- 1Integrative Omics and Molecular Modeling Laboratory, Department of Bioinformatics and Biotechnology, Government College University Faisalabad (GCUF), Faisalabad, Pakistan
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Laboratory of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, Guangxi, China
- 3Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
Arachis hypogaea (peanut) is a leading oil and protein-providing crop with a major food source in many countries. It is mostly grown in tropical regions and is largely affected by abiotic and biotic stresses. Cysteine-rich receptor-like kinases (CRKs) is a family of transmembrane proteins that play important roles in regulating stress-signaling and defense mechanisms, enabling plants to tolerate stress conditions. However, almost no information is available regarding this gene family in Arachis hypogaea and its progenitors. This study conducts a pangenome-wide investigation of A. hypogaea and its two progenitors, A. duranensis and A. ipaensis CRK genes (AhCRKs, AdCRKs, and AiCRKs). The gene structure, conserved motif patterns, phylogenetic history, chromosomal distribution, and duplication were studied in detail, showing the intraspecies structural conservation and evolutionary patterns. Promoter cis-elements, protein–protein interactions, GO enrichment, and miRNA targets were also predicted, showing their potential functional conservation. Their expression in salt and drought stresses was also comprehensively studied. The CRKs identified were divided into three groups, phylogenetically. The expansion of this gene family in peanuts was caused by both types of duplication: tandem and segmental. Furthermore, positive as well as negative selection pressure directed the duplication process. The peanut CRK genes were also enriched in hormones, light, development, and stress-related elements. MicroRNA (miRNA) also targeted the AhCRK genes, which suggests the regulatory association of miRNAs in the expression of these genes. Transcriptome datasets showed that AhCRKs have varying expression levels under different abiotic stress conditions. Furthermore, the multi-stress responsiveness of the AhCRK genes was evaluated using a machine learning-based method, Random Forest (RF) classifier. The 3D structures of AhCRKs were also predicted. Our study can be utilized in developing a detailed understanding of the stress regulatory mechanisms of the CRK gene family in peanuts and its further studies to improve the genetic makeup of peanuts to thrive better under stress conditions.
1 Introduction
Plants are exposed to a great number of biotic as well as abiotic stresses throughout their life. These include salinity, drought, cold, and pathogens. This has led to the evolution of several mechanisms in the immune system that helps them survive during these stresses. One of these mechanisms is receptor-like kinases (RLKs), which belong to transmembrane proteins, which sense external signals and send them to the intracellular environment (Tan et al., 2019). In this way, they respond to environmental stress. Depending upon their extracellular regions, RLKs are classified into various types, such as leucine-rich repeat RLKs, S-domain RLKs, cysteine-rich RLKs, and wall-associated RLKs (X. Zhao et al., 2022).
Cysteine-rich RLKs (CRKs) form a major group of RLKs having more than 40 members in Arabidopsis thaliana. Two copies of the domain of unknown function 26 (DUF26; PFAM domain PF01657), which contains four conserved cysteines, make up the protein’s extracellular region. These three cysteines together make up the motif C-8X-C-2X-C, which may play a role in the production of disulfide bridges that might be used to regulate thiol redox (Chen et al., 2004; Shiu and Bleecker, 2003; Wrzaczek et al., 2010). CRKs are linked to reactive oxygen species (ROS) signaling and cell death. In A. thaliana, the kinase CRK2 plays a crucial role in innate immunity and growth of plants by forming a complex with an NADPH oxidase respiratory burst oxidase homolog D (RBOHD), in response for extracellular ROS production. CRK2’s activity is vital for ROS bursts in response to elicitors, impacting defense against pathogens. Phosphorylation of RBOHD’s C-terminal enhances ROS production, suggesting an evolutionarily conserved mechanism, highlighting CRK2’s role in regulating ROS in response to microbial patterns (Kimura et al., 2020). In CRKs, oxidative stress, pathogen attack, and salicylic acid induction are the major causes of transcriptional induction (Chen et al., 2003). Several Arabidopsis CRK members regulate the defense response against pathogens and cause cell death in leaves. Moreover, over-expression of CRK5 causes increased resistance toward a virulent pathogen Pseudomonas syringae. Similarly, over-expression of CRK4/5/19 and 20 by a chemically inducible promoter causes cell death. Genetic analyses have suggested the involvement of CRK5 in the regulation of cell death independent of SA. However, increased resistance to Pseudomonas due to the over-expression of CRK13 requires increased levels of SA (Acharya et al., 2007).
CRKs have been identified in several plant species, and a number of them have been elucidated for their biological functions. These reports have demonstrated that they are mainly involved in hormonal signaling pathways, tolerance to environmental stresses, and plant growth. In Arabidopsis, higher expression of CRK1, also known as AtCBK3, results in an increased thermotolerance (Wei et al., 2019). Moreover, previous studies have shown the association between CRK3 and cytosolic glutamine synthetase (GLN1), which mobilized nitrogen during leaf senescence, while CRK1 and CRK5 conferred drought stress tolerance by negatively regulating ABA signaling (Li et al., 2006). Furthermore, AtCRK6,7 and HvCRK1 from Hordeum vulgare were found to enhance a regulatory response against the pathogen, powdery mildew (Rayapuram et al., 2012; Bourdais et al., 2015). The TaCRK1 gene in wheat showed an upregulated expression in response to a pathogen, Rhizoctonia cerealis (Saintenac et al., 2021). This reveals the involvement as well as the importance of CRK genes in physiological processes during plant development (Sarwar et al., 2023).
Arachis hypogaea L., also commonly known as peanut and groundnut, is an oilseed and grain legume which is extensively cultivated in the tropical and subtropical regions, with a yearly production of nearly 46 million tons. The genus Arachis is prevalent in South America and comprises mostly the diploid species (2n = 2x = 20), whereas A. hypogaea is an allotetraploid (AABB-type genome; 2n = 4x = 40) resulting from the hybridization occurrence between two diploid species, followed by polyploidization (Bertioli et al., 2016). Homologous A and B genomes Arachis duranensis (AA, 2n = 20) and Arachis ipaensis (BB, 2n = 20) contributed to the hybridization (Zhang et al., 2017). Peanut is rich in oil (40%–60%), carbohydrates, protein (10%–20%), minerals, vitamins, monosaturated fatty acids, and antioxidants. India is the largest consumer of edible oil worldwide, and it also consumes the most peanut oil (Patel et al., 2022). Semi-arid tropics (SAT) including Asia, Africa, and South and North America account for approximately 60% of peanut production worldwide. These areas have predominant extremes of drought, salinity, and temperature. These conditions such as salt and drought stress significantly affect the growth and productivity of plants. Estimates have shown that drought stress causes the loss of approximately 6 million tons of peanuts, which is worth about 250 USD. Similarly, soil salinity halts plant growth by reducing the mineral uptake by the plant (Banavath et al., 2018). Peanut production is greatly affected by heat, drought, and salt stresses as it grows in tropical and subtropical regions. Since CRKs are involved in the defense mechanism of plants against environmental stresses, the identification of peanut CRKs can help understand their interaction mechanisms. The availability of A. hypogaea, A. duranensis, and A. ipaensis genomes has facilitated genome-wide identification as well as the characterization of CRKs. The aim of studying and characterizing the inter- and intra-species diversity led to the pangenome-wide analysis of these three peanut genomes. A comprehensive structural evaluation, including gene structure, motif analysis, phylogenetics, chromosomal distribution, and gene enrichment, has been performed. In addition, the differential expression of the identified members under drought and salt stress has also been carried out to find multi-stress-related genes. Furthermore, their involvement in multi-stress responsiveness is also validated through a machine learning classifier algorithm. Hence, this study will broaden our knowledge of the CRK gene family in peanut, elucidating their contribution to conferring resistance against various environmental stresses, and also will serve as valuable insights for future researchers.
2 Materials and methods
2.1 Identification and characterization of the CRK gene family in A. hypogaea, A. duranensis, and A. ipaensis
The 44 A. thaliana CRK protein sequences were retrieved from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/). The protein sequence FASTA files of A. hypogaea, A. ipaensis, and A. duranensis were downloaded from the NCBI. NCBI command-line tool, BLAST+, was used to create a local database of these files. A BLASTp search was performed against these protein sequence databases, using Arabidopsis CRK protein sequences as queries. The resulting hits were further refined by removing duplicates and isoforms.
Furthermore, the identified proteins were searched to confirm the presence of the stress antifungal/DUF26 (PF01657) and protein kinase (PF00069 and PF07714) domains. For this purpose, NCBI conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2015), Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) (Schultz et al., 2000), and InterPro (https://www.ebi.ac.uk/interpro/) (Hunter et al., 2009) database were utilized. The proteins having no characteristic conserved domains were excluded from further analysis. Furthermore, TBtools (Chen et al., 2018) was used to construct the domain architecture.
Information on the various physicochemical properties [molecular weight, their isoelectric point (pI), instability index (II), aliphatic index (AI), and the grand average of hydropathicity (GRAVY)] was predicted by using the ExPASy ProtParam tool (https://web.expasy.org/protparam/) (Gasteiger et al., 2005). Subcellular localization for each three peanut CRKs was predicted using an online WoLF PSORT tool (https://wolfpsort.hgc.jp/) (Horton et al., 2007).
2.2 Phylogenetic, gene structure, and conserved motif analysis of AhCRKs
A phylogenetic tree was constructed to evaluate the evolutionary links among CRK proteins. A multiple sequence alignment of 71 A. hypogaea (AhCRKs) 36 A. duranensis (AdCRKs), 44 A. ipaensis (AiCRKs), 44 A. thaliana (AtCRKs) (K. Chen et al., 2004), 37 H. vulgare (HvCRKs) (Rayapuram et al., 2012), 36 Oryza sativa (OsCRKs) (Shumayla et al., 2019) and 46 Proteus vulgaris (PvCRKs) (Quezada et al., 2019) was done using ClustalW (Zameer et al., 2021). A phylogenetic tree was constructed using the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al., 2016). Using the maximum likelihood (ML) approach and 1,000 bootstrapping replicates, the reliability of the built-in tree was confirmed. Further editing of the tree was done using the Interactive Tree of Life (iTOL) (https://itol.embl.de/) (Letunic and Peer, 2021).
The conserved motif among the members of the CRK family of each three Arachis species was searched using the Multiple Expectation Maximization for Motif Elicitation (https://meme-suite.org/meme/tools/meme) tool (Bailey et al., 2015). A maximum of 20 conserved motifs were analyzed. The GFF files of each Arachis species were used to analyze the intron–exon pattern. Both the motifs and gene structures were visualized using TBtools (Zia et al., 2022).
2.3 Chromosomal location, Ka/Ks, and gene duplication analysis
The chromosomal location information for CRK genes from each of the three Arachis spp. was determined from the NCBI genomic database. The distribution of CRK genes from three Arachis species across their chromosomes was analyzed and visualized using TBtools. Duplicated pairs of AhCRK genes were identified using NCBI nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (Zia et al., 2022), based on the coverage of the aligned sequences which should be ≥ 70%. The Ka/Ks ratios (the rate of non-synonymous/synonymous substitution) for the duplicated gene pairs were also calculated using DnaSP v.6 software (Rozas et al., 2017). This was done to assess the molecular evolutionary rates of each gene pair. Moreover, the time of divergence for these gene pairs was calculated using the formula “t = Ks/2λ,” with a λ value of 1.5 × 10−8 for dicots, substitutions/synonymous site, and year representing the neutral substitution (Zameer et al., 2022). This was shown a million years ago (Mya). The linkage between chromosomes and duplicated pairs was shown using the Advanced Circos program of TBtools.
2.4 Protein–protein interaction, Gene Ontology enrichment, and miRNA prediction
Amino acid sequences of AhCRKs were subjected to the STRING database (https://string-db.org/) (von Mering et al., 2003) to analyze the interactions among peanut CRKs and other proteins. The top 10 interactions were set to be predicted, and the threshold level was kept medium (0.4). The interaction network was visualized using Cytoscape software (Shannon et al., 2003). The components considered for GO enrichment were biological processes (BPs), cellular components (CCs), and molecular functions (MFs), and these were predicted using the DAVID database (https://david.ncifcrf.gov/home.jsp) (Dennis et al., 2003). Moreover, KEGG pathways were also analyzed using the same database. The miRNAsong database (miRNAsong - A tool for microRNA sponge sequence generation and testing (muni.cz)) (Barta, Peskova, and Hampl, 2016) was used to identify the A. hypogaea miRNAs. Furthermore, the psRNATarget database [psRNATarget: A Plant Small RNA Target Analysis Server (2017 Update) (zhaolab.org)] (Dai and Patrick, 2011) was used to identify the putative miRNAs targeting the AhCRK genes using the coding sequences of AhCRKs as target sequences.
2.5 Cis-regulatory elements and expression profiling of AhCRKs in abiotic stresses
For identification of cis-regulatory elements, 2 kb sequences upstream of the translation start site of CRK genes from each of the three Arachis species were extracted and scanned using the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999). Finally, the diagram was illustrated using TBtools software.
The expression levels of all AhCRK genes in peanut leaves under drought and salt stress were evaluated using transcriptome datasets available at the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) (BioProjects; PRJNA706902: drought stress and PRJNA603232: salt stress). The genome and annotation (GFF) files were downloaded from the genome-NCBI database (https://www.ncbi.nlm.nih.gov/genome/). Indexes of the A. hypogaea genome were built by using Bowtie2 (Colling et al., 2013) and the clean paired-clean reads were then mapped to the genome. Furthermore, the gene level counts from RNA-seq providing the expression level of the genes were obtained by using featureCounts (Liao et al., 2014). The count values for each condition were used to generate a heatmap. The logarithmic transformed values were used to show the differential expression of genes.
2.6 Evaluation of multi-stress-related AhCRK genes using machine learning
DESeq2 (Love et al., 2014) was applied to both drought and salt samples to identify the genes which were significantly expressed. The genes were further divided based on their statistical significance depending on whether their p-value is less than 1 and the log2 fold-change values (log2 fold value >0.5 for upregulation and log2 fold value <0.5 for downregulation). The AhCRK genes common to both datasets were collected. Furthermore, to access the credibility of these genes, the classification algorithm—random forest (RF) was implied in R. Assessing the performance of a model is usually subjective, which involves comparing the model’s predictions to the known values of the dependent variable in a specific dataset. DESeq normalization was applied to the salts’ count datasets for the RF classifiers, in which the genes common to both stresses were incorporated. The performance of the RF classifier was evaluated using matrices including the accuracy, the area under the receiver operating characteristic curve (AUC), specificity, and sensitivity of the data with common multi-stress-related genes.
2.7 3D structure prediction of AhCRK proteins
The three-dimensional (3D) structure of a protein is necessary for its proper functioning. Based on the expression analysis, the 3D structures of three AhCRK proteins were predicted using AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) (Jumper et al., 2021). The predicted structures were validated using the SAVES (https://saves.mbi.ucla.edu) server (Elshemey et al., 2010) and MolProbity (http://molprobity.biochem.duke.edu/) (Davis et al., 2007). PyMOL (Alexander et al., 2011) was used to visualize these structures. The overview of the current study is shown in Figure 1.
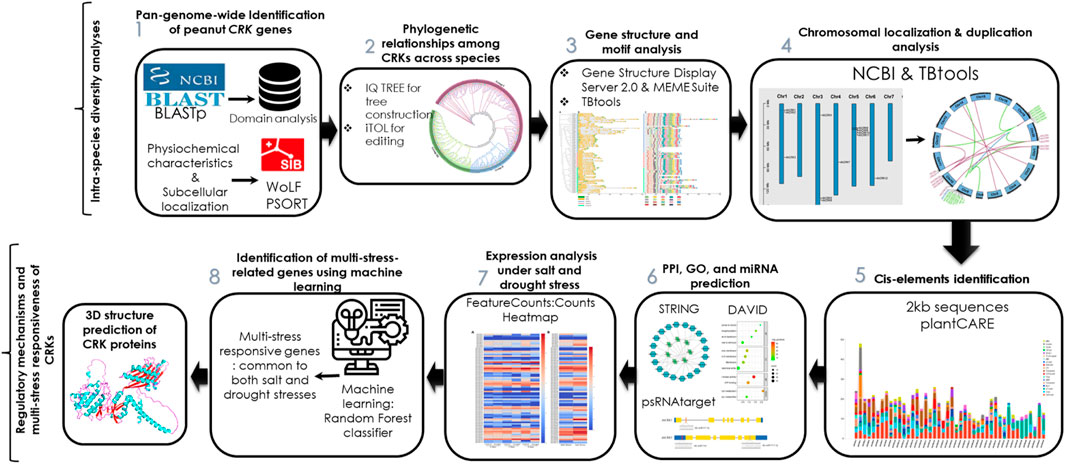
FIGURE 1. Workflow of the pangenome-wide identification of CRK genes in peanut, their structural and functional analysis, expression profiling, and multi-stress responsiveness.
3 Results
3.1 Identification of CRK genes in three Arachis species
A total of 71, 36, and 44 genes were identified in AhCRKs, AdCRKs, and AiCRKs (Supplementary Tables S1–S3). The protein domain analysis of these identified CRKs in three Arachis species confirmed the presence of the conserved stress-antifungal/DUF26 (PFAM PF01657) and Pkinases (PFAM domains PF00069 and PF07714) domains. All the proteins from these three species contained two to four PF01657 (DUF26; salt stress response/antifungal domain). For Pkinase domains, some of the genes contained PF00069 (protein kinase domain) and others had PF07714 (protein tyrosine kinase domain) conserved in them (Supplementary Tables S4). Furthermore, all the genes were named in chronological order based on their position on chromosomes.
The physicochemical properties of 71 identified Arachis CRK proteins were analyzed. There were no substantial differences in their protein length/amino acid residues, molecular weights, isoelectric point, instability index, aliphatic index, and GRAVY values among the three species. In all the three species, most of these proteins have pI values ranging between 5 and 9, indicating their acidic as well as basic behavior. The II values of most of these proteins showed that they will be unstable in the test tube. Almost all of these proteins had an AI greater than 70, which indicates that these proteins are thermally stable, and negative GRAVY values indicate that these proteins are hydrophilic (Figure 2). The determination of subcellular localization of AhCRK proteins will help understand their molecular functions. Most of the AhCRKs were localized in the plasma membrane, which was followed by the extracellular membrane, and chloroplast. In A. duranensis, AdCRKs were localized in the plasma membrane, chloroplast, and extracellular space. In A. ipaensis, these proteins were also localized in the plasma membrane, extracellular membrane, and chloroplast (Supplementary Tables S1–S3).
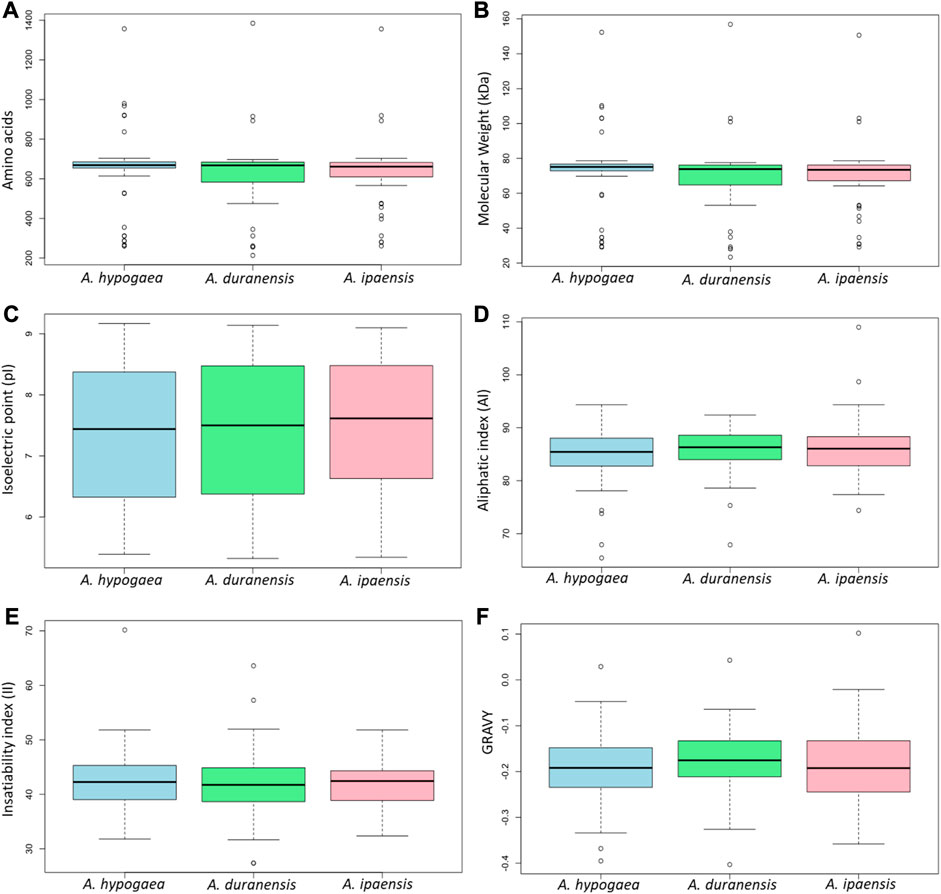
FIGURE 2. Box plots showing the physiochemical characteristics of three Arachis species: (A) amino acid residues/protein length, (B) their molecular weight, (C) their isoelectric point, (D) aliphatic index, (E) insatiability index, and (F) the grand average of hydropathicity.
3.2 Phylogenetic relations of Arachis CRK proteins
The identified protein sequences from A. hypogaea, A. duranensis, A. ipaensis, A. thaliana, O. sativa, H. vulgare, and P. vulgaris were used in the construction of the phylogenetic tree to study intra- and inter-species diversity. According to the phylogenetic tree, these protein sequences were classified into three groups, and each group contained a different number of members from each species (Figure 3). Group I had the maximum number of members present in it (166 members). Members from all seven species were present in this group, showing the shared homology among them. This group contained 30 members from A. hypogaea, 21 members from A. ipaensis, 16 members from A. duranensis, six members from A. thaliana, and 36 members from O. sativa.
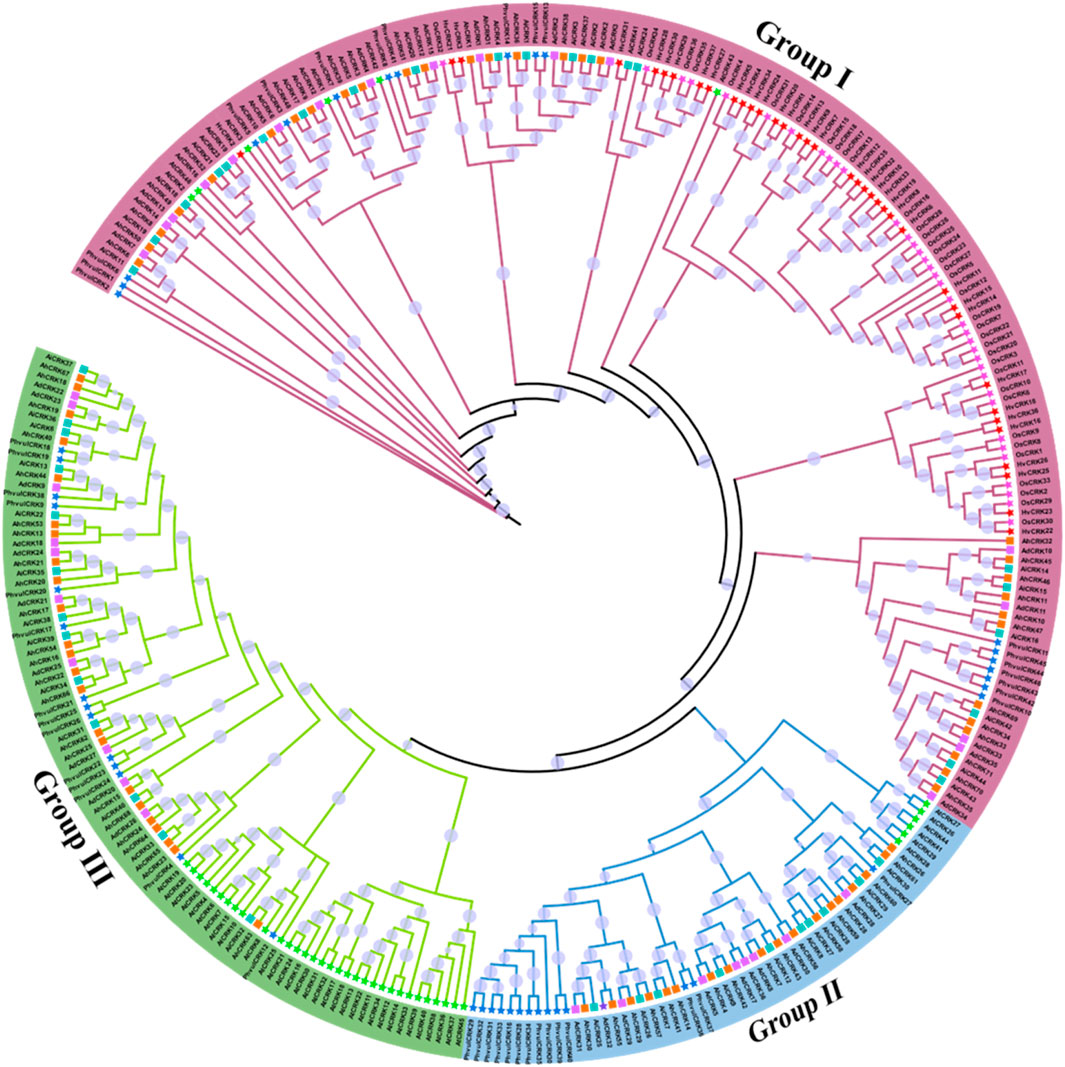
FIGURE 3. Phylogenetic tree of the CRK protein sequences from seven different plant species including three Arachis species, generated using the maximum likelihood method. Different groups are represented by specific clade and branch colors.
Group II, the smallest group with 57 members, has the following members: 18 A. hypogaea, 10 A. ipaensis, nine A. duranensis, six A. thaliana, and no members from O. sativa. Group III has 92 members: 23 A. hypogaea, 13 A. ipaensis, 10 A. duranensis, and 32 A. thaliana. This group does not have any members from Oryza sativa as well. The results suggested the close evolutionary relationships across species, which indicates structural and functional conservations as well. Members of all three Arachis species were clustered in all three groups, indicating their intraspecies conservation. The presence of members from other species in groups also indicates the orthologous relationships among species.
3.3 Gene structure and the conserved motif analysis
Gene length in A. hypogaea varied from 938 bp (AhCRK42) to 26,515 bp (AhCRK32). Exon and intron positions were associated and compared to increase our insights into the structural diversity among the CRK members of A. hypogaea, as well as intraspecies diversity among the members from three Arachis species. For A. hypogaea, the intron number varied from 1 (AhCRK63) to 16 (AhCRK35). Moreover, some members including AhCRK4, AhCRK42, and AhCRK65 contained no intronic regions. Members from each group contained a similar number of introns and exons.
In A. duranensis, the gene length ranged from 855 bp (AdCRK17) to 19,199 bp (AdCRK9). The exon–intron pattern was also conserved among members of the same group. Group I contained zero to seven introns, whereas members of Group II contained a maximum of seven introns, and the members of Group III had introns ranging from five to eleven. For A. ipaensis, the observed gene length varied from 873 bp (AiCRK8) to 10,977 bp (AiCRK1). The members from the same group showed conservation among their gene structures, while the overall number of introns ranged from 0 (AiCRK8 and AiCRK12) to 10 (AiCRK43). This variation in the number of introns and exons might have resulted in varied gene lengths among the members from the same group, as well as intraspecies (Figure 4; Supplementary Figures S1, 2).
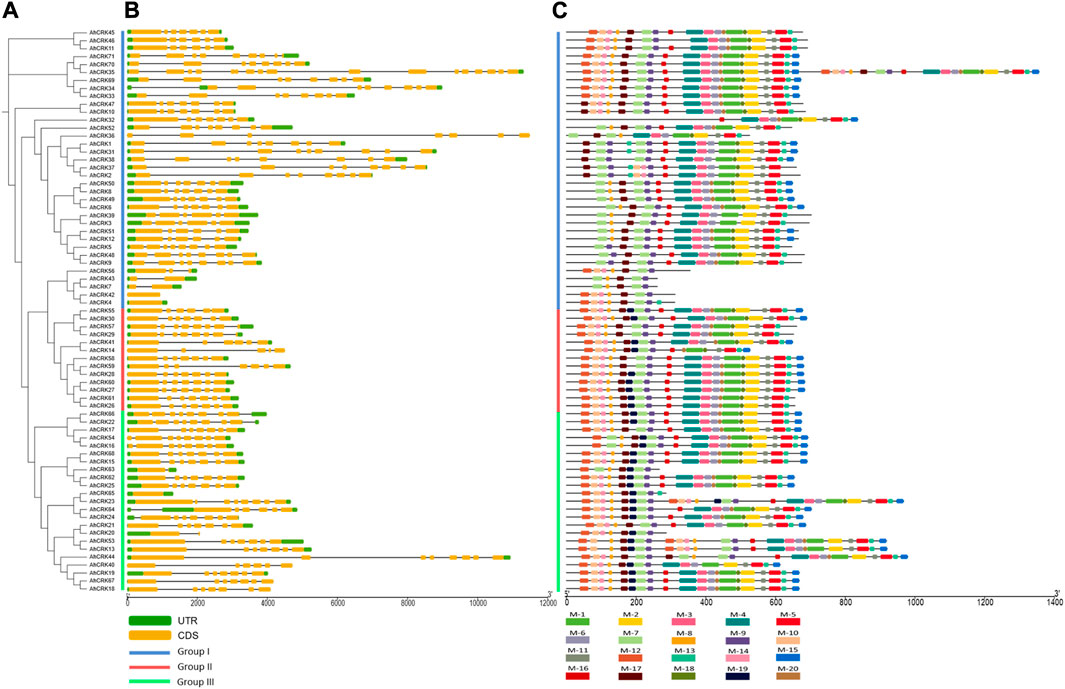
FIGURE 4. (A) Phylogenetic tree of AhCRKs, (B) structural features showing exon–intron organization, and (C) a conserved motif pattern of 71 AhCRK proteins.
The occurrence of 20 conserved motifs was analyzed among AhCRK, AdCRK, and AiCRK proteins. For AhCRKs, almost 18 motifs were conserved among the maximum members. Some members showed a fewer number of conserved motifs, whereas members of group 2 had a higher conservation of motif patterns. In A. duranensis, almost 16 motifs were conserved among most members. Members of Group I had slight variations in their conservation pattern, while those of Group II and Group III showed significant conservation of motif patterns. Members of AiCRKs showed the greatest conservation of motifs among all the members and groups with very few members having a fewer number of motifs conserved in them.
3.4 Chromosomal mapping of CRKs and their duplication analysis
To evaluate the pattern of genomic distribution of AhCRK, AdCRK, and AiCRK genes, their chromosomal gene location was identified. Furthermore, duplication events of these genes were analyzed using syntenic analysis. These analyses showed that the AhCRK genes were found to be randomly distributed on 17 out of 20 A. hypogaea chromosomes. Peanut Chr9 and Chr19 had the highest number of genes (16 members) mapped on them. Chr2, Ch7, and Chr17 had no CRK genes present in them (Figure 5). In the A. duranensis genome, eight chromosomes had AdCRK genes mapped on them, and Chr2 and Chr7 with no CRK genes mapped onto them. Consistent with the mapping pattern observed in A. hypogaea, Chr9 of the A. duranensis genome had the highest number of genes clustered on it (14 AdCRKs). Only one gene AdCRK36 was present on the scaffold. A. ipaensis also followed a similar mapping pattern, with genes mapped on every chromosome except Chr7 and the highest number of AiCRKs being clustered on Chr9 (18 genes). These results show the conservation in gene position patterns on chromosomes (Supplementary Figures S3, 4).
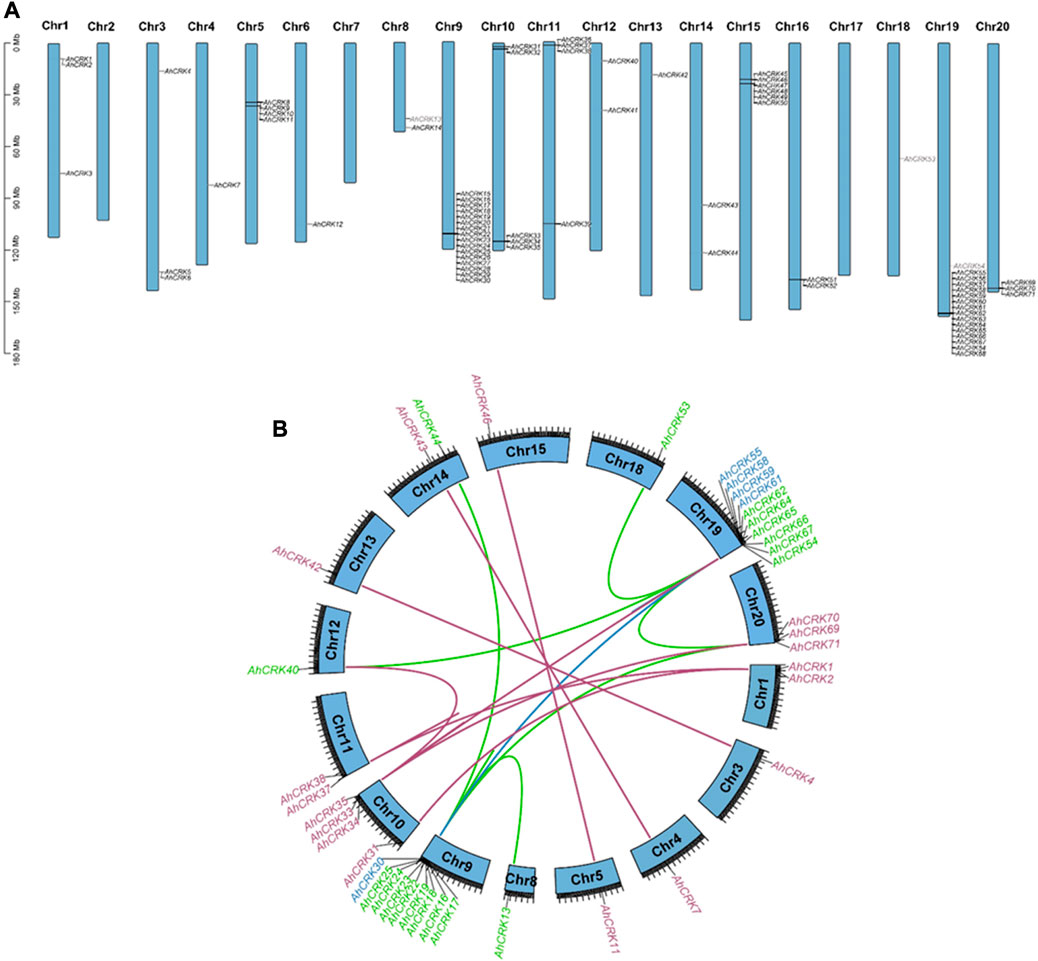
FIGURE 5. (A) Chromosomal mapping of AhCRK genes; (B) segmental and tandem duplications among the AhCRK members. Gene label colors specify the group they belong to.
Gene duplication events were also observed among AhCRK, AdCRK, and AiCRK genes, and a total of 41, 14, and 20 duplicated gene pairs were found in these members, respectively. In A. hypogaea, 10 pairs of tandem and 31 pairs of segmental duplication were observed. In A. duranensis, 11 pairs of tandemly duplicated genes were found, with three pairs being segmentally duplicated. Similarly, in A. ipaensis, 13 pairs of genes resulting from tandem duplication were analyzed with seven gene pairs being segmentally duplicated. These observations are consistent with their genomic composition; for instance, A. hypogaea have double the size of the genome as that of A. duranensis and A. ipaensis¸ indicating that the AhCRK genes are dispersed on different chromosomes. Segmental supplication is the duplication of genes from one chromosome onto another; thus, the presence of more segmental duplication pairs in A. hypogaea makes sense. To analyze the evolutionary constraints of the repeated Arachis CRK genes, the Ka, Ks, and the Ka/Ks ratios of all para-homologous gene pairs were then calculated. In A. hypogaea, almost half of the gene pairs had their Ka/Ks values ranging from 0.16 to 0.99, which were smaller than 1.0, indicating significant purification selection pressure had been applied to these gene pairs. The rest of the half pairs having Ka/Ks values more than 1.0 indicated that positive selection also occurred. As a result, the divergence time of 41 duplicated pairs was between 0.67 Mya and 117.42 Mya. In A. duranensis, three gene pairs exhibited positive selection (Ka/Ks values >1), while the rest of the duplicated pairs showed negative selection (Ka/Ks values <1). Furthermore their divergence time ranged from 1.653 to 121.91 Mya. For A. ipaensis, eight duplicated gene pairs showed positive selection, and the rest of the 12 pairs showed purifying selection, with their divergence time ranging from 0.84 to 129.4 Mya (Table 1; Supplementary Tables S5, 6).
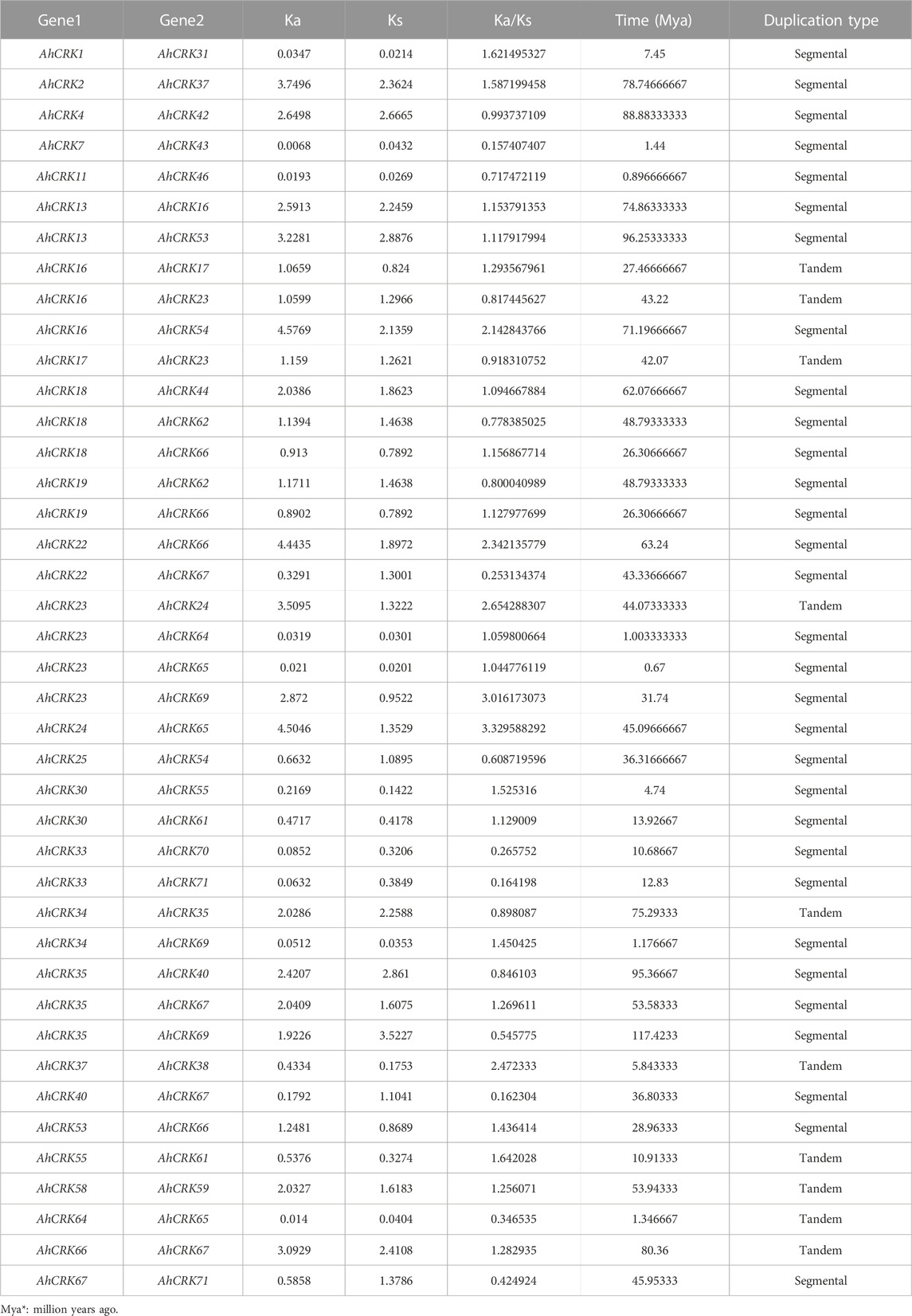
TABLE 1. Duplication data of AhCRK genes, synonymous and non-synonymous mutations, duplication time, and type of duplication between the genes.
3.5 Cis-regulatory element analysis of Arachis spp.
To get better insights and understanding regarding the functional roles of Arachis CRK genes, their upstream promoter regions were analyzed to predict the cis-acting elements present in them. Several development-related (MBSI, Circadian, O2-site, CAT-Box, and HD-Zip 1), stress-related (TC-rich repeats, MBS, LTR, GC-motif, and WUN-motif), hormone-related (P-box, TGA-element, CGTCA-motif, ABRE, and TCA-element), and light-related (GATA-motif, Box 4, GT1-motif, and G-Box) cis-regulatory elements were identified in each Arachis CRK members’ promoters (Figure 6).
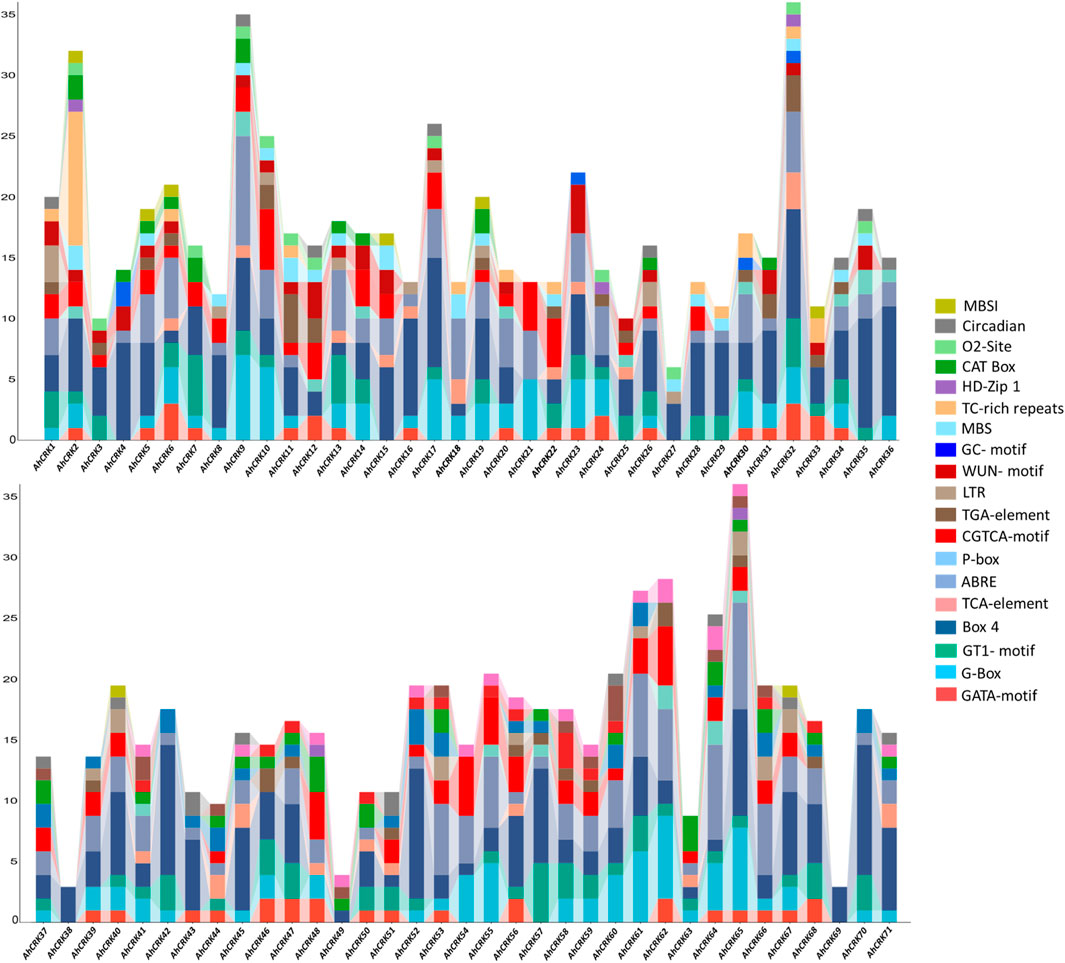
FIGURE 6. Cis-regulatory elements in the upstream promoter regions of the AhCRK genes. Each bar is representing the specific elements present in the particular gene.
In A. hypogaea, all four types of elements were observed in each AhCRK members’ promoters’ site, except for AhCRK38 and AhCRK69 which contained only one light-related element (Box 4). All these elements corresponded to gibberellin, auxin, abscisic acid, and MeJA responsiveness; endosperm and meristem expression; low-temperature responsiveness; and zein metabolism regulation. Each of the A. duranensis CRKs contain all four kinds of cis-regulatory elements. Members of each A. ipaensis CRKs contain all four types of cis-acting elements with a large number of light-related elements (GATA-motif). Moreover, AiCRK38–40 and AiCRK42 contained only light-responsive elements (Supplementary Figures S5, 6). All these results demonstrate not only the conservation of elements but also their potential involvement in growth, developmental, hormonal, and stress-related processes leading to their functional roles in the tolerance of environmental stresses.
3.6 Prediction of miRNAs, protein–protein interaction network, and Gene Ontology enrichment analysis
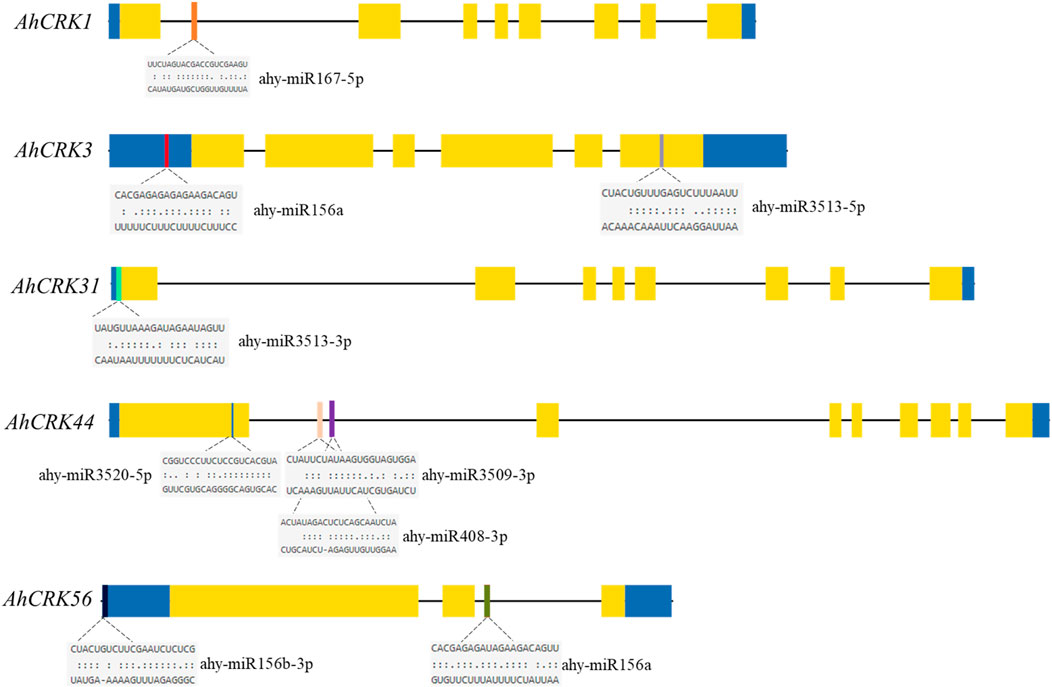
Several studies in recent years have unveiled the regulatory roles of miRNAs in the transcription and expression of genes under various developmental and stress-related conditions. Therefore, the miRNAs targeting the AhCRK genes were predicted to get insights into the miRNA-mediated post-transcriptional regulation of these genes. A total of 34 AhCRKs were targeted by miRNAs from 12 different families (Figure 7; Supplementary Table S7). Members of the miR156 family targeted AhCRK3, AhCRK20, AhCRK49, AhCRK55, and AhCRK56. miRNAs from the miR160 family targeted AhCRK32. Similarly, miRNAs of the miR167 family targeted AhCRK1 and AhCRK38. Further studies are required to determine the biological roles of these peanut miRNAs and their involvement in gene expression mechanisms.
A PPI network of the Arachis CRK proteins was also generated to understand the functional relativity among them (Figure 8A). The Arachis CRKs interacted with each other and other proteins showing connectivity as well as their functional relativity. More specifically, AhCRK59, AhCRK57, and AhCRK23 interacted with most of the other related proteins. These relative proteins were found to be mostly involved in defense and immunity response, the signaling pathway associated with transmembrane receptor protein tyrosine kinase, and the signal transduction which suggests the potential role of AhCRKs in related pathways and mechanisms.
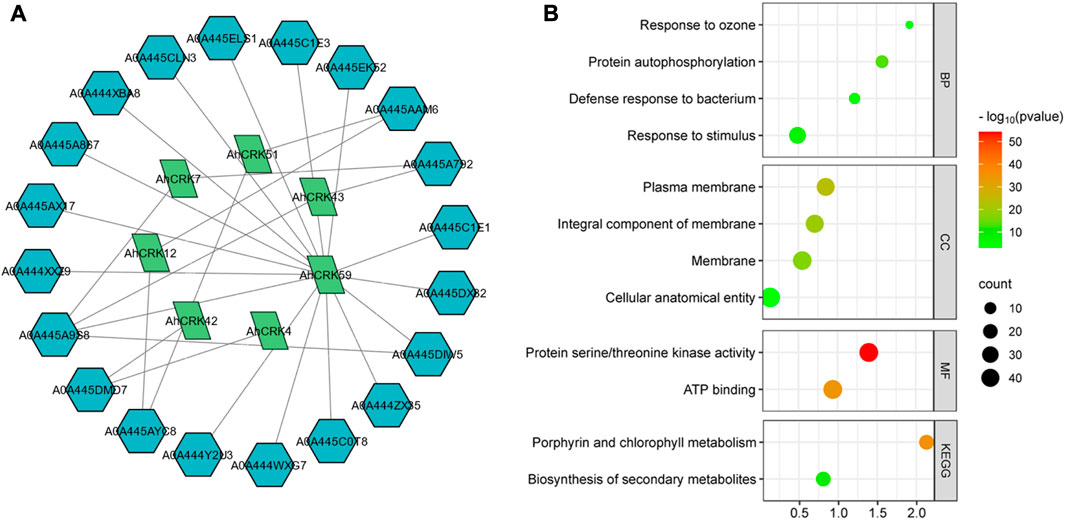
FIGURE 8. (A) Network showing the interactions among AhCRK protein members and other related proteins. The green nodes are AhCRKs, and the blue nodes are other interacting proteins. (B) GO enrichment bubble plot representing the biological processes, their cellular components, potential molecular functions, and GO and KEGG pathways in which AhCRK proteins are potentially involved.
GO enrichment analysis was then performed to further determine the dynamic roles of CRKs at the molecular level. Based on this GO analysis, AhCRK genes were classified into three different major categories: biological processes (BP), cellular components (CC), and molecular functions (MF). Biological processes in which these proteins were found to be involved included responses to stimulus and defense responses. Almost all of the proteins were found to have membranes as their cellular component. Similarly, their molecular functions included kinase activity and ATP binding. Their related KEGG pathways suggest their involvement in metabolic pathways (Figure 8B).
3.7 Expression profiling of AhCRKs under drought and salt stresses
Transcriptome expression data were used in the determination of the expression level of 71 AhCRKs in leaf tissues under drought and salt stresses. AhCRKs possessed diverse expressions under different stress conditions. Under drought stress conditions, most of the genes were highly expressed including AhCRK19, AhCRK22, AhCRK23, AhCRK24, AhCRK25, AhCRK32, AhCRK33, AhCRK34, AhCRK38, AhCRK48, AhCRK49, AhCRK62-65, AhCRK67, and AhCRK69. All these genes had the same expression under all control and treated conditions, while AhCRK1, AhCRK29, AhCRK30, AhCRK31, AhCRK57, and AhCRK70 had fluctuating expression under control and treated conditions (Figure 9A). In salt stress, an almost similar expression pattern of AhCRKs was observed as in drought stress, whereas genes including AhCRK1, AhCRK13, AhCRK29, AhCRK41, AhCRK55, AhCRK56, AhCRK57, and AhCRK68–70 had changed expression under control and treated conditions (Figure 9B).
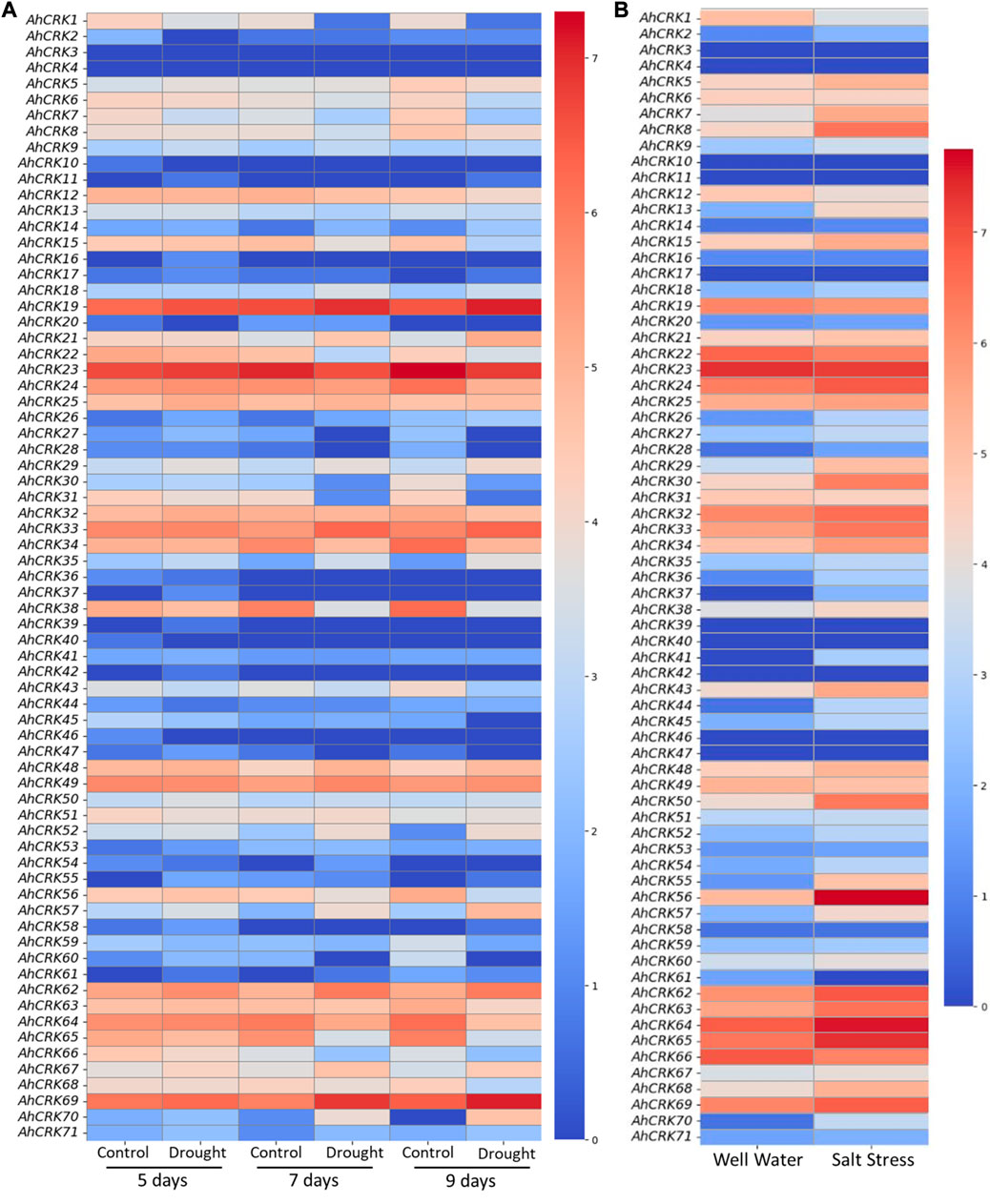
FIGURE 9. Heatmap representing the change in the expression level of AhCRKs in peanut leaves under (A) drought stress at 5, 7, and 9 days and in (B) salt stress. Blue color represents the downregulated expression, and red color represents the higher or upregulated expression.
3.8 Identification and performance evaluation of multi-stress responsive genes with a classification algorithm
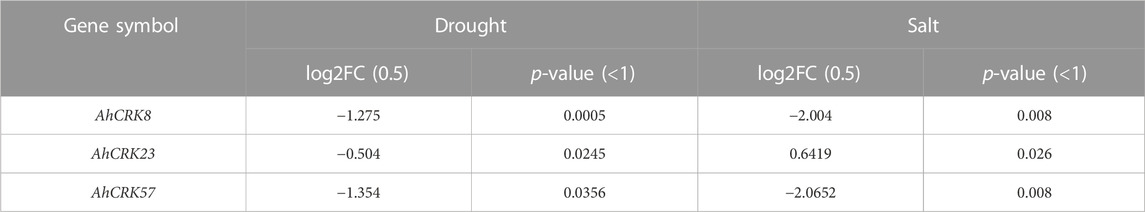
Among all the differentially expressed AhCRK genes, three genes AhCRK8 (Group I), AhCRK23 (Group III), and AhCRK57 (Group II) were found to be common in both drought and salt datasets. After the identification of these potential multi-stress responsive genes, a machine learning classifier, random forest, was implemented to evaluate the performance of these multi-stress responsive genes. To perform this task, the count data of salt stress were provided as a training dataset and multi-stress-related genes (AhCRK8, AhCRK23, and AhCRK57) were tested (Table 2). The ROC plots took into account the sensitivity, the specificity, and the false positive rate (FPR). Sensitivity = True Positives/(True Positives + False Negatives), the proportion of the actual positives that have been correctly identified by the classification model, and the specificity = True Negatives/(True Negatives + False Positives) and the FPR which is the measure of accuracy of the test; Accuracy = (True Positives + True Negatives)/(True Positives + False Positives + True Negatives + False Negatives). The ROC values observed for AhCRK8, AhCRK23, and AhCRK57 were 0.6667, 0.8333, and 0.5556, respectively. AhCRK23 was found to have acceptable ROC values, thus making it a potential multi-stress responsive gene. Supplementary Figure S7 shows the ROC plots for these genes.
3.9 3D structure prediction of AhCRK proteins
To obtain more structural and ultimately functional insights, the 3D protein structures of three multi-stress-related AhCRKs were modeled. The AhCRK8 and AhCRK23 had almost similar structures. The long spirals can be seen in both very similar structures. Similarly, the turns and loops also share similar patterns in both structures. However, the structure of AhCRK57 is significantly different, having a fewer number of helices than those of AhCRK8 and AhCRK23 with a similar number of turns and loops. The predicted similar structures suggest the potentially similar functions of these AhCRK proteins (Figure 10).
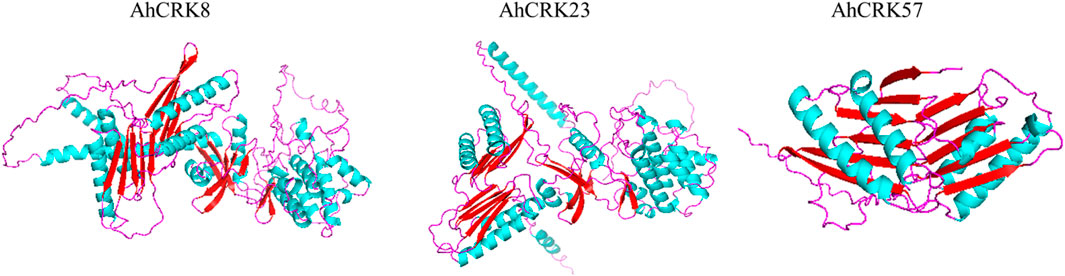
FIGURE 10. Predicted 3D structures of three multi-stress-related AhCRKs. Structures are displayed based on secondary structures: blue colors represent spirals, red shapes represent turns, and purple shapes represent loops.
4 Discussion
RLKs are perceivers of a variety of external environmental stimuli and transmit these input signals to activate the expression of specific target genes (Morris and Walker, 2003). RLKs contain an extracellular domain, an amino-terminal signal sequence, a cytoplasmic domain with serine/threonine protein kinase activity, and a single transmembrane domain. RLKs have several sub-families based on conserved residues; one of them is cysteine-rich repeat (CRR) RLKs (CRKs) (Czernic et al., 1999). CRKs have crucial roles in plants’ immunity, defense, and response to ultraviolet radiation and abiotic stresses (Liu et al., 2021). The CRK family has multiple members in different species with 44 members identified in A. thaliana (Chen et al., 2004), 36 in rice (Shumayla et al., 2019), 36 in Malus domestica (Zuo et al., 2020), and 30 in Gossypium barbadense (Li et al., 2018). However, there is no detailed study available evaluating the intraspecies diversity in Arachis members. The current study is being carried out on the CRK family in three Arachis species, namely, A. hypogaea, and its progenitors A. duranensis and A. ipaensis.
In our study, we identified 71, 36, and 44 CRK genes in A. hypogaea, A. duranensis, and A. Ipaensis, respectively. They exhibit nearly consistent molecular weight, length of amino acids, isoelectric point, aliphatic index, instability index, and GRAVY values with DUF26 and receptor-like kinase domain conserved in them.
All the Arachis CRKs have been precisely studied to understand their phylogenetic relationships. The difference in the number of CRKs across Arachis members, specifically A. hypogaea having 71 members and the other two members containing almost half the number of CRK proteins, shows their ancestral relationships. A. hypogaea originated through the combination of both A. duranensis and A. Ipaensis genomes and have double the number of chromosomes as present in each progenitor. All the CRK members are clustered on the three clades, with each clade containing Arachis and A. thaliana members. Various members of A. thaliana are shown to have roles in stress mechanisms. AtCRK1 positively regulates the heat and shock responses. AtCRK5 are the potential regulators of responses to various environmental stimuli (Baba et al., 2018). Similarly, overexpression of the CRK5 gene altered the defense and growth responses in transgenic plants (Chen et al., 2004). In O. sativa, OsCRK1, OsCRK3, and OsCRK4 were expressed in different developing stages, indicating the involvement of these genes in development mechanisms (Yadav et al., 2020). The homology and these genes with Arachis members suggest that they also have similar functional conservation and evolutionary significance.
The gene structure, number of intron–exons, and motifs can reflect the expansion, evolution, and functional relationships among the members of the gene family. These are caused mainly by three different types of mechanisms: gain/loss of intron/exon, insertion/deletion, and exonization/pseudoexonization (Wei et al., 2019). The pattern of the gene structure and motif number of CRK members is consistent across various species, indicating they are well-conserved during evolution. A similar pattern of the gene structure was observed in Arachis species and others, including watermelon (Wei et al., 2019), chili pepper (Rout et al., 2022), and Brassica napus (Sarwar et al., 2023). However, the number of introns and exons varied among the members belonging to different groups. The motif number was also highly conserved among members across CRK members from various species, such as in B. napus (Sarwar et al., 2023), hot pepper (Srideepthi et al., 2020), and in P. vulgaris (Quezada et al., 2019). Some members such as AhCRK42, AdCRK36, AdCRK5, and AdCRK9 did not contain any intron, which indicates early evolution as well, and they might have active involvement in plant development and metabolism (Chakraborty et al., 2023). All the variation in exon and intron numbers across Arachis and other species suggests the evolution of gene structures over time, which ultimately affects their functional conservation. This indicates the diversification of CRK genes. Since almost all the genes had the similar number of motifs conserved in them, it shows that their functions remained conserved during evolution. According to phylogeny analysis, it seems that CRK family members have been affected by evolutionary events that have caused their expansion, although functional diversity was observed between them. Probably, the modifications in the gene structure and regulatory regions of genes during evolution have caused the diversity of expression between members of a gene family. However, further studies are needed to understand the possible role of these modifications (Hashemipetroudi et al., 2023; Yaghobi and Heidari, 2023).
The chromosomal localization was relatively conserved among Arachis members, and Chr7 had no gene in all three Arachis species. However, the gene numbers clustered at almost the same chromosomal sites slightly varied. Evolutionary patterns are attributed to the mechanism of duplication of genes, including segmental and tandem. Segmental duplications are highly prevalent in plants, particularly in diploidized polyploids, where multiple duplicated chromosomal segments are retained, contributing to the abundance of duplicated genomic blocks within their genomes (Quezada et al., 2019). Both tandem and segmental duplication have played a significant role in the expansion of the CRK gene family. However, most plant species exhibited segmental duplication across their genomes (Wei et al., 2019; Zhao et al., 2021). However, in A. hypogaea, most CRK genes were segmentally duplicated, whereas A. duranensis and A. ipaensis CRKs observed tandem duplication. The determination of selection pressure on any protein or gene was done by utilizing the Ka/Ks ratio, where the mutation ratio was utilized. Ka/Ks greater than 1 represents positive selection, while Ka/Ks less than 1 shows purifying selection. Arachis species showed both positive and purifying selection.
Cis-regulatory elements are also one of the key players in regulating the stress-responsive activities of CRK genes and act as molecular switches, thus regulating gene expression. The cis-elements identified in the CRK gene family are related to defense-related, hormone, and abiotic stress-responsiveness. Expression analysis studies showed the involvement of AhCRK genes in drought and salt stress, which is evident by the presence of stress response elements in their promoters. Similarly, in Capsicum annuum (pepper), CaCRK5 is involved in a mechanism related to the immune response against pathogens. Various elements in this gene’s promoter region contributed to this defense response (Mou et al., 2021). The Triticum aestivum gene TaCRK68-A showed its recombinant expression in Saccharomyces cerevisiae and Escherichia coli, thereby enhancing their tolerance against drought, salinity, cold, and heat stress (Shumayla et al., 2019). This could be speculated that AhCRK genes’ expression is promoted by abiotic stresses, although further work is required to confirm this. miRNAs have received significant attention for their roles in stress tolerance and development. We identified miRNAs belonging to multiple families which targeted AhCRK genes. Other peanut genes involved in abiotic stress responses have also found miRNAs targeting them, thereby controlling their expression levels (Cai et al., 2023). The GO analysis of AhCRK genes exhibited their distinct roles in external stimulus and defense response and their involvement in functions like kinase activity. Previous studies reported the CRK genes’ roles in the positive regulation of stress responses, thus showing their involvement in various metabolic and biological pathways (Shumayla et al., 2019). The PPI analyses of these genes also showed their interaction with the other proteins involved in kinase- and stress-related activities.
The expression profile of CRK genes correlated with two abiotic stresses: drought and salt treatment in A. hypogaea. The degree of upregulation and downregulation varied in both stresses. In drought stress, almost half of the genes were upregulated (AhCRK23 and AhCRK69), and the others were downregulated, whereas some genes showed a change in expression on different days of tolerating drought stress, including AhCRK1, AhCRK21, AhCRK38, AhCRK56, AhCRK57, and AhCRK70. This indicates their crucial roles in drought stress tolerance. Moreover, in salt stress, the expression also varied under normal and treated conditions. Some genes were highly upregulated or downregulated upon exposure to salt stress (AhCRK1¸ AhCRK7, AhCRK8¸ AhCRK23, AhCRK56, AhCRK68, AhCRK69, and AhCRK70). Similar results were observed in other plants under abiotic stresses, including cold, salt (Zhang H. et al., 2017), heat, and drought (Shumayla et al., 2019). This shows that these genes would have important stress regulatory roles in real-world scenarios and fortify the foundation for future crop improvement strategies. Furthermore, the machine learning approaches were used to evaluate the genes which showed co-expression in both drought and salt stress. Three genes AhCRK8¸ AhCRK23, and AhCRK57 were found to show responsiveness under multi-stress-related conditions. The 3D structures of these three proteins were also predicted to help understand their structural and functional conservations. Hence, it can be inferred that CRK genes are regulated in abiotic stresses and help plants thrive under those conditions. In future studies, leveraging natural genetic variation within the germplasm to validate the function of identified candidate genes under specific stress conditions holds a significant confirmation value. These perspectives could contribute valuable insights toward the selection and integration of these genes in breeding and genetic engineering initiatives to enhance stress resilience in crops. Thus, these genes, most importantly the multi-stress responsive genes, can be used in future research studies on peanut.
5 Conclusion
CRKs are found to have regulatory roles in plants under various abiotic and biotic stresses. This study provides not only a systematic but also a comparative analysis of CRK genes in three nutritious and economically important peanut species. A total of 71, 36, and 44 genes were identified in A. hypogaea, A. duranensis, and A. ipaensis. The results elucidate the structural and physiochemical properties of the CRK gene family, which shows the intraspecies diversity and evolutionary conservation. The results also provide deep insights into the roles of CRK genes in the development, growth, environmental stimuli, and the mediation of abiotic stresses (salt and drought). AhCRK19, AhCRK23, AhCRK56, and AhCRK69 can potentially be candidate genes for conferring tolerance against drought stress. On the other hand, AhCRK8, AhCRK23, AhCRK24¸ AhCRK56, AhCRK65, and AhCRK69 can act as potential candidate genes in providing resistance against salt stress. Machine learning approaches were utilized to evaluate the multi-stress responsiveness of these genes. Owing to their expression on both drought and salt stress, AhCRK8, AhCRK23, and AhCRK57 can be deemed candidate genes for multi-stress responsiveness. These genes are needed to be explored further and can be used in genetic engineering research to devise multi-stress-resistant and -tolerant crops. Our study will also help further investigate the functional roles of the CRK genes in peanuts.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
KF, MS, and MJR collected the data, analyzed the data, performed all experiments, and wrote down the first draft of the manuscript. FA, NAA, AA, and MTQ planned this study, supervised the project, arranged resources, validated findings, and revised the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors are thankful to the Researchers Supporting Project number (RSPD 2023R1035) and King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1252020/full#supplementary-material
References
Acharya, B. R., Raina, S., Maqbool, S. B., Jagadeeswaran, G., Mosher, S. L., Appel, H. M., et al. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50 (3), 488–499. doi:10.1111/j.1365-313X.2007.03064.x
Alexander, N., Woetzel, N., and Meiler, J. (2011). “Bcl::Cluster: A method for clustering biological molecules coupled with visualization in the pymol molecular graphics system.” IEEE Int Conf Comput Adv. Bio Med. Sci. 2011: 13–18. doi:10.1109/ICCABS.2011.5729867
Baba, A. I., Rigó, G., Ayaydin, F., Rehman, A. U., Andrási, N., Zsigmond, L., et al. (2018). Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: atCRK1 regulates responses to continuous light. Int. J. Mol. Sci. 19 (5), 1–21. doi:10.3390/ijms19051282
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Banavath, J. N., Chakradhar, T., Pandit, V., Konduru, S., Guduru, K. K., Akila, C. S., et al. (2018). Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea L.). Front. Chem. 6, 34. doi:10.3389/fchem.2018.00034
Barta, T., Peskova, L., and Hampl, A. (2016). MiRNAsong: A web-based tool for generation and testing of MiRNA sponge constructs in silico. Sci. Rep. 6, 36625–36628. doi:10.1038/srep36625
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48 (4), 438–446. doi:10.1038/ng.3517
Bourdais, G., Burdiak, P., Gauthier, A., Nitsch, L., Salojärvi, J., Rayapuram, C., et al. (2015). Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11 (7), 10053733–e1005436. doi:10.1371/journal.pgen.1005373
Cai, T., Sharif, Y., Zhuang, Y., Yang, Q., Chen, X., Chen, K., et al. (2023). In-silico identification and characterization of O-methyltransferase gene family in peanut (Arachis hypogaea L.) reveals their putative roles in development and stress tolerance. Front. Plant Sci. 14, 1–24. doi:10.3389/fpls.2023.1145624
Chakraborty, S., Gangwar, R., Zahra, S., Poddar, N., Singh, A., and Kumar, S. (2023). Genome-wide characterization and comparative analysis of the OSCA gene family and identification of its potential stress-responsive members in legumes. Sci. Rep. 13 (1), 5914. doi:10.1038/s41598-023-33226-8
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. TBtools, a Toolkit Biol. integrating Var. HTS-data Handl. tools a user-friendly interface, 289660. doi:10.1101/289660
Chen, K., Fan, B., Du, L., and Chen, Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56 (2), 271–283. doi:10.1007/s11103-004-3381-2
Chen, K., Du, L., and Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53 (1–2), 61–74. doi:10.1023/B:PLAN.0000009265.72567.58
Colling, J., Jacob, P., Makunga, N. P., and Goossens, A. (2013). Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 1011. 30515. doi:10.3390/ijms23073945
Czernic, P., Visser, B., Sun, W., Savouré, A., Deslandes, L., Marco, Y., et al. (1999). Characterization of an Arabidopsis thaliana receptor-like protein kinase gene activated by oxidative stress and pathogen attack. Plant J. 18 (3), 321–327. doi:10.1046/j.1365-313x.1999.00447.x
Dai, X., and Patrick, X. Z. (2011). PsRNATarget: A plant small rna target analysis server. Nucleic Acids Res. 39 (2), 155–159. doi:10.1093/nar/gkr319
Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., et al. (2007). MolProbity: all-Atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35 (2), 375–383. doi:10.1093/nar/gkm216
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). David: database for annotation, visualization, and integrated discovery. Genome Biol. 4 (5), P3. doi:10.1186/gb-2003-4-5-p3
Elshemey, W. M., Elfiky, A. A., and Wissam, A. G. (2010). Correlation to protein conformation of wide-angle X-ray scatter parameters. Protein J. 29 (8), 545–550. doi:10.1007/s10930-010-9291-z
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server. proteomics Protoc. Handb., 571–607. doi:10.1385/1-59259-890-0:571
Hashemipetroudi, S. H., Arab, M., Heidari, P., and Kuhlmann, M. (2023). Genome-wide analysis of the laccase (lac) gene family in aeluropus littoralis: A focus on identification, evolution and expression patterns in response to abiotic stresses and aba treatment. Front. Plant Sci. 14, 1112354. doi:10.3389/fpls.2023.1112354
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic acids Res. 35 (2), W585–W587. doi:10.1093/nar/gkm259
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37 (1), 211–215. doi:10.1093/nar/gkn785
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kimura, S., Hunter, K., Vaahtera, L., Tran, H. C., Citterico, M., Vaattovaara, A., et al. (2020). CRK2 and C-terminal phosphorylation of NADPH oxidase RBOHD regulate reactive oxygen species production in Arabidopsis. Plant Cell 32 (4), 1063–1080. doi:10.1105/tpc.19.00525
Letunic, I., and Peer, B. (2021). Interactive tree of Life (ITOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi:10.1093/nar/gkab301
Li, R. J., Hua, W., and Lu, Y. T. (2006). Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem. Biophysical Res. Commun. 342 (1), 119–126. doi:10.1016/j.bbrc.2006.01.100
Li, T. G., Zhang, D. D., Zhou, L., Kong, Z. Q., Hussaini, A. S., Wang, D., et al. (2018). Genome-wide identification and functional analyses of the crk gene family in cotton reveals Gbcrk18 confers verticillium wilt resistance in Gossypium barbadense. Front. Plant Sci. 9, 1266–1316. doi:10.3389/fpls.2018.01266
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi:10.1093/bioinformatics/btt656
Liu, Y., Feng, Z., Zhu, W., Liu, J., and Zhang, Y. (2021). Genome-wide identification and characterization of cysteine-rich receptor-like protein kinase genes in tomato and their expression profile in response to heat stress. Diversity 13 (6), 258. doi:10.3390/d13060258
Love, M. I., Huber, W., and Anders., S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi:10.1186/s13059-014-0550-8
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). Cdd: NCBI’s conserved domain database. Nucleic acids Res. 43 (D1), D222–D226. doi:10.1093/nar/gku1221
Morris, E. R., and Walker., J. C. (2003). Receptor-like protein kinases: the keys to response. Curr. Opin. Plant Biol. 6 (4), 339–342. doi:10.1016/s1369-5266(03)00055-4
Mou, S., Meng, Q., Gao, F., Zhang, T., He, W., Guan, D., et al. (2021). A cysteine-rich receptor-like protein kinase CaCKR5 modulates immune response against ralstonia solanacearum infection in pepper. BMC Plant Biol. 21 (1), 382–415. doi:10.1186/s12870-021-03150-y
Patel, J., Khandwal, D., Choudhary, B., Ardeshana, D., Jha, R. K., Tanna, B., et al. (2022). Differential physio-biochemical and metabolic responses of peanut (Arachis hypogaea L.) under multiple abiotic stress conditions. Int. J. Mol. Sci. 23 (2), 660. doi:10.3390/ijms23020660
Quezada, E. H., García, G. X., Arthikala, M. K., Melappa, G., Lara, M., and Nanjareddy, K. (2019). Cysteine-rich receptor-like kinase gene family identification in the phaseolus genome and comparative analysis of their expression profiles specific to mycorrhizal and rhizobial symbiosis. Genes 10 (1), 59. doi:10.3390/genes10010059
Rayapuram, C., Jensen, M. K., Maiser, F., Shanir, J. V., Hornshøj, H., Rung, J. H., et al. (2012). Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol. Plant Pathol. 13 (2), 135–147. doi:10.1111/j.1364-3703.2011.00736.x
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27 (1), 295–296. doi:10.1093/nar/27.1.295
Rout, S. S., Rout, P., Uzair, M., Kumar, G., and Nanda, S. (2022). Genome-wide identification and expression analysis of CRK gene family in chili pepper (Capsicum annuum L.) in response to colletotrichum truncatum infection. J. Hortic. Sci. Biotechnol. 00 (00), 194–206. doi:10.1080/14620316.2022.2117654
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi:10.1093/molbev/msx248
Saintenac, C., Cambon, F., Aouini, L., Verstappen, E., Ghaffary, S. M. T., Poucet, T., et al. (2021). A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against septoria tritici blotch. Nat. Commun. 12 (1), 433–510. doi:10.1038/s41467-020-20685-0
Sarwar, R., Li, L., Yu, J., Zhang, Y., Geng, R., Meng, Q., et al. (2023). Functional characterization of the cystine-rich-receptor-like kinases (CRKs) and their expression response to sclerotinia sclerotiorum and abiotic stresses in Brassica napus. Int. J. Mol. Sci. 24 (1), 511. doi:10.3390/ijms24010511
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., and Bork, P. (2000). Smart: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28 (1), 231–234. doi:10.1093/nar/28.1.231
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132 (2), 530–543. doi:10.1104/pp.103.021964
Shumayla, , Tyagi, S., Sharma, A., Singh, K., and Upadhyay, S. K. (2019). Genomic dissection and transcriptional profiling of cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int. J. Biol. Macromol. 134, 316–329. doi:10.1016/j.ijbiomac.2019.05.016
Srideepthi, R., Krishna, M. S. R., Suneetha, P., Krishna, R. S., and Karthikeyan, S. (2020). Genome-wide identification, characterization and expression analysis of non-RD receptor like kinase gene family under colletotrichum truncatum stress conditions in hot pepper. Genetica 148 (5–6), 283–296. doi:10.1007/s10709-020-00104-4
Tan, S., Debellé, F., Gamas, P., Frugier, F., and Brault, M. (2019). Diversification of cytokinin phosphotransfer signaling genes in medicago truncatula and other legume genomes. BMC Genomics 20 (1), 373–419. doi:10.1186/s12864-019-5724-z
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic acids Res. 44 (W1), W232–W235. doi:10.1093/nar/gkw256
von Mering, C., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. (2003). String: A database of predicted functional associations between proteins. Nucleic Acids Res. 31 (1), 258–261. doi:10.1093/nar/gkg034
Wei, C., Zhang, R., Yang, X., Zhu, C., Li, H., Zhang, Y., et al. (2019). Direct photo transformation of tetracycline and sulfanomide group antibiotics in surface water: kinetics, toxicity and site modeling. Int. J. Mol. Sci. 20 (10), 1–9. doi:10.1016/j.scitotenv.2019.04.041
Wrzaczek, M., Brosché, M., Salojärvi, J., Kangasjärvi, S., Idänheimo, N., Mersmann, S., et al. (2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10, 95–19. doi:10.1186/1471-2229-10-95
Yadav, A., Garg, T., Singh, H., and Yadav, S. R. (2020). Tissue-specific expression pattern of calcium-dependent protein kinases-related kinases (CRKs) in rice. Plant Signal. Behav. 15 (11), 1809846. doi:10.1080/15592324.2020.1809846
Yaghobi, M., and Heidari, P. (2023). Genome-wide analysis of aquaporin gene family in Triticum turgidum and its expression profile in response to salt stress. Genes 14 (1), 202. doi:10.3390/genes14010202
Zameer, R., Fatima, K., Azeem, F., Algwaiz, H. I. M., Sadaqat, M., Rasheed, A., et al. (2022). Genome-wide characterization of superoxide dismutase (SOD) genes in daucus carota: novel insights into structure, expression, and binding interaction with hydrogen peroxide (H2O2) under abiotic stress condition. Front. Plant Sci. 13, 870241–870315. doi:10.3389/fpls.2022.870241
Zameer, R., Sadaqat, M., Fatima, K., Fiaz, S., Rasul, S., Zafar, H., et al. (2021). Two-component system genes in sorghum bicolor: genome-wide identification and expression profiling in response to environmental stresses. Front. Genet. 12, 794305. doi:10.3389/fgene.2021.794305
Zhang, H., Wei, C., Yang, X., Chen, H., Yang, Y., Mo, Y., et al. (2017a). Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (cucumis melo L.). PLoS ONE 12 (4), 01763522–e176423. doi:10.1371/journal.pone.0176352
Zhang, X., Zhang, J., He, X., Wang, Y., Ma, X., and Yin, D. (2017b). Genome-wide association study of major agronomic traits related to domestication in peanut. Front. Plant Sci. 8, 1611–1710. doi:10.3389/fpls.2017.01611
Zhao, P., Liu, Y., Kong, W., Ji, J., Cai, T., and Guo, Z. (2021). Genome-wide identification and characterization of calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in medicago truncatula. Int. J. Mol. Sci. 22 (3), 1044–1116. doi:10.3390/ijms22031044
Zhao, X., Qu, D., Wang, L., Gao, Y., Wang, A., et al. (2022). Genome-wide identification of cysteine-rich receptor-like kinases in sweet cherry reveals that PaCRK1 enhances sweet cherry resistance to salt stress. Springer Berl. Heidelb. 41, 2037–2088. doi:10.1007/s00299-022-02907-5
Zia, K., Rao, M. J., Sadaqat, M., Azeem, F., Fatima, K., Tahir Ul Qamar, M., et al. (2022). Pangenome-wide analysis of cyclic nucleotide-gated channel (CNGC) gene family in citrus Spp. Revealed their intraspecies diversity and potential roles in abiotic stress tolerance. Front. Genet. 13, 1034921. doi:10.3389/fgene.2022.1034921
Zuo, C., Liu, H., Lv, Q., Chen, Z., Tian, Y., Mao, J., et al. (2020). Genome-wide analysis of the apple (Malus domestica) cysteine-rich receptor-like kinase (CRK) family: annotation, genomic organization, and expression profiles in response to fungal infection. Plant Mol. Biol. Report. 38 (1), 14–24. doi:10.1007/s11105-019-01179-w
Keywords: peanut, pangenome-wide, receptor-like kinases, gene ontology enrichment, abiotic stress, multi-stress-related genes, random forest
Citation: Fatima K, Sadaqat M, Azeem F, Rao MJ, Albekairi NA, Alshammari A and Tahir ul Qamar M (2023) Integrated omics and machine learning-assisted profiling of cysteine-rich-receptor-like kinases from three peanut spp. revealed their role in multiple stresses. Front. Genet. 14:1252020. doi: 10.3389/fgene.2023.1252020
Received: 03 July 2023; Accepted: 05 September 2023;
Published: 20 September 2023.
Edited by:
Diaa Abd El Moneim, Arish University, EgyptReviewed by:
Balpreet Kaur Dhatt, Bayer Crop Science, United StatesParviz Heidari, Shahrood University of Technology, Iran
Copyright © 2023 Fatima, Sadaqat, Azeem, Rao, Albekairi, Alshammari and Tahir ul Qamar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Tahir ul Qamar, dGFoaXJ1bHFhbWFyQGdjdWYuZWR1LnBr
 Kinza Fatima
Kinza Fatima Muhammad Sadaqat
Muhammad Sadaqat Farrukh Azeem
Farrukh Azeem Muhammad Junaid Rao2
Muhammad Junaid Rao2 Muhammad Tahir ul Qamar
Muhammad Tahir ul Qamar