
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet., 29 August 2023
Sec. Neurogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1225197
This article is part of the Research TopicMethylation in the Human BrainView all 5 articles
 Joshua S. Danoff1,2†
Joshua S. Danoff1,2† Emma A. Page1,2†
Emma A. Page1,2† Allison M. Perkeybile1,2
Allison M. Perkeybile1,2 William M. Kenkel3
William M. Kenkel3 Jason R. Yee4
Jason R. Yee4 Craig F. Ferris5
Craig F. Ferris5 C. Sue Carter1
C. Sue Carter1 Jessica J. Connelly1,2*
Jessica J. Connelly1,2*The neurohormone oxytocin regulates many aspects of physiology primarily by binding to its receptor, the oxytocin receptor. The oxytocin receptor gene (Oxtr) has been shown to have alternative transcripts in the mouse brain which may each have different biological functions or be used in specific contexts. A popular animal model for studying oxytocin-dependent social behaviors is the prairie vole, a biparental and monogamous rodent. Alternative transcriptional capacity of Oxtr in prairie voles is unknown. We used 5′ rapid amplification of cDNA ends to identify alternative Oxtr transcription start sites in prairie vole brain tissue and uterine tissue. We then validated expression of specific transcripts in fetal brains and assessed the impact of exogenous oxytocin administration in utero on offspring brain development. We identified seven distinct Oxtr transcripts, all of which are present in both brain and uterine tissue. We then demonstrated that maternal oxytocin administration alters expression of a specific subset of Oxtr transcripts and that these different transcripts are under unique epigenetic regulation, such that in the perinatal period only one of the alternative transcripts is associated with DNA methylation in the Oxtr promoter. These data establish the existence of multiple Oxtr transcripts in prairie vole brain and uterine tissue and implicate oxytocin in the regulation of alternative transcript expression. These data have significant implications for our understanding of null mutant models in both mice and voles and translation in human birth and behavior.
Oxytocin is a neuropeptide hormone originally described for its role in promoting uterine contractions (Dale, 1906). Oxytocin signaling throughout the brain and body regulates many psychological and physiological processes including social behaviors, stress response, inflammation, and metabolism (Carter et al., 2020). Manipulation of oxytocin signaling early in life alters social behavior throughout the lifespan, including prosocial behaviors such as partner preference and parental behavior as well as aggression (Miller and Caldwell, 2015). Notably, oxytocin regulates many non-social behaviors including pain perception (Boll et al., 2018) and learning and memory (Chini et al., 2014). Physiologically, oxytocin regulates a wide array of processes, including bone development (Colaianni et al., 2014), cardiovascular development and regulation of blood pressure (Gutkowska and Jankowski, 2012), and regulation of metabolism and body weight, which is partly (but not completely) due to central effects of oxytocin in the neural control of food intake (McCormack et al., 2020). Accumulating evidence implicates aberrant oxytocin signaling in the pathophysiology of many neuropsychiatric disorders, including autism spectrum disorder, schizophrenia, mood disorders, and anxiety disorders (Cochran et al., 2013). In contemporary obstetric practice, oxytocin is used to induce and augment labor (Fox et al., 2015) and remains the first-line pharmaceutical for prevention and treatment of postpartum hemorrhage (World Health Organization, 2012). There is also interest in using oxytocin as a treatment for neuropsychiatric disorders including autism spectrum disorder, though these efforts have not been consistently successful (Huang et al., 2021; Sikich et al., 2021). Further examination of mechanism of action of oxytocin, including the genetic variability in its receptor, might improve pharmacological outcomes in behavioral disorders.
Oxytocin primarily acts by binding to its receptor, the oxytocin receptor (OXTR). In humans, OXTR is encoded by the OXTR gene, which is thought to consist of four exons and three introns (Inoue et al., 1994). To some extent, homology has been used in model organisms to define transcript structure, however, a recent study of epigenetic regulation of Oxtr in mouse hippocampus identified eight isoforms of Oxtr which originate from alternative transcription start sites, alternative splicing of exons, or both (Towers et al., 2018). Two of the isoforms have the potential to be translated into full length OXTR proteins while the remaining six would make partial proteins, if translated. An important model system for examining oxytocin-dependent social behaviors are prairie voles, which display a wide range of human-like social behaviors including pair bonding and biparental care and have a highly conserved Oxtr gene (Tabbaa et al., 2017; Perkeybile et al., 2019). In previous studies, we have shown that DNA methylation in Oxtr is predictive of gene expression in the brain (Danoff et al., 2021). However, the assay for gene expression in this study measured only two exon splicing events of Oxtr, each of which could represent one or more transcripts. We have previously identified the presence of one alternative transcript, Oxtr-H, in the prairie vole nucleus accumbens (Danoff et al., 2021). Given the findings of multiple Oxtr transcripts in the mouse brain, we sought to establish the presence of additional alternative transcripts in prairie voles.
In this study, we use 5′ rapid amplification of cDNA ends (5′ RACE) to identify Oxtr transcripts present in prairie vole nucleus accumbens and uterus. We identify seven alternative Oxtr transcripts containing the annotated fourth exon, each of which have a different transcription start site. We show that exogenous oxytocin exposure at birth differentially impacts Oxtr variant expression in the fetal brain, suggesting distinct biological functions of each of the transcripts. Finally, we provide evidence that DNA methylation in an important Oxtr regulatory region regulates only specific transcripts.
All subjects were laboratory-bred prairie voles (Microtus ochrogaster) derived from wild-caught stock near Champaign, Illinois treated as previously described (Perkeybile et al., 2019). Subjects are at least four generations removed from wild-caught stock. Breeding pairs were housed in polycarbonate cages (44 cm × 22 cm × 16 cm). Same sex sibling pairs were housed in polycarbonate cages (27 cm × 16 cm × 16 cm) after weaning on postnatal day 20 (PND20). Animals were given food (high-fiber Purina rabbit chow) and water ad libitum, cotton nestlets for nesting material, and were maintained on a 14:10 light:dark cycle.
Four nucleus accumbens RNA samples (two male and two female, aged between postnatal days 21 and 24) from our previous study of early life experience were pooled for 5′ RACE (Perkeybile et al., 2019). Uterine samples were taken from two adults: one virgin (115 days old) and one postpartum day 1 (multiparous and over a year old) from our colony at the University of Virginia. Uterine samples were collected from animals that were deeply anaesthetized with an overdose of Euthasol (Covetrus) and sacrificed via cervical dislocation. An incision was made on the abdomen and the uterus was removed and flash frozen on dry ice, then stored at −80°C until nucleic acid isolation. RNA was isolated using an AllPrep DNA/RNA Mini kit (Qiagen) according to manufacturer instructions. All procedures for collection of brain tissue were approved by the IACUC at the University of California, Davis. All procedures for collection of uterine tissue were approved by the IACUC at the University of Virginia.
5′ RACE was completed using the GeneRacer Kit with Superscript III RT (Invitrogen) according to manufacturer instructions. For uterine samples, 3 μg of RNA was used as input. For nucleus accumbens, 2.5 μg, which was pooled from four different samples, was used as input. Briefly, RNA was dephosphorylated and decapped and the GeneRacer oligo was ligated to the mRNA. mRNA was transcribed into cDNA using Superscript III RT using random primers. Touchdown PCR was performed with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) according to manufacturer instructions with an annealing temperature of 65°C, with the provided forward GeneRacer primer, and the reverse gene-specific primer 5′-CGTCTCTCCGGGCCTGCTGCCCT-3′. A nested PCR was then performed using 1 μL of initial PCR as input, the provided nested forward GeneRacer primer, and the nested reverse gene-specific primer 5′- CCGGGCCTGCTGCCCTTCAGGT-3′ according to manufacturer instructions. 20 μL of nested PCR product was run on a 1.5% agarose gel with ethidium bromide. Single bands denoted in Figure 1 were cut from the gel and DNA was extracted using the QIAquick Gel Extraction Kit (Qiagen) and resuspended in 10 μL of Buffer EB. Nonspecific PCR bands were identified by: 1) PCR controls missing either template, forward primer, or reverse primer; 2) an experiment where we denatured PCR products and slowly cooled them to prevent PCR product hybrids; and 3) sequencing. We performed PCR and downstream cloning and sequencing using 5′ RACE products on three separate occasions and present sequencing data from all three runs. However, data are compiled into one figure for visualization purposes.
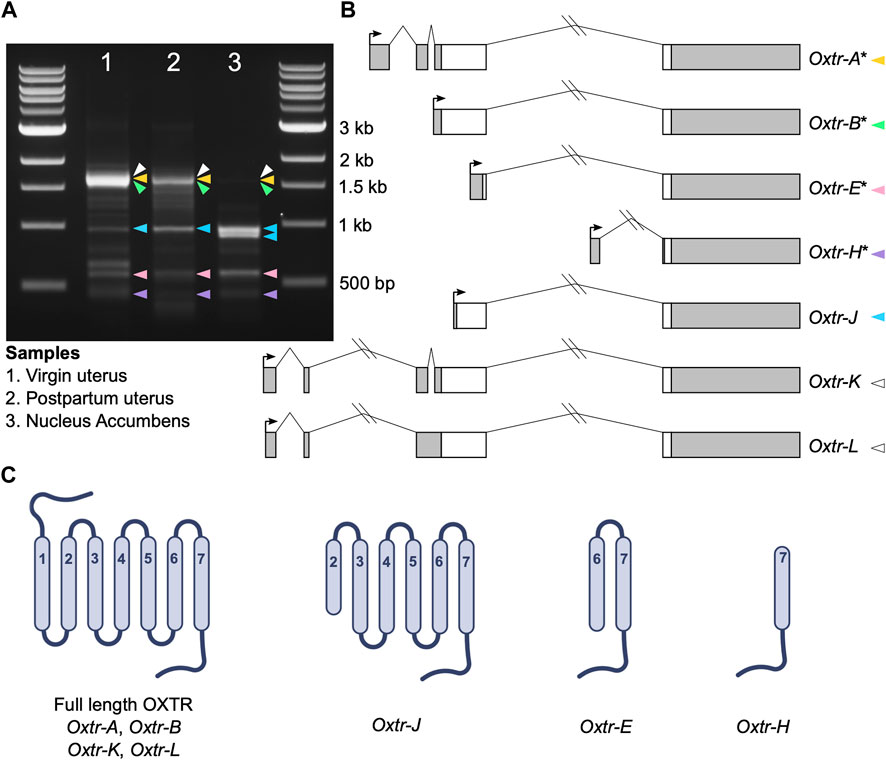
FIGURE 1. Oxtr transcripts identified in prairie vole uterus and nucleus accumbens. (A) Gel electrophoresis image showing the bands produced by 5′ RACE in adult virgin uterus (lane 1), postpartum uterus (lane 2) and juvenile nucleus accumbens (lane 3). Colored arrowheads indicate bands with specific Oxtr RACE products and match colors in Figure 1B. (B) Schematics of Oxtr transcripts identified by 5′ RACE. Exons are indicated by boxes (coding, white; untranslated, gray). Introns are shown with solid lines. Transcription start sites are indicated by arrows. Transcripts previously identified in mouse hippocampus are indicated by an asterisk. (C) Schematics showing the potential protein structures of each of the transcripts.
Each gel-extracted band represents a PCR product that was cloned using the TOPO TA Cloning Kit (Invitrogen) according to manufacturer instructions. Briefly, 4 μL of the purified PCR product was cloned into the TOPO vector. One Shot TOP10 E. coli was transformed using the heat shock method. Transformed E. coli were spread on LB-agar plates containing 50 μg/mL ampicillin for selection and incubated at 37°C for 24 h. Three single colonies from each PCR product were picked and grown overnight in liquid LB with 50 μg/mL ampicillin at 37°C shaking at 200 rpm. Cells were pelleted by spinning at 4000 rcf for 10 min and plasmids containing 5′ end clones were isolated using the PureLink Quick Plasmid Miniprep Kit (Invitrogen). Plasmids were sent to Genewiz for sequencing. Sequences were aligned to Oxtr using NCBI’s blastn. Sequence information for each transcript is provided in Supplementary Data.
Methods for generation of samples for the induction of labor study and measurement of DNA methylation in these samples were previously reported and are summarized below (Kenkel et al., 2019). Prairie voles were paired and allowed to mate using a timed-mating paradigm which enabled prediction of the day of birth. On the expected day of birth, pregnant dams were intraperitoneally injected with either saline (n = 9 dams), low dose oxytocin (0.125 mg/kg, n = 6 dams), medium dose oxytocin (0.25 mg/kg, n = 8 dams), or high dose oxytocin (0.5 mg/kg, n = 6 dams). An additional group of pregnant dams were not injected with anything and used as a no treatment control (n = 9 dams). 90 min following injection, pregnant dams were anesthetized with isoflurane and fetuses were removed and euthanized. Fetal brains were extracted and sectioned into thirds (which roughly correspond to forebrain, midbrain, and hindbrain), then they were flash frozen on dry ice. DNA and RNA were extracted from the midbrain of one male and one female fetus per mother using AllPrep mini kits (Qiagen). All procedures for the induction labor study were approved by the IACUC at Northeastern University.
Offspring used for gene expression analysis were restricted to animals with G/G genotype at SNP KLW2. This genotype which has previously been associated with expression of Oxtr-H (Danoff et al., 2021). The number of offspring included in each group following filtering for genotype are: saline treatment: 7 males, 7 females; low dose oxytocin: 6 males, 6 females; medium dose oxytocin: 6 males, 7 females; high dose oxytocin: 5 males, 6 females; no treatment control: 7 males, 8 females. 1 μg RNA from fetal midbrain was processed for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to manufacturer instruction. Reverse transcriptase quantitative PCR (RT-qPCR) was completed using the CFX966 system (Bio-Rad) with Power SYBR Green chemistry (Applied Biosystems). Primers were designed to capture distinct transcripts or sets of transcripts. We note that we attempted to generate an assay specific to transcripts Oxtr-K and Oxtr-L, and while PCR was successful and generated the correct amplicon, the assay could not be made efficient enough for quantitation, likely because of low abundance of these transcripts in brain tissue. Expression of all transcripts were normalized to Gapdh using the comparative Ct method, 2−ΔCt. See Supplementary Table S1 for all primers and PCR conditions. Specificity of products was confirmed using melt curve analysis. All RT-qPCR reactions were completed in triplicate, and the average Ct value of replicates was used for further analysis.
5 ng of DNA was used as template for PCR using a Pyromark PCR kit (Qiagen, Valencia, CA) according to manufacturer instructions using 0.2 μM forward primer 5′-GGACCCAAGGCCTATGTCA-3′ and biotinylated reverse primer 5′-ATGAGCTTGACGCTACTGACTCG-3’. The following cycling conditions were used for amplification of the target fragment: [Step 1: (95°C/15 min)/1 cycle, Step 2: (94°C/30 s, 58°C/30 s, 72°C/30 s)/45 cycles, Step 3: (72°C/10 min)/1 cycle, Step 4: 4°C hold. Pyrosequencing was completed using Pyromark Gold Q24 reagents (Qiagen) according to manufacturer instructions with sequencing primer 5′-TCAAGATCTGGCAGAAC-3’.
Methods for measurement of DNA methylation in induction of labor samples have previously been reported (Kenkel et al., 2019). Briefly, 200 ng genomic DNA were subject to bisulfite treatment (Kit MECOV50, Invitrogen, Carlsbad, CA) per manufacturer instructions. 12 ng of bisulfite converted DNA was used as a template for PCR using a Pyromark PCR kit (Qiagen) and 0.2 uM of primers 5′-GGGGATAGGATGGTTAGTTAGTATT-3′ and 5′- CCAACAACCTCAAAACTCTACT-3’. The following cycling conditions were used for amplification of the target Oxtr fragment, which included CpG sites −934_1, −934_2, −924, and −901: [Step 1: (95°C/15 min)/1 cycle, Step 2: (94°C/30 s, 58°C/30 s, 72°C/30 s)/50 cycles, Step 3: (72°C/10 min)/1 cycle, Step 4: 4°C hold. Pyrosequencing was performed using two primers: 5′-GAGGGAAGGTTTTGGAGTTTTTTATAT-3′, which covers CpG sites −934_1, −934_2, and −924, and 5′-AGGGATTGAAAAGTGA-3′, which covers CpG site −901, on a Pyromark Q24 using PyroMark Gold Q24 Reagents (Qiagen) per the manufacturer protocol. Epigenotypes reported are an average of the three replicates.
Statistical analysis was completed in R statistical software. RT-qPCR data was analyzed using mixed-effect linear models with fixed effects of a treatment by sex interaction and random effects of litter ID to account for genetic relatedness using the afex package in R (Team, 2018; Singmann et al., 2022). Post-hoc tests were completed used the emmeans R package and a modified Dunnett correction (“dunnetx”) for multiple comparisons (Lenth, 2022). Comparison of RT-qPCR data from the no treatment and saline groups were completed using the same model structure. Since no sex or treatment by sex effects were associated with expression of any transcript, the relationship between DNA methylation and Oxtr transcript expression was examined using mixed-effect linear models with fixed effects of DNA methylation at a specific CpG site and treatment and random effects of litter ID. Separate models were constructed for each CpG site measured to avoid collinearity.
Using 5′ RACE, we identified seven Oxtr transcripts with unique 5′ transcription start sites in prairie vole nucleus accumbens and uterine tissue (Figures 1A, B). Though we are interested in transcriptional products of Oxtr in the brain, we also used two uterine samples, a virgin uterus and a postpartum uterus (day of birth), because of the abundance of Oxtr expression in this tissue. Four of these transcripts were previously identified in mouse hippocampus (Towers et al., 2018) and are named according to their homology. These include Oxtr-A, which is considered the canonical transcript, consisting of four exons and three introns. Oxtr-B consists of a 5′ extended exon 3 spliced to exon 4. Oxtr-E consists of a truncated exon 3 spliced to exon 4. Oxtr-H consists of a novel exon originating from the third intron of the gene spliced to exon 4. Three novel transcripts were also identified. Oxtr-J which originates in the middle of exon 3 and is spliced to exon 4. Oxtr-K and Oxtr-L both consist of two novel exons about 4,000 basepairs upstream of the canonical transcription start site, which get spliced to exon 2. Oxtr-L differs from Oxtr-K in that first exon is 35 bp shorter and the intron between exon 2 and 3 is retained, though we cannot rule out the possibility that Oxtr-L represents a transcript similar to Oxtr-K that has not been fully spliced. Sequences of each transcript are provided in Supplementary Data.
We predicted putative translation start sites using the ATG in the transcript sequence that produces the longest peptide sequence, which was always in frame. We compared this predicted sequence to the prairie vole OXTR protein sequence (Uniprot E0V872) to generate schematics of the predicted protein. The predicted protein products of these transcripts are shown in Figure 1C. Oxtr-A, Oxtr-B, Oxtr-K, and Oxtr-L would all make a full length OXTR protein if translated. The other transcripts could be translated to truncated OXTR proteins as shown in Figure 1C. Importantly, we do not yet have evidence that these transcripts are all translated; it is possible that some transcripts are not translated and possess gene regulatory functions or do not have any biological function.
We investigated how transcriptional dynamics of Oxtr isoforms in the fetal brain are impacted by biological context using a model of labor induction (Kenkel et al., 2019). In this study, we injected pregnant dams with saline, low dose oxytocin (0.125 mg/kg), medium dose oxytocin (0.25 mg/kg), or high dose oxytocin (0.5 mg/kg) on the expected day of birth. An additional group of pregnant dams were not injected and used as a no treatment control. 90 min after the injection, fetuses were extracted and the fetal brain was removed. Our previous results indicate that maternal oxytocin treatment leads to increased Oxtr DNA methylation and impacts Oxtr expression (Kenkel et al., 2019). We suspected that a relationship between maternal oxytocin exposure and Oxtr gene expression in the fetal brain might be further clarified by examining single transcript specific expression. We investigated this using reverse transcriptase quantitative PCR (RT-qPCR) with primers that can detect distinct alternative transcripts (Oxtr-A, Oxtr-B, Oxtr-H) or groups of transcripts (Oxtr-All, Oxtr-All except H) (Figure 2A).
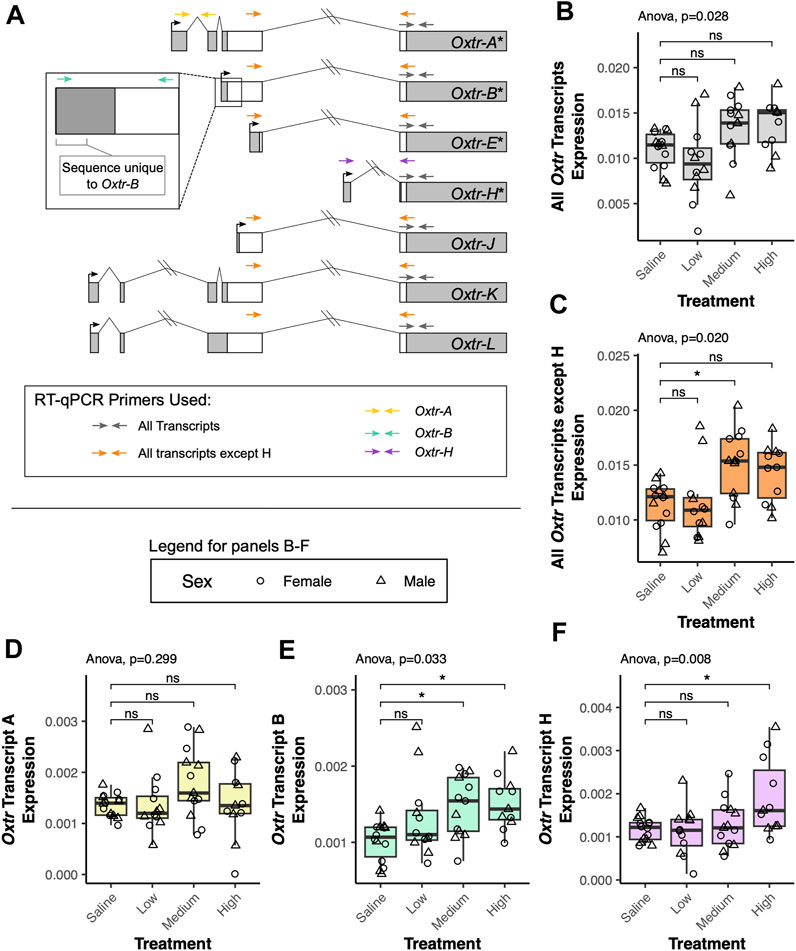
FIGURE 2. Maternal oxytocin treatment leads to increased expression of a subset of Oxtr transcripts in the fetal brain. (A) Schematic of Oxtr transcripts identified by 5′ RACE. Transcripts with asterisks indicate transcripts previously identified in mouse hippocampus. Primers used for RT-PCR are indicated by colored arrows with each color representing a different transcript or set of transcripts captured by the assay. (B) Maternal oxytocin treatment leads to differential expression of Oxtr when primers capture all transcripts [F(3,22.5) = 3.66, p = 0.028]. Post-hoc tests do not indicate significant differences between any treatment group and saline after correction for multiple comparisons. (C) Maternal oxytocin treatment leads differential expression of Oxtr when measuring all transcripts except H [F(3,22.4) = 4.03, p = 0.020]. Post-hoc tests indicate that the medium dose group differs from saline [t(21.8) = 2.80, p = 0.029]. (D) Maternal oxytocin treatment does not lead to differential expression of Oxtr transcript A [F(3,22.5) = 1.30, p = 0.229]. (E) Maternal oxytocin treatment leads differential expression of Oxtr transcript B [F(3,21.7) = 3.50, p = 0.033]. Post-hoc tests indicate that the medium dose group differs from saline [t(21.6) = 2.63, p = 0.041] and that the high dose group differs from saline [t(21.9) = 2.89, p = 0.023]. (F) Maternal oxytocin treatment leads differential expression of Oxtr transcript H [F(3,42) = 4.49, p = 0.008]. Post-hoc tests indicate that the high dose group differs from saline [t(21.6) = 3.15, p = 0.013]. * p < 0.05, ns p > 0.05.
Our results indicate that there are significant group differences in expression when using primers that capture all Oxtr transcripts, but post hoc tests do not indicate that any specific group differs from saline [F(3,22.5) = 3.66, p = 0.028, Figure 2B]. When using primers that capture all Oxtr transcripts except Oxtr-H, there is again a significant group difference, with the medium group having increased expression compared to saline [F(3,22.4) = 4.03, p = 0.020, Figure 2C]. We then examined if expression of only specific Oxtr transcripts is altered by maternally administered oxytocin. None of the treatment group expression differences in Oxtr transcripts can be attributed to Oxtr-A, which does not change expression levels with maternal oxytocin administration [F(3,22.5) = 1.30, p = 0.229, Figure 2D]. However, these changes appear to be driven by changes in expression of Oxtr-B, which does significantly differ by maternal oxytocin administration, and is elevated in the medium dose and high dose groups [F(3,21.7) = 3.50, p = 0.033, Figure 2E]. We also identify group differences in expression of Oxtr-H, where expression is elevated in the high dose group [F(3,42) = 4.49, p = 0.008, Figure 2F]. Importantly, expression of each of these transcripts or groups of transcripts does not differ between offspring of saline treated dams and offspring of dams receiving no injection (Supplementary Figure S1). These results indicate that maternal oxytocin treatment alters expression of specific Oxtr transcripts in the fetal brain in a dose-dependent manner and suggests the importance of clarifying expression of these specific transcripts in previous literature in the vole and the mouse. Moreover, these results underscore the importance of precise quantification of specific transcripts when examining how endogenous or exogenous factors impact Oxtr expression in the brain.
DNA methylation is an epigenetic modification that occurs when a methyl group is covalently added to cytosine, typically in a cytosine-phosphate-guanine (CpG) context (Bird, 2011). DNA methylation in gene promoters is generally associated with transcriptional repression (Bird, 2011). DNA methylation in a regulatory region of Oxtr called MT2, which is located within exon 1 and intron 1 of Oxtr, is associated with reduced transcription of OXTR in both humans and prairie voles (Kusui et al., 2001; Danoff et al., 2021). We tested if DNA methylation in MT2 regulates specific Oxtr transcripts. We examined DNA methylation at four CpG sites in MT2 (CpGs −934_1, −934_2, −924, and −901) which we have previously shown to be sensitive to early life experience and are among the CpG sites where methylation is most highly associated with Oxtr expression (Danoff et al., 2021). Results of all DNA methylation models are in Supplementary Table S2 and indicate that all CpG sites measured show similar relationships with Oxtr transcript expression. For simplicity, results from CpG site −924 are provided below and in Figure 3. DNA methylation at CpG site −924 is negatively associated with expression of Oxtr when the assay does not distinguish between individual transcripts [F(1,51.6) = 12.55, p < 0.001; Figure 3A] and Oxtr expression when the assay measures all transcripts except H [F(1,58.7) = 11.28, p = 0.001; Figure 3B]. We next measured the expression of specific transcripts to identify which transcripts are associated with MT2 DNA methylation. In the fetal brain, the relationship between Oxtr gene expression and DNA methylation cannot be attributed to regulation of Oxtr-A, which is not associated with DNA methylation at CpG −924 [F(1,59.0) = 1.29, p = 0.26; Figure 3C]. However, DNA methylation at CpG −924 is negatively associated with expression of Oxtr-B [F(1,54.4) = 14.15, p < 0.001; Figure 2D]. DNA methylation at CpG −924 is not associated with expression of Oxtr-H [F(1,59.0) = 2.44, p = 0.123; Figure 2E]. These results indicate that DNA methylation in MT2 impacts expression of Oxtr-B in the fetal brain.
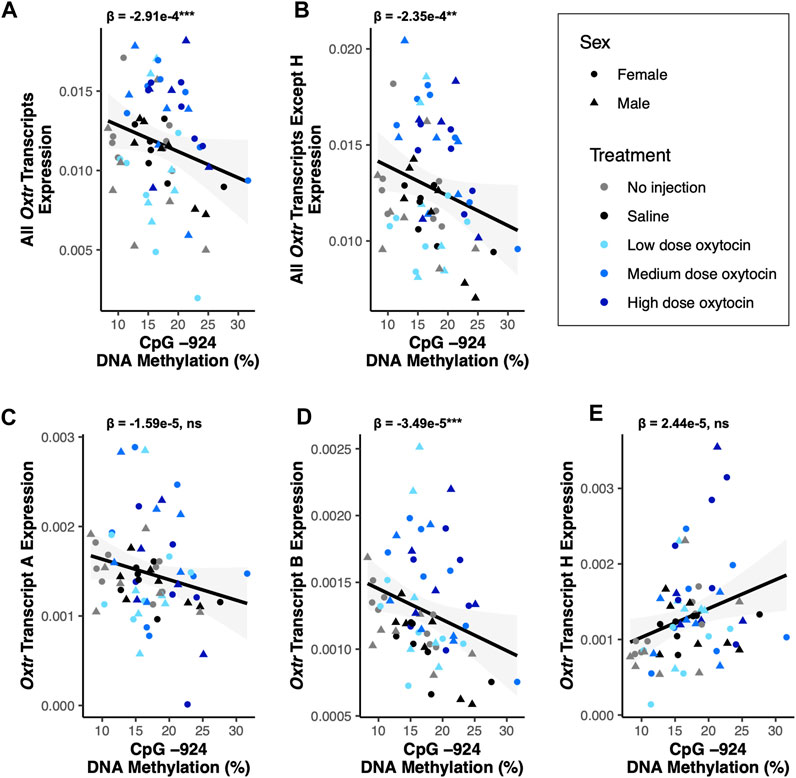
FIGURE 3. DNA methylation of MT2 in the fetal brain is associated with Oxtr-B expression but not Oxtr-A or Oxtr-H. For all plots, β values for CpG −924 DNA methylation from mixed model regressions which account for oxytocin treatment group and litter effects are shown. Regression lines are shown for visualization purposes. (A) DNA methylation at CpG −924 is negatively associated with expression of all Oxtr transcripts [F(1,51.6) = 12.55, p < 0.001]. (B) DNA methylation at CpG −924 is negatively associated with expression of all Oxtr transcripts except H [F(1,58.7) = 11.28, p = 0.001]. (C) DNA methylation at CpG −924 is not associated with expression of Oxtr transcript A [F(1,59.0) = 1.29, p = 0.26]. (D) DNA methylation at CpG −924 is negatively associated with expression of Oxtr transcript B [F(1,54.4) = 14.15, p < 0.001]. (E) DNA methylation at CpG −924 is not associated with expression of Oxtr transcript H [F(1,59.0) = 2.44, p = 0.123].
We provide evidence of seven alternative transcripts of Oxtr in prairie voles in multiple tissues. Four of these transcripts were previously identified in mouse hippocampus, indicating conservation of these transcripts across species. An additional three transcripts were identified which are novel. We confirmed the presence of five of these transcripts with RT-qPCR in the fetal brain. Putative protein product sequence-based prediction indicates that while four of these transcripts are predicted to produce full length OXTR protein, the remaining three would make partial protein products. Possible functions of these isoforms are discussed below, though we have not provided evidence that these transcripts are translated into protein. Further investigation of these transcripts is required to examine functional roles on oxytocin-dependent processes including birth and social behavior.
Four of the described isoforms contain the entire protein coding sequence and could therefore be translated into the full length OXTR protein. It is possible that there is context- or tissue-dependent expression of particular transcripts which might relate to the regulation or function of OXTR and other receptors known to bind oxytocin. In other GPCRs, it has been shown that transcripts with extended 5′ untranslated regions are translated less efficiently and may allow for a quick context-dependent change in expression of a gene that is readily reversible (Kreth et al., 2008). This mechanism may be the functional effect of Oxtr-K and Oxtr-L expression, allowing the brain and/or uterus to slow Oxtr translation while keeping pools of mRNA encoding for full length Oxtr in the cell.
If the transcripts which do not include the entire protein-coding region are indeed translated, these truncated OXTR proteins may alter the availability or activity of the full-length protein or other closely related receptors known to bind to oxytocin (e.g., AVPR1A, AVPR1B). It is well documented in other GPCRs that truncated receptors regulate both the activity and availability of full-length receptors (Markovic and Challiss, 2009; Liu et al., 2021). Crystal structure data suggest an important modulatory role for the seventh transmembrane domain of OXTR, indicating that translated Oxtr-E and Oxtr-H may regulate oxytocin binding capacity (Meyerowitz et al., 2022). OXTR homodimerizes and heterodimerizes with other GPCRs, and interacts with many G proteins and β-arrestin at its C-terminus, all of which have implications for oxytocin signaling (Oakley et al., 2001; Busnelli and Chini, 2018). Truncated forms of OXTR containing the C terminus in the membrane or cytoplasm could bind these proteins and make them unavailable to full-length OXTR and other GPCRs. Altogether, the presence of multiple truncated forms of OXTR proteins would have wide implications for the function of OXTR at the cellular level.
It is also possible that many of the alternative isoforms are not translated into protein but instead act as regulatory RNAs for the Oxtr gene. Many protein coding genes express non-coding alternative isoforms with regulatory function (Dhamija and Menon, 2018). These functions can either increase or decrease expression of the canonical transcript or be independent of the canonical transcript’s function. Given that we have previously shown correlation between the canonical Oxtr transcript and Oxtr-H, the described isoforms might increase expression of the canonical transcript (Danoff et al., 2021). Further investigation using methods to knockdown or knockout specific Oxtr transcripts will provide insight as to the function of these transcripts.
Finally, using samples from a study simulating labor induction, we show that specific Oxtr transcripts are sensitive to maternal oxytocin administration in a dose-dependent manner. Most effects in our assays which measure expression of multiple transcripts can be attributed to increased expression of Oxtr-B in the medium and high dose oxytocin administration groups. While Oxtr-B has increased expression in both the medium and high dose groups, Oxtr-H only has increased expression in the high dose group, suggesting that this transcript might be invoked in situations where the brain is responding to an abundance of oxytocin signaling. Notably, previous work in mice suggested that Oxtr-B is the transcript most tightly regulated by DNA methylation (Towers et al., 2018). Our results provide further evidence that DNA methylation in MT2 regulates expression of Oxtr-B specifically. DNA methylation in MT2 at least in the fetal period is not associated with expression of transcripts Oxtr-A and Oxtr-H. Altogether, these results suggest that Oxtr-B may be the dominant transcript expressed in the brain, and is the most sensitive to changes in the environment and local epigenetic state.
Our study does not present an exhaustive list of Oxtr transcripts. The gene-specific primer we used for amplification of 5′ RACE products binds to the fourth exon of transcript Oxtr-A. As such, we only identified transcripts containing this exon and cannot exclude the possibility that there are transcripts which do not include this exon. Additionally, we may not have sequenced enough clones to capture very rare transcripts. Nonetheless, the results presented here provide evidence of seven alternative isoforms of Oxtr in prairie voles. These isoforms represent alternative Oxtr gene products which may modulate the activity of full length OXTR in one of three ways: by producing proteins which interact with (1) the full-length OXTR, (2) other receptors that dimerize with OXTR, or (3) other proteins that interact with OXTR such as G proteins and β-arrestin. These products might also alter the transcriptional or translational ability of the canonical transcript. This has widespread implications for understanding regulation of oxytocin signaling in many tissues. Studies measuring expression of Oxtr in prairie voles should consider which of these transcripts the chosen quantification method measures for proper interpretation. Interpretation of such results should also consider that context-dependent expression of these isoforms may allow the cell to quickly and reversibly modulate oxytocin signaling in response to both internal processes such as parturition or external events such as social interaction without fully adjusting the regulation of the Oxtr gene. Further studies are necessary to examine the functional role of each of these transcripts in the cell and how the expression of each transcript is modulated by biological and environmental context. In the context of the uterus, for example, specific transcripts might be critical for contractions during birth which require oxytocin signaling, a process which might be disrupted if gene regulation goes awry. In the brain, there might be more subtle variation in transcript use which is related to specific environmental exposures, experiences, or developmental trajectories. Future studies which examine the functional characteristic of different Oxtr transcripts in the brain should also study such factors, which may explain why some transcripts are upregulated and others are not. Lastly, it has not escaped our notice that critical SNP findings in the human literature identify a prominent effect of a polymorphism in the third intron of OXTR on many behavioral and psychiatric outcomes (Wu et al., 2005; Li et al., 2015; Gong et al., 2017). Follow up work in the human to identify the presence of alternative OXTR transcripts is needed, especially transcript H, which would have a transcription start site near commonly studied OXTR SNPs if present in humans.
Transcript sequences are deposited in GenBank with the following accession numbers: OR044891, OR044892, OR044893, OR044894, OR044895, OR044896, OR044897. Sequences are also available in Supplementary Data. All other data is available upon request from the corresponding author.
The animal study was approved by the University of Virginia IACUC, University of California Davis IACUC, and Northeastern University IACUC. The study was conducted in accordance with the local legislation and institutional requirements.
Conceptualization: JD, EP, and JC; Investigation: JD, EP, AP, JY, and WK; Formal analysis: JD and EP; Visualization: JD and EP; Supervision: CF, CC, and JC; Writing—original draft: JD, EP, and JC; Writing—review and editing: AP, JY, WK, CF, and CC. All authors contributed to the article and approved the submitted version.
This work was funded by NIH grants P01HD075750 to CC, CF, and JC, R01HD098117 to CC and JC, F32HD092051 to AP, and Autism Speaks Grant 7110 to CC and JC.
We thank the animal care staff at the University of California, Davis, Northeastern University, and the University of Virginia for their contributions in maintaining our animal colonies. We also thank Dr. Krassimira Botcheva for assisting with primer design for 5′ RACE and Dr. Marta Busnelli for helpful discussions about possible functions of partial OXTR proteins.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1225197/full#supplementary-material
Bird, A. (2011). The dinucleotide CG as a genomic signalling module. J. Mol. Biol. 409, 47–53. doi:10.1016/j.jmb.2011.01.056
Boll, S., Almeida de Minas, A. C., Raftogianni, A., Herpertz, S. C., and Grinevich, V. (2018). Oxytocin and pain perception: from animal models to human research. Neuroscience 387, 149–161. doi:10.1016/j.neuroscience.2017.09.041
Busnelli, M., and Chini, B. (2018). “Molecular basis of oxytocin receptor signalling in the brain: what we know and what we need to know,” in Behavioral Pharmacology of neuropeptides: Oxytocin current topics in behavioral neurosciences. Editors R. Hurlemann, and V. Grinevich (Cham: Springer International Publishing), 3–29. doi:10.1007/7854_2017_6
Carter, C. S., Kenkel, W. M., MacLean, E. L., Wilson, S. R., Perkeybile, A. M., Yee, J. R., et al. (2020). Is oxytocin “nature’s medicine”. Pharmacol. Rev. 72, 829–861. doi:10.1124/pr.120.019398
Chini, B., Leonzino, M., Braida, D., and Sala, M. (2014). Learning about oxytocin: pharmacologic and behavioral issues. Biol. Psychiatry 76, 360–366. doi:10.1016/j.biopsych.2013.08.029
Cochran, D., Fallon, D., Hill, M., and Frazier, J. A. (2013). The role of oxytocin in psychiatric disorders: A review of biological and therapeutic research findings. Harv Rev. Psychiatry 21, 219–247. doi:10.1097/HRP.0b013e3182a75b7d
Colaianni, G., Tamma, R., Di Benedetto, A., Yuen, T., Sun, L., Zaidi, M., et al. (2014). The oxytocin–bone Axis. J. Neuroendocrinol. 26, 53–57. doi:10.1111/jne.12120
Dale, H. H. (1906). On some physiological actions of ergot. J. Physiology 34, 163–206. doi:10.1113/jphysiol.1906.sp001148
Danoff, J. S., Wroblewski, K. L., Graves, A. J., Quinn, G. C., Perkeybile, A. M., Kenkel, W. M., et al. (2021). Genetic, epigenetic, and environmental factors controlling oxytocin receptor gene expression. Clin. Epigenetics 13, 23. doi:10.1186/s13148-021-01017-5
Dhamija, S., and Menon, M. B. (2018). Non-coding transcript variants of protein-coding genes – what are they good for? RNA Biol. 15, 1025–1031. doi:10.1080/15476286.2018.1511675
Fox, K. A., Ramirez, M. M., and Ramin, S. M. (2015). “Induction and augmentation of labor,” in Management of labor and delivery (New Jersey, United States: John Wiley & Sons, Ltd), 62–85. doi:10.1002/9781118327241.ch4
Gong, P., Fan, H., Liu, J., Yang, X., Zhang, K., and Zhou, X. (2017). Revisiting the impact of OXTR rs53576 on empathy: A population-based study and a meta-analysis. Psychoneuroendocrinology 80, 131–136. doi:10.1016/j.psyneuen.2017.03.005
Gutkowska, J., and Jankowski, M. (2012). Oxytocin revisited: its role in cardiovascular regulation. J. Neuroendocrinol. 24, 599–608. doi:10.1111/j.1365-2826.2011.02235.x
Huang, Y., Huang, X., Ebstein, R. P., and Yu, R. (2021). Intranasal oxytocin in the treatment of autism spectrum disorders: A multilevel meta-analysis. Neurosci. Biobehav. Rev. 122, 18–27. doi:10.1016/j.neubiorev.2020.12.028
Inoue, T., Kimura, T., Azuma, C., Inazawa, J., Takemura, M., Kikuchi, T., et al. (1994). Structural organization of the human oxytocin receptor gene. J. Biol. Chem. 269, 32451–32456. doi:10.1016/s0021-9258(18)31656-9
Kenkel, W. M., Perkeybile, A. M., Yee, J. R., Pournajafi-Nazarloo, H., Lillard, T. S., Ferguson, E. F., et al. (2019). Behavioral and epigenetic consequences of oxytocin treatment at birth. Sci. Adv. 5, eaav2244. doi:10.1126/sciadv.aav2244
Kreth, S., Ledderose, C., Kaufmann, I., Groeger, G., and Thiel, M. (2008). Differential expression of 5′-UTR splice variants of the adenosine A2A receptor gene in human granulocytes: identification, characterization, and functional impact on activation. FASEB J. 22, 3276–3286. doi:10.1096/fj.07-101097
Kusui, C., Kimura, T., Ogita, K., Nakamura, H., Matsumura, Y., Koyama, M., et al. (2001). DNA methylation of the human oxytocin receptor gene promoter regulates tissue-specific gene suppression. Biochem. Biophysical Res. Commun. 289, 681–686. doi:10.1006/bbrc.2001.6024
Li, J., Zhao, Y., Li, R., Broster, L. S., Zhou, C., and Yang, S. (2015). Association of oxytocin receptor gene (OXTR) rs53576 polymorphism with sociality: A meta-analysis. PLOS ONE 10, e0131820. doi:10.1371/journal.pone.0131820
Liu, S., Kang, W.-J., Abrimian, A., Xu, J., Cartegni, L., Majumdar, S., et al. (2021). Alternative pre-mRNA splicing of the mu opioid receptor gene, OPRM1: insight into complex mu opioid actions. Biomolecules 11, 1525. doi:10.3390/biom11101525
Markovic, D., and Challiss, R. A. J. (2009). Alternative splicing of G protein-coupled receptors: physiology and pathophysiology. Cell. Mol. Life Sci. 66, 3337–3352. doi:10.1007/s00018-009-0093-4
McCormack, S. E., Blevins, J. E., and Lawson, E. A. (2020). Metabolic effects of oxytocin. Endocr. Rev. 41, 121–145. doi:10.1210/endrev/bnz012
Meyerowitz, J. G., Robertson, M. J., Barros-Álvarez, X., Panova, O., Nwokonko, R. M., Gao, Y., et al. (2022). The oxytocin signaling complex reveals a molecular switch for cation dependence. Nat. Struct. Mol. Biol. 29, 274–281. doi:10.1038/s41594-022-00728-4
Miller, T. V., and Caldwell, H. K. (2015). Oxytocin during development: possible organizational effects on behavior. Front. Endocrinol. 6, 76. doi:10.3389/fendo.2015.00076
Oakley, R. H., Laporte, S. A., Holt, J. A., Barak, L. S., and Caron, M. G. (2001). Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-β-arrestin complexes after receptor endocytosis. J. Biol. Chem. 276, 19452–19460. doi:10.1074/jbc.M101450200
Perkeybile, A. M., Carter, C. S., Wroblewski, K. L., Puglia, M. H., Kenkel, W. M., Lillard, T. S., et al. (2019). Early nurture epigenetically tunes the oxytocin receptor. Psychoneuroendocrinology 99, 128–136. doi:10.1016/j.psyneuen.2018.08.037
Sikich, L., Kolevzon, A., King, B. H., McDougle, C. J., Sanders, K. B., Kim, S.-J., et al. (2021). Intranasal oxytocin in children and adolescents with autism spectrum disorder. N. Engl. J. Med. 385, 1462–1473. doi:10.1056/NEJMoa2103583
Singmann, H., Bolker, B., Westfall, J., Aust, F., and Ben-Shachar, M. S. (2022). Afex: Analysis of factorial experiments. R package version 1.1-1. Available at: https://cran.r-project.org/package=afex.
Tabbaa, M., Paedae, B., Liu, Y., and Wang, Z. (2017). Neuropeptide regulation of social attachment: the prairie vole model. Compr. Physiol. 7, 81–104. doi:10.1002/cphy.c150055
Team, R. C. (2018). R: A language and environment for statistical computing. Available at: https://www.r-project.org/.
Towers, A. J., Tremblay, M. W., Chung, L., Li, X., Bey, A. L., Zhang, W., et al. (2018). Epigenetic dysregulation of Oxtr in Tet1-deficient mice has implications for neuropsychiatric disorders. JCI Insight 6, e120592. doi:10.1172/jci.insight.120592
World Health Organization (2012). WHO recommendations for the prevention and treatment of postpartum haemorrhage. Geneva: World Health Organization.
Keywords: oxytocin, alternative transcript isoforms, prairie vole (Microtus ochrogaster), DNA methyaltion, oxytocin receptor (OXTR)
Citation: Danoff JS, Page EA, Perkeybile AM, Kenkel WM, Yee JR, Ferris CF, Carter CS and Connelly JJ (2023) Transcriptional diversity of the oxytocin receptor in prairie voles: mechanistic implications for behavioral neuroscience and maternal physiology. Front. Genet. 14:1225197. doi: 10.3389/fgene.2023.1225197
Received: 18 May 2023; Accepted: 14 August 2023;
Published: 29 August 2023.
Edited by:
Jun-ichi Iga, Ehime University, JapanReviewed by:
Karolina Pierzynowska, University of Gdansk, PolandCopyright © 2023 Danoff, Page, Perkeybile, Kenkel, Yee, Ferris, Carter and Connelly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica J. Connelly, amVzc2ljYS5jb25uZWxseUB2aXJnaW5pYS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.