- 1Laboratório de Citogenética de Peixes, Departamento de Genética e Evolução, Universidade Federal de São Carlos, São Carlos, Brazil
- 2Laboratory of Fish Genetics, Institute of Animal Physiology and Genetics, Czech Academy of Sciences, Liběchov, Czechia
- 3Institut für Humangenetik, Universitätsklinikum Jena, Jena, Germany
- 4Museu de Zoologia, Universidade de São Paulo, São Paulo, Brazil
- 5Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Ponta Grossa, Brazil
The remarkable fish biodiversity encompasses also great sex chromosome variability. Harttia catfish belong to Neotropical models for karyotype and sex chromosome research. Some species possess one of the three male-heterogametic sex chromosome systems, XY, X1X2Y or XY1Y2, while other members of the genus have yet uncharacterized modes of sex determination. Particularly the XY1Y2 multiple sex chromosome system shows a relatively low incidence among vertebrates, and it has not been yet thoroughly investigated. Previous research suggested two independent X-autosome fusions in Harttia which led to the emergence of XY1Y2 sex chromosome system in three of its species. In this study, we investigated evolutionary trajectories of synteny blocks involved in this XY1Y2 system by probing six Harttia species with whole chromosome painting (WCP) probes derived from the X (HCA-X) and the chromosome 9 (HCA-9) of H. carvalhoi. We found that both painting probes hybridize to two distinct chromosome pairs in Amazonian species, whereas the HCA-9 probe paints three chromosome pairs in H. guianensis, endemic to Guyanese drainages. These findings demonstrate distinct evolutionary fates of mapped synteny blocks and thereby elevated karyotype dynamics in Harttia among the three evolutionary clades.
1 Introduction
The Loricariidae family, which includes the Neotropical armored catfishes, is a promising group for evolutionary studies. Slightly above 1,000 species are distributed among 115 genera and most of this diversity occurs in the subfamilies Hypostominae, Hypoptopomatinae, and Loricariinae (Fricke et al., 2023). Loricariids have spread to almost all freshwater environments during their evolutionary history, being thus especially valuable in the artisanal fishery and the ornamental fish trade (Novák et al., 2022). Despite their great diversity in morphology, ecology, and habitats, Loricariidae is recovered as a monophyletic group in both morphological and molecular phylogenetic reconstructions, though the relationships between the tribes and genera have not been fully resolved so far (Covain et al., 2016; Londoño-Burbano and Reis, 2021). In this context, cytogenetic research provides important landmarks for solving issues related to taxonomy, phylogeny, and biodiversity in loricariids (e.g., Rocha-Reis et al., 2018; Glugoski et al., 2020; Takagui et al., 2020; Takagui et al., 2023; de Paula et al., 2022) as well as in other teleosts (Bellafronte et al., 2005; Cioffi et al., 2018; Gavazzoni et al., 2023). Moreover, particularly in the Neotropical region, with the richest freshwater biodiversity on the planet (Albert et al., 2020), cytogenetic investigation has abundant opportunities to deeply scrutinize the relationship between karyotype and sex chromosome evolution and reproductive isolation/diversification (Cioffi et al., 2017; Cioffi et al., 2018; de Souza et al., 2022).
Armored catfishes exhibit a wide variation in diploid chromosome numbers, ranging from 2n = 34 in Rineloricaria latirostris (Giuliano-Caetano, 1998) to 2n = 96 in Hemipsilichthys gobio (published as Upsilodus sp. in Kavalco et al., 2005). Some reports also documented population polymorphisms in the karyotype organization and repetitive DNA distribution, and the presence of B and sex chromosomes, the latter in different stages of differentiation (Scavone and Julio, 1994; Centofante et al., 2006; Rosa et al., 2012; Porto et al., 2014; Glugoski et al., 2018; Marajó et al., 2023). While the plesiomorphic 2n for loricariids has been supposed to be 2n = 54 (Artoni and Bertollo, 2001), this issue is still under intense debate, particularly considering the karyotypic features of species from the early-diverging clades (Takagui et al., 2020; Sassi et al., 2021).
Teleost fishes are well known for remarkable diversity of sex determination mechanisms (Devlin and Nagahama, 2002; Godwin and Roberts, 2018; Shen and Wang, 2018; Guiguen et al., 2019; Schartl et al., 2023). To date, more than 500 species have been found to carry one of nine so far known sex chromosome systems (El Taher et al., 2021; Sember et al., 2021), five of which are so-called multiple sex chromosome systems as they involve more than two chromosomes. Among 81 currently known cases of teleost multiple sex chromosomes (Deon et al., 2020; Sember et al., 2021; Ferchaud et al., 2022; de Araújo et al., 2023; Marajó et al., 2023; Nirchio et al., 2023), only nine are of the XY1Y2 type and evolved either via Y-fission or X-autosome fusion (Deon et al., 2020; Sember et al., 2021), Intriguingly, three of these systems have been found in Harttia spp., all from the Southeastern Brazilian drainages (Centofante et al., 2006; Blanco et al., 2013; Deon et al., 2020).
The genus Harttia (Loricariidae, Loricariinae) currently harbors 28 valid species (Oyakawa et al., 2018; Caldas et al., 2022; Fricke et al., 2023) as well as three other predicted species based on the cytogenetic data (Sassi et al., 2021). Harttia catfish are organized into three phylogenetic clades grouping representatives from Guyanese, Amazonian, and Southeastern Brazilian drainages (Covain et al., 2016). Harttia spp. are excellent models for studying interplay between chromosomal and species diversity in Neotropical fishes as they underwent extensive karyotype repatterning. Their 2n varies from 2n = 52/53 in H. carvalhoi (Centofante et al., 2006) to 2n = 62 in H. absaberi and Harttia sp. 2 (Rodrigues, 2010; Deon et al., 2020) and at least three distinct sex chromosome systems have been described in some species: an XX/XY sex chromosome system in H. rondoni (Sassi et al., 2020), and the two following multiple sex chromosome systems: X1X1X2X2/X1X2Y in H. duriventris, H. punctata, and H. villasboas (Blanco et al., 2013; Sassi et al., 2020), and XX/XY1Y2 in H. carvalhoi, H. intermontana and Harttia sp. 1 (Centofante et al., 2006; Deon et al., 2020). Studies using whole chromosome painting (WCP) probes have recently demonstrated that both the XY and X1X2Y sex chromosome systems are homologous, while the XY1Y2 system is formed by different linkage groups and thus has evolved independently (Deon et al., 2022a; b). Intriguingly, the three XY1Y2-bearing species share the ancestral sex chromosomes but differ by autosomal additions. While H. carvalhoi and Harttia sp. 1 entirely share the identity of XY1Y2 chromosomes, the same system in H. intermontana involves linkage group corresponding to chromosome 9 in H. carvalhoi, which implies two independent X-autosome fusions behind the origin of known Harttia XY1Y2 sex chromosomes (Deon et al., 2022a).
Here, we investigate six Harttia species from the Guyanese (H. guianensis) and Amazonian (H. villasboas, H. duriventris, H. rondoni, Harttia sp., and H. dissidens) clades using cross-species (i.e., Zoo-FISH) WCP experiments to examine the trajectory of karyotype and sex chromosome evolution. For this, we used the probes derived from the largest chromosomes (pairs X and 9) of H. carvalhoi (2n = 52/53 XY1Y2), bearing in mind that chromosome 9 is involved in the XY1Y2 sex chromosome system of H. intermontana. The results allowed us to demonstrate distinct evolutionary fates of mapped synteny blocks and thereby elevated karyotype dynamics in Harttia.
2 Materials and methods
2.1 Sampling and chromosome preparation
Individuals belonging to seven species (including H. carvalhoi used solely for the WCP probes preparation) were collected in distinct locations in Amazonas and Tocantins-Araguaia River basins (Figure 1; Table 1), with the approval of the Brazilian environmental agencies ICMBIO/SISBIO (License 48628-14) and SISGEN (A96FF09). We used cells from the posterior kidney and spleen tissue to obtain metaphase chromosomes by the classical air-drying technique (Bertollo et al., 2015). The specimens were deposited in the fish collection of the Instituto Nacional de Pesquisa da Amazônia (INPA) and the Museu de Zoologia da Universidade de São Paulo (MZUSP) as indicated in Table 1. The anesthesia and ethical practices used during the procedures were previously approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315).
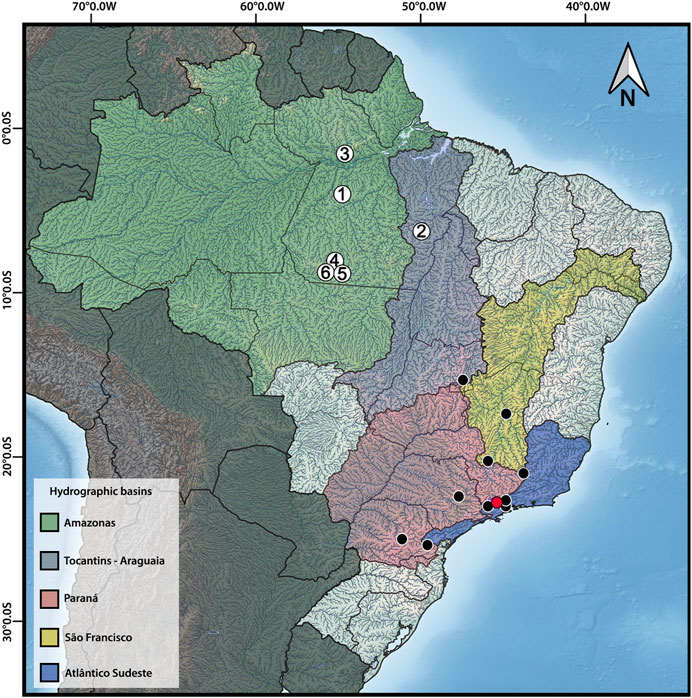
FIGURE 1. Distribution map of Harttia species with available cytogenetic data, highlighting the six species studied herein: 1 – H. dissidens, 2 – H. duriventris, 3 – H. guianensis, 4 – H. rondoni, 5 – H. villasboas, 6 – Harttia sp. 3. The red circle indicates the collection site of H. carvalhoi, a species used to construct WCP probes, while the black circles correspond to the distribution of other Harttia species which were not included in the present investigation. Major Brazilian hydrographic basins are highlighted: Amazonas in green, Tocantins-Araguaia in purple, Paraná in red, São Francisco in yellow, and Atlântico-Sudeste in blue. The map was created in QGIS 3.20 using Natural Earth package.
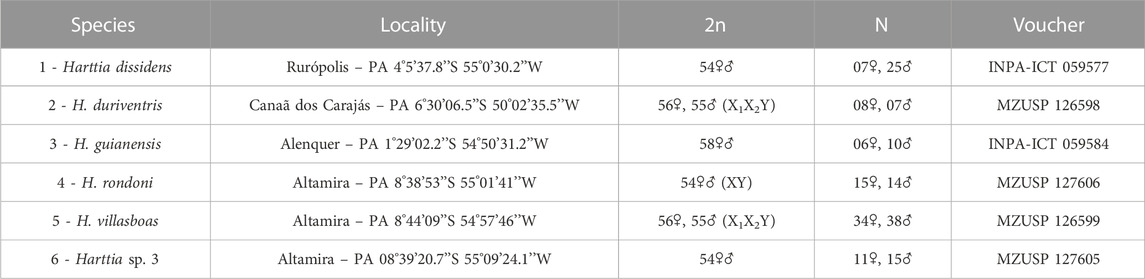
TABLE 1. Harttia species, collection sites, diploid chromosome numbers and sex chromosomes, sampling, and voucher numbers.
2.2 Microdissection and preparation of hybridization probes
We selected the largest metacentric (X) and the largest submetacentric (No. 9) chromosomes of H. carvalhoi since both chromosomes are involved in the two variant forms of the XY1Y2 system in Harttia species (Deon et al., 2022b). Glass-needle-based microdissection was used to isolate 15 copies of each target chromosome under an inverted microscope (Zeiss Axiovert 135). The obtained DNA was amplified using a degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) following Yang et al. (2009). The probes were labeled with Spectrum Green-dUTP and Spectrum Orange-dUTP (Vysis, Downers Grove, United States), in a secondary DOP-PCR reaction using 1 µL of the first amplification as template (Yang and Graphodatsky, 2009).
2.3 Whole chromosome painting (WCP)
Chromosome preparations were aged at 60 °C for 1h, then treated with RNAse solution [1.5 µL RNase A (10 mg/mL) in 1.5 mL 2×SSC] for 1 h 30min. Next, chromosomes were treated with 0.005% (v/v) pepsin solution [99 µL H2O, 10 µL 1M HCl, and 2.5 µL pepsin (20 mg/mL)]. Slides were denatured in 70% formamide in 2× SSC at 72 °C for 3 min. The probe mix per each slide contained 200 ng of each painting probe and 20 µg of blocking DNA which was obtained by DOP-PCR from H. carvalhoi genome and used as a blocker to highly and moderated repeated sequences (Yang et al., 2009). The final probe mixture was dissolved in the hybridization buffer containing 50% formamide, 2× SSC and 10% dextran sulfate, then denatured for 10 min at 86°C, cooled at 4°C for 2 min, and pre-annealed for 40 min at 37°C before being applied onto denatured chromosome slides. Hybridization took place in a dark moist chamber at 37°C for 72 h, followed by a series of post-hybridization washes in 1× SSC, 4× SSC/Tween, and 1× PBS solutions (for details, see Sassi et al., 2023). Chromosomes were finally counterstained with 4’, 6-diamidino-2-phenylindole (DAPI) 0.2 μg/mL in Vectashield mounting medium (Vector, California, United States).
2.4 Microscopic analyses and image processing
At least 30 metaphase spreads per individual were evaluated to confirm the WCP results. Metaphases were captured in an Axioplan II microscope (Carl Zeiss Jena GmbH, Germany) fluorescence microscope using the ISIS software (MetaSystems, Silver spring, MD, USA).
3 Results
Both HCA-X and HCA-9 probes hybridized to the chromosomes of all analyzed species (Figure 2). Each probe completely painted from end to end two distinct chromosome pairs (one metacentric and one submetacentric in both cases) in H. villasboas, H. duriventris, H. rondoni, and Harttia sp. 3. In addition to those pairs, H. guianensis also demonstrated a small metacentric pair probed with HCA-9 (Figure 2). In H. rondoni, additional HCA-X signals were found in pericentromeric regions of XY sex chromosomes, probably due to shared ribosomal DNA (rDNA) content which has been amplified on sex chromosomes in this species. Finally, in H. dissidens, the HCA-9 probe stained entirely one submetacentric pair but further only long arms of one metacentric pair, while the HCA-X probe revealed the same patterns as for the majority of studied species, with a metacentric and a submetacentric pair being stained. In this species, repetitive DNA sequences contained in both painting probes form distinct clusters, revealed as additional hybridization signals on several chromosomes. An ideogram compiling the present WCP results is provided in Figure 3.
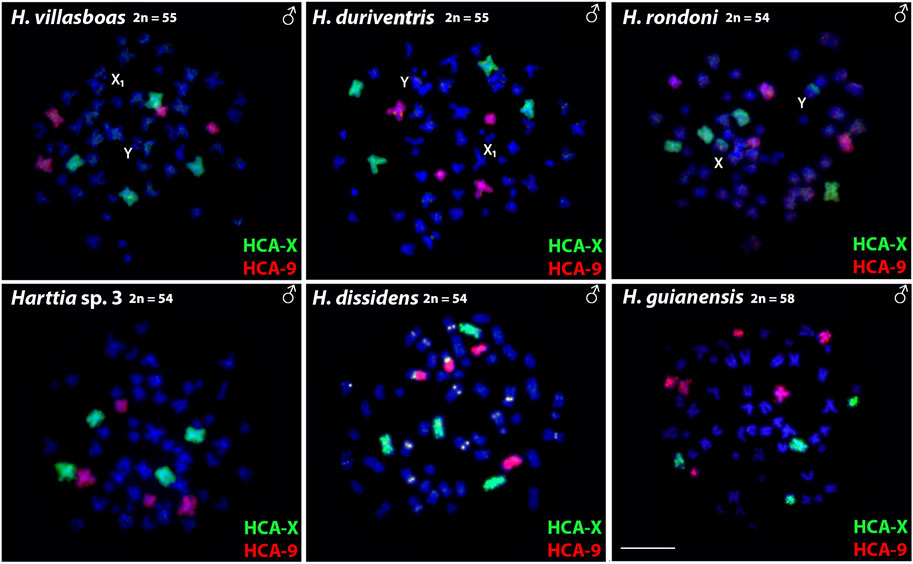
FIGURE 2. Whole chromosome painting with HCA-X (green) and HCA-9 (red) probes on the male chromosomes of six Harttia species. Sex chromosomes are indicated if present and detectable in the given species (first line). Scale Bar = 10 µm.
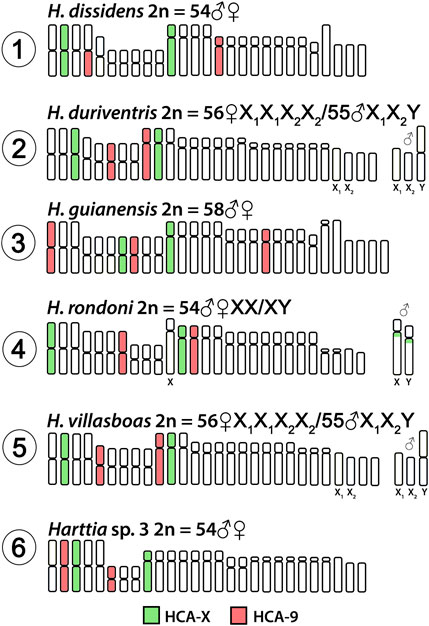
FIGURE 3. Idiogram summarizing obtained WCP/Zoo-FISH patterns by HCA-X and HCA-9 probes. HCA = Harttia carvalhoi.
4 Discussion
In the present study, we mapped two WCP probes prepared from H. carvalhoi chromosomes, where the chromosomes X and 9 were involved in rearrangements leading to emergence of the XY1Y2 sex chromosome system which is present in three Harttia species (Deon et al., 2022a). Both probes painted mostly entire chromosomes, thus pointing to preserved synteny blocks. The probes hybridized only to chromosomes without detectable sex-specific differences in size and shape (and therefore do not represent differentiated sex chromosomes) in species from the Amazonian and Guyanese clades (Figures 2, 3). These findings corroborate the view of independent events behind X1X2Y and XY1Y2 origin in Harttia (Deon et al., 2022a; b). We also show that chromosome 9 of H. carvalhoi, which has been fused to X chromosome in H. intermontana, is not involved in any other known sex chromosome system in Harttia, however, its propensity to fuse with other chromosomes has been recorded in H. dissidens.
The ancestral karyotype of Harttia is thought to be 2n = 58, with poorly differentiated or absent sex chromosomes (Blanco et al., 2017; Sassi et al., 2021). Until now, four species – H. gracilis, H. guianensis, H. kronei, H. longipinna–from the three phylogenetic clades are known to share these karyotype characteristics. In addition, H. punctata exhibits 58 chromosomes in females, but 57 in males due to the presence of X1X2Y sex chromosome system. Except for H. guianensis, which possesses three chromosome pairs labeled with the HCA-9 probe, all remaining five examined species, including H. punctata (Deon et al., 2022b), share the pattern of two distinct chromosome pairs being painted either with HCA-X or HCA-9 probe. Based on this evidence, it is possible to infer that the chromosome number reduction from the probable ancestral 2n = 58 to 2n = 56 in H. loricariformis, H. villasboas, and H. duriventris, and to 2n = 54 in H. rondoni and Harttia sp. 3 was achieved by chromosome fusions which did not involve the herein mapped chromosomes. On the other hand, in H. dissidens (2n = 54♀♂, Figure 2) the HCA-9 probe mapped to a submetacentric pair and the short arms of a metacentric pair, thus indicating that a fusion event involving the chromosome 9 of H. carvalhoi was responsible for its 2n reduction in relation to the ancestral karyotype. In support of this hypothesis, H. guianensis (2n = 58) also possesses more acrocentric chromosomes – five pairs – than the other species with lower 2n (Blanco et al., 2017; Deon et al., 2020; Sassi et al., 2020; Sassi et al., 2021). While the trajectory via centric fusions of acrocentric chromosomes seems highly probable, based on the available data we cannot entirely rule out also the possible involvement of biarmed (metacentric and submetacentric) chromosomes in the (specifically end-to-end/tandem) fusion events (cf. Schubert and Lysak, 2011) in the evolutionary history of Harttia.
It is notable that the patterns of hybridization in the Zoo-FISH experiments follows a biogeographical pattern, with H. guianensis (distributed in Guyanese drainages) showing three chromosomal pairs labeled with HCA-9, while the species from Amazonian and Tocantins-Araguaia River basins share the pattern of two chromosomal pairs being stained with the same particular WCP probe. Indeed, species from the Amazonian and Tocantins-Araguaia River basins exhibit more similar cytogenetic features when compared to H. guianensis, the only species from the Guyanese drainages karyotyped to date (discussed also in Sassi et al., 2021). Based on the cytogenetic and phylogenetic data from nuclear and mitochondrial DNA (Covain et al., 2016), it is possible to suggest that species from Guyanese drainages carry the ancestral-like karyotype arrangement (Sassi et al., 2021). As H. guianensis with the most diversified karyotype is the only species from the left bank of the Amazon River, it is tempting to speculate about the possible role of Amazon River as a barrier for gene flow. However, it is difficult to address this issue in the light of the still growing list of Harttia species which have been described within the last few years (Oyakawa et al., 2018; Caldas et al., 2022), and the existence of at least three karyotype forms proposed to represent new Harttia species (see Deon et al., 2020; Sassi et al., 2021). Moreover, at least seven Harttia species are known to occur in Guyanese drainages (Py-Daniel et al., 2001), but only H. guianensis has been investigated cytogenetically thus far. It is also noteworthy that the phylogenetic patterns of Harttia spp. are not well resolved, with the previously recognized monophyletic status of the genus (Covain et al., 2016) being questioned in a recent study (Cherobim, 2022).
Both the X and Y chromosomes of H. rondoni exhibit a positive HCA-X signal in the pericentromeric region of their long arms. As our previous study showed that pericentromeric regions of these chromosomes are enriched with tandem repeats of the major rDNA gene cluster (Sassi et al., 2020), the observed signal pattern might be attributed rather to shared repeat content between the HCA-X probe and the X and Y chromosomes and therefore does not represent homology of synteny blocks. However, the chromosome X of H. carvalhoi does not carry major rDNA site, neither do the chromosomes composing both forms of the XY1Y2 sex system of Harttia sp. 1 and H. intermontana (Blanco et al., 2017; Deon et al., 2020; 2022a). Bearing also in mind that rDNA sites outside the homeologous synteny blocks were not revealed in other herein studied species by the painting probes, it is highly probable that other repeat class interspersed with rDNA on the XY sex chromosomes of H. rondoni is responsible for the observed signal pattern. Analogous situation has been observed after application of HCA-X or HCA-9 probe in other Harttia spp. (Deon et al., 2022b). Similarly, an unknown repeat sequence contained in both painting probes demonstrated homology with certain regions on multiple chromosomes of H. dissidens.
Sex chromosomes in Loricariidae are promising targets for evolutionary research, since this family is the most diverse among Neotropical fishes (Fricke et al., 2023) and exhibits a wide range of sex chromosomes at distinct stages of differentiation (reviewed in Sember et al., 2021). Taking Harttia as an example, although inhabiting large Brazilian rivers, such as São Francisco, Paraná and Araguaia (Caldas et al., 2022), most species are found in riffle areas where they form isolated subpopulations with restricted/absent gene flow. This condition is being further pronounced by low vagility of these fishes, favoring a faster fixation of chromosomal rearrangements by genetic drift (Lande, 1977; King, 1993; Araya-Jaime et al., 2017), and thus also faster species divergence. Structuring into small, geographically isolated populations may also facilitate sex chromosome differentiation and transition between sex chromosome systems (e.g., Primo et al., 2017; Cioffi et al., 2018; Krysanov & Demidova, 2018; Glugoski et al., 2020; Štundlová et al., 2022; Marajó et al., 2023) To answer the question whether indeed genetic drift is a major force in sex chromosomes fixation in Harttia and other fish lineages with similar eco-geographical and demographic features and whether certain selective forces and ecological factors (cf. Pennell et al., 2015; Veller et al., 2017; Saunders et al., 2018; Meisel, 2022) might have played a role in this process, we have to acquire much deeper knowledge especially concerning genetic content and degree of sex chromosome differentiation, presence/absence of sex chromosomes in wide range of conspecific populations, and their phylogenetic distribution based on well-resolved phylogeny (cf. Sember et al., 2021). Particularly in Harttia, most of the phylogenetic reconstructions do not include all valid species, neither those recognized by cytogenetic studies (Covain et al., 2016; Roxo et al., 2019; Londono-Burbano and Reis, 2021; Sassi et al., 2021; Cherobim, 2022). In this context, given the collection efforts performed within the frame of the recent cytogenetic studies, distinct evolutionary units were identified in Harttia (Deon et al., 2020; Sassi et al., 2020) and their recognition as valid species remains to be investigated.
In the present study, we provide another piece of evidence strengthening the proposed view about 2n reduction in Harttia by fusions and about independent evolution of Harttia multiple sex chromosomes, with repeated origins involving different synteny blocks among species. A thorough phylogenetic reconstruction including all currently recognized representatives of Harttia is urgently needed for achieving a complete picture of sex chromosome evolution and karyotype reshaping in this noteworthy group of the Neotropical ichthyofauna.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315).
Author contributions
FS, GD, and MC conceived and designed research. FS and GD conducted experiments. FS, GD, AS, OM, LB, MV, and MC analysed the data. AS, TL, OO contributed with new methods. FS, AS, GD, LB, MV, and MC wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by São Paulo Research Foundation (FAPESP) grants 2020/02681-9 and 2022/04261-2 (FS), 2020/11772-8 (MC) and Brazilian National Council for Scientific and Technological Development (CNPq), grant number 302928/2021-9 (MC). AS was supported by Czech Academy of Sciences (RVO: 67985904 of IAPG CAS, Liběchov). We also acknowledge support for TL by the German Research Foundation Projekt-Nr. 512648189 and the Open Access Publication Fund of the Thueringer Universitaets-und Landesbibliothek Jena. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001. This study was supported by INCT - Peixes, funded by MCTIC/CNPq (proc. 405706/2022-7).
Acknowledgments
The authors are grateful to Eliana Feldberg and Lúcia Helena Rapp Py-Daniel from National Institute of Amazonian Research who provided assistance in the collection and identification of animal samples. This paper is dedicated in memory of Daniel Rodrigues Blanco, who contributed significantly to the research of Harttia chromosomes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albert, J. S., Tagliacollo, V. A., and Dagosta, F. (2020). Diversification of Neotropical freshwater fishes. Annu. Rev. Ecol. Evol. Syst. 51, 27–53. doi:10.1146/annurev-ecolsys-011620-031032
Araya-Jaime, C., Mateussi, N. T. B., Utsunomia, R., Costa-Silva, G. J., Oliveira, C., and Foresti, F. (2017). ZZ/Z0: The new system of sex chromosomes in Eigenmannia aff. Trilineata (Teleostei: Gymnotiformes: Sternopygidae) characterized by molecular cytogenetics and DNA barcoding. Zebrafish 14 (5), 464–470. doi:10.1089/zeb.2017.1422
Artoni, R. F., and Bertollo, L. A. C. (2001). Trends in the karyotype evolution of Loricariidae fish (Siluriformes). Hereditas 134 (3), 201–210. doi:10.1111/j.1601-5223.2001.00201.x
Bellafronte, E., Margarido, V. P., and Moreira-Filho, O. (2005). Cytotaxonomy of Parodon nasus and Parodon tortuosus (Pisces, Characiformes). A case of synonymy confirmed by cytogenetic analyses. Genet. Mol. Biol. 28, 710–716. doi:10.1590/S1415-47572005000500010
Bertollo, L. A. C., Cioffi, M. B., and Moreira-Filho, O. (2015). “Direct chromosome preparation from freshwater teleost fishes,” in Fish cytogenetic techniques. Editors C. Ozouf-Costaz, E. Pisano, F. Foresti, and L. F. Almeida-Toledo (Boca Raton, FL: CRC Press), 31–36.
Blanco, D. R., Vicari, M. R., Lui, R. L., Bertollo, L. A. C., Traldi, J. B., and Moreira-Filho, O. (2013). The role of the Robertsonian rearrangements in the origin of the XX/XY1Y2 sex chromosome system and in the chromosomal differentiation in Harttia species (Siluriformes, Loricariidae). Rev. Fish. Biol. Fish. 23, 127–134. doi:10.1007/s11160-012-9283-5
Blanco, D. R., Vicari, M. R., Lui, R. L., Traldi, J. B., Bueno, V., Martinez, J. D. F., et al. (2017). Karyotype diversity and evolutionary trends in armored catfish species of the genus Harttia (Siluriformes: Loricariidae). Zebrafish 14 (2), 169–176. doi:10.1089/zeb.2016.1377
Caldas, L., Cherobim, A. M., and Langeani, F. (2022). A new species of Harttia from the rio São Francisco basin (Siluriformes: Loricariidae). Neotrop. Ichthyol. 20 (4), e220051. doi:10.1590/1982-0224-2022-0051
Centofante, L., Bertollo, L. A. C., and Moreira-Filho, O. (2006). Cytogenetic characterization and description of an XX/XY1Y2 sex chromosome system in catfish Harttia carvalhoi (Siluriformes, Loricariidae). Cytogenet. Genome Res. 112 (3-4), 320–324. doi:10.1159/000089887
Cherobim, A. M. (2022). Phylogenetic analysis of Harttia Steindachner, 1877 (Siluriformes: Loricariidae: Loricariinae). [PhD Thesis]. [São José do Rio Preto (SP)]: Universidade Estadual Paulista.
Cioffi, M. B., Moreira-Filho, O., Ráb, P., Sember, A., Molina, W. F., and Bertollo, L. A. C. (2018). Conventional cytogenetic approaches—useful and indispensable tools in discovering fish biodiversity. Curr. Genet. Med. Rep. 6, 176–186. doi:10.1007/s40142-018-0148-7
Cioffi, M. B., Yano, C. F., Sember, A., and Bertollo, L. A. C. (2017). Chromosomal evolution in lower vertebrates: Sex chromosomes in neotropical fishes. Genes 8 (10), 258. doi:10.3390/genes8100258
Covain, R., Fisch-Muller, S., Oliveira, C., Mol, J. H., Montoya-Burgos, J. I., and Dray, S. (2016). Molecular phylogeny of the highly diversified catfish subfamily Loricariinae (Siluriformes, Loricariidae) reveals incongruences with morphological classification. Mol. Phylogenet. Evol. 94, 492–517. doi:10.1016/j.ympev.2015.10.018
de Araújo, L., Ramos, L. I., Vieira, M. M. D. R., Oliveira, A. V. D., Portela-Castro, A. L. D. B., Borin-Carvalho, L. A., et al. (2023). Cytogenetic and molecular characterization of Eigenmannia aff. desantanai (gymnotiformes: Sternopygidae): A first report of system of sex chromosomes ZW1W2/ZZ in Gymnotiformes. Zebrafish 20 (2), 77–85. doi:10.1089/zeb.2022.0059
de Paula, G. B., Gavazzoni, M., Zawadzki, C. H., Fernandes, C. A., Portela-Castro, A. L., Lui, R. L., et al. (2022). Identification of cryptic species in allopatric populations of Hypostomus tietensis (Siluriformes: Loricariidae) through cytogenetics analyses. Neotrop. Ichthyol. 20 (2), e210158. doi:10.1590/1982-0224-2021-0158
de Souza, F. H. S., Sassi, F. M. C., Ferreira, P. H. N., Bertollo, L. A. C., Ezaz, T., Liehr, T., et al. (2022). Integrating cytogenetics and population genomics: Allopatry and neo-sex chromosomes may have shaped the genetic divergence in the Erythrinus erythrinus species complex (Teleostei, Characiformes). Biology 11, 315. doi:10.3390/biology11020315
Deon, G. A., Glugoski, L., Hatanaka, T., Sassi, F. M. C., Nogaroto, V., Bertollo, L. A. C., et al. (2022a). Evolutionary breakpoint regions and chromosomal remodeling in Harttia (Siluriformes: Loricariidae) species diversification. Genet. Mol. Biol. 45 (2), e20210170. doi:10.1590/1678-4685-GMB-2021-0170
Deon, G. A., Glugoski, L., Sassi, F. M. C., Hatanaka, T., Nogaroto, V., Bertollo, L. A. C., et al. (2022b). Chromosomal rearrangements and origin of the multiple XX/XY1Y2 sex chromosome system in Harttia species (Siluriformes: Loricariidae). Front. Genet. 13, 877522. doi:10.3389/fgene.2022.877522
Deon, G. A., Glugoski, L., Vicari, M. R., Nogaroto, V., Sassi, F. M. C., Cioffi, M. B., et al. (2020). Highly rearranged karyotypes and multiple sex chromosome systems in armored catfishes from the genus Harttia (Teleostei, Siluriformes). Genes 11 (11), 1366. doi:10.3390/genes11111366
Devlin, R. H., and Nagahama, Y. (2002). Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 208 (3–4), 191–364. doi:10.1016/S0044-8486(02)00057-1
El Taher, A., Ronco, F., Matschiner, M., Salzburger, W., and Böhne, A. (2021). Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci. Adv. 7 (36), eabe8215. doi:10.1126/sciadv.abe8215
Ferchaud, A. L., Mérot, C., Normandeau, E., Ragoussis, J., Babin, C., Djambazian, H., et al. (2022). Chromosome-level assembly reveals a putative Y-autosomal fusion in the sex determination system of the Greenland halibut (Reinhardtius hippoglossoides). G3 (Bethesda) 12 (1), jkab376. doi:10.1093/g3journal/jkab376
Fricke, R., Eschmeyer, W. N., and Van der Laan, R. (2023). ESCHMEYER'S catalog of fishes: GENERA, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Accessed April 15, 2023).
Gavazzoni, M., Pavanelli, C. S., Graça, W. J., De Oliveira, E. A., Moreira-Filho, O., and Margarido, V. P. (2023). Species delimitation in Psalidodon fasciatus (Cuvier, 1819) complex (Teleostei: Characidae) from three hydrographic basins. Biol. J. Linn. Soc. 138 (1), 51–67. doi:10.1093/biolinnean/blac139
Giuliano-Caetano, L. (1998). Polimorfismo cromossômico Robertsoniano em populações de Rineloricaria latirostris (Pisces, Loricariinae). [Ph.D Thesis]. [São Carlos (SP)]: Universidade Federal de São Carlos.
Glugoski, L., Deon, G., Schott, S., Vicari, M. R., Nogaroto, V., and Moreira-Filho, O. (2020). Comparative cytogenetic analyses in ancistrus species (Siluriformes: Loricariidae). Neotrop. Ichthyol. 18 (2), e200013. doi:10.1590/1982-0224-2020-0013
Glugoski, L., Giuliano-Caetano, L., Moreira-Filho, O., Vicari, M. R., and Nogaroto, V. (2018). Co-located hAT transposable element and 5S rDNA in an interstitial telomeric sequence suggest the formation of Robertsonian fusion in armored catfish. Gene 650, 49–54. doi:10.1016/j.gene.2018.01.099
Godwin, J., and Roberts, R. (2018). “Environmental and genetic sex determining mechanisms in fishes,” in Transitions between sexual systems. Editor J. Leonard (Cham: Springer), 311–344. doi:10.1007/978-3-319-94139-4_11
Guiguen, Y., Fostier, A., and Herpin, A. (2019). “Sex determination and differentiation in fish: Genetic, genomic, and endocrine aspects,” in Sex control in aquaculture. Editors H. P. Wang, F. Piferrer, and S. L. Chen (Hoboken: John Wiley & Sons), 35–63. doi:10.1002/9781119127291.ch2
Kavalco, K. F., Pazza, R., Bertollo, L. A. C., and Moreira-Filho, O. (2005). Karyotypic diversity and evolution of Loricariidae (Pisces, Siluriformes). Heredity 94 (2), 180–186. doi:10.1038/sj.hdy.6800595
King, M. (1993). Species evolution: The role of chromosome change. Cambridge: Cambridge University Press.
Krysanov, E., and Demidova, T. (2018). Extensive karyotype variability of African fish genus Nothobranchius (Cyprinodontiformes). Comp. Cytogenet. 12 (3), 387–402. doi:10.3897/CompCytogen.v12i3.25092
Lande, R. (1977). The influence of the mating system on the maintenance of genetic variability in polygenic characters. Genetics 86 (2), 485–498. doi:10.1093/genetics/86.2.485
Londoño-Burbano, A., and Reis, R. E. (2021). A combined molecular and morphological phylogeny of the Loricariinae (Siluriformes: Loricariidae), with emphasis on the Harttiini and Farlowellini. PloS One 16 (3), 0247747. doi:10.1371/journal.pone.0247747
Marajó, L., Viana, P. F., Ferreira, A. M. V., Py-Daniel, L. H. R., Cioffi, M. B., Sember, A., et al. (2023). Chromosomal rearrangements and the first indication of an ♀X1X1X2X2/♂X1X2Y sex chromosome system in Rineloricaria fishes (Teleostei: Siluriformes). J. Fish. Biol. 102 (2), 443–454. doi:10.1111/jfb.15275
Meisel, R. P. (2022). Ecology and the evolution of sex chromosomes. J. Evol. Biol. 35 (12), 1601–1618. doi:10.1111/jeb.14074
Nirchio, M., Oliveira, C., Cioffi, M. B., Sassi, F. M. C., Valdiviezo, J., Paim, F. G., et al. (2023). Occurrence of sex chromosomes in fish of the genus Ancistrus with a new description of multiple sex chromosomes in the Ecuadorian endemic Ancistrus clementinae (Loricariidae). Genes 14 (2), 306. doi:10.3390/genes14020306
Novák, J., Hofmann, J., Hohl, D., Magalhães, A. L. B., and Patoka, J. (2022). Enigmatic armoured catfishes (Siluriformes: Callichthyidae and Loricariidae) in ornamental aquaculture: A new insight into neotropical fish diversity. Aquaculture 547, 737460. doi:10.1016/j.aquaculture.2021.737460
Oyakawa, O. T., Fichberg, I., and Py-Daniel, L. R. (2018). Three new species of Harttia (Loricariidae: Loricariinae) from serra do Cachimbo, rio Xingu basin, Pará, Northern Brazil. Zootaxa 4387 (1), 75–90. doi:10.11646/zootaxa.4387.1.3
Pennell, M. W., Kirkpatrick, M., Otto, S. P., Vamosi, J. C., Peichel, C. L., Valenzuela, N., et al. (2015). Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 11 (5), e1005237. doi:10.1371/journal.pgen.1005237
Porto, F. E., Gindri, B. S., Vieira, M. M. R., Borin, L. A., Portela-Castro, A. L. B., and Martins-Santos, I. C. (2014). Polymorphisms of the nucleolus organizing regions in Loricaria cataphracta (Siluriformes, Loricariidae) of the upper Paraguay River basin indicate an association with transposable elements. Genet. Mol. Res. 13, 1627–1634. doi:10.4238/2014.March.12.15
Primo, C. C., Glugoski, L., Almeida, M. C., Zawadski, C. H., Moreira-Filho, O., Vicari, M. R., et al. (2017). Mechanisms of chromosomal diversification in species of Rineloricaria (Actinopterygii: Siluriformes: Loricariidae). Zebrafish 14 (2), 161–168. doi:10.1089/zeb.2016.1386
Rocha-Reis, D. A., Brandão, K. O., Almeida-Toledo, L. F., Pasa, R., and Kavalco, K. F. (2018). The persevering cytotaxonomy: Discovery of a unique XX/XY sex chromosome system in catfishes suggests the existence of a new, endemic and rare species. Cytogenet. Genome Res. 156, 45–55. doi:10.1159/000492959
Rodrigues, R. M. (2010). Estudos cromossômicos e moleculares em Loricariinae com ênfase em espécies de Rineloricaria (Siluriformes, Loricariidae): uma perspectiva evolutiva. São Paulo: Universidade de São Paulo. [PhD Thesis]. [São Paulo (SP)].
Rosa, K. O., Ziemniczak, K., de Barros, A. V., Nogaroto, V., Almeida, M. C., Cestari, M. M., et al. (2012). Numeric and structural chromosome polymorphism in Rineloricaria lima (Siluriformes: Loricariidae): Fusion points carrying 5S rDNA or telomere sequence vestiges. Rev. Fish. Biol. Fish. 22, 739–749. doi:10.1007/s11160-011-9250-6
Roxo, F. F., Ochoa, L. E., Sabaj, M. H., Lujan, N. K., Covain, R., Silva, G. S., et al. (2019). Phylogenomic reappraisal of the Neotropical catfish family Loricariidae (Teleostei: Siluriformes) using ultraconserved elements. Mol. Phylogenet. Evol. 135, 148–165. doi:10.1016/j.ympev.2019.02.017
Sassi, F. M. C., Deon, G. A., Moreira-Filho, O., Vicari, M. R., Bertollo, L. A. C., Liehr, T., et al. (2020). Multiple sex chromosomes and evolutionary relationships in Amazonian catfishes: The outstanding model of the genus Harttia (Siluriformes: Loricariidae). Genes 11 (10), 1179. doi:10.3390/genes11101179
Sassi, F. M. C., Moreira-Filho, O., Deon, G. A., Sember, A., Bertollo, L. A. C., Liehr, T., et al. (2021). Adding new pieces to the puzzle of karyotype evolution in Harttia (Siluriformes, Loricariidae): Investigation of Amazonian species. Biology 10 (9), 922. doi:10.3390/biology10090922
Sassi, F. M. C., Toma, G. A., and Cioffi, M. B. (2023). “FISH—In fish chromosomes,” in Cytogenetics and molecular cytogenetics. Editor T. Liehr (Boca Raton, FL: CRC Press), 281–296.
Saunders, P. A., Neuenschwander, S., and Perrin, N. (2018). Sex chromosome turnovers and genetic drift: A simulation study. J. Evol. Biol. 31 (9), 1413–1419. doi:10.1111/jeb.13336
Scavone, M. D. P., and Júlio, H. F. (1994). Cytogenetic analysis and probable supernumerary chromosomes of Loricaria prolixa and Loricaria sp. females (Loricariidae-Siluriformes) from the Paraná River basin. Rev. Ictiol. 2 (3), 41–47.
Schartl, M., Georges, A., and Graves, J. A. M. (2023). Polygenic sex determination in vertebrates–is there any such thing? Trends Genet. 39, 242–250. doi:10.1016/j.tig.2022.12.002
Schubert, I., and Lysak, M. A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27 (6), 207–216. doi:10.1016/j.tig.2011.03.004
Sember, A., Nguyen, P., Perez, M. F., Altmanová, M., Ráb, P., and Cioffi, M. B. (2021). Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: State of the art and future challenges. Phil. Trans. R. Soc. B Biol. Sci. 376 (1833), 20200098. doi:10.1098/rstb.2020.0098
Shen, Z. G., and Wang, H. P. (2018). “Environmental sex determination and sex differentiation in teleosts–how sex is established,” in Sex control in aquaculture. Editors H-P. Wang, F. Piferrer, S-L. Chen, and Z-G. Shen (Hoboken, NJ: John Wiley & Sons), 85–115.
Štundlová, J., Hospodářská, M., Lukšíková, K., Voleníková, A., Pavlica, T., Altmanová, M., et al. (2022). Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci. Chromosome Res. 30 (4), 309–333. doi:10.1007/s10577-022-09707-3
Takagui, F. H., Baumgärtner, L., Venturelli, N. B., Paiz, L. M., Viana, P., Pompeo, L. R. S., et al. (2020). Unrevealing the karyotypic evolution and cytotaxonomy of armored catfishes (Loricariinae) with emphasis in Sturisoma, Loricariichthys, Loricaria, Proloricaria, Pyxiloricaria, and Rineloricaria. Zebrafish 17, 319–332. doi:10.1089/zeb.2020.1893
Takagui, F. H., Rubert, M., Dionisio, J. F., Baumgärtner, L., Cardoso, Y. P., Jerep, F. C., et al. (2023). Cytogenetic markers reinforce the redescription of the armored pleco Hypostomus spiniger (Loricariidae-Hypostominae), an endemic species in the Uruguay River basin and Patos Lagoon system. Braz. Arch. Biol. Tech. 66, e23220154. doi:10.1590/1678-4324-2023220154
Veller, C., Muralidhar, P., Constable, G. W., and Nowak, M. A. (2017). Drift-induced selection between male and female heterogamety. Genetics 207 (2), 711–727. doi:10.1534/genetics.117.300151
Yang, F., and Graphodatsky, A. S. (2009). “Animal probes and ZOO-FISH,” in Fluorescence in situ hybridization (FISH)—application guide. Editor T. Liehr (Berlin: Springer), 323–346.
Keywords: microdissection, WCP, chromosomal rearrangements, karyotype, evolution
Citation: Sassi FdMC, Deon GA, Sember A, Liehr T, Oyakawa OT, Moreira Filho O, Bertollo LAC, Vicari MR and Cioffi MdB (2023) Turnover of multiple sex chromosomes in Harttia catfish (Siluriformes, Loricariidae): a glimpse from whole chromosome painting. Front. Genet. 14:1226222. doi: 10.3389/fgene.2023.1226222
Received: 20 May 2023; Accepted: 17 July 2023;
Published: 28 July 2023.
Edited by:
Vindhya Mohindra, National Bureau of Fish Genetic Resources (ICAR), IndiaReviewed by:
Kris Christensen, Fisheries and Oceans Canada (DFO), CanadaJulio Cesar Pieczarka, Federal University of Pará, Brazil
Duílio M. Z. A. Silva, São Paulo State University, Brazil
Copyright © 2023 Sassi, Deon, Sember, Liehr, Oyakawa, Moreira Filho, Bertollo, Vicari and Cioffi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Liehr, dGhvbWFzLkxpZWhyQG1lZC51bmktamVuYS5kZQ==
 Francisco de Menezes Cavalcante Sassi
Francisco de Menezes Cavalcante Sassi Geize Aparecida Deon
Geize Aparecida Deon Alexandr Sember
Alexandr Sember Thomas Liehr
Thomas Liehr Osvaldo Takeshi Oyakawa4
Osvaldo Takeshi Oyakawa4 Luiz Antonio Carlos Bertollo
Luiz Antonio Carlos Bertollo Marcelo Ricardo Vicari
Marcelo Ricardo Vicari Marcelo de Bello Cioffi
Marcelo de Bello Cioffi