
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 19 September 2023
Sec. ELSI in Science and Genetics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1233338
This article is part of the Research Topic Bioinformatics and computational approaches for precision medicine implementation View all 5 articles
Personalized medicine has been identified as a powerful tool for addressing the myriad of health issues facing different health systems globally. Although recent studies have expanded our understanding of how different factors such as genetics and the environment play significant roles in affecting the health of individuals, there are still several other issues affecting their translation into personalizing health interventions globally. Since African populations have demonstrated huge genetic diversity, there is a significant need to apply the concepts of personalized medicine to overcome various African-specific health challenges. Thus, we review the current state, progress, and challenges facing the adoption of personalized medicine in Africa with a view to providing insights to critical stakeholders on the right approach to deploy.
All over the world today, there is focused attention on personalized medicine as a way of revolutionizing healthcare delivery. Personalized medicine is a revolutionary form of medicine that utilizes an individual’s genetic, proteomic, and environmental information in the prevention, diagnosis, monitoring and treatment of diseases (McGonigle, 2016). It holds the potential of providing tailor-made medical interventions which maximize health benefits and minimize treatments’ side effects (McGonigle, 2016). This approach to preventing and/or treating diseases is hinged on taking each individual’s peculiarities into account (Collins and Varmus, 2015). The scientific basis underlying the concept of personalized medicine is that individual-level genomic information can be investigated and used significantly in clinical usage as well as in providing strategies for public health policies, See Figure 1. The concept underlying individualized healthcare has become a source of great hope to treat many complex diseases, including cancer (Ghoorah et al., 2021; Hasanzad et al., 2021). This concept might help Africans in overcoming many diseases that are related to the African continent. For example, studies have revealed that Africans have a seemingly high risk of developing chronic kidney disease (CKD) at an early age and with a faster progression toward kidney failure (George et al., 2021). Thus, personalized medicine strategies may be adopted to mitigate this and several other health conditions.
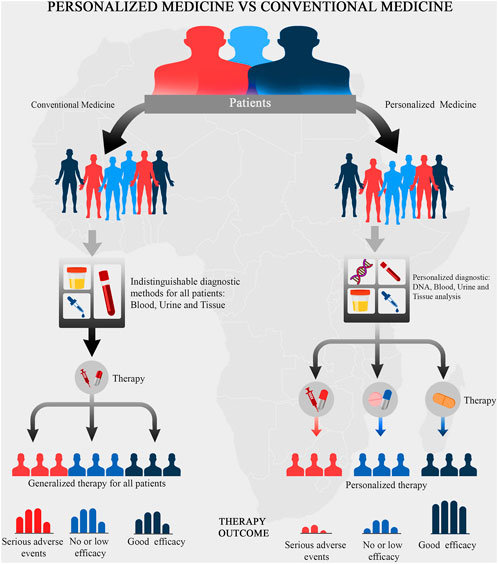
FIGURE 1. A schematic representation of the concept underlying personalized medicine. The underlying concepts of personalized medicine aims to prioritize the outcome of the medical treatment.
Personalized medicine entails optimizing drug choice, dosage, and treatment regimen while avoiding adverse drug effects for the specific patient. Its goal is to make patient-specific prescriptions of medicine and treatment based on their genetic makeup. Genetic variants could potentially play critical roles in the efficient administration of a particular drug, i.e., absorption, distribution, metabolism, and excretion of active components (ADME) (Adeyemo and Rotimi, 2014; Sigman, 2018). However, the application of personalized medicine might be limited when applied to non-genetic diseases. For instance, the burden of non-communicable diseases is largely considered to be as a result of lifestyle changes and urbanization (Collins and Varmus, 2015; Drake et al., 2018; Afolaranmi et al., 2021) rather than individualized genetic changes. Nonetheless, the implementation of personalized medicine will result in a paradigm shift from the conventional symptom-based, “trial-and-error” health system to a genome-based approach, thereby, bringing about precision and personalization in disease diagnosis, management, and treatment (Drake et al., 2018).
This review is aimed at highlighting the giant strides recorded so far in the journey to personalizing medicine in Africa as well as identifying some of the current challenges faced in its translation into routine clinical practice. It also seeks to proffer insights on how these challenges can be effectively tackled.
The term “personalized medicine” was introduced to the scientific community in 1971 (Gibson, 1971). However, the advent of personalized medicine in Africa is still evolving. For instance, by querying PubMed for personalized medicine-related terms on 15 May 2023, we obtained a total of 79,273 hits. The histogram of these hits demonstrates that there is an exponential growth of the publications in this area, see Figure 2. We used the search syntax below for querying PubMed for personalized medicine:
((“personalized medicine”) OR (“individualized medicine”) OR (“precision medicine"))
However, to restrict the hits for personalized medicine in Africa, we used the search syntax below:
(((“personalized medicine”) OR (“individualized medicine”) OR (“precision medicine")) AND (“Africa"))
Although few authors have argued about the subtle distinction in the terms ‘personalized medicine’, ‘individualized medicine’, and ‘precision medicine’, we used them synonymously in our search query since most authors on the subject have used them interchangeably (Khoury, 2016; Goetz and Schork, 2018; Sugeir and Naylor, 2018). For more details about the above PubMed query syntax, refer to Bramer et al. (2018). We obtained only 620 hits (0.78%) when we restricted our results to Africa and surprisingly the scientific effort started after 30 years, i.e., started in 2000. Moreover, when we restricted our search to Clinical Trials or Randomized Controlled Trials, we obtained only 21 hits (1.17%) for Africa out of 1,791 hits. The outcome of these 21 hits is summarized in Table 1. It is worth noting that by looking at the authors’ list of these 21 articles, we observed that there are only 46 researchers from Africa affiliated with 34 African institutes. Thus, it is safe to say that many developing countries in Africa still lack the expertise and technologies necessary to adopt genomic medicine widely, yet they continue to face challenges caused by the growing number of noncommunicable diseases (like cancer, diabetes, heart disease, and other chronic illnesses), as well as infectious diseases that are emerging and re-emerging (Kaze et al., 2018; Mulder et al., 2018; Mapesi and Paris, 2019; Mudie et al., 2019). As a result of these factors, Africa’s healthcare system is heavily burdened (Schlebusch et al., 2012).
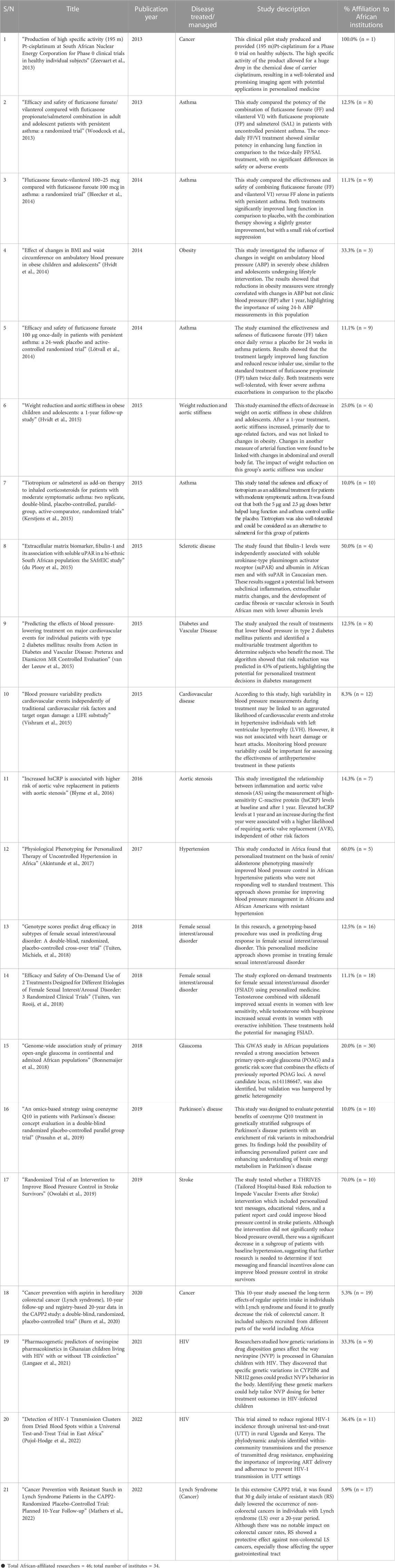
TABLE 1. Summary of the 21 PubMed hits on the Clinical Trials or Randomized Controlled Trials in Africa.
A simple breakdown of the results of the PubMed search shows that most of the hits were studies on conditions such as asthma, cancer, HIV, female sexual interest/arousal disorders, obesity, and cardiovascular diseases among others. The results obtained from these studies are summarized as follows.
A total of four studies have been done to investigate the application of personalized medicine in Asthma. The underlying samples were grouped based on disease status, i.e., persistent asthma versus intermittent asthma, or based on the patient’s reaction to the treatment. In detail, Woodcock et al. (2013) performed a randomized, double blind, double dummy, parallel group trial design. Their study involved 806 patients to assess the potency of fluticasone furoate-vilanterol combination (FF/VI) in comparison with fluticasone propionate (FP)/salmeterol (SAL) in asthmatic patients. The study concluded that the once-daily FF/VI was capable of improving lung function in the group of patients with persistent asthma. In 2014, Bleecker et al. (2014) investigated the efficacy and safety of the combination of corticosteroid fluticasone furoate and vilanterol (FF-VI) in comparison with fluticasone furoate (FF) in 609 asthmatic patients. They observed that FF-VI and FF resulted in improved lung function than placebo in the group of patients with persistent asthma. Furthermore, Lötvall et al. (2014) assessed the effectiveness and safety of the novel inhaled corticosteroids (ICS) fluticasone furoate (FF) compared to placebo. They reported that treating patients older than 12 years with FF 100 μg or 200 μg once daily will improve lung function in the group of patients with moderate-severe persistent asthma. In 2015, Kerstjens et al. (2015) investigated the safety and effectiveness of tiotropium in moderately asthmatic patients who showed symptoms despite being treated with medium-dose of ICS. They concluded that a once-daily tiotropium add-on to medium-dose inhaled corticosteroids lowers airflow obstruction and improves asthma control in the group of patients with moderate symptoms. Also, they considered Tiotropium to be a safe and potent bronchodilator and a possible substitute for salmeterol in this patient population.
Regarding cancer diseases, three studies investigated the utility of personalized medicine based on the underlying ethnic groups of the participants, i.e., participants are grouped based on their ancestry information. In detail, Zeevaart et al. (2013) performed a clinical trial for evaluating a cisplatinum product on 10 healthy volunteers. Although they used a relatively small sample size; however, they considered this product to be a promising imaging agent with potential applications in personalized medicine for the group of patients who are subject to platinum chemotherapy. In 2020, Burn et al. (2020) investigated Lynch syndrome which is linked with an increased risk of colorectal cancer, as well as with many different cancers (Burn et al., 2020). They conducted a double-blind, randomized CAPP2 trial. The total number of participants was 861 patients from 43 distinct international research centers worldwide. The participants were grouped based on their underlying ancestry as follows: 707 Europeans, 112 Australians, 38 Africans, and 4 Americans. The patients were randomly assigned to receive 600 mg aspirin (n = 427) or placebo (n = 434). The researchers monitored cancer outcomes up to 10 years from recruitment. The study’s findings reported that aspirin could be considered as a good preventive agent for colorectal cancer in group of pateints with Lynch syndrome. In 2022, Mathers et al. (2022) subjected participants with Lynch syndrome to a randomized double-blind treatment with 30 g resistant starch (RS) (n = 463) daily or placebo (n = 455) for up to 4 years. Upon following up with the participant for up to 20 years, they concluded that 30 g daily RS confers substantial protection against non–colorectal cancers for the group of patients with Lynch syndrome.
So far, five studies have applied personalized medicine strategies in understanding Diabetes and Cardiovascular Diseases by demonstrating variability in treatment response among the patients. In detail, van der Leeuw et al. (2015) attempted to identify which group of patients benefited most from blood pressure-lowering treatment in a large type 2 diabetes mellitus clinical trial. They developed a multivariable treatment model for predicting patients who benefited most from the therapy in terms of absolute risk reduction of major adverse cardiovascular events. Their findings reported the usefulness of using multivariable treatment algorithm in personalized medicine as the algorithm can be used to identify groups of patients that will be subject to blood pressure-lowering therapy. Another study, conducted by Vishram et al. (2015) used 8,505 patients randomized to two kinds of hypertension treatments: losartan and atenolol-based treatment. They tested the potential association between blood pressure (BP) variability and target organ damage. Vishram et al. (2015) concluded that the diastolic BP is associated with stroke. However, there was no statistical evidence for the association between diastolic BP and myocardial infarction. In 2016, Blyme et al. (2016) investigated connections between inflammation and aortic valve stenosis in 1,423 patients. Their findings reported that the increasement of high-sensitivity C-reactive protein (hsCRP) during the first year can be used to predict aortic valve replacement. Thus, the group of patients with increase in hsCRP are more likely to be subject to aortic valve replacement. In 2017, Akintunde et al. (2017) tested the possibility of using physiology-based individualized therapy to treat hypertension. The study sample included patients with uncontrolled hypertension from African countries, including Nigeria, Kenya, and South Africa. Their findings reported that using physiology-based individualized therapy considerably improved blood pressure control among African patients with uncontrolled hypertension. Also, they recommended applying their approach with other African ancestries, i.e., African American, or to all patients with resistant hypertension. In 2019, Owolabi et al. (2019) carried out a randomized stroke trial in Nigeria to determine the effect of a Tailored Hospital-based Risk reduction to Impede Vascular Events after Stroke intervention on blood pressure control among a group of patients with stroke. They concluded that the intervention did not significantly reduce systolic BP among groups of patients with stroke.
Regarding Female sexual interest/arousal disorder, two studies have been done so far and these studies examine the variation in response to treatment among the participating subjects. Using a double-blind, randomized, placebo-controlled crossover study, Tuiten et al. (2018) designed and tested a new genotyping procedure for predicting which drugs will yield a positive treatment response. A total of 139 female sexual interest/arousal disorder patients were treated with three different on-demand drug combinations for three 2-week periods. The study was able to predict which group of women benefits from which on-demand drug. This study could be potentially useful clinically as a companion diagnostic through a personalized medicine approach. In addition, Tuiten et al. (2018b) investigated the efficacy and safety of 2 novel on-demand drugs designed to treat 2 subgroups of patients with female sexual interest/arousal disorder (FSIAD). They used a personalized medicine approach through an allocation formula involving genetic, hormonal, and psychological variables for predicting drug effectiveness in the two subgroups. They reported that drugs under investigation were well tolerated, safe and largely resulted in more satisfying sexual events in the two subgroups.
Regarding Glaucoma disease, Bonnemaijer et al. (2018) conducted a genome-wide association study in 1,113 cases of Primary open-angle glaucoma (POAG) and 1826 controls. They studied samples from Tanzanian, South African, and African American. Their findings reported POAG loci that were significantly associated with POAG in the different groups of the studied samples.
Two studies have demonstrated the application of personalized medicine in HIV disease. The participating samples in these two studies were grouped based on disease status or based on their response to treatment. In detail, Langaee et al. (2021) investigated the relationship between genetic variations in relevant drug disposition genes and Nevirapine (NVP) pharmacokinetics parameters in children living with HIV in Ghana and eligible to receive NVP-based antiretroviral therapy. The subjects were treated with NVP plus zidovudine and lamivudine or abacavir and lamivudine twice daily, TB-coinfected patients got concurrent anti-TB therapy with NVP. Langaee et al. (2021) concluded that genotyping for SNPs encoding the transcriptional factor, pregnane X receptor, improves the prediction of NVP for individualized therapy. In addition, Pujol-Hodge et al. (2022) carried out a universal test-and-treat trial known as the Sustainable East Africa Research in Community Health (SEARCH) trial. They targeted individuals in Uganda and Kenya to reduce regional incidence of HIV-1. They investigated the distribution of HIV-1 drug resistance sequence and HIV-1 subtypes in both groups of population. The study findings emphasized the necessity of improving delivery and adherence to current antiretroviral therapy recommendations, to stop HIV-1 transmission.
Obesity-related disease studies showed how targeting medical interventions can result in changes in health outcomes. Hvidt et al. (2014) investigated the weight changes effects on ambulatory BP in 61 severely obese patients. The patients were subjected to lifestyle intervention and examined with ambulatory BP monitoring at start date and after a year’s treatment. The study concluded that changes in ambulatory BP are linked with different groups of obese patients. Moreover, Hvidt et al. (2015) investigated the impact of weight loss on aortic stiffness in 72 obese patients. However, they reported that the effect of reduction in weight on aortic stiffness was unclear in the different groups of obese patients.
Regarding Parkinson’s disease, Prasuhn et al. (2019) performed a study using a double-blind, randomized, and placebo-controlled approach focused on genetically stratified subgroups of Parkinson’s disease patients (PD) with enrichment of risk variants in mitochondrial genes. This group of patients is more likely to benefit from treatment with the coenzyme Q10, i.e., a mitochondrial enhancer. This study may be an initial step in successfully predicting treatment response on the basis of the genetic status of PD patients thereby translating progress in molecular genetics into personalized patient care.
Regarding Sclerotic disease, du Plooy et al. (2015) studied the independent relationship of fibulin-1 with the inflammatory markers of atherosclerosis. Their study sample involved 290 Africans, i.e., bi-ethnic South African population, and 343 sex- and age-matched Caucasians. This study investigated the utility of personalized medicine based on the underlying ethnic groups of the participants, i.e., participants are grouped based on their ancestry information. Their results indicated that the South African men with lower albumin levels are more likely to have cardiac fibrosis or vascular sclerosis.
Significant progress has been recorded so far in various aspects related to the implementation of personalized medicine in Africa. Some of these aspects include education system and training, infrastructure, and African-specific data generation. As a result, there has been a recent rise in the number of skilled human resources, infrastructure, and the quality of genomics research emanating from Africa and led by Africans (Uthman et al., 2015).
Since genomics is pivotal to the development of an effective system for personalized medicine (McGonigle, 2016), investments in the training and education of healthcare practitioners among other investments have been recommended as a path to harnessing the opportunities in genomics and precision medicine for the advancement of healthcare (Crellin et al., 2019). Therefore, efforts have been made to provide notable genomics initiatives in Africa. For instance, many organizations such as the Institute of Human Virology, African Collaborative Center for Microbiome and Genomics Research, African Research Group for Oncology, African Center for Translational Genomics, and Center for Genomic and Precision Medicine are among the initiatives taking bold steps in this direction. Refer to Table 2 for examples of prominent genomics initiatives in Africa that are making strong statements in genomics thereby providing frameworks and policies for personalized medicine in Africa. Noting that this list is not intended to be an exhaustive one, but rather a snapshot of a burgeoning list of genomics initiatives in Africa. Although these initiatives and several others have recorded some level of success in advancing the field of genomics in Africa, a lot still remains to be done to fully tap into the wealth of genetic diversity domiciled on the continent towards personalizing medicine for Africans and advancing global health.
Although many efforts at implementing personalized medicine in Africa have been addressed, there are still several challenges in this area, which have been summarized below.
The challenges associated with the rising incidence of non-communicable diseases, the prevalence of infectious diseases together with the widely known fact of Africa’s wide genetic diversity (Schlebusch et al., 2012), diverse environmental and climatic conditions, dietary and cultural backgrounds (Ibrahim and Bekele, 2019), require that serious focus be given to the research and application of genomics in addressing health issues in clinical settings on the continent. Despite the fact that different kinds of literature have identified the huge prospects associated with harnessing genomics in the African context (Fatumo, 2020), not so much satisfactory work, in comparison with other populations, has been done so far in this regard (Gurdasani et al., 2015; Zhang et al., 2022). This is primarily attributed to the fact that many low- and middle-income countries (LMICs), especially those in Africa, are still in the early stages of development and lack sufficient knowledge and implementation of genomics-based approaches for diagnostic and therapeutic purposes. (Drake et al., 2018).
However, research facilities such as ACEGID at the Redeemer’s University, CApIC-ACE at Covenant University, both in Nigeria, and the Medical Research Council/Uganda Virus Research Institute (MRC/UVRI) and London School of Hygiene and Tropical Medicine (LSHTM) research unit in Uganda, to mention but a few, are blazing the trail in genomics on the continent. ACEGID is particularly interested in the genomics of infectious diseases in Africa. They were instrumental in the first genome sequencing of SARS-COV-2 in Africa (Ihekweazu et al., 2020). CApIC-ACE seeks to develop new diagnostic biomarkers for prostate and breast cancers. They are also building a federated genomics (FEDGEN) infrastructure customized to process and analyze indigenous genomic data (Adetiba, E. et al., 2022). The MRC/UVRI and LSHTM Uganda Research Unit is promoting the genetic epidemiology of both communicable and non-communicable diseases (NCDs) in Uganda. Notwithstanding, more still needs to be done to set Africa in motion for large-scale genomic and personalized medicine adoption.
It is also noteworthy to mention that initiatives such as the H3Africa Consortium are currently doing a lot in bridging the genomics research gap. The consortium has invested in the establishment of high-quality biorepositories in Africa, a bioinformatic network, and a strong training program that has developed skills in genomic data analysis and interpretation among bioinformaticians, wet-lab researchers, and healthcare professionals (Mulder et al., 2018). This initiative aims to enhance genomics capacity and infrastructure across the continent by supporting numerous cutting-edge research projects into type-2 diabetes (T2D), cardiometabolic disease, rheumatic heart disease, chronic kidney disease, neurological diseases, microbiomes, various infectious diseases, and pharmacogenomics, among others—(http://h3africa.org/projects). There is also the Southern African Human Genome Programme (SAHGP) which is an “initiative that aspires to unlock the unique genetic character of southern African populations for a better understanding of human genetic diversity” (Choudhury et al., 2017). The outcomes of all these research endeavors hold significant potential to positively transform genomic medicine, not only within Africa but also on a global scale.
As high-throughput sequencing technologies become more widely accessible, genomic datasets are growing larger and more complex. Thus, there is a pressing need to train a significant mass of African researchers with the requisite skills and expertise to be able to contribute to research in this area rather than just being data generators. Knowledge deficit among healthcare professionals is also a huge barrier to the implementation of personalized genomic medicine. Most clinical practitioners are out-of-touch with the latest trends in genomics technology and many are unable to interpret genetic testing results. Also, several studies (even in developed countries) have shown that a significant number of non-genetics providers and specialists have acknowledged having limited knowledge of genomics and an insufficient capacity to interpret genomic information for their patients. (Bonter et al., 2011). Thus, in the face of the rapidly evolving body of knowledge in this field, it is imperative to recognize that 21st century physicians need more than a fundamental knowledge of human genetics, but a more robust understanding of it (Dhar et al., 2012). Apart from these, insufficient funding for health and education has been largely responsible for patients presenting at health facilities at later stages of diseases with complications that make treatment extremely difficult to manage (Drake et al., 2018).
To effectively utilize next-generation sequencing and other innovative technologies in personalized medicine, it is crucial to have a solid foundation of skills, knowledge, and infrastructure to effectively apply genetic information in healthcare (Ghoorah et al., 2021). Concerted efforts must be put in place in terms of capacity building to create a significant pool of bioinformaticians for biomedical data analysis and provision of genomic medicine training programs for healthcare professionals (Goetz and Schork, 2018). Moreover, emphasis should be placed on more training activities that are participatory in nature such as mentorships, internships, workshops, hackathons, fellowships, and conferences where early career investigators can interact with experts and learn the needed skills and knowledge through practical hands-on applications of concepts bordering around genomics and personalized medicine.
The lack of adequate infrastructures and technologies to support research and clinical translation has been identified as another major barrier to the adoption of personalized genomic medicine in Africa (Mulder, 2017; Jongeneel et al., 2022). To facilitate comprehensive DNA analysis, Africa should prioritize improving access to technology for researchers. Investment is required in various areas to support DNA sequencing and genotyping facilities, establish biobanks for sample storage along with their associated data, and develop robust data infrastructure and information management systems for efficient data generation, storage, and analysis pipelines. Additionally, there is a need to improve electronic health records, ensure reliable internet connectivity, and establish facilities for clinical action and conducting clinical trials.
Genomic medicine aims to reveal an individual’s genetic predisposition to diseases, usually by establishing a link between genotype and phenotype through genome-wide association studies (GWAS). Although prediction methods such as polygenic risk scores (PRS) hold potential for clinical applications, individuals of non-European descent may not benefit much from genomic medicine due to their current underrepresentation in extensive genetics and genomics research (Ju et al., 2022). For genomic medicine to translate into personalized care globally, as many different populations as feasible must be sufficiently represented in genomic research (Bentley et al., 2019). It has been suggested that failure to prioritize genomic studies in Africa, which harbors the highest genetic diversity among human populations, would hinder global health equity. Thus, there is a need for more data on African population genomics to address this challenge. Also, it is expedient for extensive research to be done to develop databases and other platforms for easy access to data on genomic variants and their associated genes and diseases (Bombard, 2015), especially among African populations.
Most genomic research involves the generation as well as the analysis of genomic data which can be quite expensive in terms of cost, time, and computational resources required. Most of these costs can be offset if there are huge investments in place for funding research work in these areas. Most African genomic scientists and researchers operate in relatively resource-scarce environments and largely lack the needed capacity to contribute and compete effectively with their counterparts in larger and better-resourced groups in the analysis of genomics data. Therefore, many of them resort to data generation in their quest to at least make any contribution. Thus, adequate funding will go a long way to address this challenge.
It is very crucial for deliberate efforts to be made at developing genomics research capacity in Africa. For instance, there is a serious need for collaborations among African investigators as well as with researchers in high-income settings who have the capacities, expertise, and funding to drive research. Mechanisms can also be put in place to allow government authorities, industrial partners, and/or researchers in different African countries to pool resources together in establishing and strengthening regional research centers and research networks with common interests instead of working in isolation (Tekola-Ayele and Rotimi, 2015).
Populations with non-European ancestry have not kept pace with their European ancestry counterparts, partly due to insufficient representation as both research participants and researchers in genomics research (Fatumo, 2020). More research and data that is applicable to indigenous populations in Africa is needed. This could help prevent the mistaken consideration of African individuals for disease risk variants that seem to be overrepresented in African populations (Mulder, 2017). African investigators need to be equipped to play vital roles in the global arena of genomic research so that Africa is not left out in the current waves of personalized precision medicine. Also, more public enlightenment must be done to encourage more members of the public to volunteer as subjects in genetic studies. Ethical and societal issues surrounding the use of genomic technologies should also be properly addressed through adequate education, orientation, and legislation where necessary.
The central importance of Africa to the origin of man cannot be overemphasized. The continent, which is the second largest in the world in terms of size and population, accommodates diverse ethnicities. For instance, Nigeria, which is the most populous and most diverse country in Africa and has 250 ethnic groups with over 500 different native languages, is characterized by high levels of genetic diversity and a broad population substructure (Fatumo et al., 2020). A recent study by Joshi et al. (2023) described the whole-genome sequencing of 449 Nigerian individuals across 47 unique self-reported ethnolinguistic groups in the country. Their results emphasize the value of the African genomic resource in improving our understanding of human ancestry and health.
As a matter of fact, obtaining an accurate representation of the reference genome is the cornerstone of the personalized medicine revolution. Thus, there have been discussions in the scientific community on the need for a more representative reference genome, especially in populations with wide genetic diversity. This is with the hope that such will allow for more utility and compatibility of the outcomes of whole genome sequencing in such populations and reduce biases. For instance, results from the 1,000 Genomes dataset reveal that the African super-population genomic data has the largest divergence from the GRCh38 reference (Tetikol et al., 2022). In fact, Sherman et al. (2019) also observed that the African pan-genome used in their study contains ∼10% more DNA than the current human reference genome. Therefore, different methods have been proposed to address this disparity in the reference genome and the population genomic data during large sequencing projects. Some of these approaches include: adding and extending nucleotides in the existing reference (Huang et al., 2013); generating a population-specific consensus sequence by assembling raw read data from scratch (Duan et al., 2019; Sherman et al., 2019), and graph-based references that have the ability to simultaneously represent multiple distinct populations. Tetikol et al. (2022) proposed a population-specific graph construction method to ensure that a more representative reference is used for the downstream analysis of large-scale genome sequencing projects. Since graph genomes offer more genetic diversity, they provide a viable framework for assembling African-specific references that would significantly reduce the biases of the GRCh38 reference.
In many healthcare settings in Africa, doctors and other healthcare providers still depend largely on the “one-size-fits-all” generalized approach of administering care rather than considering the individual uniqueness of each patient in care decisions. Also, most health facilities are poorly funded and understaffed. As a result, they lack the infrastructure and capacity for individualized healthcare delivery. Thus, it is important to adequately reimagine and reconfigure the health system on the continent to meet up with global best practices.
For personalized medicine to be successful, it requires sufficient infrastructure within the healthcare system to support the research and implementation of targeted diagnosis and treatment processes. For instance, a couple of health facilities across the world are using sequencing to unravel maladies that have defied diagnosis, including life-threatening conditions, particularly among children and the elderly (Worthey et al., 2011; Manolio et al., 2013; Gorzynski et al., 2022). Hence, it is necessary that national and sub-national frameworks are put in place for the adoption of routine sequencing, electronic medical records (EMR), and other standard practices to effectively improve the health outcomes of African communities. Health systems should also be ready to embrace research as there is also the possibility of key actors in the health sector failing to adopt research-proven interventions (Goetz and Schork, 2018). Resistance to change rather than lack of information or resources sometimes could be the problem impeding the deployment of certain interventions. Therefore, there must be willingness and openness to adopt current trends in clinical care.
Personalized medicine, if well implemented, holds the promise of being more cost-effective than the conventional traditional medical practices. It also has the potential of improving the quality of life of the patients. Diversity in genomics research is vital for both social justice and scientific advancement. Because of its rich genetic diversity, Africa needs to be well fortified with the right resources and technologies to be able to combat most of its health issues. However, this responsibility of adapting personalized medicine into the healthcare system in Africa should not be seen only as an “African issue to address”. It is a global endeavor that necessitates partnerships with diverse communities throughout the research process, dissemination of findings, and utilization of new technologies. Therefore, international partners (in the form of funding organizations, research collaborators, etc.), as well as individuals with African ancestry, have diverse but important roles to play in this global venture, as already highlighted in this review. Thus, all hands must be on deck to ensure its success in the African context.
Based on the reviewed scientific literature, we would like to share the following considerations to successfully apply personalized medicine concepts on a pan-African level. First, to enable personalized genomic medicine to flourish in Africa, it is essential to have inclusive discussions among stakeholders from clinical, research, industry, non-governmental, and legislative sectors. These dialogues should result in the development of comprehensive national, regional, and continental policies, as well as research infrastructure. These policies and infrastructure are crucial for advancing research, improving patient care, and ensuring fair access to new genomic technologies. The integration of genomics into the educational and health system of African communities is pivotal to the actualization of individualized medicine as it will create the human capital that will drive development in this field. Second, the implementation of personalized medicine would require the capacity building of a diverse array of skilled personnel. These include researchers, doctors, nurses, clinical geneticists, genetic counselors, pharmacists, bioinformaticians, biostatisticians, technicians, data scientists, and data security personnel. Collaborative and synergistic efforts must be made to harness the expertise of these personnel to develop personalized medicine on the continent. Also, the skilled diaspora community should be deliberately attracted to tap into their expertise by providing an enabling environment and investment in infrastructure. Third, the stakeholders, including health authorities, need to provide clear policies for personalized medicine applications to guide research in their research lines.
PO: Conceptualization, Writing–Original Draft Preparation, Writing–Review and Editing; YA: Conceptualization, Writing–Original Draft Preparation, Writing–Review and Editing; EA: Conceptualization, Funding Acquisition, Supervision, Writing–Review and Editing. All authors contributed to the article and approved the submitted version.
Research reported in this publication is supported by the National Human Genome Research Institute (NHGRI), Office Of The Director, National Institutes of Health (OD) under award numbers U24HG006941 and U2RTW010679. Also, the research reported in this publication is supported by the World Bank funding for the ACE Impact projects. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the World Bank.
The authors acknowledge the logistical support of Mr. Babajide Ayodele, Mr Promise Onyemaechi and Mr. Kehinde Philip Akinode. Covenant University provided the infrastructural support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adetiba, E., et al. (2022). “FEDGEN testbed: A federated genomics private cloud infrastructure for precision medicine and artificial intelligence research,” in Informatics and intelligent applications. ICIIA 2021. Editors S. Misra, J. Oluranti, R. Damaševičius, and R. Maskeliunas (Cham: Springer). Communications in Computer and Information Science, vol 1547. doi:10.1007/978-3-030-95630-1_6
Adeyemo, A., and Rotimi, C. (2014). What does genomic medicine mean for diverse populations? Mol. Genet. genomic Med. 2 (1), 3–6. doi:10.1002/mgg3.63
Adoga, M. P., Fatumo, S. A., and Agwale, S. M. (2014). H3Africa: A tipping point for a revolution in bioinformatics, genomics and health research in Africa. Source Code Biol. Med. 9 (1), 10. doi:10.1186/1751-0473-9-10
Afolaranmi, O., Salako, O., Okunade, K., James, A., and Fagbenro, G. (2021). Integrating genomics education into Nigerian undergraduate medical training - a narrative review. J. Clin. Sci. 18 (1), 3. doi:10.4103/jcls.jcls_6_20
Akintunde, A., Nondi, J., Gogo, K., Jones, E. S. W., Rayner, B. L., Hackam, D. G., et al. (2017). Physiological phenotyping for personalized therapy of uncontrolled hypertension in Africa. Am. J. Hypertens. 30 (9), 923–930. doi:10.1093/ajh/hpx066
Bentley, A. R., Callier, S., and Rotimi, C. (2019). The emergence of genomic research in Africa and new frameworks for equity in biomedical research. Ethn. Dis. 29, 179–186. Suppl 1. doi:10.18865/ed.29.S1.179
Bleecker, E. R., Lötvall, J., O’Byrne, P. M., Woodcock, A., Busse, W. W., Kerwin, E. M., et al. (2014). Fluticasone furoate–vilanterol 100-25 mcg compared with fluticasone furoate 100 mcg in asthma: A randomized trial. J. Allergy Clin. Immunol. Pract. 2 (5), 553–561. doi:10.1016/j.jaip.2014.02.010
Blyme, A., Asferg, C., Nielsen, O. W., Boman, K., Gohlke-Bärwolf, C., Wachtell, K., et al. (2016). Increased hsCRP is associated with higher risk of aortic valve replacement in patients with aortic stenosis. Scand. Cardiovasc. J. 50 (3), 138–145. doi:10.3109/14017431.2016.1151928
Bombard, Y. (2015). Translating personalized genomic medicine into clinical practice: evidence, values, and health policy. Genome 58 (12), 491–497. doi:10.1139/gen-2015-0145
Bonnemaijer, P. W. M., Iglesias, A. I., Nadkarni, G. N., Sanyiwa, A. J., Hassan, H. G., Cook, C., et al. (2018). Genome-wide association study of primary open-angle glaucoma in continental and admixed African populations. Hum. Genet. 137 (10), 847–862. doi:10.1007/s00439-018-1943-7
Bonter, K., Desjardins, C., Currier, N., Pun, J., and Ashbury, F. D. (2011). Personalised medicine in Canada: A survey of adoption and practice in oncology, cardiology and family medicine. BMJ Open 1 (1), e000110. doi:10.1136/bmjopen-2011-000110
Bramer, W. M., de Jonge, G. B., Rethlefsen, M. L., Mast, F., and Kleijnen, J. (2018). A systematic approach to searching: an efficient and complete method to develop literature searches. J. Med. Libr. Assoc. JMLA 106 (4), 531–541. doi:10.5195/jmla.2018.283
Burn, J., Sheth, H., Elliott, F., Reed, L., Macrae, F., Mecklin, J.-P., et al. (2020). Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: A double-blind, randomised, placebo-controlled trial. Lancet 395 (10240), 1855–1863. doi:10.1016/S0140-6736(20)30366-4
Choudhury, A., Ramsay, M., Hazelhurst, S., Aron, S., Bardien, S., Botha, G., et al. (2017). Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat. Commun. 8 (1), 2062. doi:10.1038/s41467-017-00663-9
Collins, F. S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372 (9), 793–795. doi:10.1056/nejmp1500523
Crellin, E., McClaren, B., Nisselle, A., Best, S., Gaff, C., and Metcalfe, S. (2019). Preparing medical specialists to practice genomic medicine: education an essential part of a broader strategy. Front. Genet. 10, 789. doi:10.3389/fgene.2019.00789
Dhar, S. U., Alford, R. L., Nelson, E. A., and Potocki, L. (2012). Enhancing exposure to genetics and genomics through an innovative medical school curriculum. Genet. Med. 14 (1), 163–167. doi:10.1038/gim.0b013e31822dd7d4
Drake, T. M., Knight, S. R., Harrison, E. M., and Søreide, K. (2018). Global inequities in precision medicine and molecular cancer research. Front. Oncol. 8, 346. Issue SEP. doi:10.3389/fonc.2018.00346
du Plooy, C. S., Kruger, R., Huisman, H. W., Rasmussen, L. M., Eugen-Olsen, J., and Schutte, A. E. (2015). Extracellular matrix biomarker, fibulin-1 and its association with soluble uPAR in a Bi-ethnic South African population: the safreic study. Heart, Lung Circulation 24 (3), 298–305. doi:10.1016/j.hlc.2014.09.019
Duan, Z., Qiao, Y., Lu, J., Lu, H., Zhang, W., Yan, F., et al. (2019). Hupan: A pan-genome analysis pipeline for human genomes. Genome Biol. 20 (1), 149. doi:10.1186/s13059-019-1751-y
El-Attar, E. A., Helmy Elkaffas, R. M., Aglan, S. A., Naga, I. S., Nabil, A., and Abdallah, H. Y. (2022). Genomics in Egypt: current status and future aspects. Front. Genet. 13, 797465. doi:10.3389/fgene.2022.797465
Fatumo, S., Ebenezer, T. E., Ekenna, C., Isewon, I., Ahmad, U., Adetunji, C., et al. (2020). The Nigerian bioinformatics and genomics network (NBGN): A collaborative platform to advance bioinformatics and genomics in Nigeria. Glob. Health, Epidemiol. Genomics 5, e3. doi:10.1017/gheg.2020.3
Fatumo, S. (2020). “The opportunity in African genome resource for precision medicine,” in EBioMedicine (54) (Amsterdam, Netherlands: Elsevier B.V). doi:10.1016/j.ebiom.2020.102721
George, C., Stoker, S., Okpechi, I., Woodward, M., Kengne, A., and Ckd-Africa Collaboration, (2021). The chronic kidney disease in Africa (CKD-Africa) collaboration: lessons from a new pan-african network. BMJ Glob. health 6 (8), e006454. doi:10.1136/bmjgh-2021-006454
Ghoorah, A. W., Chaplain, T., Rindra, R., Goorah, S., Chinien, G., and Jaufeerally-Fakim, Y. (2021). Population structure of the South west Indian ocean islands: implications for precision medicine. Front. Genet. 12, 758563. doi:10.3389/fgene.2021.758563
Gibson, W. M. (1971). Can personalized medicine survive? Can. family physician Med. famille Can. 17 (8), 29–88.
Goetz, L. H., and Schork, N. J. (2018). Personalized medicine: motivation, challenges, and progress. Fertil. Steril. 109 (6), 952–963. doi:10.1016/j.fertnstert.2018.05.006
Gorzynski, J. E., Goenka, S. D., Shafin, K., Jensen, T. D., Fisk, D. G., Grove, M. E., et al. (2022). Ultrarapid nanopore genome sequencing in a critical care setting. N. Engl. J. Med. 386 (7), 700–702. doi:10.1056/nejmc2112090
Gurdasani, D., Carstensen, T., Tekola-Ayele, F., Pagani, L., Tachmazidou, I., Hatzikotoulas, K., et al. (2015). The african genome variation project shapes medical genetics in Africa. Nature 517 (7534), 327–332. doi:10.1038/nature13997
Hasanzad, M., Sarhangi, N., Naghavi, A., Ghavimehr, E., Khatami, F., Ehsani Chimeh, S., et al. (2021). Genomic medicine on the frontier of precision medicine. J. Diabetes and Metabolic Disord. 21 (1), 853–861. doi:10.1007/s40200-021-00880-6
Huang, L., Popic, V., and Batzoglou, S. (2013). Short read alignment with populations of genomes. Bioinformatics 29 (13), i361–i370. doi:10.1093/bioinformatics/btt215
Hvidt, K. N., Olsen, M. H., Ibsen, H., and Holm, J.-C. (2014). Effect of changes in BMI and waist circumference on ambulatory blood pressure in obese children and adolescents. J. Hypertens. 32 (7), 1470–1477. doi:10.1097/HJH.0000000000000188
Hvidt, K. N., Olsen, M. H., Ibsen, H., and Holm, J.-C. (2015). Weight reduction and aortic stiffness in obese children and adolescents: A 1-year follow-up study. J. Hum. Hypertens. 29 (9), 535–540. doi:10.1038/jhh.2014.127
Ibrahim, M. E., and Bekele, E. (2019). “Disease, selection, and evolution in the african landscape,” in The genetics of african populations in health and disease (Cambridge, England: Cambridge University Press), 50–70. doi:10.1017/9781139680295.003
Ihekweazu, C., Happi, C., Omilabu, S., Salako, B. L., Abayomi, A., and Olaniyi, P. (2020). First African SARS-CoV-2 genome sequence from Nigerian COVID-19 case; 2020. Available at: http://virological.org/t/firstafrican-sars-cov-2-genome-sequence-from-nigerian-covid-19-case/421 (Accessed April 8, 2020).
Jongeneel, C. V., Kotze, M. J., Bhaw-Luximon, A., Fadlelmola, F. M., Fakim, Y. J., Hamdi, Y., et al. (2022). A view on genomic medicine activities in Africa: implications for policy. Front. Genet. 13, 769919. doi:10.3389/fgene.2022.769919
Joshi, E., Biddanda, A., Popoola, J., Yakubu, A., Osakwe, O., Attipoe, D., et al. (2023). “Whole-genome sequencing across 449 samples spanning 47 ethnolinguistic groups provides insights into genetic diversity in Nigeria,” in Cell genomics (Amsterdam, Netherlands: Elsevier BV). doi:10.1016/j.xgen.2023.100378
Ju, D., Hui, D., Hammond, D. A., Wonkam, A., and Tishkoff, S. A. (2022). Importance of including non-European populations in large human genetic studies to enhance precision medicine. Annu. Rev. Biomed. Data Sci. 5 (1), 321–339. doi:10.1146/annurev-biodatasci-122220-112550
Kaze, A. D., Ilori, T., Jaar, B. G., and Echouffo-Tcheugui, J. B. (2018). Burden of chronic kidney disease on the african continent: A systematic review and meta-analysis. BMC Nephrol. 19 (1), 125. doi:10.1186/s12882-018-0930-5
Kerstjens, H. A. M., Casale, T. B., Bleecker, E. R., Meltzer, E. O., Pizzichini, E., Schmidt, O., et al. (2015). Tiotropium or salmeterol as add-on therapy to inhaled corticosteroids for patients with moderate symptomatic asthma: two replicate, double-blind, placebo-controlled, parallel-group, active-comparator, randomised trials. Lancet Respir. Med. 3 (5), 367–376. doi:10.1016/S2213-2600(15)00031-4
Khoury, M. (2016). The shift from personalized medicine to precision public health: words matter! center for disease control and prevention. Available from: https://blogs.cdc.gov/genomics/2016/04/21/shift/(Accessed April 21, 2016).
Langaee, T., Al-Shaer, M. H., Gong, Y., Lima, E., Antwi, S., Enimil, A., et al. (2021). Pharmacogenetic predictors of nevirapine pharmacokinetics in Ghanaian children living with HIV with or without TB coinfection. Infect. Genet. Evol. 92, 104856. doi:10.1016/j.meegid.2021.104856
Lötvall, J., Bleecker, E. R., Busse, W. W., O’Byrne, P. M., Woodcock, A., Kerwin, E. M., et al. (2014). Efficacy and safety of fluticasone furoate 100 μg once-daily in patients with persistent asthma: A 24-week placebo and active-controlled randomised trial. Respir. Med. 108 (1), 41–49. doi:10.1016/j.rmed.2013.11.009
Manolio, T. A., Chisholm, R. L., Ozenberger, B., Roden, D. M., Williams, M. S., Wilson, R., et al. (2013). Implementing genomic medicine in the clinic: the future is here. Genet. Med. 15 (4), 258–267. doi:10.1038/gim.2012.157
Mapesi, H., and Paris, D. H. (2019). Non-communicable diseases on the rise in sub-saharan Africa, the underappreciated threat of a dual disease burden. Praxis 108 (15), 997–1005. doi:10.1024/1661-8157/a003354
Mathers, J. C., Elliott, F., Macrae, F., Mecklin, J.-P., Möslein, G., McRonald, F. E., et al. (2022). Cancer prevention with resistant starch in Lynch syndrome patients in the CAPP2-randomized placebo controlled trial: planned 10-year follow-up. Cancer Prev. Res. 15 (9), 623–634. doi:10.1158/1940-6207.CAPR-22-0044
McGonigle, I. V. (2016). The collective nature of personalized medicine. Genet. Res. 98, e3. doi:10.1017/S0016672315000270
Mudie, K., Jin, M. M., Tan Kendall, L., Addo, J., Dos-Santos-Silva, I., Quint, J., et al. (2019). Non-communicable diseases in sub-saharan Africa: A scoping review of large cohort studies. J. Glob. health 9 (2), 020409. doi:10.7189/jogh.09.020409
Mulder, N., Abimiku, A., Adebamowo, S. N., de Vries, J., Matimba, A., Olowoyo, P., et al. (2018). H3Africa: current perspectives. Pharmacogenomics personalized Med. 11, 59–66. doi:10.2147/PGPM.S141546
Mulder, N. (2017). Development to enable precision medicine in Africa. Pers. Med. 14 (6), 467–470. doi:10.2217/pme-2017-0055
Owolabi, M. O., Gebregziabher, M., Akinyemi, R. O., Akinyemi, J. O., Akpa, O., Olaniyan, O., et al. (2019). Randomized trial of an intervention to improve blood pressure control in stroke survivors. Circulation Cardiovasc. Qual. Outcomes 12 (12), e005904. doi:10.1161/CIRCOUTCOMES.119.005904
Prasuhn, J., Brüggemann, N., Hessler, N., Berg, D., Gasser, T., Brockmann, K., et al. (2019). An omics-based strategy using coenzyme Q10 in patients with Parkinson’s disease: concept evaluation in a double-blind randomized placebo-controlled parallel group trial. Neurological Res. Pract. 1 (1), 31. doi:10.1186/s42466-019-0033-1
Pujol-Hodge, E., Salazar-Gonzalez, J., Ssemwanga, D., Charlebois, E., Ayieko, J., Grant, H., et al. (2022). Detection of HIV-1 transmission clusters from dried blood spots within a universal test-and-treat trial in East Africa. Viruses 14 (8), 1673. doi:10.3390/v14081673
Schlebusch, C. M., Skoglund, P., Sjödin, P., Gattepaille, L. M., Hernandez, D., Jay, F., et al. (2012). Genomic variation in seven khoe-san groups reveals adaptation and complex african history. Science 338 (6105), 374–379. doi:10.1126/science.1227721
Sherman, R. M., Forman, J., Antonescu, V., Puiu, D., Daya, M., Rafaels, N., et al. (2019). Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 51 (1), 30–35. doi:10.1038/s41588-018-0273-y
Sigman, M. (2018). Introduction: personalized medicine: what is it and what are the challenges? Fertil. Steril. 109 (6), 944–945. doi:10.1016/j.fertnstert.2018.04.027
Sugeir, S., and Naylor, S. (2018). Critical care and personalized or precision medicine: who needs whom? J. Crit. care 43, 401–405. doi:10.1016/j.jcrc.2017.11.026
Tekola-Ayele, F., and Rotimi, C. N. (2015). Translational genomics in low- and middle-income countries: opportunities and challenges. Public Health Genomics 18 (4), 242–247. doi:10.1159/000433518
Tetikol, H. S., Turgut, D., Narci, K., Budak, G., Kalay, O., Arslan, E., et al. (2022). Pan-African genome demonstrates how population-specific genome graphs improve high-throughput sequencing data analysis. Nat. Commun. 13 (1), 4384. doi:10.1038/s41467-022-31724-3
Tuiten, A., Michiels, F., Böcker, K. B., Höhle, D., van Honk, J., de Lange, R. P., et al. (2018). Genotype scores predict drug efficacy in subtypes of female sexual interest/arousal disorder: A double-blind, randomized, placebo-controlled cross-over trial. Women’s Health 14, 1745506518788970. doi:10.1177/1745506518788970
Tuiten, A., van Rooij, K., Bloemers, J., Eisenegger, C., van Honk, J., Kessels, R., et al. (2018b). Efficacy and safety of on-demand use of 2 treatments designed for different etiologies of female sexual interest/arousal disorder: 3 randomized clinical trials. J. Sex. Med. 15 (2), 201–216. doi:10.1016/j.jsxm.2017.11.226
Uthman, O. A., Wiysonge, C. S., Ota, M. O., Nicol, M., Hussey, G. D., Ndumbe, P. M., et al. (2015). Increasing the value of health research in the WHO african region beyond 2015-reflecting on the past, celebrating the present and building the future: A bibliometric analysis. BMJ open 5 (3), e006340. doi:10.1136/bmjopen-2014-006340
van der Leeuw, J., Visseren, F. L. J., Woodward, M., Zoungas, S., Kengne, A. P., van der Graaf, Y., et al. (2015). Predicting the effects of blood pressure-lowering treatment on major cardiovascular events for individual patients with type 2 diabetes mellitus: results from action in diabetes and vascular disease: preterax and diamicron mr controlled evaluation. Hypertension 65 (1), 115–121. doi:10.1161/HYPERTENSIONAHA.114.04421
Vishram, J. K. K., Dahlöf, B., Devereux, R. B., Ibsen, H., Kjeldsen, S. E., Lindholm, L. H., et al. (2015). Blood pressure variability predicts cardiovascular events independently of traditional cardiovascular risk factors and target organ damage: A LIFE substudy. J. Hypertens. 33 (12), 2422–2430. doi:10.1097/HJH.0000000000000739
Woodcock, A., Bleecker, E. R., Lötvall, J., O’Byrne, P. M., Bateman, E. D., Medley, H., et al. (2013). Efficacy and safety of fluticasone furoate/vilanterol compared with fluticasone propionate/salmeterol combination in adult and adolescent patients with persistent asthma: A randomized trial. Chest 144 (4), 1222–1229. doi:10.1378/chest.13-0178
Worthey, E. A., Mayer, A. N., Syverson, G. D., Helbling, D., Bonacci, B. B., Decker, B., et al. (2011). Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 13 (3), 255–262. doi:10.1097/GIM.0b013e3182088158
Zeevaart, J. R., Wagener, J., Marjanovic-Painter, B., Sathekge, M., Soni, N., Zinn, C., et al. (2013). Production of high specific activity 195m Pt-cisplatinum at South African Nuclear Energy Corporation for Phase 0 clinical trials in healthy individual subjects. J. Label. Compd. Radiopharm. 56 (9–10), 495–503. doi:10.1002/jlcr.3091
Keywords: personalized medicine, individualized medicine, precision medicine, genomics, Africa
Citation: Owolabi P, Adam Y and Adebiyi E (2023) Personalizing medicine in Africa: current state, progress and challenges. Front. Genet. 14:1233338. doi: 10.3389/fgene.2023.1233338
Received: 02 June 2023; Accepted: 11 September 2023;
Published: 19 September 2023.
Edited by:
Raphael Zozimus Sangeda, Muhimbili University of Health and Allied Sciences, TanzaniaReviewed by:
Karen Y. He, Janssen Research and Development, United StatesCopyright © 2023 Owolabi, Adam and Adebiyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ezekiel Adebiyi, ZXpla2llbC5hZGViaXlpQGNvdmVuYW50dW5pdmVyc2l0eS5lZHUubmc=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.