
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet. , 10 August 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1226052
This article is part of the Research Topic Evolution of Abiotic Stress Responses in Land Plants View all 6 articles
Chañar (Geoffroea decorticans (Gill., ex Hook. & Arn.) Burkart) has been highly significant for indigenous people in the Atacama Desert for over 3,000 years. Through evolutionary processes, the G. decorticans mitogenome likely underwent changes facilitating its adaptation to the extreme conditions of the Atacama Desert. Here, we compare the mitochondrial genome of G. decorticans with those of other Papilionoideae family species. The complete mitogenome of G. decorticans was sequenced and assembled, making it the first in the genus Geoffroea. The mitogenome contained 383,963 base pairs, consisting of 33 protein coding genes, 21 transfer RNA genes, and 3 ribosomal RNA genes. The Chañar mitogenome is relatively compact, and has two intact genes (sdh4 and nad1) which were not observed in most other species. Additionally, Chañar possessed the highest amount of mitochondrial DNA of plastid origin among angiosperm species. The phylogenetic analysis of the mitogenomes of Chañar and 12 other taxa displayed a high level of consistency in taxonomic classification, when compared to those of the plastid genome. Atp8 was subjected to positive selection, while the ccmFc and rps1 were subjected to neutral selection. This study provides valuable information regarding its ability to survive the extreme environmental conditions of the Atacama Desert.
Chañar, Geoffroea decorticans (Gill., ex Hook. & Arn.) Burkart, is considered to have been one of the most important wild trees for the indigenous populations that resided in the Atacama Desert around 1000 years BP (Ugalde et al., 2021). In the present day, this species is recognized for its diverse utility as a food resource, furniture material and medicinal product (Giménez, 2004; Nuñez et al., 2009; Costamagna et al., 2013; Jiménez-Aspee et al., 2017; Cotabarren et al., 2020). Surviving and providing sustenance to local communities under such challenging conditions is a remarkable achievement for any plant. The Atacama Desert, known as the world`s oldest and driest desert, presents extreme environmental conditions including high levels of UV radiation, high temperatures, extreme aridity, and highly saline and oxidizing soils (Eshel et al., 2021; Azua-Bustos et al., 2022). Geoffroea decorticans also inhabits other arid and semi-arid regions in Bolivia, Peru, and Argentina (Contreras Díaz, Porcile Saavedra and Aguayo Cruces, 2018), which are facing increasing aridity due to climate change. Drought, salinity, and high temperatures are highly important environmental factors that severely restrict plant growth and development (Krasensky and Jonak, 2012). In response to these abiotic stresses, plants employ various mechanisms, such as the production of reactive oxygen species (ROS), which can cause oxidative damage to lipids, proteins, and nucleic acids, ultimately leading to programmed cell death (Van Aken et al., 2009; Tang and Zhu, 2023).
Mitochondria play a key role in plant responses to abiotic stress (Newton et al., 2004; Liberatore et al., 2016). They are involved in energy production, metabolism, regulation of PCD, and ROS production (Tang and Zhu, 2023). Compared to plastid genomes, mitochondrial genomes demonstrate substantial variability in terms of size, structure (Smith and Keeling, 2015), and gene content (Liberatore et al., 2016). Plant mitochondrial genomes (mitogenomes) exhibit distinctive characteristics, including high rates of point mutations and structural rearrangements, genome expansion and contraction, integration of foreign DNA, gene loss, and transfer to the nuclear genome (Palmer et al., 2000; Chevigny et al., 2020). It is highly likely that the mitogenome structure of G. decorticans has undergone changes, through evolution, enabling its adaptation to extreme conditions, and accounting for its remarkable survival capability. Studying the genetic characteristics of plants that have adapted to these harsh conditions can contribute to the preservation of this valuable genetic resource that has sustained indigenous cultures for millennia. The objective of this study is 1) to compare the structural characteristics of the mitochondrial genome of G. decorticans with other species of Papilionoideae family species, focusing on gene content, genome size, the number of protein-coding genes with RNA editing, transfer of DNA from plastid regions, and 2) to confirm its phylogeny.
Fresh leaves of Chañar were collected near Copiapó, Chile. A subsample was stored in the Index Herbariorum of Universidad de Chile, with the voucher number EIF13815, and the rest of the leaves were used for DNA isolation, using a modified cetyltrimethylammonium bromide (CTAB) protocol (Contreras et al., 2020). The concentration of the DNA was measured using a Qubit™ 3.0 fluorometer and a Qubit™ dsDNA HS Assay Kit. To verify the integrity of the DNA, an Agilent 2100 Bioanalyzer was used, prior to sequencing. The NGS library was prepared using the TruSeq DNA LT Kit and sequencing was performed on Illumina next-generation sequencing (NGS) platforms. Paired-end sequences of 150 bp were generated for both forward (R1) and reverse (R2) reads. To filter the reads we used the Trim-Galore software (Krueger, 2019), which eliminates adapter remnants and low quality sequences (phred value <25). The SPAdes 4 software, version 3.13.0 (Bankevich et al., 2012) was used to assemble the filtered reads. Additionally, we mapped the reads back to the G. decorticans mitogenome assembly to visualize the read coverage, using Geneious Prime v2022.0.1 (Supplementary Figure S1, http://www.geneious.com; Kearse et al. (2012)). The annotation of the mitogenome was performed using AGORA (Jung et al., 2018) and MITOFY (Alverson et al., 2010) software. The circular map of the mitochondrial genome, along with annotation information, was generated using OrganellarGenomeDRAW (OGDRAW) (Greiner et al., 2019). The final annotated mitogenome sequence of G. decorticans was deposited in the NCBI GenBank, with the accession number OQ707067.
The features of the G. decorticans mitogenome were compared with ten closely related species in the Papilionoideae subfamily, i.e., Dalbergia odorifera T.C. Chen (MW441235), Arachis hypogaea L. (MW448460), Lotus japonicus (Regel) K. Larsen (NC_016743), Medicago sativa L. (ON782580), Glycine max (L.) Merr., 1917 (NC_020455), Phaseolus vulgaris L. (MK176514), Vigna angularis (Willd.) Ohwi & H. Ohashi (NC_021092), Pongamia pinnata (L.) Pierre (NC_016742), Sophora koreensis Nakai (NC_072933) and Castanospermum australe A. Cunn. & C. Fraser (MK426679). Additionally, we compared the G. decorticans mitogenome with mitogenomes of angiosperm-species from non-polar desert habitats (Supplementary Table S1). The length of the plastid-derived region of the mitogenome was evaluated using BLASTN (Johnson et al., 2008) with default parameters, because plant mitogenomes contain sequence elements that originate in the plastid genome (plastome), known as mitochondrial DNA of plastid origin (MIPT). Therefore, each mitogenome was used as the query versus a database comprising the plastomes corresponding to the species: MW672397, KX257487, NC_049008, MT571487, NC_007942, NC_002694, NC_042841, JN673818, EU196765, AP012598, and MW628966.
Thirty-three protein-coding gene (PCG) sequences, i.e., nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, sdh4, cob, cox1, cox2, cox3, atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFc, ccmFn, rps1, rps3, rps4, rps10, rps12, rps14, rpl5, rpl16, matR, and mttB, were used in the phylogenetic analysis of G. decorticans along with the previously mentioned ten Papilionoideae species. In addition, two Caesalpinioideae species, Leucaena trichandra (Zucc.) Urb. (NC_039738) and Acacia ligulata A. Cunn. ex Benth. (NC_040998), were included as outgroups. The 33 PCG sequences were aligned separately using MAFFT v7 (Katoh and Standley, 2013) and any gaps in the alignment were trimmed using trimAl v1.4 (Capella-Gutiérrez et al., 2009). Subsequently, the sequences were concatenated with Mesquite 3.81 software (Maddison and Maddison, 2023). The analyses of the mitochondrial genomes’ 33 PCG sequences were conducted using the maximum likelihood (ML) method. We did the same for the complete plastid genome sequences, using the plastome accessions listed above (including the outgroup species accessions NC026134.2 and NC028733) in order to compare the resulting phylogenetic trees. The best-fitting nucleotide substitution model of sequence evolution, model TVM + G4, was determined using the Corrected Akaike Information Criterion (AICc) through Modeltest-NG on XSEDE (Darriba et al., 2020). The ML analyses were carried out using RAxML-HPC BlackBox v.8.1.12 (Stamatakis, 2014) with 1,000 bootstrap replicates, using the CIPRES Science Gateway v3.3 (Miller et al., 2010). Non-parametric bootstrap support (BS) values were used to measure the internal nodes of the resulting trees. The ratio of non-synonymous substitution (Ka) to synonymous substitution (Ks) was calculated for 25 PCGs of G. decorticans and ten Papilionoideae species, using the KaKs_Calculator tool 3.0 (Zhang, 2022) with the MA model, where Ka/Ks values of >1 signify that the gene is subjected to positive selection, Ka/Ks values equal to 1 indicate neutral selection, and Ka/Ks values < 1 signify purification.
We successfully sequenced and assembled the complete mitogenome of G. decorticans, resulting in a single circular genome with a length of 383,969 bp (Figure 1; GenBank accession number OQ707067). The mitogenome sizes of G. decorticans and ten other Papilionoideae species varied from 290,285 to 592,341 bp (Table 1). Mitogenome sizes can exhibit significant variation among plant species, for example, among angiosperm the mitogenome sizes range from 66 kb in the parasitic plant, Viscum scurruloideum (Skippingtona et al., 2015) to 11,300 kb in Silene conica (Sloan et al., 2012). According to Choi et al. (2019), the median size of seed plant mitogenomes is 476 kb. However, within the Fabaceae family, mitogenome sizes vary considerably from 271,618 to 729,504 bp. Therefore, G. decorticans possesses a relatively small mitogenome size compared to other Papilionoideae species, but it falls within an intermediate range when compared to angiosperm mitogenomes. The variations in mitogenome size among plant species can be attributed to various factors. Mitogenomic chromosome loss, gain of exogenous DNA through intracellular gene transfer and horizontal gene transfer, and the acquisition of repetitive DNA are likely explanations for the increases and decreases observed in mitogenome sizes in angiosperms (Choi et al., 2019). Additionally, some studies suggest that changes in mitochondrial genome size can be influenced by environmental stresses (Xiong et al., 2022).
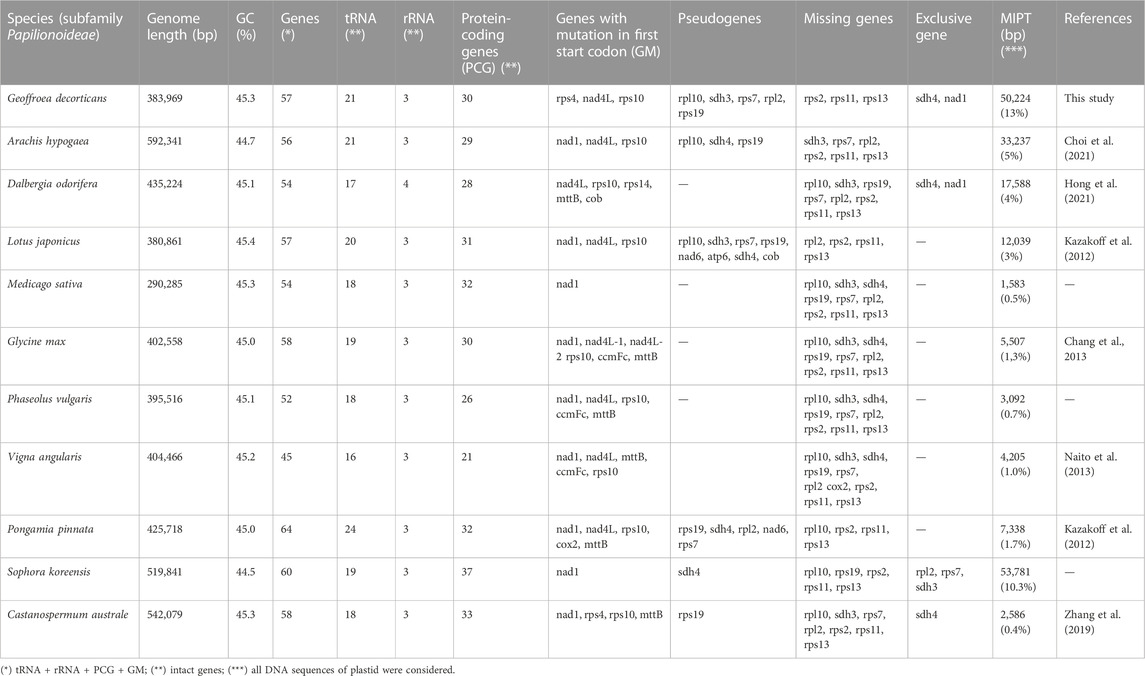
TABLE 1. General features of mitogenome of Geoffroea decorticans and other ten Papilionoideae species.
The total GC content of G. decorticans was 45.3%, which was similar to the other Papilionoideae species, ranging from 44.5% in S. koreensis to 45.4% in L. japonicus (Table 1). In the mitogenome of G. decorticans, we identified a total of 57 genes, including 33 protein-coding genes (PCG), of which 30 were intact PCGs and 3 had mutations in the first start codon (Table 1). Additionally, there were 21 tRNA genes and 3 rRNA genes (Table 1). The number of genes in the mitogenomes of other Papilionoideae species varied from 45 genes in V. angularis to 64 genes in P. pinnata (Table 1). Moreover, V. angularis had the lowest number of PCGs (21) and S. koreensis the highest (37). The number of tRNA genes ranged from 16 in V. angularis to 24 in P. pinnata, while the number of rRNA genes was consistent across most mitogenomes (3 genes), except for Dalbergia odorifera which had 4 rRNA genes (Table 1). Interestingly, G. decorticans and A. hypogaea had the second-highest number of tRNA (21) genes. In angiosperms, mitochondrial tRNA genes are known to be heterogeneous, with a variable number of native tRNA genes (typically 11–13 genes) and tRNAs acquired from different sources through intracellular and horizontal transfers (Warren et al., 2021). Several studies have suggested a link between highly accelerated rates of mitochondrial sequence evolution and a reduced number of tRNA genes. For example, species like Silene conica and Silene noctiflora and Viscum (mistletoe) have a reduced tRNA gene content (Skippingtona et al., 2015; Warren et al., 2021). In these cases, tRNA genes are replaced by nuclear-encoded homologs, leading to a gene substitution process (Warren et al., 2021). Therefore, considering the high number of tRNA genes (21) in G. decorticans, it could be hypothesized that this species exhibits a reduced rate of mitochondrial sequence evolution.
The presence of genes with mutations in the first start codon was observed in the mitogenomes of the other Papilionoideae species, ranging from 1 gene in Medicago sativa and S. koreensis to 6 genes in G. max (Table 1). These mutations, known as RNA editing (C-to-U RNA editing) occur at protein genes’ first and second codon positions. The functional significance of RNA editing is not yet fully understood (Sloan et al., 2012), but it might play a role in the maintenance and function of gene and genome architecture (Linch, 2007), as well as in gene regulation, protein isoform generation and modification of active protein complexes (Lo Giudice et al., 2019). Furthermore, Murayama et al. (2012) suggested that mitochondrial function, specifically RNA editing at the nad4 gene, interacts with and regulates the action of stress-related hormones in plants. It was found that an RNA editing site in mitochondrial nad4 transcripts was targeted by AHG11, resulting in the production of more mRNAs for oxidative stress-responsive genes (Murayama et al., 2012). In G. decorticans, as well as in the other species belonging to the Papilionoideae family, the nad4 gene remains intact, while in most of them the nad4L gene undergoes RNA editing.
Generally, vascular plants have been found to contain between 20 and 40 protein-coding genes (PCGs) in their mitogenomes (Møller et al., 2021). Mitogenomes of the species of the Fabaceae family have around 30 intact PCGs (Choi et al., 2019). In the case of the G. decorticans mitogenome, we discovered 30 intact PCGs, 3 PCGs with mutations in the first start codon, 5 pseudogenes (rpl10, sdh3, rps7, rpl2, and rps19) and 3 lost ribosomal protein genes (rps2, rps11, and rps13) (Table 1). The number of PCGs in G. decorticans (30) falls thus within the expected range for the Fabaceae species. It has been observed before that pseudogenes, truncations and deletions of the rps7, rps11, rps13, and rps2 genes were prevalent in numerous Fabaceae species (Choi et al., 2019). This observation aligns with our findings in G. decorticans and the other Papilionoideae mitogenomes studied, except for S. koreensis, which retained an intact rps7 gene. The rps19 gene was missing in all Papilionoideae species used in our study (Table 1). Similarly, Wang et al. (2023) reported that most rps genes (rps2, rps7, rps10, rps11, and rps19) were absent in the mitogenome of Photinia serratifolia, as well as in some Rosacea species. The loss of ribosomal protein genes (rps genes) and the occurrence of putative mutations in the first start codon (RNA editing) can potentially be compensated for by nuclear genes (Newton et al., 2004). In fact, nuclear genes have the ability to influence the organization of mitochondrial genomes and regulate the expression of mitochondrial genes (Newton et al., 2004). Gene loss can occur through the transfer of a gene to the nucleus, functional substitution by a related protein, or loss of the protein and its function (Adams et al., 2002). In several Fabaceae species, the presence or absence of genes such as cox2, rpl2, rpl10, rps1, sdh4, and sdh3 has been found to be variable (Choi et al., 2019). This variability in gene presence or absence was also observed in the eleven Papilionoideae species analyzed in our study (Table 1), where some species retained the genes while others exhibited pseudogenization or complete loss.
Interestingly, we discovered four Papilionoideae species that retained the intact sdh genes: D. odorifera (sdh4), Castanospermum australe (sdh4), G. decorticans (sdh4) and S. koreensis (sdh3) (Table 1). In contrast, a study by Choi et al. (2019) revealed that all Papilionoideae species had lost the rpl10, sdh3, and sdh4 genes. The exclusive conservation of functional sdh4 or sdh3 genes, without RNA editing, such as in G. decorticans, may provide an important advantage for survival in the extreme conditions of the Atacama Desert. Research has demonstrated that succinate dehydrogenase (SDH) can activate the expression of stress-related genes, thereby inducing antioxidant responses and stress tolerance in plants (Jardim-Messeder et al., 2015). The authors suggested that SDH plays a crucial role in reactive oxygen species (ROS) production and in regulating both plant development and responses to stress (Jardim-Messeder et al., 2015). It is worth noting that within angiosperms, mitochondrial rps genes (16 genes) and sdh genes (sdh3 and sdh4) have been lost from the mitochondrial genome multiple times throughout plant evolution (Adams et al., 2002). This further underscores the significance of intact genes in certain plant species.
On the other hand, we found two Papilionoideae species, D. odorifera and G. decorticans, that have the intact nad1 gene (Table 1). Similar to what was explained earlier, this gene might play a crucial role in buffering the stress conditions experienced by G. decorticans on the Atacama Desert. In fact, a study by Jethva et al. (2023) investigated the function of alternative NADH dehydrogenases (nad1) and confirmed that this gene is essential in preventing excessive ROS formation in mitochondria during reoxygenation. The absence of nad1 and nad2 led to elevated ROS production, while their overexpression limited ROS levels (Jethva et al., 2023).
Plastid-to-mitochondria transfers have been suggested to have been occurring since the colonization of land by plants. Mitochondrial DNA of plastid origin (MIPT) is present in angiosperm mitogenomes in varying amounts, representing 0.1%–10.3% of the mitogenome (Sloan and Wu, 2014). In our comparative analysis, we found that the percentage coverage of MIPTs ranged from 0.4% in C. australe to 13% in G. decorticans (Table 1). It is surprising to note that G. decorticans exhibits higher MIPT coverage than any other angiosperm species. Initially, we had doubts regarding the accuracy of our MIPT coverage values. However, when comparing our findings, such as the 1.3% coverage in G. max, with the results of other studies such as Gandini and Sanchez-Puerta, (2017), we found consistency in the values. This provides confidence in the reliability of our data. In the past MIPTs were considered as “junk” sequences and were thought to have no functional contribution to the mitogenome (Wang et al., 2007). However, recent research has revealed their significance in mitochondrial function. For instance, rice MIPTs have been found to possess promoter sequences that are utilized by the mitochondrial gene atp9 (Nakazono et al., 1996), and tRNA genes of MIPTs have also been found to contribute functionally to the mitogenome (Wang et al., 2012). The unusually high percentage of MIPTs found in G. decorticans may suggest a substantial acquisition of genes that could play important roles in mitogenome functioning. Investigating these genes and their potential contributes in future research would be highly valuable.
The mitogenomes from other angiosperm species that inhabit non-polar deserts (Table 2) varied between 339,352 and 758.210 bp. The mitogenomes contained 51 to 70 genes, 12 to 30 tRNA genes and 0.8%–10.3% MIPT, and were comparable to the mitogenome of the species in Table 1. Therefore, we did not find a common pattern that characterizes the mitogenomes of species that are able to inhabit deserts. Interestingly, the majority of the mitogenomes of the species from the desert contain an intact sdh4 gene as is observed in G. decorticans, with the exception of Phoenix dactylifera, Vigna unguiculata and Glycyrrhiza glabra where the gene is lost or present as a pseudogene. As stated before, the sdh4 gene plays an important role in the response to environmental stress. We therefore stress the importance of gaining more insight in why this gene is retained in most of the angiosperms that inhabit deserts. We observed that RNA editing had occurred in nad1 gene of the majority of the angiosperms from deserts, however, R. stricta (an extremophile plant from the desert in South-West Asia) still had the intact gene, similar as G. decorticans. Rhazya stricta, as G. decorticans, is able to survive high temperatures and high salinity (Hajrah et al., 2017). We therefore recommend to evaluate the nad1 gene in species along salinity gradients.
Previous studies have used plastid genome data to determine the molecular phylogeny and position of the genus Geoffroea Jack, including Geoffroea spinosa and G. decorticans (Lee et al., 2021; Contreras-Díaz et al., 2022). Additionally, researchers have developed SSR markers specific for G. decorticans to study the phylogeny and diversity of populations (Contreras et al., 2019; Contreras Díaz et al., 2021). However, the phylogenetic relationships of G. decorticans had not been assessed using mitogenome data. To address this, we analyzed concatenated sequences from 33 PCGs and complete plastid genome sequences, which were used in ML phylogenetic analysis. The resulting ML tree revealed two main clades: one containing the outgroup species L. trichandra and A. ligulata (Caesalpinioideae), and the other containing all 11 Papilionoideae species. Both clades were strongly supported with a bootstrap value of 100 (Figure 2). Within the Papilionoideae cluster, four subclades were identified: the Dalbergieae clade consisting of A. hypogaea, G. decorticans and D. odorifera (BP = 100); the NPAAA (non-protein–amino-acid-accumulating) clade including L. japonicus, M. sativa, G. max, P. vulgaris, V. angularis and P. pinnata (BP = 100); the Genistoids clade, represented solely by S. koreensis (BP = 100); and the ADA (Angylocalyceae, Dipterygeae, and Amburaneae) clade, which solely comprised C. australe (BP = 100) (Figure 2). These results align with previous phylogenetic studies (Cardoso et al., 2013; Choi et al., 2022). Within the Dalbergieae clade, two subclades were observed: one containing D. odorifera and the other containing A. hypogaea and G. decorticans (BP = 100) (Figure 2). This analysis strongly supported G. decorticans as a sister species of A. hypogaea (BP = 100) (Figure 2). These two species belong within the Pterocarpus clade, while D. odorifera belongs within the Dalbergia clade (Cardoso et al., 2013). Our phylogenetic analysis using the mitogenome database was backed up by the analysis using the plastid genome database (Figure 2), confirming the taxonomic classification of G. decorticans. Phylogenetic analysis of Fabaceae species, along with other angiosperms suggests that in certain legumes the presence of rpl2, rps19, and sdh3 genes can be attributed to remnants of a native ancestral gene (Choi et al., 2019). In our study, we found intact sdh4 and nad1 genes only in G. decorticans and D. odorifera but not in A. hypogaea. Although these two species do not belong to the same Pterocarpus clade (Contreras-Díaz et al., 2022), it is possible that these intact genes have been preserved from a common native ancestor.
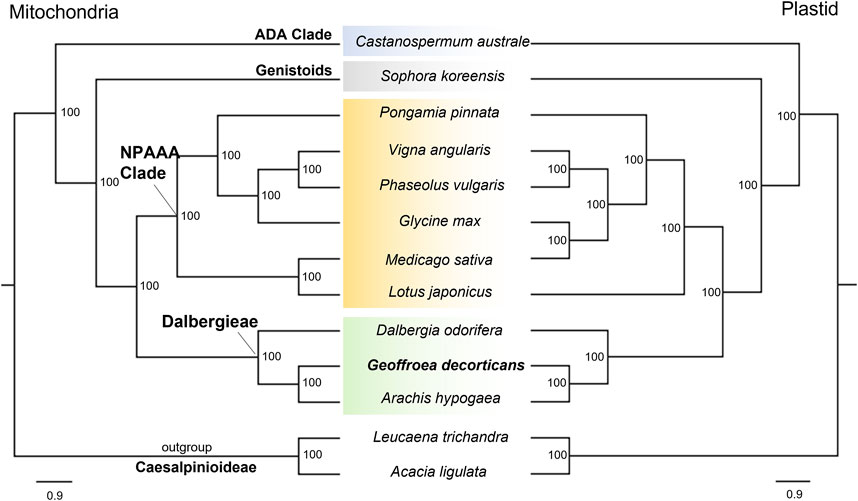
FIGURE 2. Maximum likelihood phylogeny of thirteen Fabaceae mitogenome based on nucleotide datasets of 33 protein-coding genes (left), and with plastid genomes (right). Bootstrap values are place on the nodes. Scale indicates number of nucleotide substitutions per site.
Ka/Ks ratios can be used to reflect the natural selective pressure of protein-coding genes during evolution (Feng et al., 2019). We compared the Ka/Ks ratio for 25 protein-coding genes in the mitogenomes, comparing G. decorticans and the ten Papilionoideae species that were used in our phylogenetic analysis (Figure 3). The mean Ka/Ks value in most protein-coding genes was less than 1 (Figure 3), suggesting that these genes are purified to keep the genes functional and remove deleterious mutations. However, the mean Ka/Ks value of atp8 (1.41) was greater than 1, (Figure 3), indicating that this gene was subjected to positive selection. Similarly, Ka/Ks values greater than 1 in the atp8 gene were founded in the xerophytic legume species, Ammopiptanthus mongolicus (sister of Ammopiptanthus nanus) from the desert in northwest China (Feng et al., 2019); and the authors of this study have speculated that the atp8 gene might play a role in the adaptation to dry environments. Furthermore, in the same study the evaluation of the mitogenome of A. mongolicus showed that the sdh4 gene was found to be intact and unaltered (similar to G. decorticans), while in other legumes the gene was lost or pseudogenized (Feng et al., 2019). Further research is needed to understand why these two legume species (G. decorticans and A. mongolicus) from deserts on different continents show similar positive selection of some genes (atp8) and retention of other genes (such as sdh4).
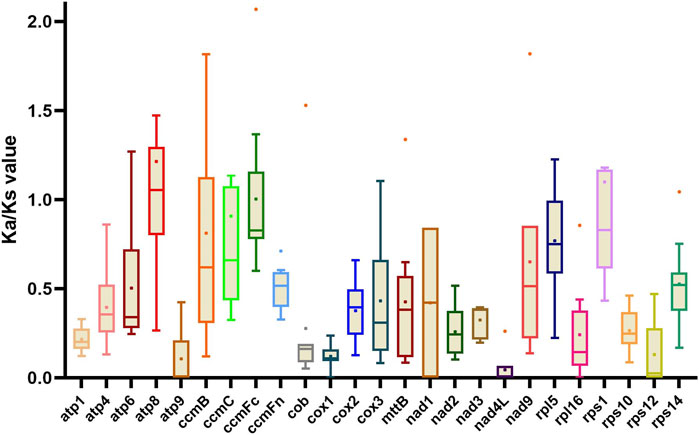
FIGURE 3. Box-and-whisker plots of Ka/Ks value of 25 protein-coding genes in G. decorticans and ten Papilionoideae species. Each box (with whiskers) shows the variation of the Ka/Ks values of a gene, among the 11 species studied using G. decorticans as a reference. Box plots show the median (central line), mean (dot on the box plot) and outliers.
Phylogenetic analysis conducted using the mitogenomes of Chañar and 12 other taxa revealed a remarkable level of consistency in taxonomic classification. When compared to other Papilionoideae species, the structure of the Geoffroea decorticans mitogenome exhibited minimal changes in terms of gene content, genome size and functional genes. However, it is important to note that the mitogenome of G. decorticans displayed distinct rearrangements, directionality, and organization in comparison to the other Papilionoideae species. One notable aspect is the conservation of native mitochondrial DNA in G. decorticans, as shown by positive selection for some genes, such as atp8, during evolution. The retention of the intact sdh4, nad1 and nad4 genes in G. decorticans suggests they might be important in drought tolerance mechanisms, and therefore in the species’ ability to cope with arid environments, as they have been lost in many plants that grow under more favorable conditions. Furthermore, Chañar stands out for possessing the highest amount of mitochondrial DNA of plastid origin (MIPTs) identified in any known mitogenome to date. MIPTs are involved in mitogenome functionality, and their abundance in Chañar is likely a result of the species’ evolutionary adaptation to the extreme environmental conditions of the Atacama Desert. The acquisition of additional DNA from other organelles, such as plastids, through horizontal gene transfer, provides Chañar with unique genetic material that potentially contributes to its survival strategies. The combination of conserved genes that facilitate drought stress responses and the acquisition of plastid material has likely contributed to the exceptional characteristics of G. decorticans. This species not only survives, but also provides sustenance to the inhabitants of the driest desert on Earth, making it an example of adaptation in challenging environments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/OQ707067.1/. The raw reads have been deposited in NCBI SRA with the number “PRJNA719569”.
RC-D conceptualized, executed the analyses and wrote the first draft. LvdB and FC provided comments and suggestions for improvement, and edited the final version. All authors contributed to the article and approved the submitted version.
This work was supported by the Universidad de Atacama (DIUDA 22423), and ANID—MILENIO—NCS 2022_024.
We thank José Luis Gutiérrez Alvarado for his picture of the Chañar tree used in Figure 1. RC-D and FC thanks to AFOREST, a Millennium Nucleus supported by ANID—MILENIO—NCS 2022_024.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1226052/full#supplementary-material
Adams, K. L., Qiu, Y. L., Stoutemyer, M., and Palmer, J. D. (2002). Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. U. S. A. 99 (15), 9905–9912. doi:10.1073/pnas.042694899
Alverson, A. J., Wei, X., Rice, D. W., Stern, D. B., Barry, K., and Palmer, J. D. (2010). Insights into the evolution of mitochondrial genome size from complete sequences of citrullus lanatus and cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 27 (6), 1436–1448. doi:10.1093/molbev/msq029
Azua-Bustos, A., González-Silva, C., and Fairén, A. G. (2022). The Atacama Desert in northern Chile as an analog model of mars. Front. Astronomy Space Sci. 8. doi:10.3389/fspas.2021.810426
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. doi:10.1089/cmb.2012.0021
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15), 1972–1973. doi:10.1093/bioinformatics/btp348
Cardoso, D., Pennington, R., de Queiroz, L., Boatwright, J., Van Wyk, B. E., Wojciechowski, M., et al. (2013). Reconstructing the deep-branching relationships of the papilionoid legumes. South Afr. J. Bot. 89, 58–75. doi:10.1016/j.sajb.2013.05.001
Chang, S., Wang, Y., Lu, J., Gai, J., Li, J., Chu, P., et al. (2013). The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS ONE 89 (2). doi:10.1371/journal.pone.0056502
Chevigny, N., Schatz-Daas, D., Lotfi, F., and Gualberto, J. M. (2020). DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 21 (1), 328. doi:10.3390/ijms21010328
Choi, I. S., Cardoso, D., de Queiroz, L. P., de Lima, H. C., Lee, C., Ruhlman, T. A., et al. (2022). Highly resolved papilionoid legume phylogeny based on plastid phylogenomics. Front. Plant Sci. 13, 823190. doi:10.3389/fpls.2022.823190
Choi, I. S., Wojciechowski, M. F., Ruhlman, T. A., and Jansen, R. K. (2021). In and out: evolution of viral sequences in the mitochondrial genomes of legumes (Fabaceae). Mol. Phylogenet. Evol. 163, 107236. doi:10.1016/j.ympev.2021.107236
Choi, I. S., Schwarz, E. N., Ruhlman, T. A., Khiyami, M. A., Sabir, J. S. M., Hajarah, N. H., et al. (2019). Fluctuations in Fabaceae mitochondrial genome size and content are both ancient and recent. BMC Plant Biol. 19 (1), 448. doi:10.1186/s12870-019-2064-8
Contreras Díaz, R., Porcile Saavedra, V., and Aguayo Cruces, F. (2018). Genetic diversity of geoffroea decorticans, a native woody leguminous species from atacama desert in Chile. Bosque 39 (2), 321–332. doi:10.4067/S0717-92002018000200321
Contreras Díaz, R., Carevic, F. S., Arias Aburto, M., Huanca Mamani, W., and Díaz Martín, B. (2021). Development of new microsatelite loci for geoffroea decorticans (gillies ex Hook. & Arn) Burkart. Bol. Soc. Argent. Bot. 56 (4), 533–545. doi:10.31055/1851.2372.V56.N4.32721
Contreras, R., Porcile, V., and Aguayo, F. (2019). Los microsatélites revelan una alta diferenciación genética entre las poblaciones nativas de Geoffroea decorticans en el desierto de Atacama chileno. Bol. Soc. Argent. Bot. 54 (2), 225–240. doi:10.31055/1851.2372.v54.n2.24367
Contreras, R., van den Brink, L., Burgos, B., González, M., and Gacitúa, S. (2020). Genetic characterization of an endangered chilean endemic species, prosopis burkartii muñoz, reveals its hybrids parentage. Plants 9 (6), 1–19. doi:10.3390/plants9060744
Contreras-Díaz, R., Carevic, F. S., Huanca-Mamani, W., Oses, R., Arias-Aburto, M., and Navarrete-Fuentes, M. (2022). Chloroplast genome structure and phylogeny of Geoffroea decorticans, a native tree from Atacama Desert. Electron. J. Biotechnol. 60, 19–25. doi:10.1016/j.ejbt.2022.09.005
Costamagna, M. S., Ordoñez, R., Zampini, I., Sayago, J., and Isla, M. (2013). Nutritional and antioxidant properties of Geoffroea decorticans, an Argentinean fruit, and derived products (flour, arrope, decoction and hydroalcoholic beverage). Food Res. Int. 54 (1), 160–168. doi:10.1016/j.foodres.2013.05.038
Cotabarren, J., Broitman, D. J., Quiroga, E., and Obregón, W. D. (2020). GdTI, the first thermostable trypsin inhibitor from Geoffroea decorticans seeds. A novel natural drug with potential application in biomedicine. Int. J. Biol. Macromol. 148, 869–879. doi:10.1016/j.ijbiomac.2020.01.214
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37 (1), 291–294. doi:10.1093/molbev/msz189
Eshel, G., Araus, V., Undurraga, S., Soto, D. C., Moraga, C., Montecinos, A., et al. (2021). Plant ecological genomics at the limits of life in the Atacama Desert. Proc. Natl. Acad. Sci. U. S. A. 118 (46), e2101177118. doi:10.1073/pnas.2101177118
Fang, Y., Wu, H., Zhang, T., Yang, M., Yin, Y., Pan, L., et al. (2012). A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS One 7 (5), e37164. doi:10.1371/journal.pone.0037164
Feng, L., Li, N., Yang, W., Li, Y., Wang, C. M., Tong, S. W., et al. (2019). Analyses of mitochondrial genomes of the genus Ammopiptanthus provide new insights into the evolution of legume plants. Plant Syst. Evol. 305 (5), 385–399. doi:10.1007/s00606-019-01578-2
Gandini, C. L., and Sanchez-Puerta, M. V. (2017). Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Sci. Rep. 7, 43402. doi:10.1038/srep43402
Giménez, A. M. (2004). Anatomía comparada de leño y corteza de Geoffroea striata y Geoffroea decorticans. Madera Bosques 10 (1), 55–68. doi:10.21829/myb.2004.1011279
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47 (W1), W59-W64–W64. doi:10.1093/nar/gkz238
Hajrah, N. H., Obaid, A. Y., Atef, A., Ramadan, A. M., Arasappan, D., Nelson, C. A., et al. (2017). Transcriptomic analysis of salt stress responsive genes in Rhazya stricta. PLoS ONE 12 (5), e0177589. doi:10.1371/journal.pone.0177589
Hong, Z., Liao, X., Ye, Y., Zhang, N., Yang, Z., Zhu, W., et al. (2021). A complete mitochondrial genome for fragrant Chinese rosewood (Dalbergia odorifera, Fabaceae) with comparative analyses of genome structure and intergenomic sequence transfers. BMC Genomics. 22 (1), 672. doi:10.1186/s12864-021-07967-7
Li, J., and Cullis, C. (2021). The Multipartite Mitochondrial Genome of Marama (Tylosema esculentum). Front. Plant Sci. 12, 787443. doi:10.3389/fpls.2021.787443
Jardim-Messeder, D., Caverzan, A., Rauber, R., de Souza Ferreira, E., Margis-Pinheiro, M., and Galina, A. (2015). Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol. 208 (3), 776–789. doi:10.1111/nph.13515
Jethva, J., Lichtenauer, S., Schmidt-Schippers, R., Steffen-Heins, A., Poschet, G., Wirtz, M., et al. (2023). Mitochondrial alternative NADH dehydrogenases NDA1 and NDA2 promote survival of reoxygenation stress in Arabidopsis by safeguarding photosynthesis and limiting ROS generation. New Phytol. 238 (1), 96–112. doi:10.1111/nph.18657
Jiménez-Aspee, F., Theoduloz, C., Soriano, M. D. P. C., Ugalde-Arbizu, M., Alberto, M. R., Zampini, I. C., et al. (2017). The native fruit geoffroea decorticans from arid Northern Chile: phenolic composition, antioxidant activities and in vitro inhibition of pro-inflammatory and metabolic syndrome-associated enzymes. Molecules 22 (9), 1565. doi:10.3390/molecules22091565
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI blast: a better web interface. Nucleic acids Res. 36, W5–W9. doi:10.1093/nar/gkn201
Jung, J., Kim, J. I., Jeong, Y. S., and Yi, G. (2018). Agora: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics 34 (15), 2661–2663. doi:10.1093/bioinformatics/bty196
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Kazakoff, S. H., Imelfort, M., Edwards, D., Koehorst, J., Biswas, B., Batley, J., et al. (2012). Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLoS One 7 (12), e51687. doi:10.1371/journal.pone.0051687
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi:10.1093/bioinformatics/bts199
Krasensky, J., and Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63 (4), 1593–1608. doi:10.1093/jxb/err460
Krueger, F. (2019). Trim galore. Available at: https://github.com/FelixKrueger/TrimGalore.
Lee, C., Choi, I. S., Cardoso, D., de Lima, H. C., de Queiroz, L. P., Wojciechowski, M. F., et al. (2021). The chicken or the egg? Plastome evolution and an independent loss of the inverted repeat in papilionoid legumes. Plant J. 107 (3), 861–875. doi:10.1111/tpj.15351
Liberatore, K. L., Dukowic-Schulze, S., Miller, M. E., Chen, C., and Kianian, S. F. (2016). The role of mitochondria in plant development and stress tolerance. Free Radic. Biol. Med. 100, 238–256. doi:10.1016/j.freeradbiomed.2016.03.033
Lo Giudice, C., Hernández, I., Ceci, L. R., Pesole, G., and Picardi, E. (2019). RNA editing in plants: a comprehensive survey of bioinformatics tools and databases. Plant Physiology Biochem. 137, 53–61. doi:10.1016/j.plaphy.2019.02.001
Maddison, W. P., and Maddison, D. R. (2023). Mesquite: a modular system for evolutionary analysis. Available at: http://www.mesquiteproject.org.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop, (GCE), New Orleans, LA, USA, November 2010. doi:10.1109/GCE.2010.5676129
Møller, I. M., Rasmusson, A. G., and Van Aken, O. (2021). Plant mitochondria – past, present and future. Plant J. 108 (4), 912–959. doi:10.1111/tpj.15495
Murayama, M., Hayashi, S., Nishimura, N., Ishide, M., Kobayashi, K., Yagi, Y., et al. (2012). Isolation of arabidopsis ahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. J. Exp. Bot. 63 (14), 5301–5310. doi:10.1093/jxb/ers188
Naito, K., Kaga, A., Tomooka, N., and Kawase, M. (2013). De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed Sci. 63 (2), 176–182. doi:10.1270/jsbbs.63.176
Nakazono, M., Nishiwaki, S., Tsutsumi, N., and Hirai, A. (1996). A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol. General Genet. MGG 252 (4), 371–378. doi:10.1007/bf02173001
Newton, K. J., Gabay-Laughnan, S., and De Paepe, R. (2004). Mitochondrial mutations in plants, 121–141. doi:10.1007/978-1-4020-2400-9_7
Nuñez, L., Mcrostie, V., and Cartajena, I. (2009). Consideraciones sobre la recolección vegetal y la horticultura durante el formativo temprano en el sureste de la cuenca de Atacama. Darwiniana 47 (1), 56–75.
Palmer, J. D., Adams, K. L., Cho, Y., Parkinson, C. L., Qiu, Y. L., and Song, K. (2000). Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. U. S. A. 97 (13), 6960–6966. doi:10.1073/pnas.97.13.6960
Park, S., Ruhlman, T. A., Sabir, J. S., Mutwakil, M. H., Baeshen, M. N., Sabir, M. J., et al. (2014). Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 15 (1), 405. doi:10.1186/1471-2164-15-405
Sanchez-Puerta, M. V., Edera, A., Gandini, C. L., Williams, A. V., Howell, K. A., Nevill, P. G., et al. (2019). Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Mol. Phylogenet. Evol. 132, 243–250. doi:10.1016/j.ympev.2018.12.006
Skippingtona, E., Barkman, T. J., Rice, D. W., and Palmer, J. D. (2015). Miniaturized mitogenome of the parasitic plant viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. U. S. A. 112 (27), E3515–E3524. doi:10.1073/pnas.1504491112
Sloan, D. B., Alverson, A. J., Chuckalovcak, J. P., Wu, M., McCauley, D. E., Palmer, J. D., et al. (2012). Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10 (1), e1001241. doi:10.1371/journal.pbio.1001241
Sloan, D. B., and Wu, Z. (2014). History of plastid DNA insertions reveals weak deletion and AT mutation biases in angiosperm mitochondrial genomes. Genome Biol. Evol. 6 (12), 3210–3221. doi:10.1093/gbe/evu253
Smith, D. R., and Keeling, P. J. (2015). Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. U. S. A. 112 (33), 10177–10184. doi:10.1073/pnas.1422049112
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi:10.1093/bioinformatics/btu033
Tang, H., and Zhu, H. (2023). Specific changes in morphology and dynamics of plant mitochondria under abiotic stress. Horticulturae 9 (1), 11. doi:10.3390/horticulturae9010011
Ugalde, P. C., McRostie, V., Gayo, E. M., García, M., Latorre, C., and Santoro, C. M. (2021). 13,000 years of sociocultural plant use in the Atacama Desert of northern Chile. Veg. Hist. Archaeobotany 30 (2), 213–230. doi:10.1007/s00334-020-00783-1
Van Aken, O., Zhang, B., Carrie, C., Uggalla, V., Paynter, E., Giraud, E., et al. (2009). Defining the mitochondrial stress response in arabidopsis thaliana. Mol. Plant 2 (6), 1310–1324. doi:10.1093/mp/ssp053
Wang, D., Rousseau-Gueutin, M., and Timmis, J. N. (2012). Plastid sequences contribute to some plant mitochondrial genes. Mol. Biol. Evol. 29 (7), 1707–1711. doi:10.1093/molbev/mss016
Wang, D., Wu, Y. W., Shih, A. C. C., Wu, C. S., Wang, Y. N., and Chaw, S. M. (2007). Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 24 (9), 2040–2048. doi:10.1093/molbev/msm133
Wang, Y., Chen, S., Chen, J., Chen, C., Lin, X., Peng, H., et al. (2023). Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Photinia serratifolia. Sci. Rep. 13 (1), 770. doi:10.1038/s41598-022-24327-x
Warren, J. M., Salinas-Giegé, T., Triant, D. A., Taylor, D. R., Drouard, L., and Sloan, D. B. (2021). Rapid shifts in mitochondrial tRNA import in a plant lineage with extensive mitochondrial tRNA gene loss. Mol. Biol. Evol. 38 (12), 5735–5751. doi:10.1093/molbev/msab255
Xiong, Y., Yu, Q., Xiong, Y., Zhao, J., Lei, X., Liu, L., et al. (2022). The complete mitogenome of elymus sibiricus and insights into its evolutionary pattern based on simple repeat sequences of seed plant mitogenomes. Front. Plant Sci. 12, 802321. doi:10.3389/fpls.2021.802321
Zhang, R., Zhang, C. Q., Gan, Y., Li, D., and Rhodes, R. E. (2019). Predicting transport-related walking in Chinese employees by integrating worksite neighbourhood walkability and social cognition. Aust. Syst. Bot. 32 (6), 484–498. doi:10.1111/aphw.12164
Keywords: Atacama Desert, Geoffroea decorticans, mitochondrial genome, stress tolerance, fabaceae, extremophiles
Citation: Contreras-Díaz R, Carevic FS and van den Brink L (2023) Comparative analysis of the complete mitogenome of Geoffroea decorticans: a native tree surviving in the Atacama Desert. Front. Genet. 14:1226052. doi: 10.3389/fgene.2023.1226052
Received: 20 May 2023; Accepted: 26 July 2023;
Published: 10 August 2023.
Edited by:
Ana Luisa Garcia-Oliveira, The International Maize and Wheat Improvement Center (CIMMYT), KenyaReviewed by:
Edi Sudianto, National Cheng Kung University, TaiwanCopyright © 2023 Contreras-Díaz, Carevic and van den Brink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Contreras-Díaz, cm9iZXJ0by5jb250cmVyYXNAdWRhLmNs
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.