- Department of Pediatrics, Texas Tech University Health Sciences Center, Amarillo, TX, United States
Fabry disease (FD) is a rare genetic condition caused by mutations in the GLA gene, located on the X chromosome in the RPL36-HNRNPH2 readthrough genomic region. This gene produces an enzyme called alpha-galactosidase A (α-Gal A). When the enzyme does not function properly due to the mutations, it causes harmful substances called globotriaosylceramide (Gb3) and globotriaosylsphingosine (lyso-Gb3) to build up in the body’s lysosomes. This accumulation can damage the kidneys, heart, eyes, and nervous system. Recent studies have shown that the RPL36A-HNRNPH2 readthrough loci, which include RPL36A and HNRNPH2 genes, as well as the regulatory sequence known as the GLA-HNRNPH2 bidirectional promoter, may also play a role in FD. However, the involvement of enhancer RNAs (eRNAs) in FD is still poorly understood despite their known role in various diseases. To investigate this further, we studied an RPL36A enhancer called GH0XJ101390 and showed its genomic setting in the RPL36-HNRNPH2 readthrough region; the eRNA is rich in Homotypic Clusters of TFBSs (HCTs) type and hosts a CpG Island (CGI). To test the functional correlation further with GLA, RPL36A, and HNRNPH2, we used siRNAs to knock down GH0XJ101390 in human kidney embryonic cells 293T. The results showed a significant decrease in RPL36A and GLA expression and a non-significant decrease in HNRNPH2 expression. These findings could have important implications for understanding the regulatory mechanisms of GH0XJ101390 and its potential role in FD. A better understanding of these mechanisms may improve diagnostic and therapeutic methods for FD, which could ultimately benefit patients with this rare condition.
Introduction
Fabry disease (FD, OMIM#301500) is a rare X-linked lysosomal storage disease. Deficiency in the activity of the α-Gal A enzyme encoded by the GLA gene, caused by pathogenic mutations that can be found on ClinVar—NCBI and have been identified in patients with Fabry disease (FD), leads to the accumulation of globotriaosylceramide (Gb3) and globotriaosylsphingosine (lyso-Gb3) in various cells throughout the body. These accumulations are associated with the development of FD (Schiffmann et al., 2000; Ortiz et al., 2018). This condition can cause damage to the kidneys, heart, and nervous system, and patients may experience various warning signs and clinical symptoms (Tuttolomondo et al., 2021). Individuals with this condition may experience a range of symptoms, including skin lesions (angiokeratomas), eye abnormalities (corneal verticillate), reduced sweating (hypohidrosis), ringing in the ears (tinnitus), gastrointestinal issues, premature aging, and other related ailments. (Dutra-Clarke et al., 2020; Burand and Stucky, 2021; Kurschat, 2021; Lo Curto et al., 2021). It is becoming increasingly evident that FD is a complex condition, as the effects of the buildup of lysosomal Gb3 and lyso-Gb3 are not yet fully comprehended. Recent findings suggest that the disease’s seriousness and the effectiveness of treatment cannot be solely attributed to the accumulation of Gb3 and lyso-Gb3 (Ortiz et al., 2018; Bichet et al., 2021; Tuttolomondo et al., 2021). Intriguingly, Ensembl (ensembl.org) and Gene-NCBI (ncbi.nlm.nih.gov/gene) databases note the potential implication of RPL36A-HNRNPH2 readthrough loci in FD. The RPL36-HNRNPH2 readthrough genomic region is located on the forward strand of chromosome X, while GLA is found on the opposite, reverse strand of the readthrough locus. According to recent experimental data, RPL36A and HNRNPH2 are suggested to be involved in FD, as reported by Al-Obaide et al., in 2021 and 2022.
While mutations in the GLA gene’s a-galactosidase-A enzyme are considered the primary cause of FD, it is also possible that mutations in noncoding genomic regions that make up 98%–99% of the human genome could have a role in the disease. The ncRNA plays an important role in regulating gene expression, and even enhancers and promoters are considered part of it (Statello et al., 2021). In particular, enhancer RNAs (eRNAs) are essential for controlling gene expression and determining cell lineage specificity (Kim et al., 2010; Arnold et al., 2020). However, when these regulatory sequences malfunction, they can be linked to cancer and other age-related diseases, making targeted cancer therapy a potential treatment option (Kim et al., 2010; Chen et al., 2018; Zhang et al., 2019; Arnold et al., 2020; Tang et al., 2020; Richart et al., 2021; Liang et al., 2022; Zhang et al., 2022). Mutations in enhancers have also been implicated in various diseases (Claringbould and Zaugg, 2021). Interestingly, research has shown that promoters with enhancer activity can also regulate gene expression (Dao and Spicuglia, 2018; Medina-Rivera et al., 2018; Tippens et al., 2018). As a result, understanding and utilizing ncRNA has become increasingly important for potential diagnostic and therapeutic applications in the medical field. Recent studies have revealed a connection between ncRNA and FD. For instance, pathogenic variants have been discovered in the GLA-HNRNPH2 bidirectional promoter (BDP), as reported by Al-Obaide et al., in 2021. Additionally, Lo Curto et al., 2021 found that premature aging in FD patients is correlated with miR-126-3p and the accumulation of lyso-Gb3 in FD patients.
The GeneHancer database (Fishilevich et al., 2017) displays enhancers, promoters, and promoters with enhancer activities and their target genes. The GeneHancer shows a predicted sequence called GH0XJ101390 with enhancer and promoter features and potential regulatory activity at the 5′-side of the RPL36A gene. The GeneHancer predicted that GH0XJ101390 can target several genes, including the RPL36A and GLA genes. This study shows that the GH0XJ101390 sequence hosts unreported CpG Island (CGI) and Homotypic Clusters of TFBSs (HCTs). To date, no experimental studies have been conducted to determine how the GH0XJ101390 enhancer affects the expression of the RPL36A-HNRNPH2 readthrough loci, RPL36A and HNRNPH2, and the GLA locus. We report that using siRNA knockdown of GH0XJ101390 decreases the GLA and RPL36A expression. Our results may suggest the potentially critical role of GH0XJ101390 enhancer in FD, considering its role in regulating GLA and RPL36A loci. The reported results could aid in better understanding the role of GH0XJ101390 in the broad range of clinical symptoms of FD, leading to improved diagnosis and treatment approaches for FD.
Materials and methods
Database search and analyses of GH0XJ101390 enhancer and related genes
The GeneHancer database, which is part of the GeneCards Suite, includes the predicted GH0XJ101390 enhancer. GeneHancer links enhancers to genes by using four methods, including tissue co-expression correlation between genes and enhancer RNAs, enhancer-targeted transcription factor genes, expression quantitative trait loci for variants within enhancers, and capture Hi-C, which is a promoter-specific genome conformation assay. GeneHancer’s combinatorial likelihood-based scores for enhancer-gene pairing are based on individual scores from each of these four methods, as well as gene-enhancer genomic distances (Fishilevich et al., 2017). The GH0XJ101390 sequence was downloaded from the UCSC Genome Browser bioinformatics website, which provides sequence data and annotations (Karolchik et al., 2003; Lee et al., 2022). Even though GeneHancer displays 211 TFBSs, it does not reveal the presence of Homotypic Clusters of TFBSs (HCTs) in the GH0XJ101390 sequence. To identify HCTs, we employed two bioinformatics tools - the JASPAR and AnimalTFDB search tools. JASPAR is supported by several open-source software tools and Application Programming Interfaces (APIs) built using various programming languages like Python/Biopython and R/Bioconductor (Castro-Mondragon et al., 2022). AnimalTFDB uses the HMMER package to search and predict transcription factors (TFs) and DNA-binding domain (DBD). Moreover, the transcription factor binding sites (TFBS) are extracted from the HOCOMOCO, TRANSFAC, JASPAR, and CISBP databases (Zhang et al., 2012; Shen et al., 2023). We utilized these tools to identify HCTs in the GH0XJ101390 sequence. We utilized the default set parameters of “EMBOSS Cpgplot” to identify the CpG Island (CGI). The search parameters we used were: Observed/Expected ratio >0.60, %G + %C > %50, and CGI length >200. To align DNA sequences, we used two alignment tools: EMBOSS Needle for end-to-end alignment and EMBOSS Matcher to identify similar regions within the sequences. We referred to several online public genomics databases including NCBI-Gene, GeneCards, UCSC Genome Browser, and Ensembl Genome Browser to define the genomic setting of GH0XJ101390 to RPL36-HNRNPH2 readthrough, GLA, RPL36A, and HNRNPH2 loci. Data mining and bioinformatics analysis were conducted from January 2021 to September 2022. Supplementary Table S1 includes URLs for genomics databases and bioinformatics tools used in the study.
Human cell line 293T [HEK-293] and RNA extraction
We acquired the Human Embryonic Kidney 293T cell line (ATCC® CRL-1573™) from the American Type Culture Collection (ATCC). The cells were grown in Eagle’s Minimum Essential Medium (EMEM), ATCC catalog number 30–2003, with 10% heat-inactivated FBS (ATCC® 30-2020TM), 100 U/mL penicillin, and 100 μg/mL streptomycin. The incubation occurred in a humidified incubator with 5% CO2 at 37°C. To extract RNA from 293T cells, we utilized a purification kit (cat. no. 48700) from Norgen Biotek Corp, Thorold, ON, Canada.
siRNA transient transfections
The siRNA-27 kit (Catalog # SR304146) was purchased from OriGene, Rockville, MD, United States. The kit includes three siRNAs targeting specific sequences in the GH0XJ101390 and RPL36A transcript. The siRNA, identified as SR304146A-GGAACCUCCUAUAUACUUCCGUUUG, targets a specific site in the enhancer GH0XJ101390 transcript upstream of the RPL36A sequence. The siRNA SR304146C-GCUCACGCAAGCAUGGUUAACGUCC targets specific sequences in the GH0XJ101390 transcript and RPL36A transcript and untranslated transcript of exon-1 and translated exon-2 transcript. Meanwhile, SR304146B-GGACUUUCUGUAAGAAGUGUGGCAA targets the RL36A exon-2 (Supplementary Table S2, Supplementary Figure S1). The 293T cultured cells underwent transient transfection following the OriGene’s protocol. To introduce siRNA pool or the SR30004-scrambled negative control siRNA into the cells, we used the Roche X-tremeGENE siRNA transfection reagent and followed the manufacturer’s instructions, using a concentration of 10 nM. Four types of treatments were done on 293T cells. The first group was the control and remained untreated. The second group was treated with an X-tremeGENE transfection reagent, the third group was treated with the siRNA pool containing three different siRNAs, and the last group was treated with scrambled control siRNA. To prepare siRNAs and the transfection reagent, an Opti-MEM medium was utilized. The cells were cultured in EMEM with the addition of 10% heat-inactivated FBS. After 48 h of transfection, the cells were trypsinized and prepared for RNA extraction or Western blotting.
Primer design and RT-qPCR analysis
We utilized the PrimerQuest Tool to design custom primers for RPL36A, HNRNPH2, and GLA (Supplementary Table S3). The Taq universal SYBR Green One-Step Kit (BIO-RAD) protocol was followed to perform RT-qPCR reactions in triplicate. The BIO-RAD iCylcer iQ system software quantified the reaction measurements. To analyze the expression of target genes GLA, HNRNPH2, and RPL36A, normalization and relative expression analysis were conducted using HPRT1 as a reference gene (Al-Obaide et al., 2021; 2022). The Cq average value to determine the 2−ΔΔCT to examine the relative gene expression data from real-time quantitative PCR (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008).
Western blotting
The protein levels of GLA, HNRNPH2, and RPL36A were assessed by Western blot analysis from three replicates. To perform the analysis, 293T cells were lysed using Pierce IP Lysis buffer from Pierce Biotechnology in Rockford, United States. The supernatants obtained after centrifugation were used for analysis, and the protein concentrations were determined using a BCA Protein Assay Kit (Pierce Biotechnology). Fifty micrograms of protein were separated on a 10% SDS-PAGE gel and then transferred to an Immobilon-P membrane (Millipore Co.) after blocking in 4% BSA solution. The blots were then incubated with one of the following primary antibodies (host: rabbit): α-galactosidase antibody (Invitogen, cat. # PA5-27349), RPL36A antibody (Invitogen, cat. # PA5-70719), anti-HNRNPH2 (Sigma, # A00956) and beta Actin antibody (Invitogen, cat. # PA5-85490) for 6 h. The blots were then incubated with goat anti-rabbit IgG, and horseradish peroxide conjugated. Positive bands were visualized by enhanced chemiluminescence (ECL, Amersham Co.). The Western blots were photographed, and the bands were quantitated by densitometry using a VersaDoc 5,000 imaging system. ImageJ software (imagej.nih.gov) was used for data analysis of the investigated protein products of GLA, HNRNPH2, and RPL36A.
Statistical analysis
The siRNA experiments were conducted thrice, and each siRNA treatment was subjected to triplicate qPCR measurements. The Cq mean data from three experiments were used for 2−ΔΔCT analysis. Microsoft Excel 365 was employed for data sorting, analysis, and drawing diagrams. The data has been presented as the mean ± SD. For analyzing two independent samples, a t-test was used, and a significant difference was considered at p < 0.05.
Results
The genomic setting of the RPL36A-HNRNPH2 readthrough locus and the predicted GH0XJ101390 enhancer
The read-through RPL36A-HNRNPH2 locus is composed of two genes, the first is upstream, RPL36A coding ribosomal protein L36a with similarity to readthrough encoded protein, as shown by the matcher and needle alignment programs. The second gene is the downstream, coding heterogeneous nuclear ribonucleoprotein H2 gene, HNRNPH2. The GLA locus, which is implicated in FD, is mapped in between the RPL36A and HNRNPH2 loci on the reverse strand of the RPL36A-HNRNPH2 readthrough locus (Figure 1A). GeneHancer identifier affiliated with the GeneCards database shows an enhancer, GH0XJ101390, associated with regulating RPL36A, GLA genes, and 13 additional loci (Figure 1B). Our analysis revealed the GH0XJ101390 enhancer’s sequence occupies a genomic region at genomic coordinates (GRCh38): chrX: 101,390,257-101,393,641, upstream of RPL36A-HNRNPH2 readthrough locus on the forward strand of the long arm of chromosome X (Figure 1A, Supplementary Figure S1A). Alignment analysis showed the enhancer sequence overlaps upstream the 5′-side of the RPL36A locus and downstream untranslated RPL36A exon-1, Intron 1, exon 2, and Intron 2; also, the GH0XJ101390 sequence overlaps the RPL36A promoter (Figure 1A). Supplementary Figure S1B shows the human RPL36A NM_021029.6 mRNA transcript used in the analysis to identify the RPL36A untranslated exon 1 and translated exon 2 in the genomic sequence of the GH0XJ101390 enhancer. Stergachis et al., 2013 showed that codons, which specify amino acids host transcription factors binding sites.
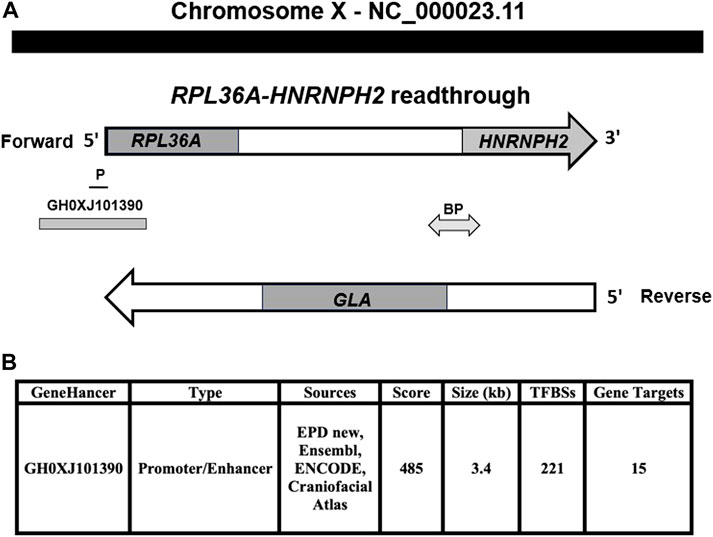
FIGURE 1. The genomic setting of GH0XJ101390 eRNA. (A) RPL36A-HNRNPH2 readthrough genomic region located on the chromosome X forward strand, RPL36A upstream, HNRNPH2 downstream, GLA gene at the reverse strand. (B) Genomic features of GH0XJ101390 enhancer and inferred number of target genes. GeneHancers are assigned a confidence score based on the number of sources, source scores, and TFBSs (from ENCODE). P: RPL36A promotor, BP: GLA, and HNRNPH2 bidirectional promoter.
In silico analysis of GH0XJ101390 sequence
Based on GeneHancer’s findings, the GH0XJ101390 spans 3.384 kb and contains 211 transcription factor binding sites (TFBSs). However, GeneHancer did not show any Homotypic Clusters of TFBSs (HCTs) in this enhancer, which are crucial components of enhancers (Gotea et al., 2010). Nonetheless, a bioinformatics analysis conducted in this study found eight HCTs types in the GH0XJ101390 sequence, which are POU3F2, NRF1, E2F4, TEF, VSX2, PDX1, PRRX2, and NKX2-5. Figure 2 provides examples of HCTs and their profiles. Furthermore, our analysis identified a CGI upstream of the GH0XJ101390 sequence. A CGI is a crucial part of an enhancer that helps to extend its long-range regulatory activity and control the responsiveness of its target genes (as discussed). The CGI sequence that we found is 428 bps in length and runs from nucleotide 537 to 964 along the GH0XJ101390 sequence, which is 3.384 kb. This CGI sequence is located at chrX: 101390737–101391163, as shown in Figure 3.
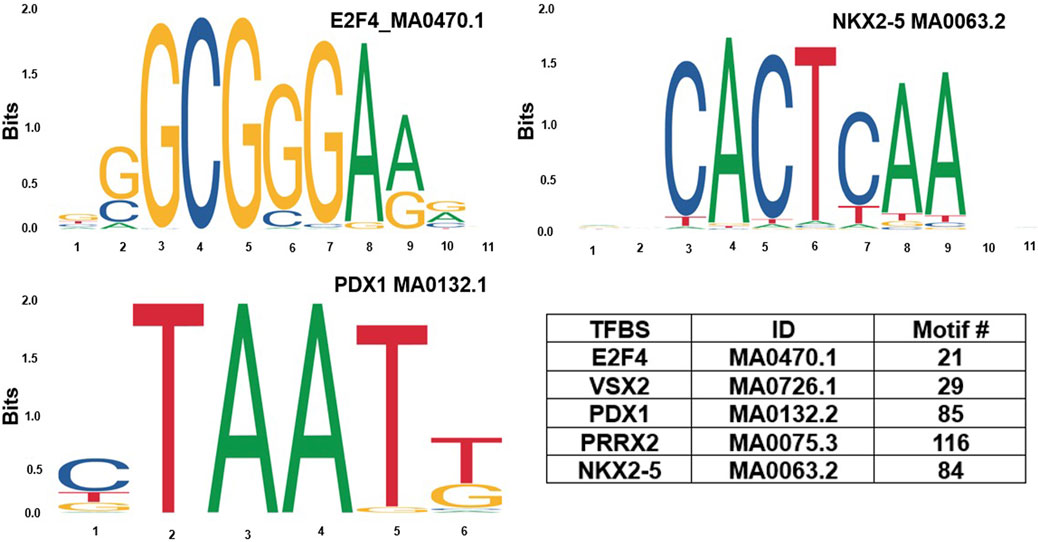
FIGURE 2. Examples of homotypic clusters of TFBSs (HCTs) profiles identified in the GH0XJ101390 eRNA sequence, and the ID and number of HCTs motifs identified in the GH0XJ101390 sequence.
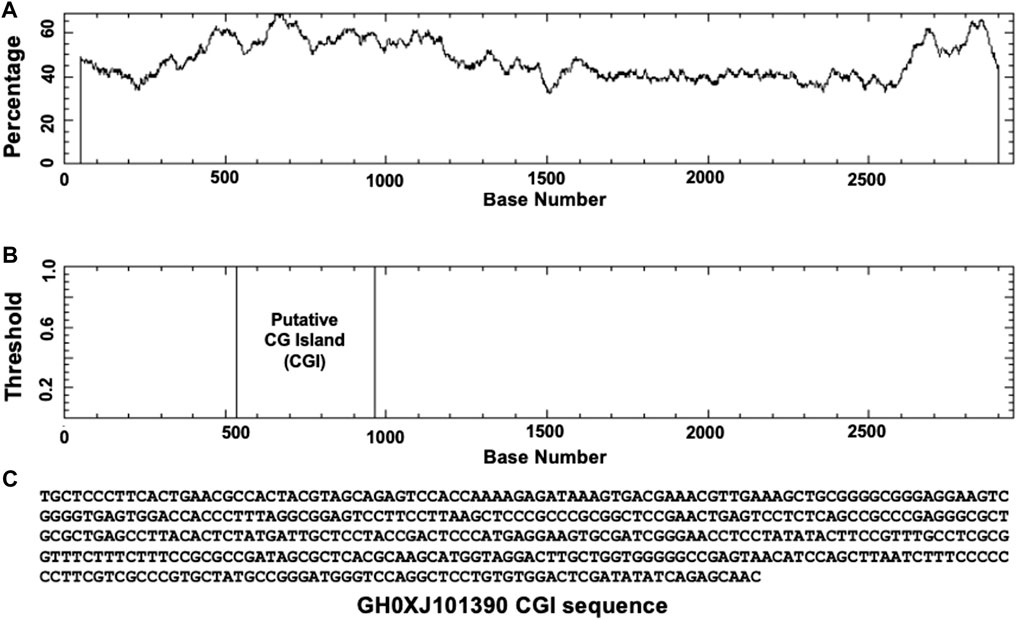
FIGURE 3. Identification of CGI in the GH0XJ101390 sequence. (A) percent C + percent G in the GH0XJ101390 sequence, (B) the position of CGI in the GH0XJ101390 nucleotide sequence, and (C) the identified CGI sequence.
Regulation of GLA, RPL36A and HNRNPH2 expressions by GH0XJ101390 eRNA
As far as we know, there have not been any experimental studies that demonstrate the effect of GH0XJ101390 enhancer on the expression of GLA, RPL36A, and HNRNPH2 genes. To explore the regulatory function of the GH0XJ101390, we conducted research on the human embryonic kidney 293T cell line. This cell line was chosen for its favorable characteristics in transfection. In order to determine the role of GH0XJ101390 in regulating GLA, and the other two loci RPL36A and HNRNPH2, we conducted a GH0XJ101390 siRNA silencing experiment on 293T cells using specific siRNAs that target the GH0XJ101390 sequence. After 48 h of transient transfection by siRNAs of 293T cells, RT-qPCR showed significant downregulation of GLA and RPL36A expressions, p < 0.05 (Figures 4A, B, Supplementary Table S4). This finding links the knocking down of GH0XJ101390 and GLA expression. On the other hand, a trend of non-significant decrease in HNRNPH2 was also observed by RT-qPCR, p > 0.05 (Figure 4C). The Western blot data indicated a decrease in GLA and RPlA36A proteins but was not observed in the HNRNPH2 protein (Figure 4D).
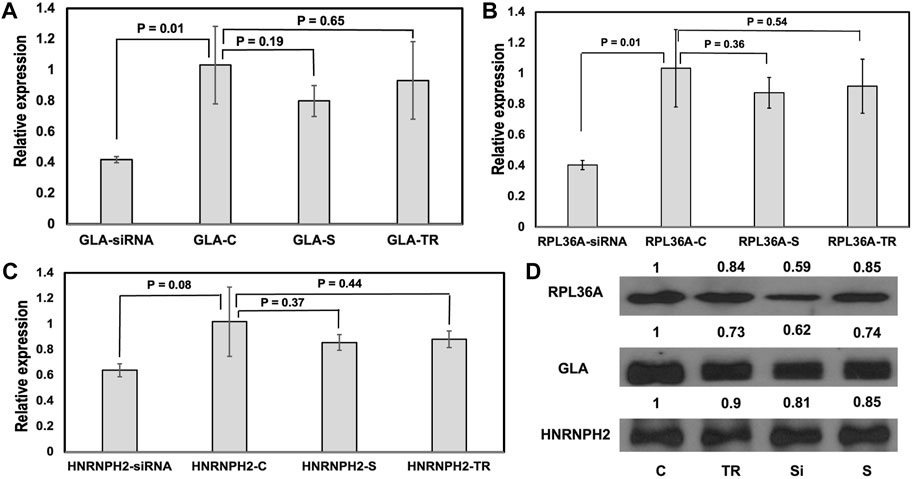
FIGURE 4. GH0XJ101390 influences the expressions of GLA, RL36A, and HNRNPH2. (A–C) RT-qPCR measured relative expressions of GLA, RPL36A, and HNRNPH2 in 293T cells treated with siRNA compared to control. *p < 0.05, ns: p > 0.05. (D) Western blot analysis of RPL36A, GLA, and HNRNPH2 proteins following siRNA- GH0XJ101390 knockdown in 293T cell line. C: untreated cells, TR: treatment by transfection reagent, S: treatment by scrambled control siRNA, siRNA: treatment by siRNA pool.
Discussion
Dysregulation of lncRNAs can lead to various diseases, including neurological and cardiovascular disorders (Esteller, 2011; Statello et al., 2021). Enhancers and promoters are examples of lncRNAs (Ørom and Shiekhattar, 2011; Ma et al., 2013; Kopp and Mendell, 2018; Han and Li, 2022; Mach and Giorgetti, 2023). Enhancer RNAs (eRNAs) are a specific type of lncRNA transcribed from enhancer regions of DNA sequences. They indicate gene expression, cell lineage specificity, and function or malfunction (Kim et al., 2010; Arnold et al., 2020). Thus, dysfunction of these regulatory sequences can result in various diseases (Izumi, 2016; Zhang et al., 2019; Gordevicius et al., 2021; Okabe and Kaneda, 2021; Zhang et al., 2022). Additionally, enhancers are linked to aging-related diseases (Chen et al., 2018). According to the findings of this study, the GH0XJ101390 eRNA may have a significant role to play in the expression of GLA. Although GLA is still a significant gene that impacts the clinical spectrum, disease progression, and treatments related to FD, there is growing suspicion that GLA pathogenic mutations alone may not be the sole cause of FD. Recent studies (Lo Curto et al., 2021; Tuttolomondo et al., 2021) suggest that FD may also occur due to the dysregulation of certain miRNAs such as miR-1307-5p and miR-126-3p levels, leading to progressive damage and premature aging of the kidneys, heart, and nervous system.
Through our study, we were able to identify the CGI present in the GH0XJ101390 sequence. The CGIs play a crucial role in enhancers, enhancing their long-range regulatory activity and controlling the responsiveness of target genes, as per Crispatzu et al. (2021). Since GH0XJ101390 modulates transcription, the identified CGI in the GH0XJ101390 sequence is likely involved in the long-range activity of GH0XJ101390, thus extending its function to GLA and HNRNPH2 expressions. Our experiments carried out in human 293T cells, were designed to provide data about potential transcriptional control of the RPL36A, GLA, and HNRNPH2 by GH0XJ101390 eRNA. The siRNA transient transfection experiments were conducted to knock down the GH0XJ101390 eRNA transcript(s), which resulted in a significant decrease in RPL36A and neighboring GLA expressions. However, HNRNPH2 expression was only insignificantly decreased. We chose to carry out our experiments in human 293T cells since it is widely utilized in transfection experiments due to its high transfectivity, rapid growth rate, and ability to grow in vitro culture (Tan et al., 2021).
Though the presence and regulatory functions of enhancers are undisputed, there is still a need for more information about their mechanisms in regulating gene expression. Our present study indicates that the GH0XJ101390 enhancer is involved in the regulation of RPL36A and GLA genes. However, the precise mechanism that regulates the GLA and RPL36A loci is yet to be understood. Our study shows that the GH0XJ101390 sequence coincides with the RPL36A exon1 and exon2 sequences. This finding sheds light on the dual purpose that human codons serve, specifying both amino acids and transcription recognition sites (Stergachis et al., 2013; Waldrop et al., 2019).
To better understand the function of GH0XJ101390 eRNA, further studies are necessary. Specifically, we should investigate the impact of specific siRNA on the GH0XJ101390 eRNA sequence upstream of RPL36A in comparison to siRNA that knockdown GH0XJ101390 sequences downstream of the RPL36A 5′-UTR region. Additionally, examining the effect of GH0XJ101390 on the RPL36A-HNRNPH2 readthrough locus is crucial. However, such a study presents a challenge due to the fact that the RPL36A-HNRNPH2 transcript spans two genes, and the NCBI-Gene reports the RPL36A-HNRNPH2 readthrough transcript produces a protein with similarity to the protein encoded by the upstream locus, RPL36A. As a result, further research is necessary to fully comprehend this critical genomic setting in readthrough transcript and protein products. In addition, it would be intriguing to explore how the secondary structure(s) of eRNAs affect their regulatory functions, as well as the impact of enhancer methylation on gene expression. It would also be worthwhile for future studies to investigate these matters in healthy individuals and compare them to GH0XJ101390 in FD patients.
To sum up, our study has uncovered an exciting possibility through analyzing the genomic setting of the GH0XJ101390 and experimental analysis. It seems that GH0XJ101390 may play a role in regulating the GLA. Furthermore, our bioinformatics analysis has identified homotypic clusters of TFBSs (HCTs) and CG Island (CGI) in the enhancer sequence of GH0XJ101390. This CGI appears to enhance the enhancer’s long-range regulatory activity, which in turn helps to modulate the expressions of GLA. This information can aid in understanding the underlying causes of Fabry disease and potentially may lead to the development of new diagnostic and treatment methods.
Data availability statement
The study’s original contributions are publicly available at [Link: https://www.ebi.ac.uk/biostudies/studies/S-BSST1240]. Supporting data and information accession number: S-BSST1240.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
TV initiated and supervised the study. MA-O conceived the study, searched, collected, curated datasets, performed bioinformatic and data analysis. MA-O, IA-O, and SI performed cell culture transfection experiments, and RT-qPCR. MA-O and TV wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Sanofi Genzyme investigator-sponsored study grant, SGZ-2019-12617.
Acknowledgments
We want to express our gratitude to Professor Kalkunte S. Srivenugopal for his support and for allowing us to utilize the equipment and facilities in his laboratory. We also appreciate the assistance provided by Dr. Viswanath Arutla in conducting the Western blot assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1229088/full#supplementary-material
References
Al-Obaide, M. A., Al-Obaidi, I. I., and Vasylyeva, T. L. (2021). Unexplored regulatory sequences of divergently paired GLA and HNRNPH2 loci pertinent to Fabry disease in human kidney and skin cells: presence of an active bidirectional promoter. Exp. Ther. Med. 21, 154. doi:10.3892/etm.2020.9586
Al-Obaide, M. A., Al-Obaidi, I. I., and Vasylyeva, T. L. (2022). The potential consequences of bidirectional promoter methylation on GLA and HNRNPH2 expression in Fabry disease phenotypes in a family of patients carrying a GLA deletion variant. Biomed. Rep. 17, 71. doi:10.3892/br.2022.1554
Arnold, P. R., Wells, A. D., and Li, X. C. (2020). Diversity and emerging roles of enhancer RNA in regulation of gene expression and cell fate. Front. Cell. Dev. Biol. 7, 377. doi:10.3389/fcell.2019.00377
Bichet, D. G., Torra, R., Wallace, E., Hughes, D., Giugliani, R., Skuban, N., et al. (2021). Long-term follow-up of renal function in patients treated with migalastat for Fabry disease. Mol. Genet. Metab. Rep. 28, 100786. doi:10.1016/j.ymgmr.2021.100786
Burand, A. J., and Stucky, C. L. (2021). Fabry disease pain: patient and preclinical parallels. Pain 162, 1305–1321. doi:10.1097/j.pain.0000000000002152
Castro-Mondragon, J. A., Riudavets-Puig, R., Rauluseviciute, I., Berhanu Lemma, R., Turchi, L., Blanc-Mathieu, R., et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. doi:10.1093/nar/gkab1113
Chen, H., Li, C., Zhou, Z., and Liang, H. (2018). Fast-evolving human-specific neural enhancers are associated with aging-related diseases. Cell Syst. 6, 604–611. doi:10.1016/j.cels.2018.04.002
Claringbould, A., and Zaugg, J. B. (2021). Enhancers in disease: molecular basis and emerging treatment strategies. Trends Mol. Med. 27, 1060–1073. doi:10.1016/j.molmed.2021.07.012
Crispatzu, G., Rehimi, R., Pachano, T., Bleckwehl, T., Cruz-Molina, S., Xiao, C., et al. (2021). The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nat. Commun. 12, 4344. doi:10.1038/s41467-021-24641-4
Dao, L. T. M., and Spicuglia, S. (2018). Transcriptional regulation by promoters with enhancer function. Transcription 9, 307–314. doi:10.1080/21541264.2018.1486150
Dutra-Clarke, M., Tapia, D., Curtin, E., Rünger, D., Lee, G. K., Lakatos, A., et al. (2020). Variable clinical features of patients with Fabry disease and outcome of enzyme replacement therapy. Mol. Genet. Metab. Rep. 26, 100700. doi:10.1016/j.ymgmr.2020.100700
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi:10.1038/nrg3074
Fishilevich, S., Nudel, R., Rappaport, N., Hadar, R., Plaschkes, I., Iny Stein, T., et al. (2017). GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017, bax028. doi:10.1093/database/bax028
Gordevicius, J., Li, P., Marshall, L. L., Killinger, B. A., Lang, S., Ensink, E., et al. (2021). Epigenetic inactivation of the autophagy-lysosomal system in appendix in Parkinson's disease. Nat. Commun. 12, 5134. doi:10.1038/s41467-021-25474-x
Gotea, V., Visel, A., Westlund, J. M., Nobrega, M. A., Pennacchio, L. A., and Ovcharenko, I. (2010). Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 20, 565–577. doi:10.1101/gr.104471.109
Han, Z., and Li, W. (2022). Enhancer RNA: what we know and what we can achieve. Cell Prolif. 55, e13202. doi:10.1111/cpr.13202
Izumi, K. (2016). Disorders of transcriptional regulation: an emerging category of multiple malformation syndromes. Mol. Syndromol. 7, 262–273. doi:10.1159/000448747
Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., et al. (2003). The UCSC genome browser database. Nucleic Acids Res. 31, 51–54. doi:10.1093/nar/gkg129
Kim, T. K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. doi:10.1038/nature09033
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Kurschat, C. E. (2021). Fabry disease-what cardiologists can learn from the nephrologist: a narrative review. Cardiovasc. Diagn. Ther. 11, 672–682. doi:10.21037/cdt-20-981
Lee, B. T., Barber, G. P., Benet-Pagès, A., Casper, J., Clawson, H., Diekhans, M., et al. (2022). The UCSC genome browser database: 2022 update. Nucleic Acids Res. 50, D1115–D1122. doi:10.1093/nar/gkab959
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lo Curto, A., Taverna, S., Costa, M. A., Passantino, R., Augello, G., Adamo, G., et al. (2021). Can be miR-126-3p a biomarker of premature aging? An ex vivo and in vitro study in Fabry disease. Cells 10, 356. doi:10.3390/cells10020356
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA. Biol. 10, 925–933. doi:10.4161/rna.24604
Mach, P., and Giorgetti, L. (2023). Integrative approaches to study enhancer-promoter communication. Curr. Opin. Genet. Dev. 80, 102052. doi:10.1016/j.gde.2023.102052
Medina-Rivera, A., Santiago-Algarra, D., Puthier, D., and Spicuglia, S. (2018). Widespread enhancer activity from core promoters. Trends biochem. Sci. 43, 452–468. doi:10.1016/j.tibs.2018.03.004
Okabe, A., and Kaneda, A. (2021). Transcriptional dysregulation by aberrant enhancer activation and rewiring in cancer. Cancer Sci. 112, 2081–2088. doi:10.1111/cas.14884
Ørom, U. A., and Shiekhattar, R. (2011). Long non-coding RNAs and enhancers. Curr. Opin. Genet. Dev. 21, 194–198. doi:10.1016/j.gde.2011.01.020
Ortiz, A., Germain, D. P., Desnick, R. J., Politei, J., Mauer, M., Burlina, A., et al. (2018). Fabry disease revisited: management and treatment recommendations for adult patients. Mol. Genet. Metab. 123, 416–427. doi:10.1016/j.ymgme.2018.02.014
Schiffmann, R., Murray, G. J., Treco, D., Daniel, P., Sellos-Moura, M., Myers, M., et al. (2000). Infusion of alpha-galactosidase A reduces tissue globotriaosylceramide storage in patients with Fabry disease. Proc. Natl. Acad. Sci. U. S. A. 97, 365–370. doi:10.1073/pnas.97.1.365
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Shen, W. K., Chen, S. Y., Gan, Z. Q., Zhang, Y. Z., Yue, T., Chen, M. M., et al. (2023). AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 51 (D1), D39–D45. doi:10.1093/nar/gkac907
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Stergachis, A. B., Haugen, E., Shafer, A., Fu, W., Vernot, B., Reynolds, A., et al. (2013). Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342, 1367–1372. doi:10.1126/science.1243490
Tan, E., Chin, C. S. H., Lim, Z. F. S., and Ng, S. K. (2021). HEK293 cell line as a platform to produce recombinant proteins and viral vectors. Front. Bioeng. Biotechnol. 9, 796991. doi:10.3389/fbioe.2021.796991
Tang, F., Yang, Z., Tan, Y., and Li, Y. (2020). Super-enhancer function and its application in cancer targeted therapy. NPJ Precis. Oncol. 4, 2. doi:10.1038/s41698-020-0108-z
Tippens, N. D., Vihervaara, A., and Lis, J. T. (2018). Enhancer transcription: what, where, when, and why? Genes Dev. 32, 1–3. doi:10.1101/gad.311605.118
Tuttolomondo, A., Simonetta, I., Riolo, R., Todaro, F., Di Chiara, T., Miceli, S., et al. (2021). Pathogenesis and molecular mechanisms of Anderson-Fabry disease and possible new molecular addressed therapeutic strategies. Int. J. Mol. Sci. 22, 10088. doi:10.3390/ijms221810088
Waldrop, E., Al-Obaide, M. A. I., and Vasylyeva, T. L. (2019). GANAB and PKD1 variations in a 12 years old female patient with early onset of autosomal dominant polycystic kidney disease. Front. Genet. 10, 44. doi:10.3389/fgene.2019.00044
Zhang, H. M., Chen, H., Liu, W., Liu, H., Gong, J., Wang, H., et al. (2012). AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40, D144–D149. doi:10.1093/nar/gkr965
Zhang, L., Ding, D., Liu, J., Liu, J., Wang, N., and Liu, J. (2022). Identification of prognostic and immunotherapy-related eRNA ID2-AS1 in bladder cancer. Med. Baltim. 10, e29759. doi:10.1097/MD.0000000000029759
Keywords: fabry disease, α-Gal A, enhancer, lncRNA, readthrough locus, bioinformatic
Citation: Al-Obaide MA, Islam S, Al-Obaidi I and Vasylyeva TL (2023) Novel enhancer mediates the RPL36A-HNRNPH2 readthrough loci and GLA gene expressions associated with fabry disease. Front. Genet. 14:1229088. doi: 10.3389/fgene.2023.1229088
Received: 25 May 2023; Accepted: 10 November 2023;
Published: 13 December 2023.
Edited by:
Bruna De Felice, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Markus Wolfien, Technical University Dresden, GermanySulev Kõks, Murdoch University, Australia
Copyright © 2023 Al-Obaide, Islam, Al-Obaidi and Vasylyeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tetyana L. Vasylyeva, dGV0eWFuYS52YXN5bHlldmFAdHR1aHNjLmVkdQ==
 Mohammed A. Al-Obaide
Mohammed A. Al-Obaide Saimul Islam
Saimul Islam Tetyana L. Vasylyeva
Tetyana L. Vasylyeva