- 1School of Life Science and Food Engineering, Huaiyin Institute of Technology, Huaian, China
- 2Department of Horticulture, Zhejiang University, Hangzhou, China
- 3National Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Key Laboratory of Horticultural Crop Biology and Genetic Improvement (East China) of MOAR, College of Horticulture, Nanjing Agricultural University, Nanjing, China
- 4Guizhou Institute of Biotechnology, Guizhou Province Academy of Agricultural Sciences, Guiyang, China
Long non-coding RNAs (lncRNAs) are increasingly recognized as cis- and trans-acting regulators of protein-coding genes in plants, particularly in response to abiotic stressors. Among these stressors, high soil salinity poses a significant challenge to crop productivity. Radish (Raphanus sativus L.) is a prominent root vegetable crop that exhibits moderate susceptibility to salt stress, particularly during the seedling stage. Nevertheless, the precise regulatory mechanisms through which lncRNAs contribute to salt response in radish remain largely unexplored. In this study, we performed genome-wide identification of lncRNAs using strand-specific RNA sequencing on radish fleshy root samples subjected to varying time points of salinity treatment. A total of 7,709 novel lncRNAs were identified, with 363 of them displaying significant differential expression in response to salt application. Furthermore, through target gene prediction, 5,006 cis- and 5,983 trans-target genes were obtained for the differentially expressed lncRNAs. The predicted target genes of these salt-responsive lncRNAs exhibited strong associations with various plant defense mechanisms, including signal perception and transduction, transcription regulation, ion homeostasis, osmoregulation, reactive oxygen species scavenging, photosynthesis, phytohormone regulation, and kinase activity. Notably, this study represents the first comprehensive genome-wide analysis of salt-responsive lncRNAs in radish, to the best of our knowledge. These findings provide a basis for future functional analysis of lncRNAs implicated in the defense response of radish against high salinity, which will aid in further understanding the regulatory mechanisms underlying radish response to salt stress.
Introduction
Soil salinity is one of the major damaging issues to cultivable land, resulting in an inevitable slowdown to plant growth and yields (Safdar et al., 2019). It is estimated that approximately 20% of global irrigated land is affected by salinization, which negatively affects land productivity (Islam et al., 2022). Hyper-ionic and hyper-osmotic injuries, as well as oxidative damage caused by prolonged saline conditions, are the main effects of salt stress in plants. To a certain degree, plants have evolved adaptive mechanisms to ensure their survival under high salinity through molecular, biochemical, and physiological adjustments. Hence, it is imperative to unravel the molecular and physiological mechanisms associated with salt stress tolerance in order to address the substantial agronomic challenges posed by salinization.
Long non-coding RNAs (lncRNAs) refer to a type of non-coding RNAs (ncRNAs) greater than 200 nucleotides (nt) in length without discernible coding potential in eukaryotes (Kapranov et al., 2007; Wang and Chekanova, 2017). Similar to classical messenger RNAs (mRNAs), lncRNAs are primarily transcribed by RNA Pol II, capped, polyadenylated, and usually spliced, but generally exhibit lower expression levels (Chekanova, 2015; Quan et al., 2015). In plants, a subset of lncRNAs is also transcribed by two other plant-specific RNA polymerases, namely, Pol IV and V, and is usually synthesized from transposable elements (St Laurent et al., 2015; Wierzbicki et al., 2021). Based on their genomic location and cellular function, lncRNAs can be classified into sense and antisense lncRNAs, intronic lncRNAs, and intergenic lncRNAs (lincRNAs) (Ma et al., 2013). Substantial evidence supports the notion that lncRNAs play crucial roles as regulators in various biological processes, exerting their influence on gene expression in both cis- and trans-acting manners (Gil and Ulitsky, 2020; Jha et al., 2020); they can serve as molecular signals, decoys, guides, and scaffolds (Wang and Chang, 2011).
Initially, lncRNAs were neglected as a component of transcriptional noise (Chen et al., 2018); nevertheless, accumulating evidence has manifested that lncRNAs play crucial roles in multiple plant biological events, including photomorphogenesis (Wang et al., 2014; Sun et al., 2020), senescence (Huang et al., 2021), reproduction (Zhang et al., 2014; Zhou et al., 2022a), seed aging (Zhang et al., 2022), and fruit ripening (Tian et al., 2019; Zhou et al., 2022b). To date, there have been experimental investigations into the functions of specific plant lncRNAs. For instance, lncRNA39026 in tomato may possess the capability to impact decoy miR168a, leading to an increase in the expression of pathogenesis-related genes, thereby improving disease resistance (Hou et al., 2020). In rice, an intronic lncRNA known as RICE FLOWERING ASSOCIATED (RIFLA) plays an essential role in the flowering process mediated by OsMADS56, achieved through the formation of a complex with OsiEZ1 (Shin et al., 2022). Recent research has hinted that lncRNAs exhibit responsiveness to various stressors, including salt stress (Baruah et al., 2021; Cao et al., 2021; Liu et al., 2022a; Hu et al., 2022). For example, a total of 742 salt-responsive lncRNAs were identified in maize (Liu et al., 2022a). Similarly, in Spirodela polyrhiza, a total of 2,815 lncRNAs were discovered under salt stress conditions, out of which 185 exhibited differential expression in response to salinity (Fu et al., 2020). In the context of cotton, the functional role of lncRNA973 in fine-tuning salt stress response has been experimentally verified (Zhang et al., 2019). Additionally, the overexpression of lncRNA77580 in Glycine max has been found to regulate the response to both salt and drought stress by modulating the transcription of distinct sets of stress-related genes (Chen et al., 2023). Overall, these reports provide substantial evidence to affirm the significant role of lncRNAs in the regulation of plant stress tolerance.
Radish (Raphanus sativus L.), an essential root vegetable belonging to the Brassicaceae family, is cultivated globally owing to its high nutritional and medicinal values. Among vegetable crops, radish exhibits moderate sensitivity to salt, with its edible fleshy taproots displaying notable responsiveness to salt stimulus up to a maximum soil salinity threshold of 1.2 dS/m (Grattan et al., 2016). The yield and quality of radish taproots are significantly impacted by salt stress, as it has become the major limiting factor due to soil salinization and secondary salinization. Investigating the response of radish to salt stress holds potential for the advancement of salt-tolerant radish lines in the field of radish breeding. In radish, a substantial number of miRNAs and protein-coding genes implicated in salt adaptation have been reported in our previous studies (Sun et al., 2015; 2016). Nevertheless, the reports on lncRNAs involved in salt stress response remain elusive in radish. Fortunately, with the notable advancements in deep transcriptome sequencing technology and associated bioinformatics methodologies, it is now easier to comprehensively mine novel non-coding RNA molecules. In this study, strand-specific RNA sequencing (ssRNA-seq) was utilized to identify and characterize the salt-responsive lncRNAs in radish. Furthermore, the function of differentially expressed lncRNAs (DE-lncRNAs) was investigated by examining their position or co-expression associations with target genes. These findings could serve as a starting point for discerning the role of lncRNAs in the regulatory mechanisms governing salt stress response in radish, thereby offering potential benefits for the development of salt-tolerant radish cultivars.
Materials and methods
Plant materials and salt treatments
The radish advanced inbred line, ‘YH’, was used in this study. The seeds were rinsed and surface-sterilized using a 1.2% NaClO solution before germinating on moist filter paper and further incubated at 25°C in darkness for 2 days. Subsequently, the seedlings were transferred to plastic pots and grown at 25°C/18°C with a relative humidity of 60% in a 14 h light/10 h dark cycle. Uniform four-leaf-old seedlings were transferred into hydroponic conditions with half-strength Hoagland’s solution. After acclimating for a 1-week slow seeding period, the plants were stressed with 200 mM NaCl. The materials for transcriptome sequencing were collected at 0 h (control), 6 h, 12 h, 24 h, 48 h, and 96 h under salt treatment with two biological replicates at each time point, and the sample for each replicate contained an equal amount of fleshy taproot from three individual seedlings. Additionally, the samples were prepared for qualitative real-time polymerase chain reaction (qRT-PCR) analysis in triplicate at every time point during salt treatments, with three individual seedlings per replicate. Last, the samples were frozen in liquid nitrogen immediately and stored at −80°C until use.
RNA extraction, quality control, and cDNA library preparation
The construction of transcriptome libraries and deep sequencing were implemented at the Novogene Bioinformatics Institute (Beijing, China). The isolation of total RNAs was performed following the manufacturer’s recommended protocol (Invitrogen, Carlsbad, CA, United States). RNA quality and integrity were monitored using the Agilent 2100 Bioanalyzer. Ribosomal RNA was filtered using the Ribo-Zero™ rRNA Removal Kit (Epicentre, United States). The cDNA libraries were generated using the NEBNext® Ultra™ II RNA Kits (NEB, United States).
Sequencing and bioinformatic discovery of lncRNAs
The constructed libraries underwent sequencing using the Illumina NovaSeq PE150 platform. To obtain clean reads, the raw reads were filtered with fastp to eliminate adapter sequences and reads containing poly-N and low-quality reads. Meanwhile, the Q20, Q30, and GC contents were calculated. The clean reads were then aligned to the radish reference genome Rs1.0 assembly (http://radish-genome.org/) using HISAT2 (version 2.1.0) (Kim et al., 2015). Based on the assembled transcripts, the candidate lncRNAs were identified using a rigorous set of criteria. Initially, transcripts possessing a class_code of “i”, “u”, “x”, and “o” were reserved. Subsequently, transcripts exceeding a length of 200 bp and containing a minimum of two exons were retained. Finally, transcripts exhibiting overlap with established protein-coding domains were removed, while those overlapping with annotated lncRNAs were preserved. The coding potential of the retained transcripts was assessed using the Coding Potential Calculator (CPC2), Coding-Non-Coding-Index (CNCI), and Pfam. Non-coding transcripts shared by these three predictive software packages were identified as novel lncRNAs.
Differential expression analysis
The StringTie program (version 1.3.4) using the “-G option” (Pertea et al., 2015), in conjunction with the calculated FPKM values, was performed to estimate the expression level of both lncRNAs and coding genes in each sample. The DESeq2 R package (version 1.10.1) (Love et al., 2014) was applied to identify differentially expressed genes (DEGs) and DE-lncRNAs between the control and salinity treatments. Genes or lncRNAs with adjusted p-values (padj) ≤ 0.05 and log2 (|fold change|) ≥ 1 were determined as DEGs and DE-lncRNAs, respectively.
Potential target gene prediction and enrichment analysis of DE-lncRNAs
In order to enhance the understanding of lncRNA functions in radish, the potential cis- and trans-target mRNAs of DE-lncRNAs were predicted. According to the genomic location of lncRNAs relative to the neighboring genes, the target mRNAs in the 100-kb region upstream or downstream of DE-lncRNAs were regarded as potential cis-target genes. Pearson’s correlation coefficient (PCC) between DE-lncRNAs and the corresponding transcripts was calculated based on their co-expression to predict the trans-target genes of lncRNAs. The transcripts up to the strict standards of (|PCC| > 0.8, p-value < 0.01) were defined as the trans-target genes of DE-lncRNAs. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were carried out to further annotate the functions of cis- and trans-target genes for DE-lncRNAs. The GO terms and KEGG pathways with corrected p-values ≤ 0.05 were recognized as significantly enriched.
Validation of DE-lncRNAs by qRT-PCR analysis
In order to validate the findings from RNA-seq analysis, a total of six DE-lncRNAs, namely, TCONS_00053136, TCONS_00062931, TCONS_00081369, TCONS_00101865, TCONS_00122106, and TCONS_00159796, were randomly selected for qRT-PCR analysis based on their significant differential expression observed in the RNA-seq data under salt stress conditions. The specific primers used for qRT-PCR analysis are listed in Supplementary Table S1. The expression of the DE-lncRNAs was normalized using RsActin as the internal reference. qRT-PCRs were operated on a LightCycler R 480 System (Roche, Mannheim, and Germany) using the 2−ΔΔCT method.
Results
Overview of whole-transcriptome sequencing in radish
To comprehensively identify the lncRNAs responsive to salt stress in radish, high-throughput ssRNA-seq of twelve libraries derived from samples of both the control (CK) and salinity treatment groups (NA_1-NA_5) were analyzed. The raw sequencing data have been deposited in the Sequence Read Archive (SRA) at the NCBI, with BioProject No. PRJNA930138. Following quality control measures on the raw data, a total of 1.11 billion clean reads (with an average of 92.71 million reads) were obtained. The Q20 and Q30 scores exceeded 90%, and the GC content ranged from 45.31% to 48.98%. All clean reads were aligned to the radish reference genome, with the alignment ratio ranging from 70.35% to 79.85%. The statistics of sequencing and mapping for each library are summarized in Supplementary Table S2.
Identification of novel lncRNAs in radish
After transcriptome assembly using the StringTie software, a total of 708,647 assembled transcripts were obtained from the mapped reads of the twelve libraries. To predict the coding potential of these novel assembled transcripts, two prediction software programs (CPC2 and CNCI) and one database (Pfam) were employed. Consequently, 7,709 transcripts were identified as novel lncRNAs (Supplementary Table S3). Meanwhile, 8,288 transcripts were determined to be novel mRNAs (Supplementary Table S4). The lncRNAs were characterized based on their relative proximity to the nearest protein-coding gene neighbor, with all lncRNAs located as lincRNAs in the intergenic region. Approximately two-thirds of the lncRNAs were distributed across all the radish chromosomes, leaving more than one-third of the lncRNAs unmapped to any specific chromosome (Figure 1A; Supplementary Table S3). In addition, a comparative analysis was conducted to assess the fundamental characteristics of lncRNAs and mRNAs, encompassing sequence length, open reading frames (ORFs), and exon numbers. The length distribution of lncRNAs in radish predominantly fell within the range of 200–7,000 bp, with approximately 93.4% of lncRNAs falling within the 200–2,000 bp range (Figure 1B). Additionally, radish lncRNAs also harbored shorter ORFs compared to mRNAs, with the majority of novel lncRNAs having ORFs of less than 150 nucleotides (Figure 1C). Structural analysis further indicated that a significant proportion (67.4%) of novel lncRNAs possessed fewer than three exons, whereas mRNAs displayed a higher exon count (Figure 1D).
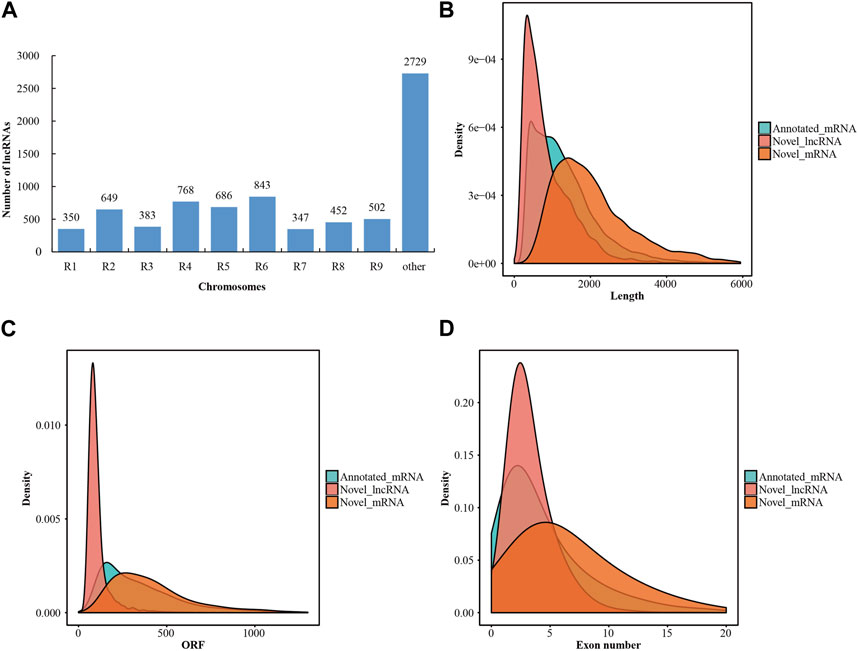
FIGURE 1. Characteristics of lncRNAs in radish. (A) Distribution of lncRNAs on different chromosomes. (B) Statistical results of length distribution between the lncRNAs and mRNAs. (C) Statistical results of ORF distribution between lncRNAs and mRNAs. (D) Statistical results of exon number percentages between lncRNAs and mRNAs.
Identification and analysis of DE-lncRNAs in radish
In order to investigate the changes in transcription of lncRNAs during salt stress, we conducted a comprehensive analysis using the edgeR package (version 3.12.1). DE-lncRNAs were filtered based on the conditions padj ≤ 0.05 and log2(|fold change|) ≥ 1. In total, 363 DE-lncRNAs were identified, with 107, 126, 160, 122, and 72 members detected in the NA_1_vs._CK, NA_2_vs._CK, NA_3_vs._CK, NA_4_vs._CK, and NA_5_vs._CK comparisons, respectively (Supplementary Table S5). Only seven common DE-lncRNAs were traced in five comparisons (Figure 2A). In the comparison between NA_1_vs._CK, a total of 73 lncRNAs were found to be upregulated, while 34 lncRNAs were downregulated. Similarly, in the comparison between NA_2_vs._CK, 80 lncRNAs were identified as upregulated and 46 lncRNAs were downregulated. Furthermore, in the comparison between NA_3_vs._CK, a total of 118 lncRNAs were upregulated and 42 lncRNAs were downregulated. Additionally, in the comparison between NA_4_vs._CK, 63 lncRNAs were discovered to be upregulated and 59 lncRNAs were downregulated. Last, in the comparison between NA_5_vs._CK, 42 lncRNAs were found to be upregulated and 30 lncRNAs were downregulated. Consequently, the prevalence of upregulated lncRNAs was observed in all comparisons, as depicted in Figure 2B. Furthermore, a heat map was employed to aggregate the DE-lncRNAs, enabling the visualization of expression patterns across all six time points of salt treatments (Figure 2C).
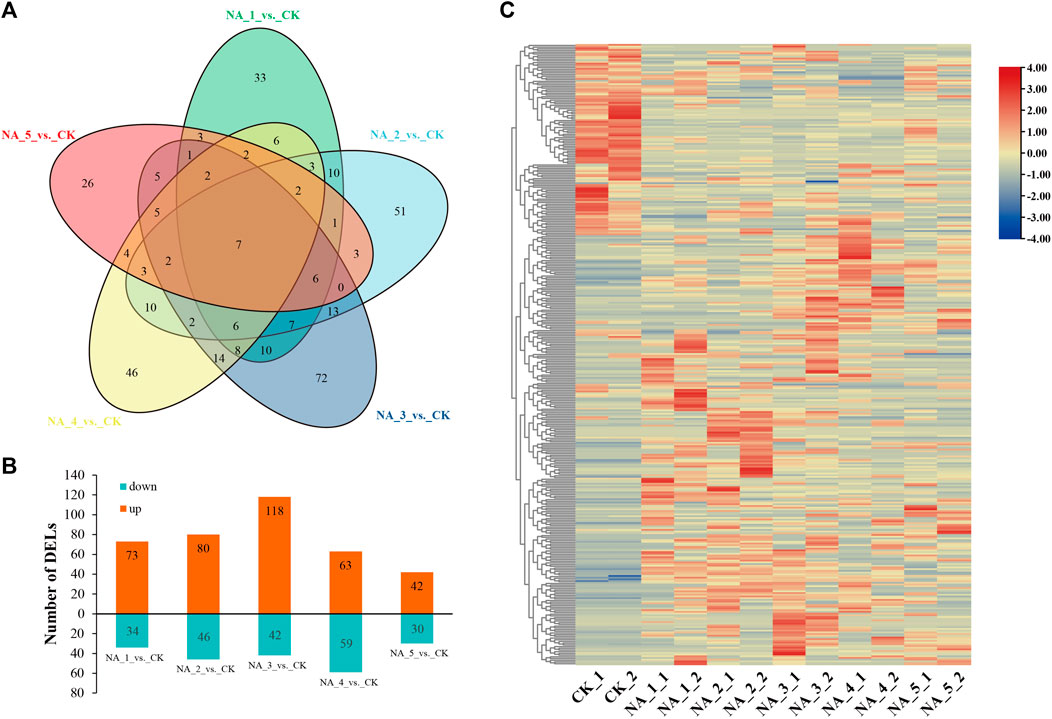
FIGURE 2. Overall view of DE-lncRNAs under different salt conditions in radish. (A) Venn diagram of DE-lncRNAs. (B) Number of upregulated/downregulated lncRNAs. (C) Hierarchical clustering plot of all DE-lncRNAs based on the average FPKM value of each set of replicates.
Analysis of potential target genes of DE-lncRNAs in radish
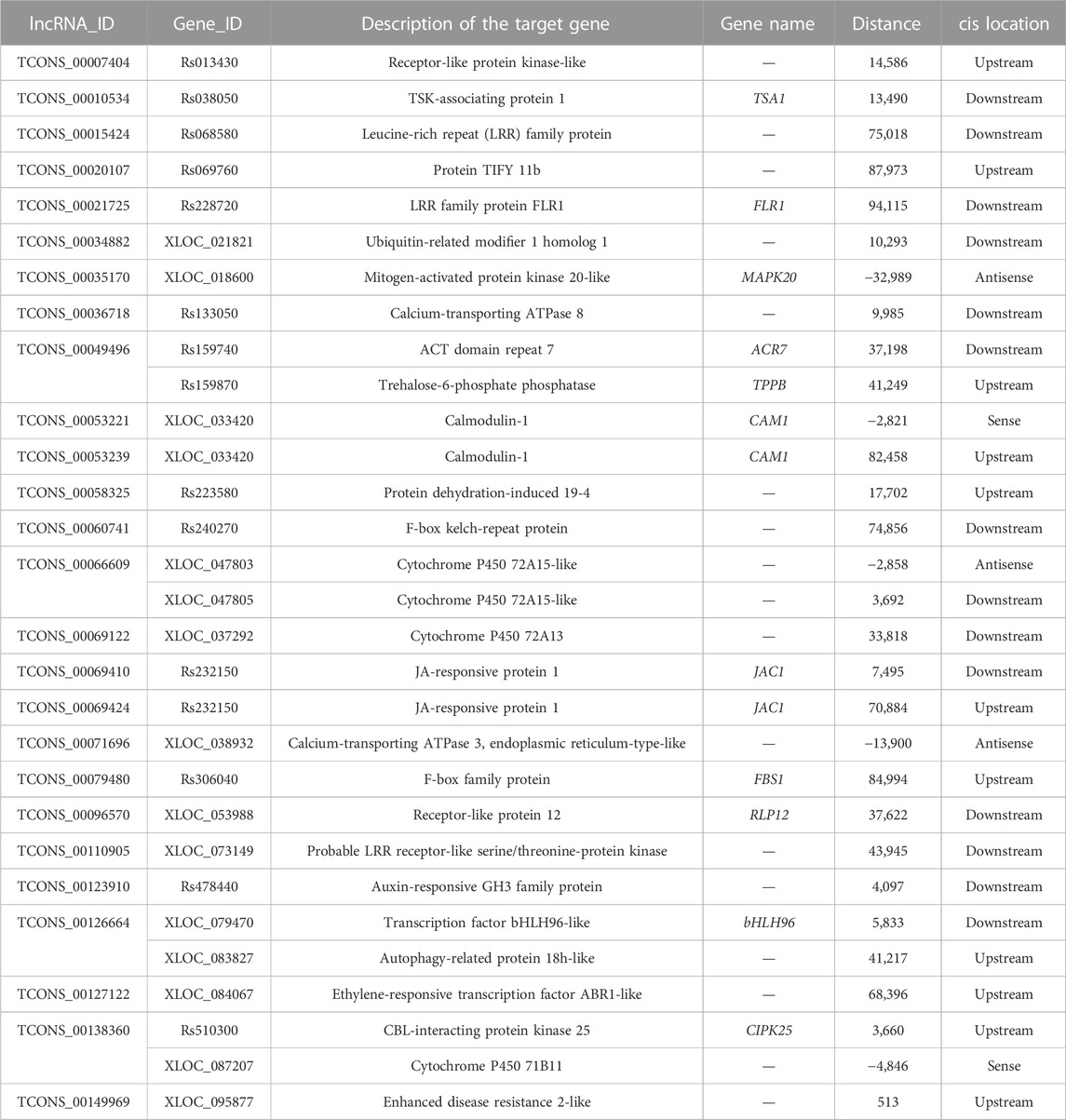
To explore the potential biological roles of lncRNAs in response to salinity stressors in radish, the target genes fine-tuned by DE-lncRNAs were identified using both cis- and trans-acting approaches (Supplementary Table S6). In terms of cis-regulation, a total of 5,006 target genes were observed for 303 DE-lncRNAs depending on their genomic location, resulting in 6,774 lncRNA–mRNA interaction pairs. Additionally, 5,983 trans-target genes were recognized for 249 DE-lncRNAs according to their co-expression relationships, resulting in 24,059 lncRNA–mRNA interaction pairs. Notably, 599 target genes were found to be jointly regulated by DE-lncRNAs via both cis- and trans-acting approaches. Moreover, the study found that 89 target genes (1.78%) exhibited differential expression under salt stress with 121 cis-regulatory pairs, and 229 target genes (3.83%) showed differential expression with 1,712 trans-regulatory pairs (Tables 1, 2; Supplementary Table S7). To further distinguish the potential key target genes responsive to salt stress, further annotation and analysis were conducted on these 121 cis-regulatory pairs and 1,712 trans-regulatory pairs. Among the differentially expressed targeted genes, there were crucial genes encoding for transcription factors (TFs) (e.g., bHLH, WRKY, MYB, and MADS-box), transport-related proteins (e.g., calcium-transporting ATPase, vacuolar glucose transporter, annexin, sugar transporter, and sulfate transporter), cytochrome P450, receptor-like protein kinases, enzymes associated with signal transduction (e.g., mitogen-activated protein kinase, calmodulin, CBL-interacting protein kinase, and CBL-interacting serine/threonine-protein kinase), enzymes related to photosynthesis (e.g., water-soluble chlorophyll protein), plant hormone regulation (e.g., ACC oxidase, auxin-responsive GH3 family protein, and proline dehydrogenase), osmoregulatory factors like trehalose-6-phosphate phosphatase, and enzymes implicated in mitigating oxidative damage (e.g., superoxide dismutase, peroxidase, and thioredoxin). All in all, these findings hinted that the identified DE-lncRNA-target pairs may play essential roles in regulating the significant biological processes related to the response of radish to salt stress.
Enrichment analysis for cis-acting target genes of DE-lncRNAs
GO analysis exhibited that the cis-target genes of DE-lncRNAs were enriched in 2,961 GO terms, including 1,677 biological processes (BPs), 376 molecular functions (MFs), and 908 cellular components (CCs) (Supplementary Table S8). Increasingly, GO analysis of cis-acting targets responsive to salt stress was closely enriched in BP terms related to photosynthesis (e.g., GO: 0015979: photosynthesis and GO: 0015994: chlorophyll metabolic process), signal transduction (e.g., GO: 0009966: regulation of signal transduction; GO: 0023051: regulation of signaling; GO: 0009755: hormone-mediated signaling pathway; and GO: 0006465: signal peptide processing), and antioxidant regulation (e.g., GO: 0000302: response to reactive oxygen species and GO: 0006979: response to oxidative stress). In the MF category, specific GO terms included numerous ion transport activities (e.g., GO: 0005216: ion channel activity; GO: 0005262: calcium channel activity; GO: 0005272: sodium channel activity; and GO: 0015079: potassium ion transmembrane transporter activity). Regarding the CC category, GO terms associated with the photosynthetic components (e.g., GO: 0009507: chloroplast; GO: 0009521: photosystem; GO: 0009522: photosystem I; and GO: 0009523: photosystem II) and ion transport channels (e.g., GO: 0034702: ion channel complex; GO: 0034703: cation channel complex; and GO: 0034706: sodium channel complex) were enriched. Nevertheless, only one MF term “glutamate-cysteine ligase activity (GO: 0004357)” was significantly enriched in the NA_3_vs._CK comparison group for cis-acting targets.
The analysis conducted using KEGG suggested that the cis-acting target genes of DE-lncRNAs were highly represented in 119 KEGG pathways (Figure 3; Supplementary Table S9). Especially, the top 20 enriched pathways in each comparison group demonstrated significant enrichment of key pathways such as plant hormone signal transduction, homologous recombination, ubiquitin-mediated proteolysis, and zeatin biosynthesis.
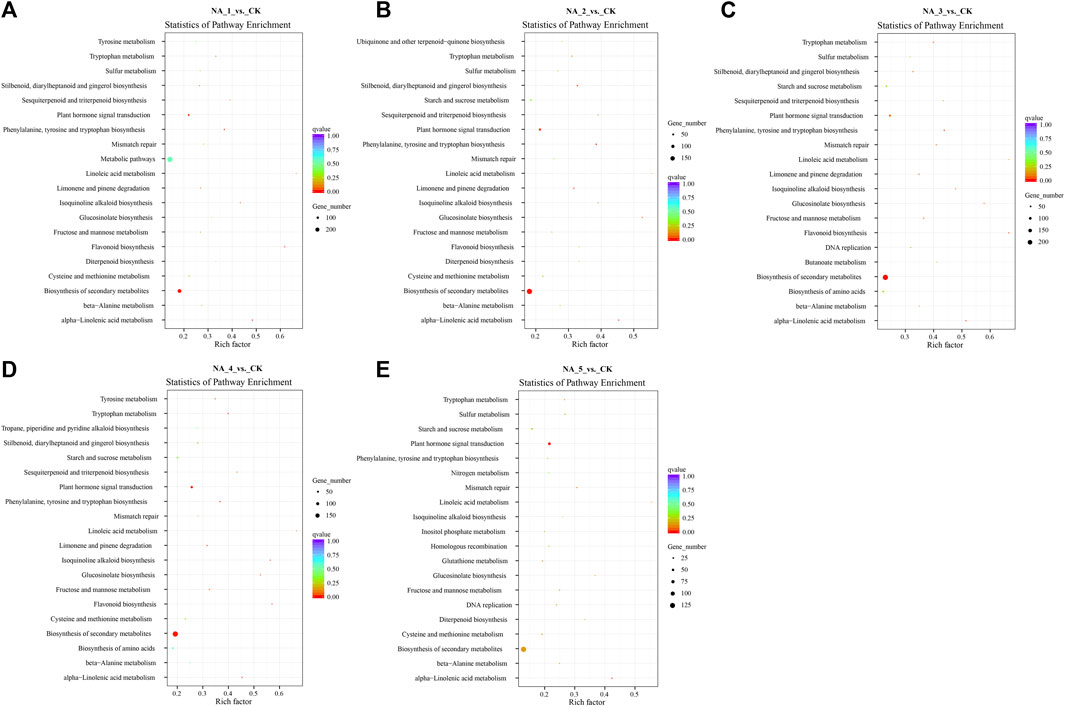
FIGURE 3. Top 20 enriched KEGG pathways for the cis-targets of DE-lncRNAs in the NA_1_vs._CK (A), NA_2_vs._CK (B), NA_3_vs._CK (C), NA_4_vs._CK (D), and NA_5_vs._CK (E) comparisons, respectively.
Enrichment analysis for trans-acting target genes of DE-lncRNAs
Functional analysis suggested that the trans-acting target genes of DE-lncRNAs were enriched in 3,051 GO terms, including 1,710 BP terms, 418 MF terms, and 923 CC terms (Supplementary Table S10). As anticipated, the trans-targets of DE-lncRNAs exhibited a significant overlap in GO terms pertaining to essential biological processes (e.g., photosynthesis, signal transduction, and antioxidant regulation), molecular functions (e.g., ion transport activity), and cellular components (e.g., photosynthetic component and ion transport channel) with the cis-acting targets. Furthermore, a notable enrichment of 11, 14, 18, 15, and 7 GO terms was observed in the NA_1_vs._CK, NA_2_vs._CK, NA_3_vs._CK, NA_4_vs._CK, and NA_5_vs._CK comparisons, respectively (Supplementary Table S10). This demonstrates that the lncRNA-trans-regulatory target genes may play prior roles in response to salt stress in radish.
The trans-acting target genes of DE-lncRNAs exhibited enrichment in 118 KEGG pathways (the top 20 enriched pathways of each group are shown in Figure 4; Supplementary Table S11). Notably, several pathways, including α-linolenic acid metabolism, β-alanine metabolism, biosynthesis of secondary metabolites, fructose and mannose metabolism, glucosinolate biosynthesis, isoquinoline alkaloid biosynthesis, plant hormone signal transduction, and tryptophan metabolism, were significantly enriched across all comparison groups. Meanwhile, certain pathways associated with salt stress defense, such as plant hormone signal transduction, glutathione metabolism, ubiquinone, and other terpenoid-quinone biosynthesis, were also enriched for some target genes.
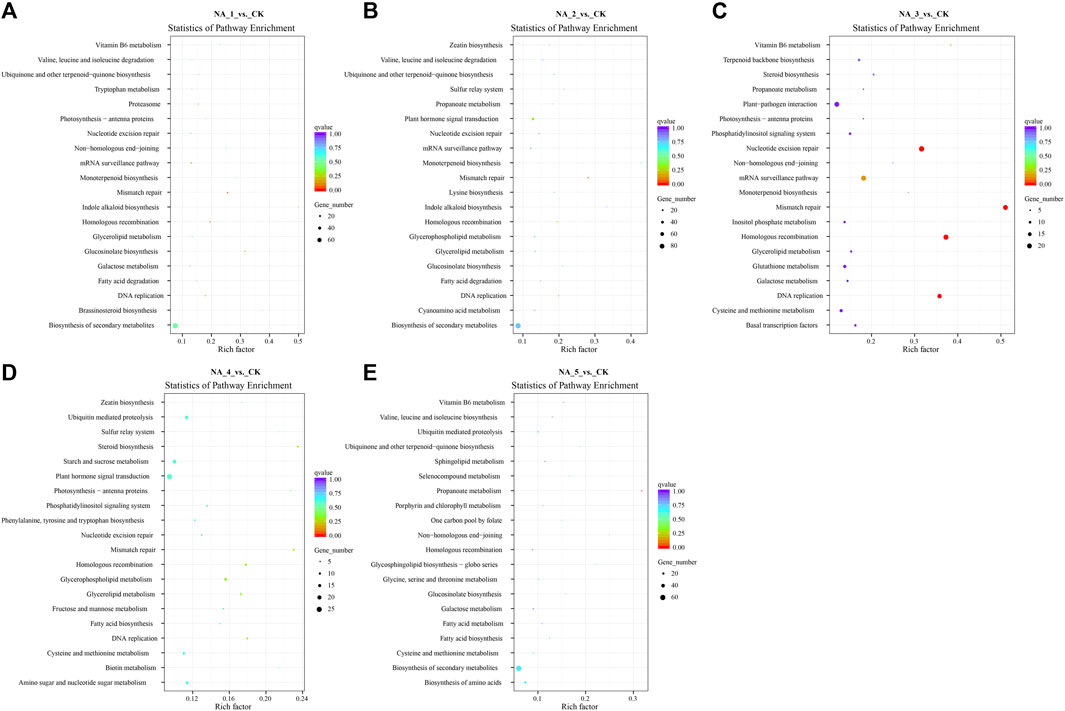
FIGURE 4. Top 20 enriched KEGG pathways for the trans-targets of DE-lncRNAs in the NA_1_vs._CK (A), NA_2_vs._CK (B), NA_3_vs._CK (C), NA_4_vs._CK (D), and NA_5_vs._CK (E) comparisons, respectively.
Experimental validation of DE-lncRNAs
To validate the analysis of the data obtained from RNA-seq, six lncRNA transcripts were examined by qRT-PCR (Figure 5). In general, the expression patterns of four salt-responsive lncRNAs (TCONS_00053136, TCONS_00062931, TCONS_00101865, and TCONS_00122106) were relatively coincident with similar trends between qRT-PCR and RNA-seq, indicating the reliability of expression profiling based on RNA-seq data. However, the expression of TCONS_00081369 and TCONS_00159796 exhibited contrasting changes at certain time points during salt treatments when comparing the two methods. For instance, the upregulation of TCONS_00081369 at 12 h and 96 h of salt treatment was observed in the RNA-seq data, while a slight downregulation was detected in the qRT-PCR analysis. Conversely, TCONS_00159796 exhibited an opposite change in expression at 96 h under 200 mM NaCl. The observed disparity in expression levels between qRT-PCR and RNA-seq could be partially attributed to variations in sensitivity and algorithmic methodologies employed by these two approaches (Xu et al., 2015).
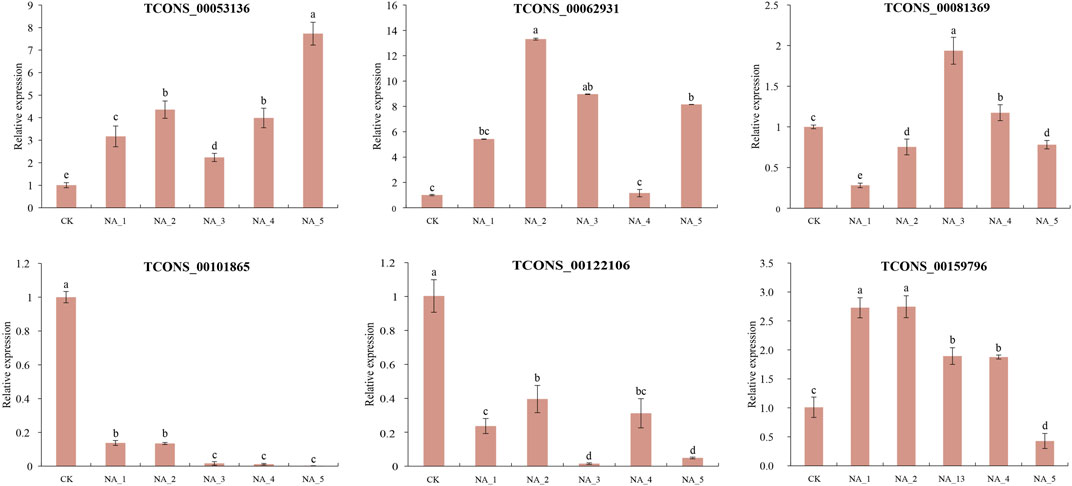
FIGURE 5. Expression validation of randomly selected DE-lncRNAs by the qRT-PCR assay. The significant difference was analyzed based on one-way ANOVA and Duncan’s multiple range tests with p < 0.05. Each bar shows the mean ± SE.
Discussion
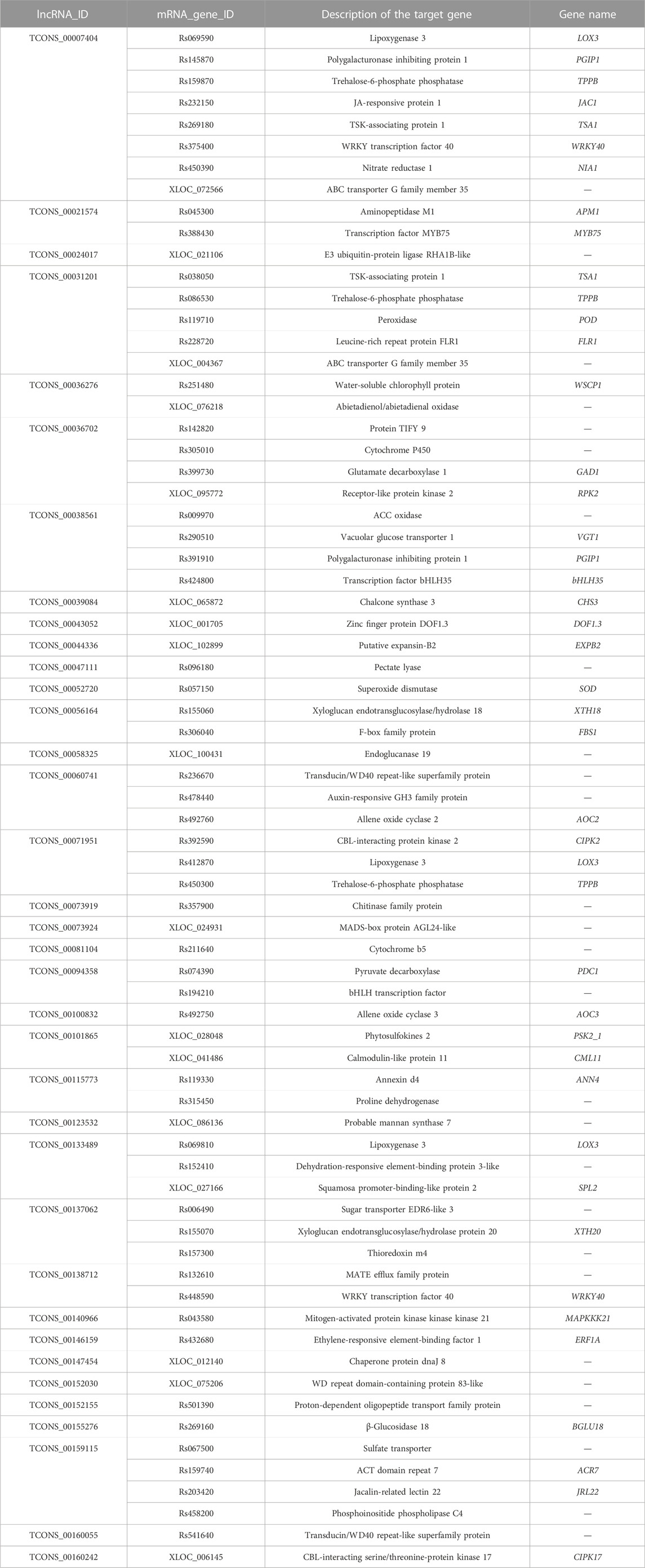
Salt stress is considered to be one of the main constraints on radish yield and quality. Recently, numerous investigations have highlighted the primary correlation between regulatory function and salt stress-responsive lncRNAs in plants (Zhang et al., 2019; Fu et al., 2020; Liu et al., 2022a; Chen et al., 2023). However, the potential role of lncRNAs during salt stress conditions in radish remains unexplored. In the current research, a total of 7,709 novel lncRNAs were systematically identified based on publicly accessible radish genome data using ssRNA-seq. Yet, in comparison to other plant species such as maize (Liu et al., 2022a), sesame (Gong et al., 2021), and peanut (Ma et al., 2020), only lincRNAs were observed in radish, manifesting that the lncRNA types may be highly specific to the context and species. Furthermore, it was also noticed that certain lncRNAs were not distributed on any chromosome, suggesting that these lncRNAs may be transcriptional noise (Nojima and Proudfoot, 2022). As anticipated, lncRNAs identified in radish were found to be shorter in total length and ORFs, and they also harbored fewer exons compared to protein-coding transcripts, which coincided with previous results in other plant species (Lv et al., 2016; Yan et al., 2020; Hu et al., 2022). We also recognized 363 lncRNAs that exhibit differential expression patterns in response to salt treatments (Supplementary Table S5). Notably, among these DE-lncRNAs, 33, 51, 72, 46, and 26 were exclusively detected at 6 h, 12 h, 24 h, 48 h, and 96 h of salt stress, respectively, when compared to the control (Figure 2A). These findings strongly suggested that the expression of salt-responsive lncRNAs was tightly regulated in a time-dependent manner, consistent with previous reports (Fu et al., 2020; Lee et al., 2022). A prior investigation has indicated that lncRNAs can exert their functions by enhancing gene expression at the transcriptional level (Ørom et al., 2010). Particularly, numerous upregulated DE-lncRNAs were dominant among all salinity conditions, implying a significant impact of high salinity on lncRNA expression.
Plants possess a diverse array of signaling molecules that facilitate the activation of appropriate defense responses in the presence of stress stimuli (Yu et al., 2017). The salt overly sensitive (SOS) signaling pathway, consisting of SOS3, SOS2, and SOS1 from the Na+/H+ exchanger (NHX), CBL-interacting protein kinase (CIPK), and calcineurin B-like (CBL) gene families, respectively, has been proposed to play a specific role in facilitating cellular signaling and maintaining ion homeostasis in response to salt stress in plants (Ji et al., 2013; Acharya et al., 2023). The CBL-CIPK signaling pathway, in particular, is a crucial component of the SOS pathway. In this work, although we did not directly predict SOS3, SOS2, and SOS1 as the lncRNA targets, the identification of salt-inducible DE-lncRNA-targeted CBL-interacting protein kinase 2 (CIPK2) and calcineurin B-like protein 1 (CBL1) might signify that lncRNAs play a role in regulating the SOS pathway in response to salt stress in radish. Annexin d4 (ANN4), a highly effective endogenous immunomodulatory protein, has been implicated in the cytosolic calcium signaling pathway in response to diverse stressors (Huh et al., 2010; Liao et al., 2017). In our experiment, the expression of TCONS_00115773-regulated ANN4 was significantly changed under salinity conditions (Table 2; Supplementary Table S7), indicating that lncRNA-ANN4-mediated calcium signaling may have a crucial function in enhancing radish resistance to high salinity. Phytohormones extensively participate in the adjustment of plants to unfavorable environmental elicitations. On the other hand, the intricate hormone signaling networks facilitate extensive crosstalk, thereby exerting crucial functions in the mediation of plant defense responses (Verma et al., 2016). Our analysis revealed the identification of DE-target genes involved in hormone signaling and responsive proteins, such as JA-responsive protein 1, and auxin-responsive GH3 family proteins (Tables 1, 2; Supplementary Table S7). Furthermore, we also noted that DE-lncRNA targeted several genes encoding mitogen-activated protein kinase (MAPK) cascades, including MAPK20 and MAPKKK21 (Tables 1, 2; Supplementary Table S7). MAPK cascades are a set of vital signaling kinases that participate in the intracellular transmission of extracellular signals via fine-tuning specific TFs, functional proteins, and transporters (Zhang and Zhang, 2022). Importantly, it is suggested that the interplay between second messengers, hormones, and MAPK modules is instrumental in adapting to diverse environmental cues (Smékalová et al., 2014), suggesting the essential involvement of lncRNAs in sensing and transducing stress signals to activate the defense mechanisms of radish under salt stress conditions.
The interactions between stress-specific TFs and lncRNAs were hypothesized to play a significant role in response to various abiotic stresses, including salt stress (Mirdar Mansuri et al., 2022). For example, in wheat, Shumayla et al. (2017) demonstrated that lncRNAs were co-expressed with the TFs related to WRKY, NAC, MYB, ERF, C3H, C2H2, bZIP, and bHLH families under salt stress conditions; several lncRNAs associated with TFs belonging to ARF, C2C2(Zn), and HSF families were found in duckweed (Fu et al., 2020); in maize, 11 target transcripts of the salt-responsive lncRNAs belonging to seven TF families, including bHLH, C2H2, Hap3/NF-YB, HAS, MYB, WD40, and WRKY, were predicted (Liu et al., 2022a). In this research, a specific group of genes related to TFs were also found in salt-responsive lncRNA–mRNA pairs in radish (Tables 1, 2; Supplementary Table S7). Specifically, the bHLH gene, bHLH96, was identified as a cis-regulated target gene of TCONS_00126664; a radish gene homologous to the Arabidopsis gene, namely, WRKY transcription factor 40 (WRKY40), was predicted to be targeted by TCONS_00007404 and TCONS_00138712. Moreover, TCONS_00146159 was identified to target a TF homologous to Arabidopsis ERF1A. The ERF gene TdERF1 has been found to potentially play a role in the mechanism of salt susceptibility/tolerance by regulating multiple hormone signaling pathways in wheat (Makhloufi et al., 2014), indicating its potential involvement in modulating plant responses to salt constraints. In addition, a considerable number of genes from the TIFY transcription factor family were found to be targeted by DE-lncRNAs. A recent study has reported that TIFYs play an indispensable role in various aspects of plant biology, including growth, development, signal transduction, and response to stress stimuli in plants (Liu et al., 2022b).
A set of evidence also supported the fact that high salinity induces oxidative and osmotic limitations, leading to the accumulation of ROS and dehydration (Cabot et al., 2014; Yang and Guo, 2018; Xiao and Zhou, 2023). Our results indicated that certain targets of DE-lncRNAs related to antioxidants exhibited significant changes in expression levels when subjected to salt conditions (Table 2 and S6). For instance, TCONS_00052720 and TCONS_00031201 were found to fine-tune superoxide dismutase (SOD) and peroxidase (POD), respectively, which was consistent with the report in cotton (Zhang et al., 2019), suggesting the essential role of lncRNAs in scavenging excess ROS during salt stress. Salt stress also triggers reduced water availability for plants. In this study, several DE-lncRNAs were identified to target transcripts encoding proteins analogous to potato DREB1 with dehydration-responsive element-binding protein 3-like properties (Table 2; Supplementary Table S7), which had been demonstrated to enhance salinity tolerance in transgenic potato plants (Bouaziz et al., 2013).
In addition, several functional proteins and transporters may play considerable roles in adjusting plant responses to salt stress. Chalcone synthase (CHS), a key enzyme in the flavonoid biosynthesis pathway, exerts a significant role in regulating plant growth, development, and defense against abiotic stress (Dao et al., 2011; Hou et al., 2022). The gene encoding chalcone synthase 3 (CHS3) was identified as a trans-acting target gene for four DE-lncRNAs (Table 2; Supplementary Table S7), suggesting that the lncRNA-mediated flavonoid biosynthesis pathway may play an essential role in response to salt stress in radish. Allene oxide cyclases (AOCs) were the pivotal genes in the JA biosynthetic pathway involved in regulating plant responses to developmental cues and environmental stresses. Numerous prior studies have demonstrated that the overexpression of allene oxide cyclase (AOC) enhances salinity tolerance in plants through the activation of jasmonate signaling (Pi et al., 2009; Zhao et al., 2014). In our study, a considerable number of DEGs encoding allene oxide cyclase 2 (AOC2) and allene oxide cyclase 3 (AOC3) were predicted to be potential targets of DE-lncRNAs (Table 2; Supplementary Table S7). Furthermore, the role of lipoxygenase 3 (LOX3) in Arabidopsis has been uncovered as a significant enzyme implicated in JA synthesis in response to salt stress (Ding et al., 2016). Our analysis differentiated that numerous DE-lncRNA-targeted genes encoding LOX3 displayed altered expression levels under salt stress conditions in radish (Table 2; Supplementary Table S7). Together, these findings might imply that the lncRNA-mediated regulation of the JA signaling pathway contributed to radish tolerance to salt stress in a complex and efficient way.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XS conceived and designed the research. MT and YS performed the experiments. WD and GC contributed powerful analytical tools. XS, LX, and XL analyzed data. XS wrote the manuscript. MT, LX, ZH, and CJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was, in part, supported by grants from the National Natural Science Foundation of China (32102399) and the Natural Science Foundation of Jiangsu Province (BK20181062).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1232363/full#supplementary-material
References
Acharya, B. R., Zhao, C., Reyes, L. A. R., Ferreira, J. F. S., and Sandhu, D. (2023). Understanding the salt overly sensitive pathway in Prunus: identification and characterization of NHX, CIPK, and CBL genes. Plant Genome, e20371. doi:10.1002/tpg2.20371
Baruah, P. M., Kashyap, P., Krishnatreya, D. B., Bordoloi, K. S., and Agarwala, N. (2021). Identification and functional analysis of drought responsive lncRNAs in tea plant. Plant gene. 27, 100311. doi:10.1016/j.plgene.2021.100311
Bouaziz, D., Pirrello, J., Charfeddine, M., Hammami, A., Jbir, R., Dhieb, A., et al. (2013). Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol. Biotechnol. 54, 803–817. doi:10.1007/s12033-012-9628-2
Cabot, C., Sibole, J. V., Barceló, J., and Poschenrieder, C. (2014). Lessons from crop plants struggling with salinity. Plant Sci. 226, 2–13. doi:10.1016/j.plantsci.2014.04.013
Cao, W., Gan, L., Wang, C., Zhao, X., Zhang, M., Du, J., et al. (2021). Genome-wide identification and characterization of potato long non-coding RNAs associated with Phytophthora infestans resistance. Front. Plant Sci. 12, 619062. doi:10.3389/fpls.2021.619062
Chekanova, J. A. (2015). Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 27, 207–216. doi:10.1016/j.pbi.2015.08.003
Chen, X., Jiang, X., Niu, F., Sun, X., Hu, Z., Gao, F., et al. (2023). Overexpression of lncRNA77580 regulates drought and salinity stress responses in soybean. Plants (Basel). 12, 181. doi:10.3390/plants12010181
Chen, X., Sun, Y., Cai, R., Wang, G., Shu, X., and Pang, W. (2018). Long noncoding RNA: multiple players in gene expression. BMB Rep. 51, 280–289. doi:10.5483/bmbrep.2018.51.6.025
Dao, T. T., Linthorst, H. J., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi:10.1007/s11101-011-9211-7
Ding, H., Lai, J., Wu, Q., Zhang, S., Chen, L., Dai, Y. S., et al. (2016). Jasmonate complements the function of Arabidopsis lipoxygenase 3 in salinity stress response. Plant Sci. 244, 1–7. doi:10.1016/j.plantsci.2015.11.009
Fu, L., Ding, Z., Tan, D., Han, B., Sun, X., and Zhang, J. (2020). Genome-wide discovery and functional prediction of salt-responsive lncRNAs in duckweed. BMC Genomics 21, 212. doi:10.1186/s12864-020-6633-x
Gil, N., and Ulitsky, I. (2020). Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi:10.1038/s41576-019-0184-5
Gong, H., You, J., Zhang, X., Liu, Y., Zhao, F., Cui, X., et al. (2021). Genome-wide identification and functional analysis of long non-coding RNAs in sesame response to salt stress. J. Plant Biol. 64, 555–565. doi:10.1007/s12374-021-09324-3
Grattan, S. R. (2016). Drought tip: crop salt tolerance. Richmond, CA, USA: University of California, Agriculture and Natural Resources.
Hou, Q., Li, S., Shang, C., Wen, Z., Cai, X., Hong, Y., et al. (2022). Genome-wide characterization of chalcone synthase genes in sweet cherry and functional characterization of CpCHS1 under drought stress. Front. Plant Sci. 13, 989959. doi:10.3389/fpls.2022.989959
Hou, X., Cui, J., Liu, W., Jiang, N., Zhou, X., Qi, H., et al. (2020). LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression. Phytopathology 110, 873–880. doi:10.1094/PHYTO-12-19-0445-R
Hu, X., Wei, Q., Wu, H., Huang, Y., Peng, X., Han, G., et al. (2022). Identification and characterization of heat-responsive lncRNAs in maize inbred line CM1. BMC Genomics 23, 208. doi:10.1186/s12864-022-08448-1
Huang, X., Zhang, H., Wang, Q., Guo, R., Wei, L., Song, H., et al. (2021). Genome-wide identification and characterization of long non-coding RNAs involved in flag leaf senescence of rice. Plant Mol. Biol. 105, 655–684. doi:10.1007/s11103-021-01121-3
Huh, S. M., Noh, E. K., Kim, H. G., Jeon, B. W., Bae, K., Hu, H. C., et al. (2010). Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant Cell Physiol. 51, 1499–1514. doi:10.1093/pcp/pcq111
Islam, W., Waheed, A., Naveed, H., and Zeng, F. (2022). MicroRNAs mediated plant responses to salt stress. Cells 11, 2806. doi:10.3390/cells11182806
Jha, U. C., Nayyar, H., Jha, R., Khurshid, M., Zhou, M., Mantri, N., et al. (2020). Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 20, 466. doi:10.1186/s12870-020-02595-x
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi:10.1093/mp/sst017
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi:10.1126/science.1138341
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Lee, M. H., Kim, K. M., Sang, W. G., Kang, C. S., and Choi, C. (2022). Comparison of gene expression changes in three wheat varieties with different susceptibilities to heat stress using RNA-Seq analysis. Int. J. Mol. Sci. 23, 10734. doi:10.3390/ijms231810734
Liao, C., Zheng, Y., and Guo, Y. (2017). MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 216, 163–177. doi:10.1111/nph.14679
Liu, P., Zhang, Y., Zou, C., Yang, C., Pan, G., Ma, L., et al. (2022a). Integrated analysis of long non-coding RNAs and mRNAs reveals the regulatory network of maize seedling root responding to salt stress. BMC Genomics 23, 50. doi:10.1186/s12864-021-08286-7
Liu, X., Yu, F., Yang, G., Liu, X., and Peng, S. (2022b). Identification of TIFY gene family in walnut and analysis of its expression under abiotic stresses. BMC Genomics 23, 190. doi:10.1186/s12864-022-08416-9
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Lv, Y., Liang, Z., Ge, M., Qi, W., Zhang, T., Lin, F., et al. (2016). Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genomics 17, 350. doi:10.1186/s12864-016-2650-1
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi:10.4161/rna.24604
Ma, X., Zhang, X., Traore, S. M., Xin, Z., Ning, L., Li, K., et al. (2020). Genome-wide identification and analysis of long noncoding RNAs (lncRNAs) during seed development in peanut (Arachis hypogaea L.). BMC Plant Biol. 20, 192. doi:10.1186/s12870-020-02405-4
Makhloufi, E., Yousfi, F. E., Marande, W., Mila, I., Hanana, M., Bergès, H., et al. (2014). Isolation and molecular characterization of ERF1, an ethylene response factor gene from durum wheat (Triticum turgidum L. subsp. durum), potentially involved in salt-stress responses. J. Exp. Bot. 65, 6359–6371. doi:10.1093/jxb/eru352
Mirdar Mansuri, R., Azizi, A. H., Sadri, A. H., and Shobbar, Z. S. (2022). Long non-coding RNAs as the regulatory hubs in rice response to salt stress. Sci. Rep. 12, 21696. doi:10.1038/s41598-022-26133-x
Nojima, T., and Proudfoot, N. J. (2022). Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 23, 389–406. doi:10.1038/s41580-021-00447-6
Ørom, U. A., Derrien, T., Guigo, R., and Shiekhattar, R. (2010). Long noncoding RNAs as enhancers of gene expression. Quant. Biol. 75, 325–331. doi:10.1101/sqb.2010.75.058
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi:10.1038/nbt.3122
Pi, Y., Jiang, K., Cao, Y., Wang, Q., Huang, Z., Li, L., et al. (2009). Allene oxide cyclase from Camptotheca acuminata improves tolerance against low temperature and salt stress in tobacco and bacteria. Mol. Biotechnol. 41, 115–122. doi:10.1007/s12033-008-9106-z
Quan, M., Chen, J., and Zhang, D. (2015). Exploring the secrets of long noncoding RNAs. Int. J. Mol. Sci. 16, 5467–5496. doi:10.3390/ijms16035467
Safdar, H., Amin, A., Shafiq, Y., Ali, A., and Yasin, R. (2019). A review: impact of salinity on plant growth. Nat. Sci. 17, 34–40. doi:10.7537/marsnsj170119.06
Shin, W. J., Nam, A. H., Kim, J. Y., Kwak, J. S., Song, J. T., and Seo, H. S. (2022). Intronic long noncoding RNA, RICE FLOWERING ASSOCIATED (RIFLA), regulates OsMADS56-mediated flowering in rice. Plant Sci. 320, 111278. doi:10.1016/j.plantsci.2022.111278
Shumayla, S., Sharma, S., Taneja, M., Tyagi, S., Singh, K., and Upadhyay, S. K. (2017). Survey of high throughput RNA-Seq data reveals potential roles for lncRNAs during development and stress response in bread wheat. Front. Plant Sci. 8, 1019. doi:10.3389/fpls.2017.01019
Smékalová, V., Doskočilová, A., Komis, G., and Samaj, J. (2014). Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 32, 2–11. doi:10.1016/j.biotechadv.2013.07.009
St Laurent, G., Wahlestedt, C., and Kapranov, P. (2015). The Landscape of long noncoding RNA classification. Trends Genet. 31, 239–251. doi:10.1016/j.tig.2015.03.007
Sun, X., Xu, L., Wang, Y., Luo, X., Zhu, X., Kinuthia, K. B., et al. (2016). Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 35, 329–346. doi:10.1007/s00299-015-1887-5
Sun, X., Xu, L., Wang, Y., Yu, R., Zhu, X., Luo, X., et al. (2015). Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genomics 16, 197. doi:10.1186/s12864-015-1416-5
Sun, Z., Huang, K., Han, Z., Wang, P., and Fang, Y. (2020). Genome-wide identification of Arabidopsis long noncoding RNAs in response to the blue light. Sci. Rep. 10, 6229. doi:10.1038/s41598-020-63187-1
Tian, Y., Bai, S., Dang, Z., Hao, J., Zhang, J., and Hasi, A. (2019). Genome-wide identification and characterization of long non-coding RNAs involved in fruit ripening and the climacteric in Cucumis melo. BMC Plant Biol. 19, 369. doi:10.1186/s12870-019-1942-4
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi:10.1186/s12870-016-0771-y
Wang, H., Chung, P. J., Liu, J., Jang, I. C., Kean, M. J., Xu, J., et al. (2014). Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 24, 444–453. doi:10.1101/gr.165555.113
Wang, H. V., and Chekanova, J. A. (2017). Long noncoding RNAs in plants. Adv. Exp. Med. Biol. 1008, 133–154. doi:10.1007/978-981-10-5203-3_5
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi:10.1016/j.molcel.2011.08.018
Wierzbicki, A. T., Blevins, T., and Swiezewski, S. (2021). Long noncoding RNAs in plants. Annu. Rev. Plant Biol. 72, 245–271. doi:10.1146/annurev-arplant-093020-035446
Xiao, F., and Zhou, H. (2023). Plant salt response: perception, signaling, and tolerance. Front. Plant Sci. 13, 1053699. doi:10.3389/fpls.2022.1053699
Xu, L., Wang, Y., Liu, W., Wang, J., Zhu, X., Zhang, K., et al. (2015). De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci. 236, 313–323. doi:10.1016/j.plantsci.2015.04.015
Yan, X., Ma, L., and Yang, M. (2020). Identification and characterization of long non-coding RNA (lncRNA) in the developing seeds of Jatropha curcas. Sci. Rep. 10, 10395. doi:10.1038/s41598-020-67410-x
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi:10.1111/nph.14920
Yu, X., Feng, B., He, P., and Shan, L. (2017). From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. doi:10.1146/annurev-phyto-080516-035649
Zhang, M., and Zhang, S. (2022). Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64, 301–341. doi:10.1111/jipb.13215
Zhang, X., Dong, J., Deng, F., Wang, W., Cheng, Y., Song, L., et al. (2019). The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 19, 459. doi:10.1186/s12870-019-2088-0
Zhang, Y., Fan, F., Zhang, Q., Luo, Y., Liu, Q., Gao, J., et al. (2022). Identification and functional analysis of long non-coding RNA (lncRNA) in response to seed aging in rice. Plants (Basel). 24 (11), 3223. doi:10.3390/plants11233223
Zhang, Y. C., Liao, J. Y., Li, Z. Y., Yu, Y., Zhang, J. P., Li, Q. F., et al. (2014). Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 15, 512. doi:10.1186/s13059-014-0512-1
Zhao, Y., Dong, W., Zhang, N., Ai, X., Wang, M., Huang, Z., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164, 1068–1076. doi:10.1104/pp.113.227595
Zhou, D., Chen, C., Jin, Z., Chen, J., Lin, S., Lyu, T., et al. (2022a). Transcript profiling analysis and ncRNAs' identification of male-sterile systems of Brassica campestris reveal new insights into the mechanism underlying anther and pollen development. Front. Plant Sci. 13, 806865. doi:10.3389/fpls.2022.806865
Keywords: lncRNA, salt stress, Raphanus sativus L., RNA-seq, target genes, Gene Ontology and pathway analyses
Citation: Sun X, Tang M, Xu L, Luo X, Shang Y, Duan W, Huang Z, Jin C and Chen G (2023) Genome-wide identification of long non-coding RNAs and their potential functions in radish response to salt stress. Front. Genet. 14:1232363. doi: 10.3389/fgene.2023.1232363
Received: 31 May 2023; Accepted: 17 October 2023;
Published: 02 November 2023.
Edited by:
Amaranatha Reddy Vennapusa, Delaware State University, United StatesReviewed by:
Aamir W. Khan, University of Missouri, United StatesAarthy Thiagaraya Selvam, Texas A&M University, United States
Shailesh Sharma, National Institute of Animal Biotechnology (NIAB), India
Copyright © 2023 Sun, Tang, Xu, Luo, Shang, Duan, Huang, Jin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaochuan Sun, eGNoc3VuMTk4N0AxNjMuY29t
†These authors have contributed equally to this work
 Xiaochuan Sun
Xiaochuan Sun Mingjia Tang
Mingjia Tang Liang Xu
Liang Xu Xiaobo Luo
Xiaobo Luo Yutong Shang4
Yutong Shang4 Guodong Chen
Guodong Chen