- Department of Medical Sciences, iBiMED–Institute of Biomedicine, University of Aveiro, Aveiro, Portugal
Emerging evidence highlights the multifaceted roles of the RNA epitranscriptome during viral infections. By modulating the modification landscape of viral and host RNAs, viruses enhance their propagation and elude host surveillance mechanisms. Here, we discuss how specific RNA modifications, in either host or viral RNA molecules, impact the virus-life cycle and host antiviral responses, highlighting the potential of targeting the RNA epitranscriptome for novel antiviral therapies.
Introduction
Viruses are completely dependent on their host cells’ machinery to replicate and form new infectious virus particles, employing different strategies to co-opt, hijack, or inhibit cellular processes. To restrict viral propagation and coordinate the immune response, different host cell receptors sense viral cues and trigger distinct antiviral signaling pathways driving the production of interferon (IFN), IFN-stimulated genes (ISG’s), and proinflammatory cytokines (Kreijtz et al., 2011; Chen et al., 2018). To usurp these antiviral pathways, viruses can exploit different characteristics of RNA molecules, including their chemical modifications. These RNA modifications, also known as the epitranscriptome, are catalyzed by different classes of RNA-modifying enzymes, occur in both host and viral RNAs, and have a direct impact on RNA maturation, stability, transport, and translation (Shelton et al., 2016; Boo and Kim, 2020; Xu L. et al., 2021; Boccaletto et al., 2022; Cui et al., 2022; Wilkinson et al., 2022; Zhang Y. et al., 2023). Notably, impairment of RNA modifications due to disruption of RNA modifiers has been shown to rewire the cellular epitranscriptome, and thus gene expression, being implicated in different pathologies (Huang et al., 2020). Although most studies are centred on ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA) modifications, emerging evidence also emphasizes their relevance in other non-coding (nc) and long-nc (lnc) RNAs (Squires et al., 2012; Amort et al., 2013; Sun et al., 2021a; Ge et al., 2021). Several epitranscriptomic marks identified on viral RNAs facilitate viral propagation (Gokhale et al., 2016; Courtney et al., 2017; McIntyre et al., 2018; Ringeard et al., 2019; Tsai et al., 2020). On the other hand, reprogramming of host RNA modifications is correlated with coordination of antiviral responses (Rubio et al., 2018; Winkler et al., 2019; Lou et al., 2021). However, despite the growing understanding of the relevance of the RNA epitranscriptome during viral infections, and its potential as antiviral target, the field is still largely underestimated. In this review, we delve into how viral and host RNA epitranscriptomes impact infectious disease pathogenicity by shaping virus-host interactions.
Unveiling the role of viral mRNA modifications during virus infection
To facilitate their replication and elude host immune surveillance, viruses evolved several tactics, including the modification of its own viral RNA molecules. The repertoire of epitranscriptomic modifications in viral mRNAs is expanding and, in the next sections, we discuss how viruses introduce epitranscriptomic marks into their own mRNA to potentiate their replication.
2′-O-methylation and 7-methyguanosine
mRNAs shield their 5′ end with an inverted N-7 methyl guanosine nucleoside (m7GpppN, cap1), along with an 2′-O-methyl group (cap2) within the first and second nucleosides downstream of the m7G cap (Pestova et al., 2001; Gebauer and Hentze, 2004; Chu et al., 2011; Varela et al., 2014; Hyde and Diamond, 2015; Devarkar et al., 2016). This capping event regulates several RNA functions, including metabolism, stability, and discrimination between self and foreign RNA (Hyde and Diamond, 2015; Ramanathan et al., 2016; Schlee and Hartmann, 2016; Decombe et al., 2023). Indeed, while cellular mRNAs with caps evade immune responses, pathogenic RNAs lacking caps are recognized by cellular sensors, triggering the activation of the IFN response (Züst et al., 2011; Mersinoglu et al., 2022).
Viruses evolved different strategies to incorporate a modified cap onto viral mRNAs (Hyde and Diamond, 2015; Sacco and Horner, 2021; Decombe et al., 2023). Influenza A virus (IAV), for instance, utilizes a cap-snatching mechanism to hijack m7G caps from host RNAs, allowing it to compete with host mRNAs for the translation machinery (Gu et al., 2015; De Vlugt et al., 2018). In contrast, certain viruses have evolved cap-independent modes of translation, utilizing internal ribosome entry sites (IRES) or other RNA structural elements that foster translation initiation (Hao et al., 2022). Several viruses encode specific methyltransferases that methylate the 2′-O-position of the ribose sugar of viral RNAs (Sorokin et al., 2021). Coronaviruses exploit their own NSP14 and NSP16 activities to introduce N-7 and 2′-O-methylation, respectively, into the viral mRNA (Chen et al., 2011; Chen et al., 2013), whereas dengue virus (DENV) and Ebola virus cap methylation relies on the activity of NS3, NS5, and L proteins, respectively (Daffis et al., 2010; Züst et al., 2011). Importantly, defects in the methyltransferase activity of these proteins is described to enhance the IFN response (Züst et al., 2011; Chang et al., 2016; Ringeard et al., 2019). The VP39 protein of vaccinia virus methylates viral 5′ caps to escape recognition by IFN-induced RNA binding protein 1 (IFIT1) (Daffis et al., 2010; Züst et al., 2011; Kumar et al., 2014; Menachery et al., 2014). Besides this, the human immunodeficiency virus 1 (HIV-1) was shown to hijack the host 2′O-MTase RNA 2′-O-methyltransferase 3 (FTSJ3) - Transactivation response RNA binding protein (TRBP) complex to modify its own RNA genome and escape recognition by the ISG 20-kDa (ISG20) protein (Ringeard et al., 2019; El Kazzi et al., 2023).
N4-acetylcytidine
In humans, N4-acetylcytidine (ac4C) marks are found within tRNA, rRNA, and mRNA molecules (Thomas et al., 1978; Boccaletto et al., 2022), to enhance their stability and function in translation (Arango et al., 2018). ac4C is introduced onto RNA molecules under the actions of N-acetyltransferase 10 (NAT10), which requires the assistance of THUMP domain-containing 1 (THUMPD1) (Sharma et al., 2015; Broly et al., 2022) and box C/D snoRNA U13 (Sharma et al., 2015), to introduce the mark into tRNAs and rRNAs, respectively (Xie et al., 2023).
ac4C was identified in the genomes of Zika virus (ZIKV), DENV, hepatitis C virus (HCV), poliovirus (PV), HIV-1, enterovirus-71 (EV71) and IAV (McIntyre et al., 2018; Tsai et al., 2020; Furuse, 2021). HIV-1 and EV71, for instance, co-opt the host’s NAT10 to acetylate their transcripts at multiple sites to enhance viral gene expression (Tsai et al., 2020; Hao et al., 2022). The ac4C modification of EV71 allows the recruitment of poly (rC)-binding protein 2 (PCBP2) to the EV71 IRES, enhancing transcript stability and the interaction with RNA-dependent RNA polymerase (3D) (Hao et al., 2022).
Pseudouridine
Pseudouridine (Ψ) is one of the most prevalent RNA modifications, and a substantial amount of this modification has been found in the genomes of several viruses, including ZIKV, DENV, HCV, PV, IAV and HIV-1 (Durbin et al., 2016; McIntyre et al., 2018; Furuse, 2021; Martinez Campos et al., 2021). Notably, Ψ-modified small RNAs derived from HCV were shown to bind to RIG-I with high affinity, but failed to trigger the canonical RIG-I conformational changes associated with its activation (Durbin et al., 2016). Furthermore, in a CRISPR screening seeking for host cell factors targeting HCV and DENV, several Ψ synthases were identified (Marceau et al., 2016).
Recent research has further revealed the involvement of Ψ in the regulation of alternative splicing (Karijolich et al., 2015). Notably, in betacoronaviruses such as SARS-CoV-2, Ψ was shown to significantly impact the splicing patterns of betacoronavirus-associated genes (Karlebach et al., 2022). Moreover, Ψ has also been showcased as an important element in Epstein-Barr virus (EBV) infections, with one of its non-coding RNAs, EBV-encoded RNA 2 (EBER2), being significantly marked by pseudouridylation (Henry et al., 2022). Disruption of pseudouridylation in EBER2 has been associated with decreased RNA levels and reduced efficiency in viral infection and the viral lifecycle (Henry et al., 2022).
5-methylcytidine
5-methycytidine (m5C) is found within multiple RNA classes and is catalyzed by enzymes belonging to the NOL1/NOP2/SUN domain (NSUN) family and DNA methyltransferase family protein (DNMT2) in eukaryotes (Bohnsack et al., 2019).
Accumulating evidence indicates that several viruses harbor m5C on their genomic RNA (Eckwahl et al., 2020; Cristinelli et al., 2022). For instance, m5C marks on murine leukemia virus (MLV) transcripts are recognized by the m5C reader ALYREF to promote their nuclear export (Eckwahl et al., 2020). The host’s RNA methyltransferase NSUN2 was shown to introduce m5C in HIV-1 genomic RNA to regulate viral gene expression. In accordance, depletion of NSUN2 reduced the abundance of m5C in HIV-1 transcripts and inhibited viral propagation by disturbing the splicing and translation of viral mRNAs (Courtney et al., 2019; Kong et al., 2020). Defects in NSUN2 activity also reduced the levels of m5C-enhancer RNA (eRNA), a transcriptional activator that, upon methylation, contributes to the metabolic reprogramming of HCV-infected cells (Shlomai et al., 2012).
Another RNA methyltransferase, DNMT2, has been shown to be translocated to stress granules to methylate HIV-1 RNAs (Dev et al., 2017) and to be involved in the fruit fly response to Drosophila C virus (DSV) (Durdevic et al., 2013). NSUN1 was also shown to restrict HIV-1 replication by inducing m5C methylation of TAR RNA (Kong et al., 2020).
N6-methyladenosine
N6-methyladenosine (m6A) plays a role in regulating mRNA stability, splicing, and translation (Shi et al., 2022; Xue et al., 2022). This modification is catalyzed by a complex of proteins, including methyltransferase-like 3 (METTL3), METTL14, and RNA-binding motif protein 15 (RBM15), among others, while eraser enzymes fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) revert m6A marks (Xue et al., 2022).
Several m6A marks have been identified in the genomes of several viruses, potentially serving as a shield to avoid recognition by the immune system and IFN production (Li et al., 2017; Ye et al., 2017; Hesser et al., 2018; Imam et al., 2018; Gokhale et al., 2020; Tsai et al., 2020; Lu et al., 2021; Zhuang et al., 2023). For instance, during vesicular stomatitis virus (VSV) infection, METTL3-mediated m6A modification reduced production of viral dsRNAs to elude RIG-I or MDA5 detection (Qiu et al., 2021). Similarly, rotavirus (RV) infection potentiates m6A modifications on mRNAs by downregulating the levels of the m6A eraser ALKBH5 (Wang et al., 2022). In HBV infection, the viral protein HBx interacted with the METTL3/14 complex to affect the m6A content of viral RNAs, with m6A modification of the HBV epsilon stem-loop of pgRNA preventing its ISG20-mediated degradation (Imam et al., 2018; Kim and Siddiqui, 2021). In this context, 3-deazaadenosine (DAA), an m6A inhibitor, or inactivation of METTL3, hindered IAV replication (Bader et al., 1978; Fischer et al., 1990; Fustin et al., 2013; Courtney et al., 2017).
The cellular m6A machinery can have diverse regulatory roles during viral infections. For instance, the m6A reader YTHDC1 was shown to bind to HIV-1 transcripts in a METTL3-dependent manner to ensure effective splicing and viral production (N’Da Konan et al., 2022). YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) was also suggested to facilitate EBV viral RNA decapping and promote its RNA decay by recruiting RNA degradation complexes (Xia et al., 2021).
A-to-I RNA editing
A-to-I RNA editing is a post-transcriptional modification involving the conversion of adenosine (A) to inosine (I) within RNA transcripts (Pfaller et al., 2021) by a family of enzymes known as ADARs (adenosine deaminases acting on RNA) encoded by three genes in mammals: ADAR1 (ADAR), ADAR2 (ADARB1), and ADAR3 (ADARB2) (Slotkin and Nishikura, 2013; Zhu et al., 2023). A-to-I RNA editing can be found in both viral and host RNAs, shaping diverse aspects of virus-host interaction. Regarding viral RNA, editing can lead to sequence variations that impact viral replication, translation, and immune evasion (Pfaller et al., 2021; Zhu et al., 2023). ADAR1 is believed to be co-opted by several viruses, including measles virus (Cattaneo et al., 1988), hepatitis D virus (HDV) (Poison et al., 1996), HIV (Doria et al., 2009), IAV (de Chassey et al., 2013; Furuse, 2021), DENV (de Chassey et al., 2013), ZIKV (Khrustalev et al., 2017), and SARS-CoV-2 (Di Giorgio et al., 2020), to edit their own viral RNA. For instance, through an ADAR-1 mediated RNA editing event, an UAG stop codon in HDV RNA is recoded to UIG. As the latter is read as UGG, this recoding phenomenon leads to a translation read-through, resulting in the production of a protein involved in virus replication. ADAR1 has been described to edit RNA from HIV (Phuphuakrat et al., 2008) and measles virus (Ward et al., 2011), promoting and restricting their replication, respectively. Additionally, ADAR1 was shown to enhance DENV propagation by facilitating the translation of its non-structural proteins (de Chassey et al., 2013). The roles of A-to-I editing during virus infection has been extensively reviewed by others (Samuel, 2011; Thompson et al., 2021; Vlachogiannis et al., 2021; Song et al., 2022; Zhu et al., 2023).
The host mRNA epitranscriptome: a new player regulating antiviral responses?
The host mRNA modification landscape undergoes substantial alterations during virus infection (Figure 1A). It is likely that viruses manipulate host mRNA modification patterns to rewire gene expression and hence modulate key biological processes undermining viral infections. For instance, it has been noted that m6A methylations are a known mark used by pattern recognition receptors (PRRs), to differentiate between host and foreign/pathogenic RNA. m6A-methylated RNA binds poorly to RIG-I, in contrast to RNA with Ψ which binds with high affinity, with both failing to initiate RIG-I downstream immune activation. In DNA viruses infections, disturbing hnRNPA2B1-FT0 interaction elevated the levels of cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), and interferon gamma inducible protein 16 (IFI16) m6A-modified mRNA to bolster the antiviral response (Wang et al., 2019) (Figure 1A). Conversely, METTL3/14 depletion boosted the IFN-response (Rubio et al., 2018; Winkler et al., 2019; Li et al., 2021), whereas ALKBH5 was shown to demethylate the mRNAs of MAVS, TRAF3, and TRAF5 to limit IFN production (Zheng et al., 2017) (Figure 1A). The YTH N6-Methyladenosine RNA Binding Protein F2 (YTHDF2) reader enzyme binds to methylated RNA, reducing RIG-I binding, blocking RIG-I conformational changes and IFN transcription (Lou et al., 2021). However, the exact mechanism by which this suppression occurs remains mostly unknown (Durbin et al., 2016; Lou et al., 2021).
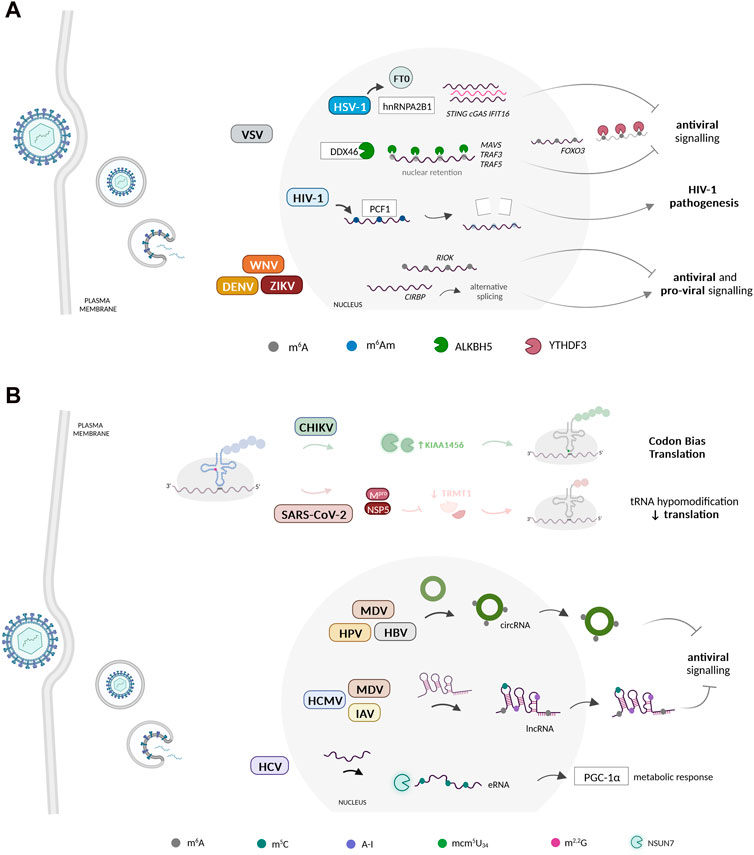
FIGURE 1. Dynamic changes in the host RNA epitranscriptomic landscape during virus infection. (A) Viruses reprogram host mRNA modification patterns to enhance their propagation. Several viruses have been shown to benefit from manipulating the m6A epitranscriptome of several host mRNAs. The host mRNA m6Am methylome was also recently proposed to play important regulatory roles during HIV-1 infection. These modifications are manipulated by viruses to regulate host gene expression during the infectious process. (B) Host non-coding RNAs undergo reprogramming at the epitranscriptome level during virus infection. Several viruses dynamically regulate the host ncRNA epitranscriptome to potentiate their replication, translation, and evasion from host immune surveillance. CHIKV induces a specific reprogramming of the host tRNA epitranscriptome via KIAA1456 and mcm5U34 to facilitate the decoding of its codon-biased transcripts. During SARS-CoV-2 infections, the levels of the tRNA-modifying enzyme TRMT1 and its modifications are significantly reduced. Beyond the tRNA, epitranscriptomic modification of other types of ncRNA molecules, including lncRNAs, circRNAs, and eRNAs, have been shown to play essential regulatory roles during virus infection, namely in the inhibition of the host antiviral response. Created with BioRender.com.
m6A also seems to play a role in regulating the translation of ISGs during antiviral response. Studies have shown that a subset of ISGs, including interferon induced transmembrane protein (IFITM1), appear to have their translation enhanced by m6A and its machinery, in addition to the fact that YTHDF1 also increases the expression of IFITM1 in an m6A-dependent manner (McFadden et al., 2021). Similarly, YTHDF3 hampered ISG expression by enhancing FOXO3 translation, an inhibitor of VSV replication (Zhang et al., 2019) (Figure 1A). Still within VSV infection, a TBK1-METTL3 axis enhanced IRF3 mRNA stability and translation through its m6A modification (Chen et al., 2022). The latter was also shown to regulate IRF7 and TLR9 mRNA stability to amplify the IFN-response to RV and EBV infection, respectively (Zheng et al., 2021; Wang et al., 2022). The m6A content of two specific host transcripts, RIOK3 and CIRBP, was reprogrammed in response to DENV, ZIKV, WNV and HCV. Functionally, m6A gain or loss in RIOK and CIRBP mRNAs facilitated their translation and splicing, respectively, to regulate virus infection (Gokhale et al., 2020) (Figure 1A). Moreover, in response to IFN stimulation, m6A and YTHDF1 were shown to increase ADAR1 levels (Terajima et al., 2021). Knockdown of YTHDF1 reduces the levels of IFN-induced A-to-I RNA editing, leading to the activation of dsRNA-sensing pathways and increasing the expression of various ISGs (Terajima et al., 2021).
Besides the immune response, m6A can regulate other biological processes during viral infections. For example, pseudorabies (PRV) exploits its US3 protein to reduce the m6A methylome as part of the PRV-induced metabolic dysfunction (Jansens et al., 2022; Yu et al., 2023). Reduced m6A on a-ketoglutarate dehydrogenase transcripts also hindered itaconate accumulation and contributed to mRNA decay during VSV infection (Liu et al., 2019). Furthermore, HBV infection altered the levels of m6A of PTEN transcripts and inhibited the IRF3 nuclear export (Kim and Siddiqui, 2021). The m6A methylation of the Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF50 RNA enabled its binding to the m6A reader SND1 for ORF50 transcript stability (Baquero-Perez et al., 2019).
Apart from m6A, HIV-1 infection decreased the amount of N6,2′-O-dimethyladenosine (m6Am) modified host mRNAs by degrading the phosphorylated CTD Interacting Factor 1 (PCIF1), an inhibitor of HIV-1 transcription (Zhang et al., 2021) (Figure 1A). Additionally, in Newcastle disease virus (NDV) infections, expression of two accessory non-structural proteins, V and W, relies on RNA editing (Jadhav et al., 2020). In the case of ZIKV, host-induced RNA editing has a pro-viral effect (Zhou et al., 2019; Zhu et al., 2023). Notably, both isoforms of ADAR1 (p110 and p150) promote ZIKV replication by inhibiting the eukaryotic translation initiation factor 2 a (eIF2α) phosphorylation and IFN during immune responses (Zhou et al., 2019; Zhu et al., 2023).
The non-coding RNA epitranscriptome landscape during viral infections
Viruses have been shown to manipulate and exploit several host ncRNAs to successfully propagate and evade the host immune response (Figure 1B). As previously mentioned, viruses manipulate host gene expression to maximize their replication and elude the antiviral response. Though most studies focus on mRNA, recent findings showcase that viruses also impact host ncRNAs, and even encode their own to perturb host antiviral responses. Next, we explore what is known regarding the importance of the ncRNA epitranscriptome in the context of viral infections.
The role of the host cell tRNA epitranscriptome during viral infections
Viruses heavily rely on the host cell translation machinery, including the host tRNAs, to effectively translate their genomes (Pavon-Eternod et al., 2013; Stern-Ginossar et al., 2019; Nunes et al., 2020). Chemical modification of tRNAs, catalyzed by several tRNA-modifying enzymes, are essential for translation efficiency, namely when occurring within the tRNA anticodon loop region (Nedialkova and Leidel, 2015; Pereira et al., 2018; Tavares et al., 2021). Although the role of the tRNA epitranscriptome in viral infections remains largely unexplored, Chikungunya virus (CHIKV) was shown to induce changes in tRNA mcm5 wobble modification levels, through increased expression of the tRNA modifying enzyme KIAA1456, which facilitated the decoding of CHIKV-biased transcripts (Jungfleisch et al., 2022) (Figure 1B).
tRNALysUUU is used by HIV as a primer for reverse transcription. It has been found that both mcm5s2 wobble modification and ms2t6A modification at position 37 of tRNALysUUU favor its interaction with HIV’s nucleocapsid protein NCp7, which is essential for the primer recognition (Graham et al., 2011). These tRNALys3UUU modifications directly impact NCp7 binding and remodeling of the human anticodon stem and loop domain (hASLLys3), as this protein exhibits a higher affinity for the modified hASLLys3UUU compared to the unmodified human tRNA (Graham et al., 2011). This shows that modifications of htRNALys3UUU play a critical role in determining the recognition by NCp7 before the annealing of tRNALys3UUU to the viral genome as the primer for reverse transcription (Graham et al., 2011).
Reprogramming of tRNA modifications was also observed during Shewanella phage 1/4 infection of the marine bacterium Shewanella glacialimarina (Lampi et al., 2023). Late-infection viral transcripts favoured GUA codons, which correlated with increased queuosine (Q) modification at the wobble position of the corresponding tRNATyrUAC. This suggests a potential correlation between phage codon bias and tRNA modification content (Lampi et al., 2023). On the other hand, TRMT1 and its modifications were reduced in response to SARS-CoV-2 infection, likely due to the activity of specific viral proteases (D’Oliviera et al., 2023; Zhang K. et al., 2023) (Figure 1B). Interestingly, a multiplex small RNA sequencing analysis of nasopharyngeal swabs showed significant variation of tRNA modification patterns among patients with distinct clinical manifestations of COVID-19 (Katanski et al., 2022).
Also, some tRNA modifications have been found in viral RNAs (McIntyre et al., 2018), raising the question of whether tRNA modifying enzymes also play a role in catalyzing viral RNA modifications that may be important for viral replication. Specifically, mcm5s2U modification was found in PV, whereas ncm5U was identified in RNAs of ZIKV, DENV, HCV and PV (McIntyre et al., 2018).
Though a direct link between viral infections and host antiviral responses is still missing, emerging data suggests that tRNA modifications may regulate the immune response. In fact, naïve T cells increase TRMT61A and TRMT6 levels to methylate specific tRNAs during transition to an active state (Liu et al., 2022). At the peak of proliferation, wybutosine and ms2t6a also decrease drastically to promote ribosomal frameshifting (Rak et al., 2021). As loss of wybutosine increases ribosomal frameshifting, this may explain the HIV-1 preference for proliferating T cells (Rak et al., 2021).
It is worth mentioning that tRNAs comprise an abundant source of tRNA-derived small RNAs (tsRNAs) that serve a variety of cellular regulatory functions (Oberbauer and Schaefer, 2018; Liu et al., 2021). Alterations in tRNA pools are often associated with tRNA-derived fragments (tRFs) generation in response to viral infections (Wang et al., 2013; Zhou et al., 2017; Choi et al., 2020). For instance, the tRNA demethylase ALKBH1 mediated the cleavage of tRNA-GluCTC to promote RSV replication (Choi et al., 2022). Then, it is plausible that the tRNA epitranscriptome influences tRF biogenesis during viral infections; host cells may counteract viruses by changing tRF patterns, or these are exploited by viruses to empower their own spread. Interestingly, tRFs also harbor chemical modifications, but in what extent these regulate tRF functions is unclear (Guzzi et al., 2018; Su et al., 2022).
Beyond tRNAs: impact of epitranscriptomic marks on other ncRNA molecules
The influence of epitranscriptomic marks on various ncRNA molecules, besides tRNAs, is becoming a relevant focus of investigation. Studies on miRNAs, lncRNAs and circRNAs have shown that ncRNAs play important roles in the regulation of immune function and the occurrence and development of viral infections. For instance, loss of m5C in the EBV ncRNA 7SL has been shown to enhance its binding to RIG-I and, thus the IFN response (Zhang Y. et al., 2022). Furthermore, during EBV infection, m5C loss increased the levels of another EBV ncRNA, EBV-encoded RNA 1 (EBER1), likely indicating that this ncRNA is an Angiogenin target for m5C-dependent cleavage (Henry et al., 2020). On the other hand, introduction of Ψ into EBER2 enhanced its stability and promoted EBV lytic replication (Henry et al., 2022).
The function of miRNAs during virus infections has gathered significant attention (Girardi et al., 2018; Letafati et al., 2022; Ostrycharz and Hukowska-Szematowicz, 2022). While some viruses encode their own miRNAs, others can either suppress or hijack host miRNAs to disturb host immune-miRNA translation to potentiate propagation (Mishra et al., 2020). Conversely, hosts can also exploit their miRNAs to suppress viral replication. Despite the increasing evidence demonstrating the crosstalk between viruses and miRNAs upon infection, the relevance of the epitranscriptome for some of the observed miRNA alterations is still not known. However, different studies linked RNA modifications to miRNA biogenesis and degradation in cancer and plants, respectively (Yu and Chen, 2010; Shelton et al., 2016; Li et al., 2018; Han et al., 2021; Marceca et al., 2021; Mei et al., 2023). Although there is not enough evidence to back up this hypothesis, it is tempting to speculate that a crosstalk between the epitranscriptome and miRNAs may also occur during viral infections to, for instance, induce the degradation of antiviral miRNAs.
lncRNAs are generated by the host to counteract infection, but viruses themselves can also encode lncRNAs to counteract host activities, thereby regulating host immune responses, viral gene expression, and viral replication (Rossetto and Pari, 2012; Rossetto et al., 2013; Xu J. et al., 2021; Li et al., 2022). Some lncRNAs can act as molecular decoys, sequestering viral proteins or miRNAs, hence preventing their interaction with host factors pivotal for viral replication (Liu and Ding, 2017). Interestingly, the expression and tissue specificity of lncRNAs is regulated by their epitranscriptome (Jacob et al., 2017). Specific HCMV and MDV lncRNAs have been shown to harbor m6A marks to increase stability (Sun et al., 2021b; Lee et al., 2022). The amount of m6A-modified lncRNAs increases with MDV infection and is accompanied by increased expression of METTL14 and ALKBH5 (Sun et al., 2021b). Similarly, an hyperediting of the viral edited repeat-long (ERL) lncRNA occurred during MDV infection to downregulate IFN, particularly at the MDV lytic phase, and correlated with increased ADAR1 activity (Figueroa et al., 2016). Recently, m5C peaks were identified, following IAV infection, in several host lncRNAs, mostly associated with immune recognition and disease pathogenesis, possibly to regulate host responses to IAV by influencing the expression and stability of specific lncRNAs (Jiang et al., 2023) (Figure 1B).
Circular RNAs (circRNAs) have recently emerged as a class of ncRNAs involved in viral infection. Several viruses can alter the circRNA landscape of infected cells to modulate host gene expression (Xie et al., 2021). Notably, viruses have been shown to exploit the cellular machinery to generate viral circRNAs for their own profit, while hosts may also use these molecules to suppress viral replication (Zhang X. et al., 2022). The circRNAs epitranscriptome has also been linked to viral infections. For instance, the m6A methylome of specific circRNAs, allied to ErbB and insulin pathways, was altered by MDV infection and enabled immune surveillance escape (Sun et al., 2021b). The HBV HBx protein upregulates METTL3 to introduce m6A onto circ-ARL3 to assist its reverse splicing and biogenesis (Figure 1B). Ultimately, circ-ARL3 acts as a sponge for miR-1303, counteracting its inhibitory effect on specific oncogenes (Rao et al., 2021). Besides that, the oncogenic human papillomavirus (HPV) was shown to form circRNAs, including the m6A-modified E7 oncogene (circE7), which associated with polysomes and was translated into the E7 oncoprotein (Zhao et al., 2019).
Viruses can also affect the expression and function of other ncRNAs, such as small nucleolar RNAs (snoRNAs) (Stamm and Lodmell, 2019), piwi-interacting RNAs (piRNAs) (Wang et al., 2018; Joosten et al., 2021; Corsello et al., 2022; Wang X. et al., 2023) and eRNAs (Perez et al., 2012), to establish a favorable environment for viral propagation. Although not much is known regarding this altered expression and putative epitranscriptome reprograming, loss of m5C via NSUN7 depletion leads to hypomethylation of eRNAs, a transcriptional coactivator that interacts with proliferator-activated receptor-gamma co-activator 1 alpha (PGC-1α) to modulate cellular metabolic responses. As the metabolic response is reprogrammed during HCV infection, and accompanied by PGC-1α induction, NSUN7 may be involved in this process (Shlomai et al., 2012).
Final remarks
The RNA epitranscriptome plays a critical role in shaping viral infections and host antiviral responses. It affects viral RNA stability, translation, and recognition by the host, hindering the host’s ability to detect viruses and mount a response (Gokhale et al., 2016; Courtney et al., 2017; Imam et al., 2018; Courtney et al., 2019; McFadden et al., 2021). Conversely, host RNA modification reprograming upon infection affects the expression of antiviral genes that suppress viral replication or facilitate viral gene expression (Rubio et al., 2018; Wang et al., 2019; Winkler et al., 2019; Wang H. et al., 2023).
A comprehensive understanding of the enzymes responsible for writing and erasing RNA modifications, as well as the reader proteins that interpret these modifications, presents exciting opportunities for developing more effective antiviral therapies. The development of small-molecule inhibitors, RNA-targeting therapies, and epitranscriptomic editing tools, holds promise in defeating viral resistance to antiviral therapies. While the field of RNA modification-targeted antiviral drugs is still in its early stages, several studies have shown promising results in potential targets. For example, in HSV-1 infection, inhibition of m6A by 3-DAA significantly decreased viral replication (Feng et al., 2022). Conversely, YTHDF1 recognition and destabilization of m6A-modified EBV transcripts, suppresses EBV infection, showing that this enzyme induces RNA deterioration (Xia et al., 2021). Nevertheless, several challenges lie ahead.
To grasp the crosstalk between viruses and the RNA epitranscriptome, a detailed mapping and characterization of RNA modifications during virus infection is required. Additionally, unraveling the crosstalk between different RNA modifications, their modifying enzymes, and their interplay with viruses will deepen our understanding of the complex dynamics at the virus-host cell interface. Further research efforts and technological advancements are crucial for fully harnessing the therapeutical potential of the RNA epitranscriptome.
Author contributions
Conceptualization, DRR, AN, DR, and ARS; writing-original draft preparation, DRR and AN; writing-review and editing, DR and ARS; visualization, DRR; supervision, DR and ARS; project administration, DR and ARS; funding acquisition, DR and ARS. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Portuguese Foundation for Science and Technology (FCT), POCH, FEDER, and COMPETE2020, through the grants SFRH/BD/146703/2019 (DRR), UI/BD/151372/2021 (AN), UIDB/04501/2020 (iBiMED), under the scope of the Operational Program “Competitiveness and internationalization”, in its FEDER/FNR component, and by Centro 2020 program, Portugal 2020. It was furthermore supported by the European Union thought the Horizon 2020 program: H2020-WIDESPREAD-2020-5 ID-952373. DR and ARS are supported by individual CEEC auxiliary research contracts CEECIND/03747/2017 and CEECIND/00284/2018, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amort, T., Soulière, M. F., Wille, A., Jia, X. Y., Fiegl, H., Wörle, H., et al. (2013). Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 10, 1003–1008. doi:10.4161/rna.24454
Arango, D., Sturgill, D., Alhusaini, N., Dillman, A. A., Sweet, T. J., Hanson, G., et al. (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24. doi:10.1016/j.cell.2018.10.030
Bader, J. P., Brown, N. R., Chiang, P. K., and Cantoni, G. L. (1978). 3-Deazaadenosine, an inhibitor of adenosylhomocysteine hydrolase, inhibits reproduction of Rous sarcoma virus and transformation of chick embryo cells. Virology 89, 494–505. doi:10.1016/0042-6822(78)90191-5
Baquero-Perez, B., Antanaviciute, A., Yonchev, I. D., Carr, I. M., Wilson, S. A., and Whitehouse, A. (2019). The Tudor SND1 protein is an m(6)A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. Elife 8, e47261. doi:10.7554/eLife.47261
Boccaletto, P., Stefaniak, F., Ray, A., Cappannini, A., Mukherjee, S., Purta, E., et al. (2022). Modomics: A database of RNA modification pathways 2021 update. Nucleic Acids Res. 50, D231–D235. doi:10.1093/nar/gkab1083
Bohnsack, K. E., Höbartner, C., and Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m⁵C) RNA methyltransferases: Mechanisms, cellular functions, and links to disease. Genes 10, 102. doi:10.3390/genes10020102
Boo, S. H., and Kim, Y. K. (2020). The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 52, 400–408. doi:10.1038/s12276-020-0407-z
Broly, M., Polevoda, B. V., Awayda, K. M., Tong, N., Lentini, J., Besnard, T., et al. (2022). THUMPD1 bi-allelic variants cause loss of tRNA acetylation and a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 109, 587–600. doi:10.1016/j.ajhg.2022.02.001
Cattaneo, R., Schmid, A., Eschle, D., Baczko, K., ter Meulen, V., and Billeter, M. A. (1988). Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55, 255–265. doi:10.1016/0092-8674(88)90048-7
Chang, D. C., Hoang, L. T., Mohamed Naim, A. N., Dong, H., Schreiber, M. J., Hibberd, M. L., et al. (2016). Evasion of early innate immune response by 2’-O-methylation of dengue genomic RNA. Virology 499, 259–266. doi:10.1016/j.virol.2016.09.022
Chen, J., Wei, X., Wang, X., Liu, T., Zhao, Y., Chen, L., et al. (2022). TBK1-METTL3 axis facilitates antiviral immunity. Cell Rep. 38, 110373. doi:10.1016/j.celrep.2022.110373
Chen, X., Liu, S., Goraya, M. U., Maarouf, M., Huang, S., and Chen, J. L. (2018). Host immune response to influenza A virus infection. Front. Immunol. 9, 1–13. doi:10.3389/fimmu.2018.00320
Chen, Y., Su, C., Ke, M., Jin, X., Xu, L., Zhang, Z., et al. (2011). Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2’-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 7, e1002294. doi:10.1371/journal.ppat.1002294
Chen, Y., Tao, J., Sun, Y., Wu, A., Su, C., Gao, G., et al. (2013). Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 87, 6296–6305. doi:10.1128/JVI.00061-13
Choi, E.-J., Wu, W., Zhang, K., Lee, I., Kim, I. H., Lee, Y. S., et al. (2020). ELAC2, an enzyme for tRNA maturation, plays a role in the cleavage of a mature tRNA to produce a tRNA-derived RNA fragment during respiratory syncytial virus infection. Front. Mol. Biosci. 7, 609732. doi:10.3389/fmolb.2020.609732
Choi, E.-J., Wu, W., Zhang, K., Yuan, X., Deng, J., Ismail, D., et al. (2022). Parent tRNA modification status determines the induction of functional tRNA-derived RNA by respiratory syncytial virus infection. Viruses 15, 57. doi:10.3390/v15010057
Chu, C., Das, K., Tyminski, J. R., Bauman, J. D., Guan, R., Qiu, W., et al. (2011). Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. U. S. A. 108, 10104–10108. doi:10.1073/pnas.1106610108
Corsello, T., Kudlicki, A. S., Liu, T., and Casola, A. (2022). Respiratory syncytial virus infection changes the piwi-interacting RNA content of airway epithelial cells. Front. Mol. Biosci. 9, 931354. doi:10.3389/fmolb.2022.931354
Courtney, D. G., Kennedy, E. M., Dumm, R. E., Bogerd, H. P., Tsai, K., Heaton, N. S., et al. (2017). Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22, 377–386.e5. doi:10.1016/j.chom.2017.08.004
Courtney, D. G., Tsai, K., Bogerd, H. P., Kennedy, E. M., Law, B. A., Emery, A., et al. (2019). Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26, 217–227.e6. doi:10.1016/j.chom.2019.07.005
Cristinelli, S., Angelino, P., and Ciuffi, A. (2022). Exploring m<sup>6</sup>A and m<sup>5</sup>C epitranscriptomes upon viral infection: An example with HIV. J. Vis. Exp. 2022. doi:10.3791/62426
Cui, L., Ma, R., Cai, J., Guo, C., Chen, Z., Yao, L., et al. (2022). RNA modifications: Importance in immune cell biology and related diseases. Signal Transduct. Target. Ther. 7, 334. doi:10.1038/s41392-022-01175-9
de Chassey, B., Aublin-Gex, A., Ruggieri, A., Meyniel-Schicklin, L., Pradezynski, F., Davoust, N., et al. (2013). The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 9, e1003440. doi:10.1371/journal.ppat.1003440
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., et al. (2010). 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. doi:10.1038/nature09489
De Vlugt, C., Sikora, D., and Pelchat, M. (2018). Insight into influenza: A virus cap-snatching. Viruses 10, 641. doi:10.3390/v10110641
Decombe, A., El Kazzi, P., and Decroly, E. (2023). Interplay of RNA 2’-O-methylations with viral replication. Curr. Opin. Virol. 59, 101302. doi:10.1016/j.coviro.2023.101302
Dev, R. R., Ganji, R., Singh, S. P., Mahalingam, S., Banerjee, S., and Khosla, S. (2017). Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem. J. 474, 2009–2026. doi:10.1042/BCJ20170258
Devarkar, S. C., Wang, C., Miller, M. T., Ramanathan, A., Jiang, F., Khan, A. G., et al. (2016). Structural basis for m7G recognition and 2’-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U. S. A. 113, 596–601. doi:10.1073/pnas.1515152113
Di Giorgio, S., Martignano, F., Torcia, M. G., Mattiuz, G., and Conticello, S. G. (2020). Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 6, 5813–6428. doi:10.1126/sciadv.abb5813
D’Oliviera, A., Dai, X., Mottaghinia, S., Geissler, E. P., Etienne, L., Zhang, Y., et al. (2023). Recognition and cleavage of human tRNA methyltransferase TRMT1 by the SARS-CoV-2 main protease. bioRxiv 20, 529306. doi:10.1101/2023.02.20.529306
Doria, M., Neri, F., Gallo, A., Farace, M. G., and Michienzi, A. (2009). Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 37, 5848–5858. doi:10.1093/nar/gkp604
Durbin, A. F., Wang, C., Marcotrigiano, J., and Gehrke, L. (2016). RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. MBio 7, e00833. doi:10.1128/mBio.00833-16
Durdevic, Z., Mobin, M. B., Hanna, K., Lyko, F., and Schaefer, M. (2013). The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 4, 931–937. doi:10.1016/j.celrep.2013.07.046
Eckwahl, M., Xu, R., Michalkiewicz, J., Zhang, W., Patel, P., Cai, Z., et al. (2020). 5-Methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J. Virol. 94, e00544. doi:10.1128/JVI.00544-20
El Kazzi, P., Rabah, N., Chamontin, C., Poulain, L., Ferron, F., Debart, F., et al. (2023). Internal RNA 2’O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res. 51, 2501–2515. doi:10.1093/nar/gkac996
Feng, Z., Zhou, F., Tan, M., Wang, T., Chen, Y., Xu, W., et al. (2022). Targeting m6A modification inhibits herpes virus 1 infection. Genes Dis. 9, 1114–1128. doi:10.1016/j.gendis.2021.02.004
Figueroa, T., Boumart, I., Coupeau, D., and Rasschaert, D. (2016). Hyperediting by ADAR1 of a new herpesvirus lncRNA during the lytic phase of the oncogenic Marek’s disease virus. J. Gen. Virol. 97, 2973–2988. doi:10.1099/jgv.0.000606
Fischer, A. A., Müller, K., and Scholtissek, C. (1990). Specific inhibition of the synthesis of influenza virus late proteins and stimulation of early, M2, and NS2 protein synthesis by 3-deazaadenosine. Virology 177, 523–531. doi:10.1016/0042-6822(90)90517-u
Furuse, Y. (2021). RNA modifications in genomic RNA of influenza A virus and the relationship between RNA modifications and viral infection. Int. J. Mol. Sci. 22, 9127. doi:10.3390/ijms22179127
Fustin, J.-M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. doi:10.1016/j.cell.2013.10.026
Ge, Y., Liu, T., Wang, C., Zhang, Y., Xu, S., Ren, Y., et al. (2021). N6-methyladenosine RNA modification and its interaction with regulatory non-coding RNAs in colorectal cancer. RNA Biol. 18, 551–561. doi:10.1080/15476286.2021.1974749
Gebauer, F., and Hentze, M. W. (2004). Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835. doi:10.1038/nrm1488
Girardi, E., López, P., and Pfeffer, S. (2018). On the importance of host MicroRNAs during viral infection. Front. Genet. 9, 439. doi:10.3389/fgene.2018.00439
Gokhale, N. S., McIntyre, A. B. R., Mattocks, M. D., Holley, C. L., Lazear, H. M., Mason, C. E., et al. (2020). Altered m(6)A modification of specific cellular transcripts affects flaviviridae infection. Mol. Cell 77, 542–555.e8. doi:10.1016/j.molcel.2019.11.007
Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A., et al. (2016). N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665. doi:10.1016/j.chom.2016.09.015
Graham, W. D., Barley-Maloney, L., Stark, C. J., Kaur, A., Stolarchuk, C., Sproat, B., et al. (2011). Functional recognition of the modified human tRNALys3UUU anticodon domain by HIV’s nucleocapsid protein and a peptide mimic. J. Mol. Biol. 410, 698–715. doi:10.1016/j.jmb.2011.04.025
Gu, W., Gallagher, G. R., Dai, W., Liu, P., Li, R., Trombly, M. I., et al. (2015). Influenza A virus preferentially snatches noncoding RNA caps. RNA 21, 2067–2075. doi:10.1261/rna.054221.115
Guzzi, N., Cieśla, M., Ngoc, P. C. T., Lang, S., Arora, S., Dimitriou, M., et al. (2018). Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26. doi:10.1016/j.cell.2018.03.008
Han, X., Guo, J., and Fan, Z. (2021). Interactions between m6A modification and miRNAs in malignant tumors. Cell Death Dis. 12, 598. doi:10.1038/s41419-021-03868-5
Hao, H., Liu, W., Miao, Y., Ma, L., Yu, B., Liu, L., et al. (2022). N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. Nucleic Acids Res. 50, 9339–9354. doi:10.1093/nar/gkac675
Henry, B. A., Kanarek, J. P., Kotter, A., Helm, M., and Lee, N. (2020). 5-methylcytosine modification of an Epstein-Barr virus noncoding RNA decreases its stability. RNA 26, 1038–1048. doi:10.1261/rna.075275.120
Henry, B. A., Marchand, V., Schlegel, B. T., Helm, M., Motorin, Y., and Lee, N. (2022). Pseudouridylation of Epstein-Barr virus noncoding RNA EBER2 facilitates lytic replication. RNA 28, 1542–1552. doi:10.1261/rna.079219.122
Hesser, C. R., Karijolich, J., Dominissini, D., He, C., and Glaunsinger, B. A. (2018). N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 14, e1006995. doi:10.1371/journal.ppat.1006995
Huang, H., Weng, H., Deng, X., and Chen, J. (2020). RNA modifications in cancer: Functions, mechanisms, and therapeutic implications. Annu. Rev. Cancer Biol. 4, 221–240. doi:10.1146/annurev-cancerbio-030419-033357
Hyde, J. L., and Diamond, M. S. (2015). Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation. Virology 479–480, 66–74. doi:10.1016/j.virol.2015.01.019
Imam, H., Khan, M., Gokhale, N. S., McIntyre, A. B. R., Kim, G. W., Jang, J. Y., et al. (2018). N6-methyladenosine modification of Hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U. S. A. 115, 8829–8834. doi:10.1073/pnas.1808319115
Jacob, R., Zander, S., and Gutschner, T. (2017). The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 18, 2387. doi:10.3390/ijms18112387
Jadhav, A., Zhao, L., Ledda, A., Liu, W., Ding, C., Nair, V., et al. (2020). Patterns of RNA editing in Newcastle disease virus infections. Viruses 12, 1249. doi:10.3390/v12111249
Jansens, R. J. J., Verhamme, R., Mirza, A. H., Olarerin-George, A., Van Waesberghe, C., Jaffrey, S. R., et al. (2022). Alphaherpesvirus US3 protein-mediated inhibition of the m6A mRNA methyltransferase complex. Cell Rep. 40:111107. doi:10.1016/j.celrep.2022.111107
Jiang, S., Hu, J., Bai, Y., Hao, R., Liu, L., and Chen, H. (2023). Transcriptome-wide 5-methylcytosine modification profiling of long non-coding RNAs in A549 cells infected with H1N1 influenza A virus. BMC Genomics 24, 316. doi:10.1186/s12864-023-09432-z
Joosten, J., Overheul, G. J., Van Rij, R. P., and Miesen, P. (2021). Endogenous piRNA-guided slicing triggers responder and trailer piRNA production from viral RNA in Aedes aegypti mosquitoes. Nucleic Acids Res. 49, 8886–8899. doi:10.1093/nar/gkab640
Jungfleisch, J., Böttcher, R., Talló-Parra, M., Pérez-Vilaró, G., Merits, A., Novoa, E. M., et al. (2022). CHIKV infection reprograms codon optimality to favor viral RNA translation by altering the tRNA epitranscriptome. Nat. Commun. 13, 4725. doi:10.1038/s41467-022-31835-x
Karijolich, J., Yi, C., and Yu, Y.-T. (2015). Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 16, 581–585. doi:10.1038/nrm4040
Karlebach, G., Aronow, B., Baylin, S. B., Butler, D., Foox, J., Levy, S., et al. (2022). Betacoronavirus-specific alternate splicing. Genomics 114, 110270. doi:10.1016/j.ygeno.2022.110270
Katanski, C. D., Alshammary, H., Watkins, C. P., Huang, S., Gonzales-Reiche, A., Sordillo, E. M., et al. (2022). tRNA abundance, modification and fragmentation in nasopharyngeal swabs as biomarkers for COVID-19 severity. Front. Cell Dev. Biol. 10, 999351. doi:10.3389/fcell.2022.999351
Khrustalev, V. V., Khrustaleva, T. A., Sharma, N., and Giri, R. (2017). Mutational pressure in Zika virus: Local ADAR-editing areas associated with pauses in translation and replication. Front. Cell. Infect. Microbiol. 7, 44–17. doi:10.3389/fcimb.2017.00044
Kim, G.-W., and Siddiqui, A. (2021). Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc. Natl. Acad. Sci. U. S. A. 118, e2019455118. doi:10.1073/pnas.2019455118
Kong, W., Biswas, A., Zhou, D., Fiches, G., Fujinaga, K., Santoso, N., et al. (2020). Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog. 16, e1008430. doi:10.1371/journal.ppat.1008430
Kreijtz, J. H. C. M., Fouchier, R. A. M., and Rimmelzwaan, G. F. (2011). Immune responses to influenza virus infection. Virus Res. 162, 19–30. doi:10.1016/j.virusres.2011.09.022
Kumar, P., Sweeney, T. R., Skabkin, M. A., Skabkina, O. V., Hellen, C. U. T., and Pestova, T. V. (2014). Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5’-terminal regions of cap0-cap1- and 5’ppp-mRNAs. Nucleic Acids Res. 42, 3228–3245. doi:10.1093/nar/gkt1321
Lampi, M., Gregorova, P., Qasim, M. S., Ahlblad, N. C. V., and Sarin, L. P. (2023). Bacteriophage infection of the marine bacterium Shewanella glacialimarina induces dynamic changes in tRNA modifications. Microorganisms 11, 355. doi:10.3390/microorganisms11020355
Lee, S., Kim, H., Hong, A., Song, J., and Kim, M. (2022). Functional and molecular dissection of HCMV long non-coding RNAs. Sci. Rep. 12, 19303. doi:10.1038/s41598-022-23317-3
Letafati, A., Najafi, S., Mottahedi, M., Karimzadeh, M., Shahini, A., Garousi, S., et al. (2022). MicroRNA let-7 and viral infections: Focus on mechanisms of action. Cell. Mol. Biol. Lett. 27, 14. doi:10.1186/s11658-022-00317-9
Li, H.-B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017). m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342. doi:10.1038/nature23450
Li, L., Song, Y., Shi, X., Liu, J., Xiong, S., Chen, W., et al. (2018). The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 28, 132–143. doi:10.1101/gr.224386.117
Li, N., Hui, H., Bray, B., Gonzalez, G. M., Zeller, M., Anderson, K. G., et al. (2021). METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 35, 109091. doi:10.1016/j.celrep.2021.109091
Li, Z., Gao, J., Xiang, X., Deng, J., Gao, D., and Sheng, X. (2022). Viral long non-coding RNA regulates virus life-cycle and pathogenicity. Mol. Biol. Rep. 49, 6693–6700. doi:10.1007/s11033-022-07268-6
Liu, B., Cao, J., Wang, X., Guo, C., Liu, Y., and Wang, T. (2021). Deciphering the tRNA-derived small RNAs: Origin, development, and future. Cell Death Dis. 13, 24. doi:10.1038/s41419-021-04472-3
Liu, W., and Ding, C. (2017). Roles of LncRNAs in viral infections. Front. Cell. Infect. Microbiol. 7, 205. doi:10.3389/fcimb.2017.00205
Liu, Y., You, Y., Lu, Z., Yang, J., and Liu, L. (2019). N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176. doi:10.1126/science.aax4468
Liu, Y., Zhou, J., Li, X., Zhang, X., Shi, J., Wang, X., et al. (2022). tRNA-m1A modification promotes T cell expansion via efficient MYC protein synthesis. Nat. Immunol. 23, 1433–1444. doi:10.1038/s41590-022-01301-3
Lou, X., Wang, J.-J., Wei, Y.-Q., and Sun, J.-J. (2021). Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis. 12, 300. doi:10.1038/s41419-021-03585-z
Lu, M., Xue, M., Wang, H. T., Kairis, E. L., Ahmad, S., Wei, J., et al. (2021). Nonsegmented negative-sense RNA viruses utilize N(6)-methyladenosine (m(6)A) as a common strategy to evade host innate immunity. J. Virol. 95, e01939. doi:10.1128/JVI.01939-20
Marceau, C. D., Puschnik, A. S., Majzoub, K., Ooi, Y. S., Brewer, S. M., Fuchs, G., et al. (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163. doi:10.1038/nature18631
Marceca, G. P., Distefano, R., Tomasello, L., Lagana, A., Russo, F., Calore, F., et al. (2021). MiREDiBase, a manually curated database of validated and putative editing events in microRNAs. Sci. data 8, 199. doi:10.1038/s41597-021-00979-8
Martinez Campos, C., Tsai, K., Courtney, D. G., Bogerd, H. P., Holley, C. L., and Cullen, B. R. (2021). Mapping of pseudouridine residues on cellular and viral transcripts using a novel antibody-based technique. RNA 27, 1400–1411. doi:10.1261/rna.078940.121
McFadden, M. J., McIntyre, A. B. R., Mourelatos, H., Abell, N. S., Gokhale, N. S., Ipas, H., et al. (2021). Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep. 34, 108798. doi:10.1016/j.celrep.2021.108798
McIntyre, W., Netzband, R., Bonenfant, G., Biegel, J. M., Miller, C., Fuchs, G., et al. (2018). Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 46, 5776–5791. doi:10.1093/nar/gky029
Mei, Z., Mou, Y., Zhang, N., Liu, X., He, Z., and Gu, S. (2023). Emerging mutual regulatory roles between m6A modification and microRNAs. Int. J. Mol. Sci. 24, 773. doi:10.3390/ijms24010773
Menachery, V. D., Yount, B. L., Josset, L., Gralinski, L. E., Scobey, T., Agnihothram, S., et al. (2014). Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 88, 4251–4264. doi:10.1128/JVI.03571-13
Mersinoglu, B., Cristinelli, S., and Ciuffi, A. (2022). The impact of epitranscriptomics on antiviral innate immunity. Viruses 14, 1666. doi:10.3390/v14081666
Mishra, R., Kumar, A., Ingle, H., and Kumar, H. (2020). The interplay between viral-derived miRNAs and host immunity during infection. Front. Immunol. 10, 3079. doi:10.3389/fimmu.2019.03079
N’Da Konan, S., Ségéral, E., Bejjani, F., Bendoumou, M., Ait Said, M., Gallois-Montbrun, S., et al. (2022). YTHDC1 regulates distinct post-integration steps of HIV-1 replication and is important for viral infectivity. Retrovirology 19, 4. doi:10.1186/s12977-022-00589-1
Nedialkova, D. D., and Leidel, S. A. (2015). Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618. doi:10.1016/j.cell.2015.05.022
Nunes, A., Ribeiro, D. R., Marques, M., Santos, M. A. S., Ribeiro, D., and Soares, A. R. (2020). Emerging roles of tRNAs in RNA virus infections. Trends biochem. Sci. 45, 794–805. doi:10.1016/j.tibs.2020.05.007
Oberbauer, V., and Schaefer, M. R. (2018). tRNA-derived small RNAs: Biogenesis, modification, function and potential impact on human disease development. Genes (Basel). 9, 607. doi:10.3390/genes9120607
Ostrycharz, E., and Hukowska-Szematowicz, B. (2022). Micro-players of great significance—host microRNA signature in viral infections in humans and animals. Int. J. Mol. Sci. 23, 10536. doi:10.3390/ijms231810536
Pavon-Eternod, M., David, A., Dittmar, K., Berglund, P., Pan, T., Bennink, J. R., et al. (2013). Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic Acids Res. 41, 1914–1921. doi:10.1093/nar/gks986
Pereira, M., Francisco, S., Varanda, A. S., Santos, M., Santos, M. A. S., and Soares, A. R. (2018). Impact of tRNA modifications and tRNA-modifying enzymes on proteostasis and human disease. Int. J. Mol. Sci. 19, 3738. doi:10.3390/ijms19123738
Perez, J. T., Zlatev, I., Aggarwal, S., Subramanian, S., Sachidanandam, R., Kim, B., et al. (2012). A small-RNA enhancer of viral polymerase activity. J. Virol. 86, 13475–13485. doi:10.1128/JVI.02295-12
Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I., et al. (2001). Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 98, 7029–7036. doi:10.1073/pnas.111145798
Pfaller, C. K., George, C. X., and Samuel, C. E. (2021). Adenosine deaminases acting on RNA (ADARs) and viral infections. Annu. Rev. Virol. 8, 239–264. doi:10.1146/annurev-virology-091919-065320
Phuphuakrat, A., Kraiwong, R., Boonarkart, C., Lauhakirti, D., Lee, T. H., and Auewarakul, P. (2008). Double-Stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J. Virol. 82, 10864–10872. doi:10.1128/JVI.00238-08
Poison, A. G., Bass, B. L., and Casey, J. L. (1996). RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380, 454–456. doi:10.1038/380454a0
Qiu, W., Zhang, Q., Zhang, R., Lu, Y., Wang, X., Tian, H., et al. (2021). N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 12, 1582. doi:10.1038/s41467-021-21904-y
Rak, R., Polonsky, M., Eizenberg-Magar, I., Mo, Y., Sakaguchi, Y., Mizrahi, O., et al. (2021). Dynamic changes in tRNA modifications and abundance during T cell activation. Proc. Natl. Acad. Sci. U. S. A. 118, e2106556118. doi:10.1073/pnas.2106556118
Ramanathan, A., Robb, G. B., and Chan, S.-H. (2016). mRNA capping: Biological functions and applications. Nucleic Acids Res. 44, 7511–7526. doi:10.1093/nar/gkw551
Rao, X., Lai, L., Li, X., Wang, L., and Yang, Q. (2021). N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life 73, 408–417. doi:10.1002/iub.2438
Ringeard, M., Marchand, V., Decroly, E., Motorin, Y., and Bennasser, Y. (2019). FTSJ3 is an RNA 2’-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504. doi:10.1038/s41586-018-0841-4
Rossetto, C. C., and Pari, G. (2012). KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 8, e1002680. doi:10.1371/journal.ppat.1002680
Rossetto, C. C., Tarrant-Elorza, M., Verma, S., Purushothaman, P., and Pari, G. S. (2013). Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 87, 5540–5553. doi:10.1128/JVI.03111-12
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L., and Mohr, I. (2018). RNA m(6) A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 32, 1472–1484. doi:10.1101/gad.319475.118
Sacco, M. T., and Horner, S. M. (2021). Flipping the script: Viral capitalization of RNA modifications. Brief. Funct. Genomics 20, 86–93. doi:10.1093/bfgp/elaa025
Samuel, C. E. (2011). Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411, 180–193. doi:10.1016/j.virol.2010.12.004
Schlee, M., and Hartmann, G. (2016). Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580. doi:10.1038/nri.2016.78
Sharma, S., Langhendries, J. L., Watzinger, P., Kötter, P., Entian, K. D., and Lafontaine, D. L. J. (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 43, 2242–2258. doi:10.1093/nar/gkv075
Shelton, S. B., Reinsborough, C., and Xhemalce, B. (2016). Who watches the watchmen: Roles of RNA modifications in the RNA interference pathway. PLoS Genet. 12, 6139–6210. doi:10.1371/journal.pgen.1006139
Shi, H., Xu, Y., Tian, N., Yang, M., and Liang, F.-S. (2022). Inducible and reversible RNA N6-methyladenosine editing. Nat. Commun. 13, 1958. doi:10.1038/s41467-022-29665-y
Shlomai, A., Rechtman, M. M., Burdelova, E. O., Zilberberg, A., Hoffman, S., Solar, I., et al. (2012). The metabolic regulator PGC-1α links hepatitis C virus infection to hepatic insulin resistance. J. Hepatol. 57, 867–873. doi:10.1016/j.jhep.2012.06.021
Slotkin, W., and Nishikura, K. (2013). Adenosine-to-inosine RNA editing and human disease. Genome Med. 5, 105. doi:10.1186/gm508
Song, B., Shiromoto, Y., Minakuchi, M., and Nishikura, K. (2022). The role of RNA editing enzyme ADAR1 in human disease. WIREs RNA 13, 16655–e1735. doi:10.1002/wrna.1665
Sorokin, I. I., Vassilenko, K. S., Terenin, I. M., Kalinina, N. O., Agol, V. I., and Dmitriev, S. E. (2021). Non-canonical translation initiation mechanisms employed by eukaryotic viral mRNAs. Biochem. (Mosc). 86, 1060–1094. doi:10.1134/S0006297921090042
Squires, J. E., Patel, H. R., Nousch, M., Sibbritt, T., Humphreys, D. T., Parker, B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033. doi:10.1093/nar/gks144
Stamm, S., and Lodmell, J. S. (2019). C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks. Non-coding RNA Res. 4, 46–53. doi:10.1016/j.ncrna.2019.02.001
Stern-Ginossar, N., Thompson, S. R., Mathews, M. B., and Mohr, I. (2019). Translational control in virus-infected cells. Cold Spring Harb. Perspect. Biol. 11, a033001. doi:10.1101/cshperspect.a033001
Su, Z., Monshaugen, I., Wilson, B., Wang, F., Klungland, A., Ougland, R., et al. (2022). TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat. Commun. 13, 2165. doi:10.1038/s41467-022-29790-8
Sun, A., Wang, R., Yang, S., Zhu, X., Liu, Y., Teng, M., et al. (2021a). Comprehensive profiling analysis of the N6-methyladenosine-modified circular RNA transcriptome in cultured cells infected with Marek’s disease virus. Sci. Rep. 11, 11084. doi:10.1038/s41598-021-90548-1
Sun, A., Zhu, X., Liu, Y., Wang, R., Yang, S., Teng, M., et al. (2021b). Transcriptome-wide N6-methyladenosine modification profiling of long non-coding RNAs during replication of Marek’s disease virus in vitro. BMC Genomics 22, 296. doi:10.1186/s12864-021-07619-w
Tavares, J. F., Davis, N. K., Poim, A., Reis, A., Kellner, S., Sousa, I., et al. (2021). tRNA-modifying enzyme mutations induce codon-specific mistranslation and protein aggregation in yeast. RNA Biol. 18, 563–575. doi:10.1080/15476286.2020.1819671
Terajima, H., Lu, M., Zhang, L., Cui, Q., Shi, Y., Li, J., et al. (2021). N6-methyladenosine promotes induction of ADAR1-mediated A-to-I RNA editing to suppress aberrant antiviral innate immune responses. PLOS Biol. 19, e3001292. doi:10.1371/journal.pbio.3001292
Thomas, G., Gordon, J., and Rogg, H. (1978). N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J. Biol. Chem. 253, 1101–1105. doi:10.1016/s0021-9258(17)38117-6
Thompson, M. G., Sacco, M. T., and Horner, S. M. (2021). How RNA modifications regulate the antiviral response. Immunol. Rev. 304, 169–180. doi:10.1111/imr.13020
Tsai, K., Jaguva Vasudevan, A. A., Martinez Campos, C., Emery, A., Swanstrom, R., and Cullen, B. R. (2020). Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28, 306–312.e6. doi:10.1016/j.chom.2020.05.011
Varela, M., Diaz-Rosales, P., Pereiro, P., Forn-Cuní, G., Costa, M. M., Dios, S., et al. (2014). Interferon-induced genes of the expanded IFIT family show conserved antiviral activities in non-mammalian species. PLoS One 9, e100015. doi:10.1371/journal.pone.0100015
Vlachogiannis, N. I., Verrou, K., Stellos, K., Sfikakis, P. P., and Paraskevis, D. (2021). The role of A-to-I RNA editing in infections by RNA viruses: Possible implications for SARS-CoV-2 infection. Clin. Immunol. 226, 108699. doi:10.1016/j.clim.2021.108699
Wang, A., Tao, W., Tong, J., Gao, J., Wang, J., Hou, G., et al. (2022). m6A modifications regulate intestinal immunity and rotavirus infection. Elife 11, e73628. doi:10.7554/eLife.73628
Wang, H., Feng, J., Zeng, C., Liu, J., Fu, Z., Wang, D., et al. (2023b). NSUN2-mediated M(5)c methylation of IRF3 mRNA negatively regulates type I interferon responses during various viral infections. Emerg. Microbes Infect. 12, 2178238. doi:10.1080/22221751.2023.2178238
Wang, L., Wen, M., and Cao, X. (2019). Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Sci. (80-. ) 365, 0758–818. doi:10.1126/science.aav0758
Wang, Q., Lee, I., Ren, J., Ajay, S. S., Lee, Y. S., and Bao, X. (2013). Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 21, 368–379. doi:10.1038/mt.2012.237
Wang, X., Huang, P., Lei, M., Ma, Y., Chen, H., Sun, J., et al. (2023a). Global expression and functional analysis of human piRNAs during HSV-1 infection. Virus Res. 328, 199087. doi:10.1016/j.virusres.2023.199087
Wang, Y., Jin, B., Liu, P., Li, J., Chen, X., and Gu, J. (2018). piRNA profiling of dengue virus type 2-infected asian tiger mosquito and midgut tissues. Viruses 10, 213. doi:10.3390/v10040213
Ward, S. V., George, C. X., Welch, M. J., Liou, L. Y., Hahm, B., Lewicki, H., et al. (2011). RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. 108, 331–336. doi:10.1073/pnas.1017241108
Wilkinson, E., Cui, Y. H., and He, Y. Y. (2022). Roles of RNA modifications in diverse cellular functions. Front. Cell Dev. Biol. 10, 828683–828726. doi:10.3389/fcell.2022.828683
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019). m(6 A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182. doi:10.1038/s41590-018-0275-z
Xia, T.-L., Li, X., Wang, X., Zhu, Y. J., Zhang, H., Cheng, W., et al. (2021). N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep. 22, e50128. doi:10.15252/embr.202050128
Xie, H., Sun, H., Mu, R., Li, S., Li, Y., Yang, C., et al. (2021). The role of circular RNAs in viral infection and related diseases. Virus Res. 291, 198205. doi:10.1016/j.virusres.2020.198205
Xie, L., Zhong, X., Cao, W., Liu, J., Zu, X., and Chen, L. (2023). Mechanisms of NAT10 as ac4C writer in diseases. Mol. Ther. Nucleic Acids 32, 359–368. doi:10.1016/j.omtn.2023.03.023
Xu, J., Wang, P., Li, Z., Li, Z., Han, D., Wen, M., et al. (2021b). IRF3-binding lncRNA-ISIR strengthens interferon production in viral infection and autoinflammation. Cell Rep. 37, 109926. doi:10.1016/j.celrep.2021.109926
Xu, L., Zhang, C., Yin, H., Gong, S., Wu, N., Ren, Z., et al. (2021a). RNA modifications act as regulators of cell death. RNA Biol. 18, 2183–2193. doi:10.1080/15476286.2021.1925460
Xue, C., Chu, Q., Zheng, Q., Jiang, S., Bao, Z., Su, Y., et al. (2022). Role of main RNA modifications in cancer: N6-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct. Target. Ther. 7, 142. doi:10.1038/s41392-022-01003-0
Ye, F., Chen, E. R., and Nilsen, T. W. (2017). Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N(6)-adenosine methylation to promote lytic replication. J. Virol. 91, e00466. doi:10.1128/JVI.00466-17
Yu, B., and Chen, X. (2010). Analysis of miRNA modifications. Methods Mol. Biol. 592, 137–148. doi:10.1007/978-1-60327-005-2_10
Yu, P.-L., Wu, R., Cao, S. J., Wen, Y. P., Huang, X. B., Zhao, S., et al. (2023). Pseudorabies virus exploits N(6)-methyladenosine modification to promote viral replication. Front. Microbiol. 14, 1087484. doi:10.3389/fmicb.2023.1087484
Zhang, K., Ciesla, J. H., Eldin, P., Briant, L., Lentini, J. M., Ramos, J., et al. (2023b). Proteolytic cleavage and inactivation of the TRMT1 tRNA modification enzyme by SARS-CoV-2 main protease. bioRxiv 10, 527147. doi:10.1101/2023.02.10.527147
Zhang, Q., Kang, Y., Wang, S., Gonzalez, G. M., Li, W., Hui, H., et al. (2021). HIV reprograms host m(6)Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nat. Commun. 12, 5543. doi:10.1038/s41467-021-25683-4
Zhang, X., Liang, Z., Wang, C., Shen, Z., Sun, S., Gong, C., et al. (2022b). Viral circular RNAs and their possible roles in virus-host interaction. Front. Immunol. 13, 939768. doi:10.3389/fimmu.2022.939768
Zhang, Y., Jiang, J., Ma, J., Wei, Z., Wang, Y., Song, B., et al. (2023a). DirectRMDB: A database of post-transcriptional RNA modifications unveiled from direct RNA sequencing technology. Nucleic Acids Res. 51, D106–D116. doi:10.1093/nar/gkac1061
Zhang, Y., Wang, X., Zhang, X., Wang, J., Ma, Y., Zhang, L., et al. (2019). RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U. S. A. 116, 976–981. doi:10.1073/pnas.1812536116
Zhang, Y., Zhang, L. S., Dai, Q., Chen, P., Lu, M., Kairis, E. L., et al. (2022a). 5-methylcytosine (m(5)C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. Proc. Natl. Acad. Sci. U. S. A. 119, e2123338119. doi:10.1073/pnas.2123338119
Zhao, J., Lee, E. E., Kim, J., Yang, R., Chamseddin, B., Ni, C., et al. (2019). Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10, 2300. doi:10.1038/s41467-019-10246-5
Zheng, Q., Hou, J., Zhou, Y., Li, Z., and Cao, X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103. doi:10.1038/ni.3830
Zheng, X., Wang, J., Zhang, X., Fu, Y., Peng, Q., Lu, J., et al. (2021). RNA m(6) A methylation regulates virus-host interaction and EBNA2 expression during Epstein-Barr virus infection. Immun. Inflamm. Dis. 9, 351–362. doi:10.1002/iid3.396
Zhou, J., Liu, S., Chen, Y., Fu, Y., Silver, A. J., Hill, M. S., et al. (2017). Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J. Gen. Virol. 98, 1600–1610. doi:10.1099/jgv.0.000852
Zhou, S., Yang, C., Zhao, F., Huang, Y., Lin, Y., Huang, C., et al. (2019). Double-stranded RNA deaminase ADAR1 promotes the Zika virus replication by inhibiting the activation of protein kinase PKR. J. Biol. Chem. 294, 18168–18180. doi:10.1074/jbc.RA119.009113
Zhu, T., Tang, M., Gao, M., Bi, X., Cao, J., Che, H., et al. (2023). Recent progress in atmospheric chemistry research in China: Establishing a theoretical framework for the "air pollution complex". Biol. Direct 18, 1–23. doi:10.1007/s00376-023-2379-0
Zhuang, G., Zhao, X., Jin, J., Zhu, X., Wang, R., Zhai, Y., et al. (2023). Infection phase-dependent dynamics of the viral and host N6-methyladenosine epitranscriptome in the lifecycle of an oncogenic virus in vivo. J. Med. Virol. 95, e28324. doi:10.1002/jmv.28324
Keywords: RNA epitranscriptome, virus-host cell interactions, host antiviral response, coding-RNAs, non-coding RNAs
Citation: Ribeiro DR, Nunes A, Ribeiro D and Soares AR (2023) The hidden RNA code: implications of the RNA epitranscriptome in the context of viral infections. Front. Genet. 14:1245683. doi: 10.3389/fgene.2023.1245683
Received: 23 June 2023; Accepted: 19 July 2023;
Published: 01 August 2023.
Edited by:
Margarida Gama-Carvalho, University of Lisbon, PortugalReviewed by:
Jaunius Urbonavičius, Vilnius Gediminas Technical University, LithuaniaCopyright © 2023 Ribeiro, Nunes, Ribeiro and Soares. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Raquel Soares, YW5hLnIuc29hcmVzQHVhLnB0
†These authors share first authorship
‡These authors share senior authorship
 Diana Roberta Ribeiro†
Diana Roberta Ribeiro† Daniela Ribeiro
Daniela Ribeiro Ana Raquel Soares
Ana Raquel Soares