- 1Department of Thoracic Surgery, Xiangya Hospital of Central South University, Changsha, Hunan, China
- 2The Second Clinical Medical College of Nanchang University, The Second Affiliated Hospital of Nanchang University, Nanchang, China
Non-small cell lung cancer (NSCLC) is one of the most common types of malignant tumors as well as the leading cause of cancer-related deaths in the world. The application of epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) has dramatically improved the prognosis of NSCLC patients who harbor EGFR mutations. However, despite an excellent initial response, NSCLC inevitably becomes resistant to EGFR-TKIs, leading to irreversible disease progression. Hence, it is of great significance to shed light on the molecular mechanisms underlying the EGFR-TKI resistance in NSCLC. Long non-coding RNAs (lncRNAs) are critical gene modulators that are able to act as oncogenes or tumor suppressors that modulate tumorigenesis, invasion, and metastasis. Recently, extensive evidence demonstrates that lncRNAs also have a significant function in modulating EGFR-TKI resistance in NSCLC. In this review, we present a comprehensive summary of the lncRNAs involved in EGFR-TKI resistance in NSCLC and focus on their detailed mechanisms of action, including activation of alternative bypass signaling pathways, phenotypic transformation, intercellular communication in the tumor microenvironment, competing endogenous RNAs (ceRNAs) networks, and epigenetic modifications. In addition, we briefly discuss the limitations and the clinical implications of current lncRNAs research in this field.
1 Introduction
Lung cancer is the second most frequent type of malignant tumor that causes the highest cancer-related mortalities globally (Sung et al., 2021; Siegel et al., 2023). Lung cancer is categorized into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) according to the cell morphology. NSCLC, which primarily includes lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), and large cell lung cancer, makes up almost 85% of lung cancer, while SCLC accounts for the remaining cases (Sung et al., 2021). Since NSCLC patients are not discovered and diagnosed until their symptoms are apparent in advanced stages, the prognosis of NSCLC patients is very poor, with the 5-year survival rate less than 20% (Choi and Mazzone, 2022; Ettinger et al., 2022).
EGFR mutations, which are revealed as drivers of NSCLC in 2004 (Lynch et al., 2004; Paez et al., 2004), are the most prevalent somatic genetic alterations in NSCLC that can be used as therapeutic targets. Approximately 20% of Caucasians and up to 40% of East Asians with NSCLC harbor EGFR mutations (Yatabe et al., 2015; Kobayashi and Mitsudomi, 2016). The advent of EGFR-TKIs has completely transformed the treatments for NSCLC, substantially prolonging the progression-free survival (PFS) as well as overall survival (OS) of NSCLC patients harboring EGFR mutations (Zhou et al., 2011). However, due to the development of EGFR-TKI resistance, most advanced NSCLC patients will inevitably experience disease progression around 10 months after treatment with EGFR-TKIs (Mok et al., 2009). The mechanisms of EGFR-TKI resistance in NSCLC include secondary or tertiary mutations of EGFR sites, upregulation of alternative bypass signaling pathways, phenotypic transformations, alterations in the tumor microenvironment, etc (Gomatou et al., 2023).
LncRNAs are non-coding RNAs (ncRNAs) that are more than 200 nt in length. Although lncRNAs were once regarded as transcriptional noise, they have a wide range of functions, interoperating with DNAs, RNAs, and proteins to regulate cellular genetic expression and diverse signaling pathways (Mercer et al., 2009). Many investigations have demonstrated that dysregulated lncRNAs are capable of acting as oncogenes or tumor suppressors to control the resistance to EGFR-TKIs in NSCLC through multiple mechanisms. Consequently, lncRNAs are likely to be new targets to overcome the resistance to EGFR-TKIs. In this review, we provide a brief overview of the biological occurrence and action mechanisms of lncRNAs. Furthermore, we synthetically summarize the various roles of lncRNAs in the mechanism of EGFR-TKI resistance. And finally, the current limitations and possible future trends in this field are also discussed.
2 An overview of lncRNAs
According to the Human Genome Project, about 2% of human genes can encode proteins, while over 90% of human genes are eventually transcribed into ncRNAs (Carninci et al., 2005; Birney et al., 2007; Ezkurdia et al., 2014). Initially, ncRNAs were regarded as junk fragments or transcriptional by-products with no practical role. However, new investigations have indicated that ncRNAs are critical for the biological function of cells (Prasanth and Spector, 2007; Statello et al., 2021). Based on their length, ncRNAs are divided in two groups: small non-coding RNAs (sncRNAs), which are less than 200 nt in length, and long non-coding RNAs (lncRNAs), whose lengths are more than 200 nt (Dinger et al., 2008). The majority of lncRNAs are transcribed from different genomic areas by RNA polymerase II as well as polymerase III (Dieci et al., 2007; Derrien et al., 2012). According to their transcriptional source areas, lncRNAs can be broadly classified into five types: intergenic lncRNAs, intronic lncRNAs, sense lncRNAs, antisense lncRNAs, and bidirectional lncRNAs. It is worth mentioning that some lncRNAs can form closed-loop structures, which are called circular RNAs (circRNAs) (St Laurent et al., 2015).
The action mechanisms of lncRNAs are complex and can be summarized into four types (Siegel et al., 2023): signals: lncRNAs are transcribed and act as signaling molecules to regulate the transcription of downstream genes under specific stimulation conditions (Sung et al., 2021); decoys: lncRNAs bind proteins or RNAs and block their actions (Choi and Mazzone, 2022); guides: lncRNAs incorporate and direct transcription factors to specific DNA sequences (Ettinger et al., 2022); scaffolds: lncRNAs are capable of serving as center platforms for multiple proteins to assemble into complexes (Rinn and Chang, 2012; Fang and Fullwood, 2016). Through these action modes, lncRNAs in the nucleus, cytoplasm, and exosomes can regulate gene expression on a variety of levels such as epigenetic, transcriptional as well as post-transcriptional levels, etc. As an example, in the nucleus, lncRNAs recruit histone modification enzymes to regulate histone modification (Li et al., 2016); in the cytoplasm, lncRNAs sponge miRNAs or form double strands with specific mRNAs to improve the stability of mRNAs (Rashid et al., 2016); and lncRNAs in exosomes can participate in intercellular communication (Zhang et al., 2017). Thus, as key genetic regulators, lncRNAs play comprehensive and pivotal roles in cellular physiological and pathological processes.
Emerging evidence suggests that lncRNAs are capable of acting as oncogenes or tumor suppressors to regulate oncogenesis, tumor progression, metastasis, recurrence, and drug resistance (Isin and Dalay, 2015). Additionally, the expression of lncRNAs exhibits high tissue specificity and tumor cell specificity (Yan et al., 2015). For instance, lncRNAs expression differs significantly in the EGFR TKI-resistant and EGFR TKI-sensitive NSCLC (Cheng et al., 2015a; Shi et al., 2020). Therefore, lncRNAs are probably valuable tumor diagnostic signatures as well as tumor treatment targets with great potential for clinical applications (Qi and Du, 2013), and this potential is even more prominent for exosomal lncRNAs (Tao et al., 2020; Han et al., 2021). Despite the huge amount of lncRNAs are already been found (Li et al., 2023), only a small fraction of lncRNAs have been elucidated regarding their mechanism of action and function. Moreover, the broad prospect of clinical applications based on lncRNAs has not yet become a reality. Thus, more research on lncRNAs, including the refinement of molecular mechanisms, the adjustment of research strategies, and the innovation of related technologies, is necessary.
3 LncRNAs and EGFR-TKI resistance in NSCLC
EGFR-TKIs are successful as first-line treatments for advanced NSCLC patients who harbor EGFR mutations. However, most patients develop drug resistance, which significantly diminishes the therapeutic efficacy of EGFR-TKIs treatments (Mok et al., 2009; Zhou et al., 2011). Cumulative evidence suggests that the mechanisms of EGFR-TKI resistance in NSCLC can be broadly divided into EGFR-dependent as well as EGFR-independent mechanisms (Gomatou et al., 2023). It is well known that the EGFR-dependent resistance mechanisms mainly refer to EGFR site mutations, among which T790M, a secondary mutation in EGFR site, is found to be the leading cause for resistance to the first- and second-generation EGFR-TKIs, such as gefitinib and erlotinib (Mok et al., 2017). And the EGFR C797S tertiary mutation is the most frequent EGFR-dependent resistance mechanism of third-generation EGFR-TKIs such as osimertinib (Oxnard et al., 2018). EGFR-independent resistance mechanisms, which can co-exist with EGFR site mutations, mainly include aberrant activation of alternative bypass signaling pathways, phenotypic transformation, alterations in the tumor microenvironment, etc. Aberrant activation in cellular signaling pathways, for example, the PI3K/AKT and classical MAPK pathways, dominates EGFR-independent resistance mechanisms, of which the common ones are mesenchymal-epithelial transition factor (MET) amplification, human epidermal growth factor receptor 2 (HER2) amplification, and oncogenic fusions (Liu et al., 2018).
Despite lacking the ability to encode proteins (Niazi and Valadkhan, 2012), lncRNAs are responsible for a wide range of biological processes. Aberrant lncRNAs expression is involved with many diseases, notably cancer (Bhan et al., 2017; Cao et al., 2022). In the setting of EGFR TKI-resistant NSCLC, lncRNAs are already been demonstrated to have significant roles (Wang Q. et al., 2020; Hu J. et al., 2021). There is not enough proof to support the role of lncRNAs in EGFR-dependent resistance mechanisms. However, numerous investigations have shown that lncRNAs have a broad control over EGFR-independent resistance mechanisms in NSCLC. After summarizing the large number of reported lncRNAs that are related to EGFR-TKI resistance, we have identified that in terms of activation of signaling pathways, lncRNAs mainly regulate three signaling pathways, which are the PI3K/AKT, classical MAPK, as well as JAK/STAT pathways; in terms of phenotypic transformation, epithelial to mesenchymal transition (EMT) is the main type of phenotypic transformation regulated by lncRNAs; in terms of the tumor microenvironment, exosomal lncRNAs affect EGFR-TKI response by participating in intercellular communication. Additionally, some lncRNAs modulate EGFR-TKI resistance via ceRNA networks and epigenetic modifications. In the following, we describe the above mechanisms through which lncRNAs mediate the EGFR-TKI resistance in NSCLC. The lncRNAs involved in EGFR-TKI resistance in NSCLC are listed in Table 1.
4 Modulation of alternative bypass signaling pathways
4.1 The PI3K/AKT signaling pathway
Survival signals induced by multiple receptors are mediated mainly through the PI3K/AKT signaling pathway, which has pivotal functions for the regulation of cellular growth, cell cycle progression, and cell survival (Fresno Vara et al., 2004). Abnormal upregulation of the PI3K/AKT signaling pathway facilitates the resistance to EGFR-TKIs in NSCLC (Liu et al., 2018). Extensive investigations have already documented that the upstream receptors, regulatory factors, and downstream molecules of this pathway can be regulated by lncRNAs to modulate EGFR-TKI resistance in NSCLC (Figure 1).
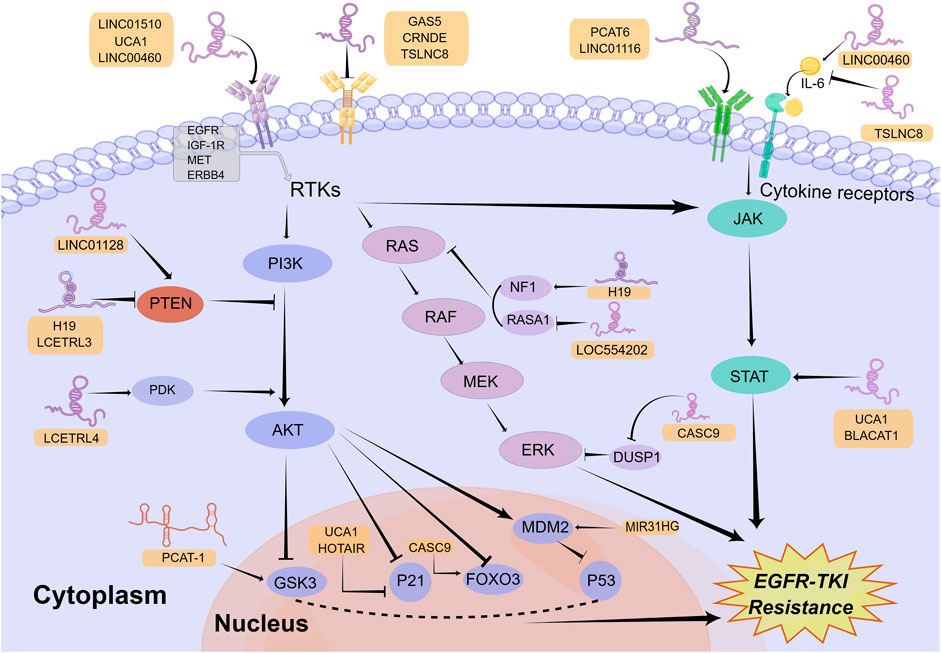
FIGURE 1. Mechanisms of lncRNAs involved in EGFR-TKI resistance in NSCLC by modulating signaling pathways. LncRNAs can regulate multiple signaling pathways to affect EGFR-TKI resistance in NSCLC, including the PI3K/AKT, classical MAPK, and JAK/STAT signaling pathways. Mechanistically, lncRNAs activate or inactivate these pathways by regulating their upstream receptors, regulatory factors, and downstream molecules. The Figure was drawn by Figdraw.
A number of receptors of the PI3K/AKT signaling pathway, such as insulin-like growth factor 1 receptor (IGF-1R), MET and EGFR, are regulated by lncRNAs to affect EGFR-TKI response in NSCLC. For example, in an investigation by Dong and collaborators, lncRNA GAS5 was revealed to be lowly expressed in gefitinib-resistant NSCLC cell lines. Overexpression of GAS5 reversed gefitinib resistance in vitro and in vivo through downregulating IGF-1R expression and inhibiting PI3K/AKT pathway (Dong et al., 2015). In another recent study, investigators found that loss of KMT5C in erlotinib-resistant cell lines resulted in enhanced transcription of lncRNA LINC01510, which could lead to upregulation of MET expression, activation of PI3K/AKT signaling pathway, and ultimately erlotinib resistance (Pal et al., 2022). The highlight in this study is the elucidation of the upstream mechanisms of LINC01510 dysregulation. In addition, since MET amplification is the most common EGFR-independent resistance mechanism (Qin et al., 2023), KMT5C/LINC01510/MET axis is of great importance in overcoming EGFR-TKI resistance and might become a potential target. In addition to IGF-1R and MET, lncRNAs also regulate other receptors in the PI3K/AKT pathway. LncRNA TRPM2-AS was reported to facilitate NSCLC cell proliferation, migration, and invasion by upregulation of EGFR and activation of the PI3K/AKT pathway via functioning as a ceRNA of miR-138-5p (Cui et al., 2020). However, whether lncRNA TRPM2-AS promotes EGFR-TKI resistance in NSCLC is still unclear, which urgently requires further exploration. Similarly, lncRNA UCA1 functioned as the ceRNA of miR-193a-3p to upregulate ERBB4 (Nie et al., 2016), which can activate the PI3K/AKT pathway (Yu et al., 2015). Moreover, UCA1 was revealed to induce EGFR-TKI resistance in non-T790M NSCLC cells via activating the AKT/mTOR pathway in another study (Cheng et al., 2015b). Thus, the miR-193a-3p/ERBB4 axis may be the mechanism by which lncRNA UCA1 leads to EGFR-TKI resistance in NSCLC.
Among numerous regulatory factors in the PI3K/AKT pathway, Phosphatase and Tensin Homolog (PTEN), one well-known inhibitor in this pathway (Kim et al., 2010), is modulated by lncRNAs to affect EGFR-TKIs response in NSCLC. Mainly through bioinformatics analysis, Ding and collaborators discovered LINC01128 was low expressed in gefitinib-resistant NSCLC cells and LINC01128/miR-25-3p/PTEN pathway was likely to enhance the resistance to EGFR-TKIs in NSCLC through mediating the PI3K/AKT pathway (Ding et al., 2022). However, due to the lack of sufficient experiments in this study, additional in vitro and in vivo experiments will be needed for the confirmation of their conclusions. In a recent investigation, Li et al. first identified lncRNAs LCETRL3 and LCETRL4 as new oncogenic genes that were highly expressed in NSCLC tissues and can reduce gefitinib efficiency against NSCLC in vitro and in vivo. Mechanistically, LCETRL3 activates NOTCH1-PTEN-AKT signaling by stabilizing TDP43 and LCETRL4 activates PDK1-AKT signaling by stabilizing EIF2S1 (Li et al., 2022). It was revealed that lncRNA H19 was upregulated in lung cancer cells and H19 knockdown augmented the gefitinib sensitivity by enhancing PTEN and PDCD4 and decreasing NFIB (Zhou and Zhang, 2020). Paradoxically, in another study, H19 was identified as significantly downregulated in clinical samples and in vitro models of resistance to EGFR-TKIs. H19 knockdown contributed to erlotinib resistance by upregulating PKM2 and activating the AKT pathway (Wang C. et al., 2017; Chen C. et al., 2020). Consequently, the exact effect of H19 within the mechanism of EGFR-TKI resistance in NSCLC still needs to be further explored.
In addition to modulating upstream receptors and regulatory factors, lncRNAs also regulate downstream molecules of the PI3K/AKT pathway, such as glycogen synthase kinase-3 (GSK3), mouse double minute 2 homolog (MDM2), FOXO3 and P21, to influence EGFR-TKI resistance in NSCLC. For example, in a study by Wang et al., lncRNA PCAT-1 was found to be highly expressed in EGFR TKI-resistant NSCLC and its knock-down inhibited the phosphorylation of AKT and GSK3, thus improving gefitinib sensitivity in NSCLC (Wang S. et al., 2021). But the more detailed mechanism of GSK3 regulation by PCAT-1 is unclear and requires further exploration. Similarly, the knockdown of lncRNA MIR31HG inhibited the PI3K/AKT signaling pathway and MDM2, thereby increasing p53 expression and enhancing gefitinib sensitivity in PC9-R cells (Wang B. et al., 2017). Interestingly, Bing et al. identified the lncRNA CASC9/miR-195-5p/FOXO3 positive feed-back loops that can enhance gefitinib resistance in NSCLC (Bing et al., 2021). In addition, both lncRNAs UCA1 and HOTAIR promoted gefitinib resistance in NSCLC through epigenetically silencing p21 expression by interacting with EZH2, a methyltransferase responsible for the trimethylation of H3K27 (Xu et al., 2020; Li et al., 2021). According to the large number of lncRNAs reported above, the regulation of the PI3K/AKT signaling pathway by lncRNAs has a significant impact in EGFR-TKI resistance.
4.2 The classical MAPK signaling pathway
The classical MAPK signaling pathway, which is generally called the RAS/MEK/ERK pathway, is one of the key signaling pathways downstream of EGFR that modulates cancer growth, survival, migration, and angiogenesis (Martinelli et al., 2017). It has already been shown that the dysregulation of this pathway was related to EGFR-TKI resistance in NSCLC (Coco et al., 2015). In a similar way to the PI3K/AKT signaling pathway, lncRNAs can activate or inhibit the classical MAPK signaling pathway by regulating its upstream receptors and regulatory factors (Figure 1).
EGFR is the primary upstream receptor in the classical MAPK pathway that is regulated through lncRNAs. For example, lncRNA LINC00460 was demonstrated to promote gefitinib resistance in NSCLC via sponging miR-769-5p and facilitating EGFR expression (Ma et al., 2019). Nevertheless, lncRNA CRNDE downregulated eIF4A3, mucin 1 (MUC1), as well as p-EGFR, thereby increasing EGFR-TKI resistance (Takahashi et al., 2021). Therefore, the relationship between EGFR expression and EGFR-TKI resistance requires further investigation. Furthermore, lncRNA DUXAP9-206 was found to interact with Cbl-b, one of the E3 ubiquitin ligases, to partially reduce EGFR degradation and activate the RAS/MEK/ERK and PI3K/AKT pathways, resulting in a degree of promotion of the malignant phenotype in NSCLC cells (Zhu et al., 2019). It indicates that DUXAP9-206 may be involved in EGFR-TKI resistance. In addition to activating the PI3K/AKT signaling pathway, as mentioned above, lncRNA LINC01510 can activate the classical MAPK signaling pathway by upregulating MET to promote EGFR-TKI resistance (Pal et al., 2022). Moreover, other lncRNAs, such as Linc00284, FAM83A-AS1, and LINC00857, were revealed to mediate lung cancer progression through the regulation of MET (Su et al., 2020; Sheng et al., 2021; Zhao et al., 2022), but their roles in EGFR-TKI resistance need to be further explored.
The modulation of regulators in the classical MAPK pathway, including DUSP1 as well as RASA, by lncRNAs contributes to EGFR-TKI resistance. Mainly through bioinformatics analysis, Ma et al. revealed that lncRNA CASC9 mediated EGFR-TKI resistance via interacting with some protein-coding genes (PCGs) (Ma et al., 2017). In a subsequent study, Chen et al. revealed that CASC9 epigenetically silenced DUSP1 and activated the ERK pathway by recruiting EZH2, resulting in enhancing gefitinib resistance in NSCLC (Chen Z. et al., 2020). Considering that the CASC9/miR-195-5p/FOXO3 regenerative feed-back loops described above can also enhance gefitinib resistance (Bing et al., 2021), CASC9 may be a target with great promise for surmounting EGFR-TKI resistance in NSCLC. Moreover, lncRNA LOC554202 promoted gefitinib resistance by upregulating the expression of miR-31, which can directly repress RASA1 and Hypoxia Inducible Factor 1 Subunit Alpha Inhibitor (FIH-1) expression, resulting in partial upregulation of the classical MAPK and PI3K/AKT pathways (He et al., 2019). Similar to DUSP1 and RASA1, as a negative regulatory factor in the classical MAPK pathway, NF1 is also capable of affecting EGFR-TKI resistance in NSCLC (de Bruin et al., 2014). One investigation has reported lncRNA H19 increased NF1 by competitively combining with microRNA-107, thus promoting NSCLC progression (Qian et al., 2018). In theory, the upregulation of NF1 could inhibit the classical MAPK signaling pathway, thereby inhibiting tumor development, which is contrary to the results of this study. Therefore, the mechanism by which H19 regulates NF1 and the classical MAPK signaling pathway needs to be further explored. In addition, whether H19 promotes EGFR-TKI resistance in NSCLC by regulating NF1 is also unknown and needs further investigation.
4.3 The JAK/STAT signaling pathway
The JAK/STAT signaling pathway, which is mainly composed of ligands, receptors, JAKs and STATs, plays a crucial role in mediating immune adaptation, tissue repair, inflammatory response, and cell apoptosis (Owen et al., 2019). The abnormal upregulation of this pathway is related to tumor progression as well as drug resistance (Jin, 2020; Hu X. et al., 2021). LncRNAs are able to mediate EGFR-TKI resistance in NSCLC via impacting the ligands, receptors, and regulatory factors in the JAK/STAT pathway (Figure 1).
More than 50 cytokines have been identified in the JAK/STAT signaling pathway (Morris et al., 2018). Interleukin 6 (IL-6), an important cytokine associated with chronic inflammatory diseases, was engaged in the progression of lung cancer (Gao et al., 2007). Researchers discovered that lncRNA LINC00460 can competitively bind with miR-149-5p to facilitate IL-6 expression, thus promoting gefitinib resistance of NSCLC cells. In mechanism, IL-6 can activate the JAK/STAT and PI3K/AKT pathways, thereby inducing an EMT-like phenotype, which is one of the mechanisms responsible for EGFR-TKI resistance in NSCLC (Nakano et al., 2020). Similarly, lncRNA TSLNC8 was downregulated in NSCLC and its over-expression suppressed the development of NSCLC by modulating the IL-6/STAT3/HIF-1α pathway (Fan et al., 2019). It is noteworthy that in another study, upregulation of TSLNC8 also markedly augmented the antitumor activity of osimertinib on NSCLC by repressing the EFGR/STAT3 signaling pathway (Zhou et al., 2020). Therefore, TSLNC8 is able to impact the JAK/STAT pathway not only by regulating IL-6 but also by EGFR, thus mediating the development as well as EGFR-TKI resistance of NSCLC. In addition, lncRNA PCAT6 was revealed to augment gefitinib resistance by serving as a competitive endogenous RNA for miR-326 in increasing interferon-alpha receptor 2 (IFNAR2) expression, an upstream receptor in the JAK/STAT pathway (Zheng et al., 2022).
Similar to the PI3K/AKT and classical MAPK signaling pathways, lncRNAs are also able to modulate the JAK/STAT pathway via affecting its regulatory factors. For example, interferon-induced protein 44 (IFI44), an upstream regulator in the JAK/STAT signaling pathway, was found to be inhibited by LINC01116 to promote gefitinib resistance in NSCLC (Wang H. et al., 2020). In addition, both lncRNA UCA1 and lncRNA BLACAT1 have been reported to mediate EGFR-TKI resistance of NSCLC cells through targeting the JAK/STAT signaling pathway (Zhang et al., 2019; Shu et al., 2020). The fundamental mechanisms through which these lncRNAs modulate the JAK/STAT signaling pathway, however, are still unknown and require additional investigation.
5 Modulation of phenotypic transformation
Phenotype transformations, including EMT, transformations from LUAD to LUSC as well as NSCLC to SCLC, are considered to be essential mechanisms of EGFR-TKI resistance in NSCLC (Rubin et al., 2020; Shaurova et al., 2020). EMT was found to be a reversible procedure in which cancer cells lose epithelial features as well as gain mesenchymal phenotypes, resulting in the acquisition of invasiveness, metastasis, and drug resistance (Witta et al., 2006; Singh and Settleman, 2010). The molecular mechanisms underlying EMT are not well studied. Current evidence suggests that this process is accompanied by abnormal expression of a wide range of molecules, including reduced expression of epithelial markers, for example E-cadherin, as well as upregulation of mesenchymal markers, for example vimentin (Sinha et al., 2020). Furthermore, alterations of multiple transcription factors (Yuan et al., 2019) and cellular signaling pathways, such as transforming growth factor-β (TGF-β), Wnt/β-catenin and PI3K/AKT signaling pathways, were found to be involved in EMT (Xu et al., 2015; Hao et al., 2019; Liu L. et al., 2020). According to recent studies, some lncRNAs associated with EGFR-TKI resistance can mediate EMT mainly by regulating transcription factors and some signaling pathways. In addition, SCLC transformation and LUSC transformation are also implicated with EGFR-TKI resistance (Shaurova et al., 2020). Nevertheless, the mechanism by which lncRNAs in regulating SCLC and LUSC transformation is ambiguous and urgently needs to be further explored.
It is revealed that silencing lncRNA CASC8 promoted osimertinib sensitivity in NSCLC cells through downregulation of Forkhead box M1 (FOXM1) (Jiang et al., 2021), one of the critical transcription factors for EMT (Balli et al., 2013; Wei et al., 2015). Researchers in this study speculated that CASC8 acted as the competitive endogenous RNA for miR-671-5P to enhance the transcription of FOXM1. However, the exact mechanism is not explored and needs further validation. Similarly, lncRNA OSER1-AS1 served as a sponge for miR-612 to enhance FOXM1 transcription, resulting in promoting gefitinib resistance (Shi et al., 2021). Zinc finger E-box binding homeobox 1 (ZEB1) has also been proven to be a transcription factor that can induce EMT in carcinoma cells (Zhang et al., 2015). LncRNA MALAT1 can promote gefitinib resistance through the miR-200a/ZEB1 axis (Feng et al., 2019). In another study, it was demonstrated that MALAT1 can promote EMT by sponging miR-124 (Wu et al., 2018). Therefore, the MALAT1/ZEB1 pathway may affect EGFR-TKI resistance by promoting EMT. LncRNA linc00673 can upregulate ZEB1 by sponging miR-150-5p to promote EMT (Lu et al., 2017). Nevertheless, the association of linc00673 with EGFR-TKI resistance needs to be further studied. Furthermore, through in vitro and in vivo experiments, lnc-ABCA12-8 was revealed to promote gefitinib resistance in NSCLC via interacting with alternative splicing factor/splicing factor 2 (ASF/SF2) and enhancing the expression of IIICS region of fibronectin 1 (FN1) (He et al., 2022), an important marker of EMT (Liu et al., 2020b).
In terms of signaling pathways, lncRNA LINC00460 was shown to serve as a sponge of miR-149-5p to activate the JAK/STAT and PI3K/AKT signaling pathways to induce EMT, thus promoting EGFR-TKI resistance in NSCLC (Nakano et al., 2020). Researchers revealed the low expression of lncRNA WT1-AS rendered lncRNA UCA1 highly expressed, thereby promoting EMT (Wan et al., 2021). Interestingly, WT1-AS also functioned as a competitive endogenous RNA of miR-494-3p to activate the PI3K/AKT pathway, thus facilitating the proliferation, migration, and invasion in NSCLC (Wu C. et al., 2021). Moreover, UCA1 was revealed to regulate gefitinib resistance through activating the PI3K/AKT pathway and mediating EMT (Cheng et al., 2015b). For this reason, the mechanism of the regulatory relationship between WT1-AS and UCA1 in EGFR-TKI resistance in NSCLC deserves further exploration. Additionally, NOTCH-1, the primary upstream receptor of NOTCH signaling pathway that regulates EMT (Xie et al., 2013), conferred gefitinib resistance of NSCLC via the lncRNA SNHG15/miR-451/ZEB1 feedback loop (Huang J. et al., 2020). Moreover, lncRNA XIST and HCP5 can promote EMT mediated by TGF-β through modulating miR-367/miR-141-ZEB2 or miR-203/SNAI axis, respectively (Li et al., 2018; Jiang et al., 2019). Nevertheless, it is not clear whether lncRNA XIST and HCP5 can mediate EGFR-TKI resistance of NSCLC via promoting EMT, which warrants further investigation.
6 Exosomal lncRNAs in the tumor microenvironment
Tumor formation results from the interaction of tumor cells with the extracellular matrix, tumor vasculature, and a variety of immune cells, so the development, metastasis, and drug resistance of tumor depend not only on genetic alterations within the tumor cells but also on the external tumor microenvironment (Junttila and de Sauvage, 2013). The tumor microenvironment, mainly including the vascular network, catabolic cancer-associated fibroblasts (CAFs), immune-related cells, as well as extracellular matrix (Klemm and Joyce, 2015), has a pivotal function in EGFR-TKI resistance in NSCLC (Jia et al., 2019; Dzul Keflee et al., 2022). Currently, many studies on the tumor microenvironment of lung cancer focus on immune cells and immunotherapy; nevertheless, the roles of exosomes from the tumor microenvironment for resistance to EGFR-TKIs in NSCLC are also significant (Liu et al., 2020c; Yuan et al., 2022). Released by eukaryotic cells, exosomes, a type of extracellular vesicles that mainly contain proteins, lipids and genetic materials, play essential roles in the intracellular communication network in the tumor microenvironment (Mostafazadeh et al., 2021). Substantial evidence indicates that lncRNAs in tumor cells or other cells can be transported as well as secreted into the tumor microenvironment via exosomes to regulate cell function and promote drug resistance (Pathania and Challagundla, 2021). Exosomal lncRNAs in the tumor microenvironment affect EGFR-TKI resistance of NSCLC mainly by regulating EGFR TKI-sensitive NSCLC cells or other non-tumor cells (Figure 2).
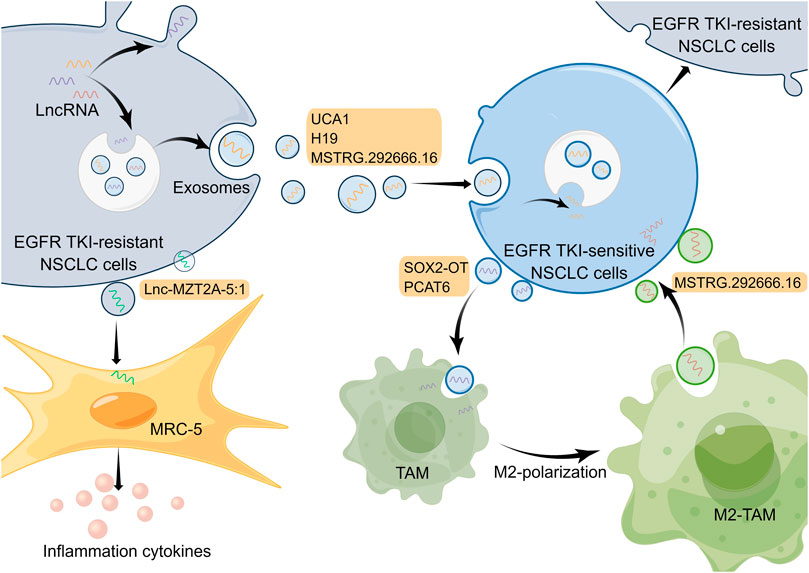
FIGURE 2. Mechanisms of lncRNAs involved in EGFR-TKI resistance in NSCLC by modulating tumor microenvironment. LncRNAs in cells can be transported and secreted into the tumor microenvironment via exosomes to participate in intercellular communication. And exosomal lncRNAs are able to affect EGFR-TKI resistance in NSCLC by regulating EGFR TKI-sensitive NSCLC cells and other non-tumor cells. The Figure was drawn by Figdraw.
6.1 Regulation of EGFR TKI-sensitive NSCLC cells
Exosomes in the tumor microenvironment can be taken up by adjacent EGFR TKI-sensitive NSCLC cells, which results in NSCLC cells becoming resistant to EGFR-TKIs (Wu S. et al., 2021). For example, lncRNA UCA1 can be delivered to gefitinib-sensitive recipient cells by exosomes isolated from gefitinib-resistant cell lines. And it promoted gefitinib resistance in NSCLC in vitro and in vivo through functioning as a ceRNA for miR-143 and modulating FOSL2 expression (Chen X. et al., 2020). It has been reported that hnRNPA2B1, an RNA-binding protein that regulates RNA loading into exosomes, mediated the packaging of lncRNA H19 into exosomes, which promoted gefitinib resistance in NSCLC cells (Lei et al., 2018). Additionally, another research has demonstrated that exosomal lncRNA H19 could enhance erlotinib resistance in NSCLC cells through the regulation of miR-615-3p/ATG7 axis (Pan and Zhou, 2020), indicating that exosomal lncRNA H19 may be a potential treatment target for NSCLC. Moreover, Deng et al. discovered exosomal lncRNA MSTRG.292666.16 was able to be uptaken by osimertinib-sensitive NSCLC cells, which led to these cells becoming resistant to osimertinib (Deng et al., 2020). However, given that this study only used one cell line for in vitro experiments, the mechanism by which this exosomal lncRNA modulates EGFR-TKI resistance needs to be explored in more NSCLC cell lines and in vivo experiments. In a subsequent study, investigators isolated exosomal lncRNA MSTRG.292666.16 from M2 type tumor-associated macrophages (TAMs) and demonstrated that it could promote osimertinib resistance of NSCLC through modulating miR-6836-5p/MAPK8IP3 axis (Wan et al., 2022). Thus, exosomal lncRNA MSTRG.292666.16 may be used as a promising therapeutic target to overcome osimertinib resistance in NSCLC. These studies illustrate that exosomal lncRNAs associated with EGFR-TKI resistance can be derived not only from EGFR TKI-resistant NSCLC cells but also from non-tumor cells, such as M2 type TAMs, which fully exemplifies the complexity of cellular communication in the tumor microenvironment.
6.2 Regulation of other non-tumor cells
Besides regulating tumor cells, exosomes in the tumor microenvironment can act on other non-tumor cells and, in turn, influence the drug resistance in tumor cells. For instance, in an investigation conducted by Zhou and collaborators, exosomal lncRNA SOX2 overlapping transcript (SOX2-OT) was delivered from NSCLC cell line H1975 to THP-1 cells and it was able to enhance macrophage M2 polarization as well as inhibit macrophage M1 polarization by regulating miR-627-3p/Smad axis, thereby promoting EGFR-TKI resistance (Zhou et al., 2021). Similarly, exosomal lncRNA PCAT6 derived from NSCLC cells also could promote macrophage M2 polarization, which enhances metastasis and EMT processes in NSCLC through modulating miR-326/KLF1 pathway (Chen et al., 2022). In addition, the phenomenon of macrophage M2 polarization in the tumor microenvironment is able to promote the EGFR-TKI resistance of NSCLC cells (Xiao et al., 2020). So exosomal lncRNAs may serve as targets to inhibit macrophage M2 polarization to suppress NSCLC. Furthermore, exosomal lnc-MZT2A-5:1 from AZD9291-resistant NSCLC cells was significantly upregulated compared with wild-type NSCLC cells, and it was able to enhance the activation of lung fibroblasts (MRC-5 cells) (Song et al., 2021). Nevertheless, whether and how activated MRC-5 cells can further influence the tumor microenvironment and EGFR-TKI resistance of NSCLC urgently requires in-depth exploration.
7 LncRNAs and other mechanisms
7.1 CeRNA networks
The intricate interplay among diverse RNA species critically contributes to the regulation of cellular functions. Different RNA transcripts that have the same miRNA response elements (MREs) and constitute a competitive relationship can act as competitive endogenous RNAs (ceRNAs), the so-called miRNA sponges (Tay et al., 2014). In recent years, there is accumulating evidence suggesting that lncRNAs and their constituent ceRNA networks are implicated with EGFR-TKI resistance of NSCLC (Kong et al., 2020; Wang T. et al., 2021). Many lncRNAs that act as ceRNAs to regulate EGFR-TKI resistance in NSCLC have been mentioned above. However, because ceRNA networks are unique and important for drug resistance, there are some points to be emphasized and elaborated. LncRNA PCAT6 has been reported to enhance gefitinib resistance of NSCLC through modulation of miR-326/IFNAR2 pathway (Zheng et al., 2022). Another study found PCAT6 promoted EMT process in NSCLC through miR-326/KLF1 pathway (Chen et al., 2022). Moreover, PCAT6 can enhance NSCLC proliferation, migration and invasion via acting as ceRNA of miR-330-5p (Cui et al., 2018). Accordingly, one lncRNA can competitively bind to different miRNAs, and one miRNA can also target different mRNAs, illustrating that ceRNA networks are heterogeneous.
In the field of lncRNAs and ceRNA networks research, bioinformatics prediction tools are used to predict and construct lncRNA-miRNA-mRNA Networks (Rincón-Riveros et al., 2021), which could subsequently be validated with dual luciferase reporter or RNA immunoprecipitation assays (Huang et al., 2019). For example, Wu et al. predicted and validated that lncRNA SNHG14 could sponge to miR-206-3p through online software starbase v2.0 and luciferase reporter analysis. Then similarly, they predicted and validated ABCB1, a member in the ATP-binding cassette (ABC) transporter family, as the target of miR-206-3p. Eventually, they demonstrated that SNHG14 promoted gefitinib resistance of NSCLC via the regulation of miR-206-3p/ABCB1 axis (Wu et al., 2019). In similar ways, it is proved lncRNA human ovarian cancer-specific transcript 2 (HOST2) facilitated gefitinib resistance via functioning as ceRNA for miRNA-621 to upregulate SYF2, a cell cycle-associated protein (Chen et al., 2019). In addition, CCAT1 and lnc-TMEM132D-AS1 were demonstrated to regulate EGFR-TKI resistance in NSCLC through their corresponding ceRNA networks (Jin et al., 2020; Wang et al., 2023). This evidence demonstrates the significance of bioinformatics prediction tools for ceRNA networks research. However, due to the wide variability in the timing, space, as well as the abundance of expression of diverse RNA species in cells, the prediction strategies of bioinformatic tools need to be continuously refined (Ebert and Sharp, 2010; Tay et al., 2014).
7.2 Epigenetic modifications
Cancer is both a multi-genic as well as an epigenetic disease, as there are not only genetic mutations but also a great number of epigenetic alterations in tumor cells (Iacobuzio-Donahue, 2009). Epigenetic modifications, for example, DNA methylation and histone modification, are critical in tumor growth, metastasis, and drug resistance (Gao et al., 2017; Bajbouj et al., 2021). LncRNAs can affect EGFR-TKI resistance in NSCLC through epigenetic modifications. For instance, lncRNAs MIAT recruited Dnmt3a, a DNA methyltransferase, to methylate the miR-34a promotor, leading to silencing miR-34a expression and finally conferring gefitinib resistance through the PI3K/AKT signaling pathway (Fu et al., 2018). Furthermore, lncRNA ferritin heavy chain 1 pseudogene 3 (FTH1P3) accelerated gefitinib resistance of NSCLC by recruiting lysine-specific demethylase 1 (LSD1), a histone-modifying enzyme, and epigenetically inhibiting the tissue inhibitor of metalloproteinase 3 (TIMP3) (Zheng G. et al., 2020). Likewise, lncRNA HAS2-AS1 can recruit LSD1 to the EphB3 promoter area and repress EphB3 transcription, thus promoting gefitinib resistance (Sun et al., 2020). EZH2, one of the key subunits in the polycomb repressive complex 2 (PRC2) complex, is the histone methyltransferase that can catalyze the trimethylation of H3K27me3 (Chase and Cross, 2011). It was reported that LINC00665 can mediate gefitinib resistance in NSCLC via interacting with EZH2 and activating the PI3K/AKT pathway (Geng et al., 2015; Liu et al., 2019). In addition, lncRNA UCA1 and HOTAIR are also able to facilitate gefitinib resistance through modification of EZH2 (Xu et al., 2020; Li et al., 2021). Therefore, EZH2 modified by lncRNAs plays a great role in EGFR-TKI resistance in NSCLC and is a possible therapeutic target for overcoming EGFR-TKI resistance in NSCLC, which deserves further exploration.
7.3 Epitranscriptomic modifications
In addition to epigenetic modifications, RNA modifications, called epitranscriptomic modifications, are also one of the mechanisms by which lncRNAs regulate EGFR-TKI resistance in NSCLC. Epitranscriptomic modifications modulate the function of mRNAs or ncRNAs by adding functional groups to them, of which methylation modifications are the most common type, such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyladenosine (m1A), etc (Barbieri and Kouzarides, 2020). Epitranscriptomic modifications can act not only as the upstream mechanism for the dysregulation of lncRNAs but also as the downstream mechanism for lncRNAs to function. For example, Zhang et al. found that METTL3-mediated m6A modification upregulated lncRNA SNHG17 by stabilizing its transcription, thereby promoting gefitinib resistance in NSCLC via the EZH2/LATS2 pathway (Zhang et al., 2022). LINC00969 was found to interact with both EZH2 and METTL3 to regulate histone methylation levels in the promoter region of NLRP3 and the m6A levels of NLRP3 mRNA to inhibit the expression of NLRP3, which promotes gefitinib resistance (Dai et al., 2023). In addition, lncRNA DGUOK-AS1 can promote the malignant phenotype of NSCLC by regulating the m6A modification of TRPM7 (Feng et al., 2023). However, whether DGUOK-AS1 can promote EGFR-TKI resistance still needs to be further investigated. This review focuses on the mechanisms of lncRNAs in EGFR-TKI resistance in NSCLC, but in fact, circRNAs, a subclass of lncRNAs, are also involved in regulating EGFR-TKI resistance (Zhou et al., 2019; Huang Y. et al., 2020; Ma et al., 2020; Yang et al., 2021; Dai et al., 2022; Fan et al., 2022; Niu et al., 2022; Wang et al., 2022; Pan et al., 2023; Wen et al., 2023). The relevant circRNAs are listed in Table 2.
8 Conclusion and perspective
In recent years, EGFR-TKI treatment for NSCLC has shown promising efficacy, but the development of drug resistance is unavoidable and significantly limits the therapeutic potency of EGFR-TKI treatments (Mok et al., 2009; Zhou et al., 2011). Therefore, in order to overcome EGFR-TKI resistance and improve patients’ prognosis, researchers have conducted numerous in-depth investigations into the mechanism of EGFR-TKI resistance. Due to the rapid development of RNA sequencing technology and bioinformatics analysis, many lncRNAs were found to participate in the regulation of EGFR-TKI resistance in NSCLC (Cheng et al., 2015a; Shi et al., 2020). The evidence described in this review suggests that lncRNAs can influence EGFR-TKI resistance by regulating aberrant activation of signaling pathways, phenotypic transformation, tumor microenvironment, and other modalities.
However, the research of lncRNAs in EGFR TKI-resistant NSCLC is still in its infancy, and some limitations exist. To start with, most studies concentrated on the downstream mechanisms underlying dysregulated lncRNAs in EGFR TKI-resistant NSCLC, while the upstream mechanism of dysregulation of lncRNA is unclear. Second, there are many lncRNAs that have been demonstrated to be implicated in NSCLC progression, but their relationship with EGFR-TKI resistance was not further explored (Lv et al., 2021; Xu et al., 2021). In third place, despite the overwhelming evidence that lncRNAs have a crucial role in EGFR-TKI resistance, to date, lncRNAs have not been clinically validated for their potential value. In terms of diagnosis and prediction, exosomal lncRNAs have great clinical application value and may be the focus of clinical application exploration due to their natural advantages of wide distribution and easy access to materials (Tao et al., 2020; Han et al., 2021). On the therapeutic side, lncRNAs targeting therapies, which rely on technologies such as RNAi, antisense oligonucleotides (ASOs), adenoviral/lentiviral vectors and CRISPR-Cas9, face many challenges (Goyal et al., 2017; Verdera et al., 2020; Kara et al., 2022), particularly safety concerns caused by off-target effects, which still need to be overcome (Toden et al., 2021). Intriguingly, many studies have demonstrated that certain drugs can regulate lncRNAs to reverse EGFR-TKI resistance in NSCLC by targeting upstream or downstream molecules of lncRNAs. Examples include the upregulation of GAS5 by ecto-ATP synthase inhibitor (Chang et al., 2020), the downregulation of MALAT1 by polyphylin I (Yang et al., 2018), the downregulation of HOTAIR by berberine (Zheng F. et al., 2020), and the downregulation of CCAT1 by hyperoside (Hu et al., 2020), all of which shed fresh light on dealing with EGFR-TKI resistance, that is, targeting upstream or downstream molecules of lncRNAs is also a possible alternative therapeutic approach.
In conclusion, lncRNAs are crucial in EGFR-TKI resistance of NSCLC via a variety of mechanisms. More in-depth investigations are warranted to further uncover the regulatory role of lncRNAs in EGFR-TKI resistance and apply lncRNAs to clinical practices to promote the prognosis of NSCLC patients.
Author contributions
WZ directed this work. DL consulted relevant literature and wrote the paper. XL modified the figures. WH revised and refined the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Hunan Provincial Science and Technology Department [2022JJ30995] to WZ.
Acknowledgments
Figures were created by Figdraw (www.figdraw.com). The authors are grateful to Figdraw.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bajbouj, K., Al-Ali, A., Ramakrishnan, R. K., Saber-Ayad, M., and Hamid, Q. (2021). Histone modification in NSCLC: Molecular mechanisms and therapeutic targets. Int. J. Mol. Sci. 22 (21), 11701. doi:10.3390/ijms222111701
Balli, D., Ustiyan, V., Zhang, Y., Wang, I. C., Masino, A. J., Ren, X., et al. (2013). Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. Embo J. 32 (2), 231–244. doi:10.1038/emboj.2012.336
Barbieri, I., and Kouzarides, T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20 (6), 303–322. doi:10.1038/s41568-020-0253-2
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77 (15), 3965–3981. doi:10.1158/0008-5472.CAN-16-2634
Bing, Z., Han, J., Zheng, Z., and Liang, N. (2021). FOXO3-induced oncogenic lncRNA CASC9 enhances gefitinib resistance of non-small-cell lung cancer through feedback loop. Life Sci. 287, 120012. doi:10.1016/j.lfs.2021.120012
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigó, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 (7146), 799–816. doi:10.1038/nature05874
Cao, Z., Oyang, L., Luo, X., Xia, L., Hu, J., Lin, J., et al. (2022). The roles of long non-coding RNAs in lung cancer. J. Cancer 13 (1), 174–183. doi:10.7150/jca.65031
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309 (5740), 1559–1563. doi:10.1126/science.1112014
Chang, Y. W., Hsu, C. L., Tang, C. W., Chen, X. J., Huang, H. C., and Juan, H. F. (2020). Multiomics reveals ectopic ATP synthase blockade induces cancer cell death via a lncRNA-mediated phospho-signaling network. Mol. Cell Proteomics 19 (11), 1805–1825. doi:10.1074/mcp.RA120.002219
Chase, A., and Cross, N. C. (2011). Aberrations of EZH2 in cancer. Clin. Cancer Res. 17 (9), 2613–2618. doi:10.1158/1078-0432.CCR-10-2156
Chen, C., Liu, W. R., Zhang, B., Zhang, L. M., Li, C. G., Liu, C., et al. (2020a). LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 and phosphorylation of AKT in EGFR-mutant lung cancers. Cancer Lett. 486, 58–70. doi:10.1016/j.canlet.2020.05.009
Chen, X., Wang, Z., Tong, F., Dong, X., Wu, G., and Zhang, R. (2020c). lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol. Ther. Nucleic Acids 19, 643–653. doi:10.1016/j.omtn.2019.10.047
Chen, Y., Hong, C., Qu, J., Chen, J., and Qin, Z. (2022). Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered 13 (5), 12834–12846. doi:10.1080/21655979.2022.2076388
Chen, Z., Chen, Q., Cheng, Z., Gu, J., Feng, W., Lei, T., et al. (2020b). Long non-coding RNA CASC9 promotes gefitinib resistance in NSCLC by epigenetic repression of DUSP1. Cell Death Dis. 11 (10), 858. doi:10.1038/s41419-020-03047-y
Chen, Z. Y., Liu, H. Y., Jiang, N., and Yuan, J. M. (2019). LncRNA HOST2 enhances gefitinib-resistance in non-small cell lung cancer by down-regulating miRNA-621. Eur. Rev. Med. Pharmacol. Sci. 23 (22), 9939–9946. doi:10.26355/eurrev_201911_19560
Cheng, N., Cai, W., Ren, S., Li, X., Wang, Q., Pan, H., et al. (2015b). Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 6 (27), 23582–23593. doi:10.18632/oncotarget.4361
Cheng, N., Li, X., Zhao, C., Ren, S., Chen, X., Cai, W., et al. (2015a). Microarray expression profile of long non-coding RNAs in EGFR-TKIs resistance of human non-small cell lung cancer. Oncol. Rep. 33 (2), 833–839. doi:10.3892/or.2014.3643
Choi, H. K., and Mazzone, P. J. (2022). Lung cancer screening. Med. Clin. North Am. 106 (6), 1041–1053. doi:10.1016/j.mcna.2022.07.007
Coco, S., Truini, A., Alama, A., Dal Bello, M. G., Venè, R., Garuti, A., et al. (2015). Afatinib resistance in non-small cell lung cancer involves the PI3K/AKT and MAPK/ERK signalling pathways and epithelial-to-mesenchymal transition. Target Oncol. 10 (3), 393–404. doi:10.1007/s11523-014-0344-7
Cui, D., Feng, Y., Shi, K., Zhang, H., and Qian, R. (2020). Long non-coding RNA TRPM2-AS sponges microRNA-138-5p to activate epidermal growth factor receptor and PI3K/AKT signaling in non-small cell lung cancer. Ann. Transl. Med. 8 (20), 1313. doi:10.21037/atm-20-6331
Cui, L. H., Xu, H. R., Yang, W., and Yu, L. J. (2018). lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 11, 7715–7724. doi:10.2147/OTT.S178597
Dai, C., Ma, Z., Si, J., An, G., Zhang, W., Li, S., et al. (2022). Hsa_circ_0007312 promotes third-generation epidermal growth factor receptor-tyrosine kinase inhibitor resistance through pyroptosis and apoptosis via the MiR-764/MAPK1 Axis in lung adenocarcinoma cells. J. Cancer 13 (9), 2798–2809. doi:10.7150/jca.72066
Dai, J., Qu, T., Yin, D., Cui, Y., Zhang, C., Zhang, E., et al. (2023). LncRNA LINC00969 promotes acquired gefitinib resistance by epigenetically suppressing of NLRP3 at transcriptional and posttranscriptional levels to inhibit pyroptosis in lung cancer. Cell Death Dis. 14 (5), 312. doi:10.1038/s41419-023-05840-x
de Bruin, E. C., Cowell, C., Warne, P. H., Jiang, M., Saunders, R. E., Melnick, M. A., et al. (2014). Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 4 (5), 606–619. doi:10.1158/2159-8290.CD-13-0741
Deng, Q., Fang, Q., Xie, B., Sun, H., Bao, Y., and Zhou, S. (2020). Exosomal long non-coding RNA MSTRG.292666.16 is associated with osimertinib (AZD9291) resistance in non-small cell lung cancer. Aging (Albany NY) 12 (9), 8001–8015. doi:10.18632/aging.103119
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22 (9), 1775–1789. doi:10.1101/gr.132159.111
Dieci, G., Fiorino, G., Castelnuovo, M., Teichmann, M., and Pagano, A. (2007). The expanding RNA polymerase III transcriptome. Trends Genet. 23 (12), 614–622. doi:10.1016/j.tig.2007.09.001
Ding, D., Zhang, J., Luo, Z., Wu, H., Lin, Z., Liang, W., et al. (2022). Analysis of the lncRNA-miRNA-mRNA network reveals a potential regulatory mechanism of EGFR-TKI resistance in NSCLC. Front. Genet. 13, 851391. doi:10.3389/fgene.2022.851391
Dinger, M. E., Pang, K. C., Mercer, T. R., and Mattick, J. S. (2008). Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 4 (11), e1000176. doi:10.1371/journal.pcbi.1000176
Dong, S., Qu, X., Li, W., Zhong, X., Li, P., Yang, S., et al. (2015). The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J. Hematol. Oncol. 8, 43. doi:10.1186/s13045-015-0140-6
Dzul Keflee, R., Leong, K. H., Ogawa, S., Bignon, J., Chan, M. C., and Kong, K. W. (2022). Overview of the multifaceted resistances toward EGFR-TKIs and new chemotherapeutic strategies in non-small cell lung cancer. Biochem. Pharmacol. 205, 115262. doi:10.1016/j.bcp.2022.115262
Ebert, M. S., and Sharp, P. A. (2010). Emerging roles for natural microRNA sponges. Curr. Biol. 20 (19), R858–R861. doi:10.1016/j.cub.2010.08.052
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J. R., Bharat, A., et al. (2022). Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 20 (5), 497–530. doi:10.6004/jnccn.2022.0025
Ezkurdia, I., Juan, D., Rodriguez, J. M., Frankish, A., Diekhans, M., Harrow, J., et al. (2014). Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 23 (22), 5866–5878. doi:10.1093/hmg/ddu309
Fan, D., Yang, Y., and Zhang, W. (2022). A novel circ_MACF1/miR-942-5p/TGFBR2 axis regulates the functional behaviors and drug sensitivity in gefitinib-resistant non-small cell lung cancer cells. BMC Pulm. Med. 22 (1), 27. doi:10.1186/s12890-021-01731-z
Fan, H., Li, J., Wang, J., and Hu, Z. (2019). Long non-coding RNAs (lncRNAs) tumor-suppressive role of lncRNA on chromosome 8p12 (TSLNC8) inhibits tumor metastasis and promotes apoptosis by regulating interleukin 6 (IL-6)/Signal transducer and activator of transcription 3 (STAT3)/Hypoxia-Inducible factor 1-alpha (HIF-1α) signaling pathway in non-small cell lung cancer. Med. Sci. Monit. 25, 7624–7633. doi:10.12659/MSM.917565
Fang, Y., and Fullwood, M. J. (2016). Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinforma. 14 (1), 42–54. doi:10.1016/j.gpb.2015.09.006
Feng, C., Zhao, Y., Li, Y., Zhang, T., Ma, Y., and Liu, Y. (2019). LncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge. Arch. Bronconeumol (Engl Ed. 55 (12), 627–633. doi:10.1016/j.arbres.2019.03.026
Feng, Y., Wu, F., Wu, Y., Guo, Z., and Ji, X. (2023). LncRNA DGUOK-AS1 facilitates non-small cell lung cancer growth and metastasis through increasing TRPM7 stability via m6A modification. Transl. Oncol. 32, 101661. doi:10.1016/j.tranon.2023.101661
Fresno Vara, J. A., Casado, E., de Castro, J., Cejas, P., Belda-Iniesta, C., and González-Barón, M. (2004). PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30 (2), 193–204. doi:10.1016/j.ctrv.2003.07.007
Fu, Y., Li, C., Luo, Y., Li, L., Liu, J., and Gui, R. (2018). Silencing of long non-coding RNA MIAT sensitizes lung cancer cells to gefitinib by epigenetically regulating miR-34a. Front. Pharmacol. 9, 82. doi:10.3389/fphar.2018.00082
Gao, S. P., Mark, K. G., Leslie, K., Pao, W., Motoi, N., Gerald, W. L., et al. (2007). Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Invest. 117 (12), 3846–3856. doi:10.1172/JCI31871
Gao, X., Reid, M. A., Kong, M., and Locasale, J. W. (2017). Metabolic interactions with cancer epigenetics. Mol. Asp. Med. 54, 50–57. doi:10.1016/j.mam.2016.09.001
Geng, J., Li, X., Zhou, Z., Wu, C. L., Dai, M., and Bai, X. (2015). EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 359 (2), 275–287. doi:10.1016/j.canlet.2015.01.031
Gomatou, G., Syrigos, N., and Kotteas, E. (2023). Osimertinib resistance: Molecular mechanisms and emerging treatment options. Cancers (Basel) 15 (3), 841. doi:10.3390/cancers15030841
Goyal, A., Myacheva, K., Groß, M., Klingenberg, M., Duran Arqué, B., and Diederichs, S. (2017). Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45 (3), e12. doi:10.1093/nar/gkw883
Han, P., Zhao, J., and Gao, L. (2021). Increased serum exosomal long non-coding RNA SNHG15 expression predicts poor prognosis in non-small cell lung cancer. J. Clin. Lab. Anal. 35 (11), e23979. doi:10.1002/jcla.23979
Hao, Y., Baker, D., and Ten Dijke, P. (2019). TGF-β-Mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20 (11), 2767. doi:10.3390/ijms20112767
He, J., Jin, S., Zhang, W., Wu, D., Li, J., Xu, J., et al. (2019). Long non-coding RNA LOC554202 promotes acquired gefitinib resistance in non-small cell lung cancer through upregulating miR-31 expression. J. Cancer 10 (24), 6003–6013. doi:10.7150/jca.35097
He, S., Shi, J., Zhou, H., Li, Q., and Wu, L. (2022). Lnc-ABCA12-8 confers acquired resistance to gefitinib in non-small cell lung cancer by regulating the alternative splicing of fibronectin 1 in the IIICS region. Cancer Gene Ther. 29 (11), 1686–1696. doi:10.1038/s41417-022-00483-0
Hu, J., Dong, S. W., Pei, Y., Wang, J., Zhang, J., and Wei, X. P. (2021a). LncRNA MITA1 promotes gefitinib resistance by inducing autophagy in lung cancer cells. Biochem. Biophys. Res. Commun. 551, 21–26. doi:10.1016/j.bbrc.2021.02.130
Hu, X., Li, J., Fu, M., Zhao, X., and Wang, W. (2021b). The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target Ther. 6 (1), 402. doi:10.1038/s41392-021-00791-1
Hu, Z., Zhao, P., and Xu, H. (2020). Hyperoside exhibits anticancer activity in non-small cell lung cancer cells with T790M mutations by upregulating FoxO1 via CCAT1. Oncol. Rep. 43 (2), 617–624. doi:10.3892/or.2019.7440
Huang, J., Pan, B., Xia, G., Zhu, J., Li, C., and Feng, J. (2020a). LncRNA SNHG15 regulates EGFR-TKI acquired resistance in lung adenocarcinoma through sponging miR-451 to upregulate MDR-1. Cell Death Dis. 11 (7), 525. doi:10.1038/s41419-020-2683-x
Huang, N., Guo, W., Ren, K., Li, W., Jiang, Y., Sun, J., et al. (2019). LncRNA AFAP1-AS1 supresses miR-139-5p and promotes cell proliferation and chemotherapy resistance of non-small cell lung cancer by competitively upregulating RRM2. Front. Oncol. 9, 1103. doi:10.3389/fonc.2019.01103
Huang, Y., Dai, Y., Wen, C., He, S., Shi, J., Zhao, D., et al. (2020b). circSETD3 contributes to acquired resistance to gefitinib in non-small-cell lung cancer by targeting the miR-520h/ABCG2 pathway. Mol. Ther. Nucleic Acids 21, 885–899. doi:10.1016/j.omtn.2020.07.027
Iacobuzio-Donahue, C. A. (2009). Epigenetic changes in cancer. Annu. Rev. Pathol. 4, 229–249. doi:10.1146/annurev.pathol.3.121806.151442
Isin, M., and Dalay, N. (2015). LncRNAs and neoplasia. Clin. Chim. Acta 444, 280–288. doi:10.1016/j.cca.2015.02.046
Jia, Y., Li, X., Jiang, T., Zhao, S., Zhao, C., Zhang, L., et al. (2019). EGFR-targeted therapy alters the tumor microenvironment in EGFR-driven lung tumors: Implications for combination therapies. Int. J. Cancer 145 (5), 1432–1444. doi:10.1002/ijc.32191
Jiang, L., Wang, R., Fang, L., Ge, X., Chen, L., Zhou, M., et al. (2019). HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 9 (9), 2460–2474. doi:10.7150/thno.31097
Jiang, X., Guan, J., Xu, Y., Ren, H., Jiang, J., Wudu, M., et al. (2021). Silencing of CASC8 inhibits non-small cell lung cancer cells function and promotes sensitivity to osimertinib via FOXM1. J. Cancer 12 (2), 387–396. doi:10.7150/jca.47863
Jin, W. (2020). Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells 9 (1), 217. doi:10.3390/cells9010217
Jin, X., Liu, X., Zhang, Z., and Guan, Y. (2020). lncRNA CCAT1 acts as a MicroRNA-218 sponge to increase gefitinib resistance in NSCLC by targeting HOXA1. Mol. Ther. Nucleic Acids 19, 1266–1275. doi:10.1016/j.omtn.2020.01.006
Junttila, M. R., and de Sauvage, F. J. (2013). Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501 (7467), 346–354. doi:10.1038/nature12626
Kara, G., Calin, G. A., and Ozpolat, B. (2022). RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 182, 114113. doi:10.1016/j.addr.2022.114113
Kim, S. M., Kim, J. S., Kim, J. H., Yun, C. O., Kim, E. M., Kim, H. K., et al. (2010). Acquired resistance to cetuximab is mediated by increased PTEN instability and leads cross-resistance to gefitinib in HCC827 NSCLC cells. Cancer Lett. 296 (2), 150–159. doi:10.1016/j.canlet.2010.04.006
Klemm, F., and Joyce, J. A. (2015). Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 25 (4), 198–213. doi:10.1016/j.tcb.2014.11.006
Kobayashi, Y., and Mitsudomi, T. (2016). Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 107 (9), 1179–1186. doi:10.1111/cas.12996
Kong, X., Hu, S., Yuan, Y., Du, Y., Zhu, Z., Song, Z., et al. (2020). Analysis of lncRNA, miRNA and mRNA-associated ceRNA networks and identification of potential drug targets for drug-resistant non-small cell lung cancer. J. Cancer 11 (11), 3357–3368. doi:10.7150/jca.40729
Lei, Y., Guo, W., Chen, B., Chen, L., Gong, J., and Li, W. (2018). Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol. Rep. 40 (6), 3438–3446. doi:10.3892/or.2018.6762
Li, C., Wan, L., Liu, Z., Xu, G., Wang, S., Su, Z., et al. (2018). Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 418, 185–195. doi:10.1016/j.canlet.2018.01.036
Li, W., Li, Y., Zhang, H., Liu, M., Gong, H., Yuan, Y., et al. (2021). HOTAIR promotes gefitinib resistance through modification of EZH2 and silencing p16 and p21 in non-small cell lung cancer. J. Cancer 12 (18), 5562–5572. doi:10.7150/jca.56093
Li, W., Sun, M., Zang, C., Ma, P., He, J., Zhang, M., et al. (2016). Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 7 (5), e2225. doi:10.1038/cddis.2016.126
Li, Y., Shen, Y., Xie, M., Wang, B., Wang, T., Zeng, J., et al. (2022). LncRNAs LCETRL3 and LCETRL4 at chromosome 4q12 diminish EGFR-TKIs efficiency in NSCLC through stabilizing TDP43 and EIF2S1. Signal Transduct. Target Ther. 7 (1), 30. doi:10.1038/s41392-021-00847-2
Li, Z., Liu, L., Feng, C., Qin, Y., Xiao, J., Zhang, Z., et al. (2023). LncBook 2.0: Integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res. 51 (D1), D186–D191. doi:10.1093/nar/gkac999
Liu, L., Zhu, H., Liao, Y., Wu, W., Liu, L., Liu, L., et al. (2020a). Inhibition of Wnt/β-catenin pathway reverses multi-drug resistance and EMT in Oct4(+)/Nanog(+) NSCLC cells. Biomed. Pharmacother. 127, 110225. doi:10.1016/j.biopha.2020.110225
Liu, Q., Yu, S., Zhao, W., Qin, S., Chu, Q., and Wu, K. (2018). EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol. Cancer 17 (1), 53. doi:10.1186/s12943-018-0793-1
Liu, X., Jiang, T., Li, X., Zhao, C., Li, J., Zhou, F., et al. (2020c). Exosomes transmit T790M mutation-induced resistance in EGFR-mutant NSCLC by activating PI3K/AKT signalling pathway. J. Cell Mol. Med. 24 (2), 1529–1540. doi:10.1111/jcmm.14838
Liu, X., Lu, X., Zhen, F., Jin, S., Yu, T., Zhu, Q., et al. (2019). LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol. Ther. Nucleic Acids 16, 155–161. doi:10.1016/j.omtn.2019.02.010
Liu, X., Meng, L., Li, X., Li, D., Liu, Q., Chen, Y., et al. (2020b). Regulation of FN1 degradation by the p62/SQSTM1-dependent autophagy-lysosome pathway in HNSCC. Int. J. Oral Sci. 12 (1), 34. doi:10.1038/s41368-020-00101-5
Lu, W., Zhang, H., Niu, Y., Wu, Y., Sun, W., Li, H., et al. (2017). Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer 16 (1), 118. doi:10.1186/s12943-017-0685-9
Lv, X., Lian, Y., Liu, Z., Xiao, J., Zhang, D., and Yin, X. (2021). Exosomal long non-coding RNA LINC00662 promotes non-small cell lung cancer progression by miR-320d/E2F1 axis. Aging (Albany NY) 13 (4), 6010–6024. doi:10.18632/aging.202522
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350 (21), 2129–2139. doi:10.1056/NEJMoa040938
Ma, G., Zhu, J., Liu, F., and Yang, Y. (2019). Long noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through epidermal growth factor receptor by sponging miR-769-5p. DNA Cell Biol. 38 (2), 176–183. doi:10.1089/dna.2018.4462
Ma, J., Qi, G., and Li, L. (2020). A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 13, 5293–5307. doi:10.2147/OTT.S243214
Ma, P., Zhang, M., Nie, F., Huang, Z., He, J., Li, W., et al. (2017). Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed. Pharmacother. 87, 20–26. doi:10.1016/j.biopha.2016.12.079
Martinelli, E., Morgillo, F., Troiani, T., and Ciardiello, F. (2017). Cancer resistance to therapies against the EGFR-RAS-RAF pathway: The role of MEK. Cancer Treat. Rev. 53, 61–69. doi:10.1016/j.ctrv.2016.12.001
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 10 (3), 155–159. doi:10.1038/nrg2521
Mok, T. S., Wu, Y. L., Ahn, M. J., Garassino, M. C., Kim, H. R., Ramalingam, S. S., et al. (2017). Osimertinib or platinum-pemetrexed in EGFR t790m-positive lung cancer. N. Engl. J. Med. 376 (7), 629–640. doi:10.1056/NEJMoa1612674
Mok, T. S., Wu, Y. L., Thongprasert, S., Yang, C. H., Chu, D. T., Saijo, N., et al. (2009). Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361 (10), 947–957. doi:10.1056/NEJMoa0810699
Morris, R., Kershaw, N. J., and Babon, J. J. (2018). The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27 (12), 1984–2009. doi:10.1002/pro.3519
Mostafazadeh, M., Samadi, N., Kahroba, H., Baradaran, B., Haiaty, S., and Nouri, M. (2021). Potential roles and prognostic significance of exosomes in cancer drug resistance. Cell Biosci. 11 (1), 1. doi:10.1186/s13578-020-00515-y
Nakano, Y., Isobe, K., Kobayashi, H., Kaburaki, K., Isshiki, T., Sakamoto, S., et al. (2020). Clinical importance of long non-coding RNA LINC00460 expression in EGFR-mutant lung adenocarcinoma. Int. J. Oncol. 56 (1), 243–257. doi:10.3892/ijo.2019.4919
Niazi, F., and Valadkhan, S. (2012). Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3' UTRs. Rna 18 (4), 825–843. doi:10.1261/rna.029520.111
Nie, W., Ge, H. J., Yang, X. Q., Sun, X., Huang, H., Tao, X., et al. (2016). LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 371 (1), 99–106. doi:10.1016/j.canlet.2015.11.024
Niu, R., Li, D., Chen, J., and Zhao, W. (2022). Circ_0014235 confers Gefitinib resistance and malignant behaviors in non-small cell lung cancer resistant to Gefitinib by governing the miR-146b-5p/YAP/PD-L1 pathway. Cell Cycle 21 (1), 86–100. doi:10.1080/15384101.2021.2009986
Owen, K. L., Brockwell, N. K., and Parker, B. S. (2019). JAK-STAT signaling: A double-edged sword of immune regulation and cancer progression. Cancers (Basel) 11 (12), 2002. doi:10.3390/cancers11122002
Oxnard, G. R., Hu, Y., Mileham, K. F., Husain, H., Costa, D. B., Tracy, P., et al. (2018). Assessment of resistance mechanisms and clinical implications in patients with EGFR t790m-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 4 (11), 1527–1534. doi:10.1001/jamaoncol.2018.2969
Paez, J. G., Jänne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., et al. (2004). EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304 (5676), 1497–1500. doi:10.1126/science.1099314
Pal, A. S., Agredo, A., Lanman, N. A., Son, J., Sohal, I. S., Bains, M., et al. (2022). Loss of KMT5C promotes EGFR inhibitor resistance in NSCLC via linc01510-mediated upregulation of MET. Cancer Res. 82 (8), 1534–1547. doi:10.1158/0008-5472.CAN-20-0821
Pan, J., Xing, J., Yu, H., Wang, Z., Wang, W., and Pan, Y. (2023). CircRBM33 promotes migration, invasion and mediates osimertinib resistance in non-small cell lung cancer cell line. Ann. Transl. Med. 11 (6), 252. doi:10.21037/atm-22-6346
Pan, R., and Zhou, H. (2020). Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 Axis. Cancer Manag. Res. 12, 4283–4297. doi:10.2147/CMAR.S241095
Pathania, A. S., and Challagundla, K. B. (2021). Exosomal long non-coding RNAs: Emerging players in the tumor microenvironment. Mol. Ther. Nucleic Acids 23, 1371–1383. doi:10.1016/j.omtn.2020.09.039
Prasanth, K. V., and Spector, D. L. (2007). Eukaryotic regulatory RNAs: An answer to the 'genome complexity' conundrum. Genes. Dev. 21 (1), 11–42. doi:10.1101/gad.1484207
Qi, P., and Du, X. (2013). The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 26 (2), 155–165. doi:10.1038/modpathol.2012.160
Qian, B., Wang, D. M., Gu, X. S., Zhou, K., Wu, J., Zhang, C. Y., et al. (2018). LncRNA H19 serves as a ceRNA and participates in non-small cell lung cancer development by regulating microRNA-107. Eur. Rev. Med. Pharmacol. Sci. 22 (18), 5946–5953. doi:10.26355/eurrev_201809_15925
Qin, K., Hong, L., Zhang, J., and Le, X. (2023). MET amplification as a resistance driver to TKI therapies in lung cancer: Clinical challenges and opportunities. Cancers (Basel) 15 (3), 612. doi:10.3390/cancers15030612
Rashid, F., Shah, A., and Shan, G. (2016). Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinforma. 14 (2), 73–80. doi:10.1016/j.gpb.2016.03.005
Rincón-Riveros, A., Morales, D., Rodríguez, J. A., Villegas, V. E., and López-Kleine, L. (2021). Bioinformatic tools for the analysis and prediction of ncRNA interactions. Int. J. Mol. Sci. 22 (21), 11397. doi:10.3390/ijms222111397
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Rubin, M. A., Bristow, R. G., Thienger, P. D., Dive, C., and Imielinski, M. (2020). Impact of lineage plasticity to and from a neuroendocrine phenotype on progression and response in prostate and lung cancers. Mol. Cell 80 (4), 562–577. doi:10.1016/j.molcel.2020.10.033
Shaurova, T., Zhang, L., Goodrich, D. W., and Hershberger, P. A. (2020). Understanding lineage plasticity as a path to targeted therapy failure in EGFR-mutant non-small cell lung cancer. Front. Genet. 11, 281. doi:10.3389/fgene.2020.00281
Sheng, W., Guo, W., Lu, F., Liu, H., Xia, R., and Dong, F. (2021). Upregulation of Linc00284 promotes lung cancer progression by regulating the miR-205-3p/c-Met Axis. Front. Genet. 12, 694571. doi:10.3389/fgene.2021.694571
Shi, J., Huang, Y., Wen, C., He, S., Wu, L., and Zhou, H. (2020). Genome-wide identification and characterization of long non-coding RNAs involved in acquired resistance to gefitinib in non-small-cell lung cancer. Comput. Biol. Chem. 87, 107288. doi:10.1016/j.compbiolchem.2020.107288
Shi, T., Sun, W., Shi, Y. L., Wang, Q., Yan, Z. X., and Zhang, M. (2021). LncRNA OSER1-AS1 interacts with miR-612/FOXM1 axis to modulate gefitinib resistance of lung adenocarcinoma. Am. J. Transl. Res. 13 (3), 1365–1376.
Shu, D., Xu, Y., and Chen, W. (2020). Knockdown of lncRNA BLACAT1 reverses the resistance of afatinib to non-small cell lung cancer via modulating STAT3 signalling. J. Drug Target 28 (3), 300–306. doi:10.1080/1061186X.2019.1650368
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73 (1), 17–48. doi:10.3322/caac.21763
Singh, A., and Settleman, J. (2010). EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 29 (34), 4741–4751. doi:10.1038/onc.2010.215
Sinha, D., Saha, P., Samanta, A., and Bishayee, A. (2020). Emerging concepts of hybrid epithelial-to-mesenchymal transition in cancer progression. Biomolecules 10 (11), 1561. doi:10.3390/biom10111561
Song, L., Qian, G., Huang, J., Chen, T., and Yang, Y. (2021). AZD9291-resistant non-small cell lung cancer cell-derived exosomal lnc-MZT2A-5:1 induces the activation of fibroblasts. Ann. Transl. Med. 9 (20), 1593. doi:10.21037/atm-21-5186
St Laurent, G., Wahlestedt, C., and Kapranov, P. (2015). The Landscape of long noncoding RNA classification. Trends Genet. 31 (5), 239–251. doi:10.1016/j.tig.2015.03.007
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Su, W., Wang, L., Zhao, H., Hu, S., Zhou, Y., Guo, C., et al. (2020). LINC00857 interacting with YBX1 to regulate apoptosis and autophagy via MET and phosphor-AMPKa signaling. Mol. Ther. Nucleic Acids 22, 1164–1175. doi:10.1016/j.omtn.2020.10.025
Sun, P., Sun, L., Cui, J., Liu, L., and He, Q. (2020). Long noncoding RNA HAS2-AS1 accelerates non-small cell lung cancer chemotherapy resistance by targeting LSD1/EphB3 pathway. Am. J. Transl. Res. 12 (3), 950–958.
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Takahashi, S., Noro, R., Seike, M., Zeng, C., Matsumoto, M., Yoshikawa, A., et al. (2021). Long non-coding RNA CRNDE is involved in resistance to EGFR tyrosine kinase inhibitor in EGFR-mutant lung cancer via eIF4A3/MUC1/EGFR signaling. Int. J. Mol. Sci. 22 (8), 4005. doi:10.3390/ijms22084005
Tao, Y., Tang, Y., Yang, Z., Wu, F., Wang, L., Yang, L., et al. (2020). Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int. J. Biol. Sci. 16 (3), 471–482. doi:10.7150/ijbs.39123
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 (7483), 344–352. doi:10.1038/nature12986
Toden, S., Zumwalt, T. J., and Goel, A. (2021). Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta Rev. Cancer 1875 (1), 188491. doi:10.1016/j.bbcan.2020.188491
Verdera, H. C., Kuranda, K., and Mingozzi, F. (2020). AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 28 (3), 723–746. doi:10.1016/j.ymthe.2019.12.010
Wan, X., Xie, B., Sun, H., Gu, W., Wang, C., Deng, Q., et al. (2022). Exosomes derived from M2 type tumor-associated macrophages promote osimertinib resistance in non-small cell lung cancer through MSTRG.292666.16-miR-6836-5p-MAPK8IP3 axis. Cancer Cell Int. 22 (1), 83. doi:10.1186/s12935-022-02509-x
Wan, Y., Yao, D., Fang, F., Wang, Y., Wu, G., and Qian, Y. (2021). LncRNA WT1-AS downregulates lncRNA UCA1 to suppress non-small cell lung cancer and predicts poor survival. BMC Cancer 21 (1), 104. doi:10.1186/s12885-020-07767-4
Wang, B., Jiang, H., Wang, L., Chen, X., Wu, K., Zhang, S., et al. (2017b). Increased MIR31HG lncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3K/AKT signaling pathway. Oncol. Lett. 13 (5), 3494–3500. doi:10.3892/ol.2017.5878
Wang, C., Jiang, J., Ji, J., Cai, Q., Chen, X., Yu, Y., et al. (2017a). PKM2 promotes cell migration and inhibits autophagy by mediating PI3K/AKT activation and contributes to the malignant development of gastric cancer. Sci. Rep. 7 (1), 2886. doi:10.1038/s41598-017-03031-1
Wang, H., Lu, B., Ren, S., Wu, F., Wang, X., Yan, C., et al. (2020b). Long noncoding RNA LINC01116 contributes to gefitinib resistance in non-small cell lung cancer through regulating IFI44. Mol. Ther. Nucleic Acids 19, 218–227. doi:10.1016/j.omtn.2019.10.039
Wang, L., Li, M., and Lian, R. (2022). Depleting hsa_circ_0000567 suppresses acquired gefitinib resistance and proliferation of lung adenocarcinoma cells through regulating the miR-377-3p/ZFX axis: An in vitro and in vivo study. Histol. Histopathol. 37 (7), 637–654. doi:10.14670/HH-18-431
Wang, N., Zhao, Q., Huang, Y., Wen, C., Li, Y., Bao, M., et al. (2023). Lnc-TMEM132D-AS1 as a potential therapeutic target for acquired resistance to osimertinib in non-small-cell lung cancer. Mol. Omics 19, 238–251. doi:10.1039/d2mo00261b
Wang, Q., Li, X., Ren, S., Su, C., Li, C., Li, W., et al. (2020a). HOTAIR induces EGFR-TKIs resistance in non-small cell lung cancer through epithelial-mesenchymal transition. Lung Cancer 147, 99–105. doi:10.1016/j.lungcan.2020.06.037
Wang, S., Liu, C., Lei, Q., Wu, Z., Miao, X., Zhu, D., et al. (2021a). Relationship between long non-coding RNA PCAT-1 expression and gefitinib resistance in non-small-cell lung cancer cells. Respir. Res. 22 (1), 146. doi:10.1186/s12931-021-01719-7
Wang, T., Yang, C., Li, B., Xing, Y., Huang, J., Zhang, Y., et al. (2021b). Identification of lncRNA-miRNA-mRNA networks linked to non-small lung cancer resistance to inhibitors of epidermal growth factor receptor. Front. Genet. 12, 758591. doi:10.3389/fgene.2021.758591
Wei, P., Zhang, N., Wang, Y., Li, D., Wang, L., Sun, X., et al. (2015). FOXM1 promotes lung adenocarcinoma invasion and metastasis by upregulating SNAIL. Int. J. Biol. Sci. 11 (2), 186–198. doi:10.7150/ijbs.10634
Wen, C., Li, Y., Huang, Y., Wang, N., He, S., Bao, M., et al. (2023). CircSETD3 mediates acquired resistance to gefitinib in non-small lung cancer cells by FXR1/ECT2 pathway. Int. J. Biochem. Cell Biol. 154, 106344. doi:10.1016/j.biocel.2022.106344
Witta, S. E., Gemmill, R. M., Hirsch, F. R., Coldren, C. D., Hedman, K., Ravdel, L., et al. (2006). Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 66 (2), 944–950. doi:10.1158/0008-5472.CAN-05-1988
Wu, C., Yang, J., Li, R., Lin, X., Wu, J., and Wu, J. (2021a). LncRNA WT1-AS/miR-494-3p regulates cell proliferation, apoptosis, migration and invasion via PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer. Onco Targets Ther. 14, 891–904. doi:10.2147/OTT.S278233
Wu, J., Weng, Y., He, F., Liang, D., and Cai, L. (2018). LncRNA MALAT-1 competitively regulates miR-124 to promote EMT and development of non-small-cell lung cancer. Anticancer Drugs 29 (7), 628–636. doi:10.1097/CAD.0000000000000626
Wu, K., Li, J., Qi, Y., Zhang, C., Zhu, D., Liu, D., et al. (2019). SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed. Pharmacother. 116, 108995. doi:10.1016/j.biopha.2019.108995
Wu, S., Luo, M., To, K. K. W., Zhang, J., Su, C., Zhang, H., et al. (2021b). Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Mol. Cancer 20 (1), 17. doi:10.1186/s12943-021-01307-9
Xiao, F., Liu, N., Ma, X., Qin, J., Liu, Y., and Wang, X. (2020). M2 macrophages reduce the effect of gefitinib by activating AKT/mTOR in gefitinib-resistant cell lines HCC827/GR. Thorac. Cancer 11 (11), 3289–3298. doi:10.1111/1759-7714.13670
Xie, M., He, C. S., Wei, S. H., and Zhang, L. (2013). Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur. J. Cancer 49 (16), 3559–3572. doi:10.1016/j.ejca.2013.07.007
Xu, S. H., Bo, Y. H., Ma, H. C., Zhang, H. N., and Shao, M. J. (2021). lncRNA LINC00473 promotes proliferation, migration, invasion and inhibition of apoptosis of non-small cell lung cancer cells by acting as a sponge of miR-497-5p. Oncol. Lett. 21 (6), 429. doi:10.3892/ol.2021.12690
Xu, T., Yan, S., Wang, M., Jiang, L., Ma, P., Lu, B., et al. (2020). LncRNA UCA1 induces acquired resistance to gefitinib by epigenetically silencing CDKN1A expression in non-small-cell lung cancer. Front. Oncol. 10, 656. doi:10.3389/fonc.2020.00656
Xu, W., Yang, Z., and Lu, N. (2015). A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 9 (4), 317–324. doi:10.1080/19336918.2015.1016686
Yan, X., Hu, Z., Feng, Y., Hu, X., Yuan, J., Zhao, S. D., et al. (2015). Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28 (4), 529–540. doi:10.1016/j.ccell.2015.09.006
Yang, B., Teng, F., Chang, L., Wang, J., Liu, D. L., Cui, Y. S., et al. (2021). Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging (Albany NY) 13 (9), 13264–13286. doi:10.18632/aging.203011
Yang, Q., Chen, W., Xu, Y., Lv, X., Zhang, M., and Jiang, H. (2018). Polyphyllin I modulates MALAT1/STAT3 signaling to induce apoptosis in gefitinib-resistant non-small cell lung cancer. Toxicol. Appl. Pharmacol. 356, 1–7. doi:10.1016/j.taap.2018.07.031
Yatabe, Y., Kerr, K. M., Utomo, A., Rajadurai, P., Tran, V. K., Du, X., et al. (2015). EGFR mutation testing practices within the asia pacific region: Results of a multicenter diagnostic survey. J. Thorac. Oncol. 10 (3), 438–445. doi:10.1097/JTO.0000000000000422
Yu, T., Li, J., Yan, M., Liu, L., Lin, H., Zhao, F., et al. (2015). MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 34 (4), 413–423. doi:10.1038/onc.2013.574
Yuan, S., Chen, W., Yang, J., Zheng, Y., Ye, W., Xie, H., et al. (2022). Tumor-associated macrophage-derived exosomes promote EGFR-TKI resistance in non-small cell lung cancer by regulating the AKT, ERK1/2 and STAT3 signaling pathways. Oncol. Lett. 24 (4), 356. doi:10.3892/ol.2022.13476
Yuan, S., Norgard, R. J., and Stanger, B. Z. (2019). Cellular plasticity in cancer. Cancer Discov. 9 (7), 837–851. doi:10.1158/2159-8290.CD-19-0015
Zhang, B., Wang, H., Wang, Q., Xu, J., Jiang, P., and Li, W. (2019). Knockout of lncRNA UCA1 inhibits drug resistance to gefitinib via targeting STAT3 signaling in NSCLC. Minerva Med. 110 (3), 273–275. doi:10.23736/S0026-4806.19.05979-2
Zhang, H., Wang, S. Q., Wang, L., Lin, H., Zhu, J. B., Chen, R., et al. (2022). m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death Dis. 13 (7), 657. doi:10.1038/s41419-022-05050-x
Zhang, P., Sun, Y., and Ma, L. (2015). ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 14 (4), 481–487. doi:10.1080/15384101.2015.1006048
Zhang, R., Xia, Y., Wang, Z., Zheng, J., Chen, Y., Li, X., et al. (2017). Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 490 (2), 406–414. doi:10.1016/j.bbrc.2017.06.055
Zhao, H., Wang, Y., Wu, X., Zeng, X., Lin, B., Hu, S., et al. (2022). FAM83A antisense RNA 1 (FAM83A-AS1) silencing impairs cell proliferation and induces autophagy via MET-AMPKɑ signaling in lung adenocarcinoma. Bioengineered 13 (5), 13312–13327. doi:10.1080/21655979.2022.2081457
Zheng, F., Li, J., Ma, C., Tang, X., Tang, Q., Wu, J., et al. (2020b). Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J. Cell Mol. Med. 24 (10), 5578–5592. doi:10.1111/jcmm.15214
Zheng, G., Chen, W., Li, W., Ding, Y., Tu, P., and Chen, W. (2020a). E2F1-induced ferritin heavy chain 1 pseudogene 3 (FTH1P3) accelerates non-small cell lung cancer gefitinib resistance. Biochem. Biophys. Res. Commun. 530 (4), 624–631. doi:10.1016/j.bbrc.2020.07.044
Zheng, Y., Guo, Z., and Li, Y. (2022). Long non-coding RNA prostate cancer-associated transcript 6 inhibited gefitinib sensitivity of non-small cell lung cancer by serving as a competing endogenous RNA of miR-326 to up-regulate interferon-alpha receptor 2. Bioengineered 13 (2), 3785–3796. doi:10.1080/21655979.2022.2031416
Zhou, C., Wu, Y. L., Chen, G., Feng, J., Liu, X. Q., Wang, C., et al. (2011). Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12 (8), 735–742. doi:10.1016/S1470-2045(11)70184-X
Zhou, D., Xia, Z., Xie, M., Gao, Y., Yu, Q., and He, B. (2021). Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975. Hum. Cell 34 (5), 1478–1489. doi:10.1007/s13577-021-00572-6
Zhou, S. Z., Li, H., Wang, Z. W., Wang, M. H., Li, N., and Wang, Y. F. (2020). LncRNA TSLNC8 synergizes with EGFR inhibitor osimertinib to inhibit lung cancer tumorigenesis by blocking the EGFR-STAT3 pathway. Cell Cycle 19 (21), 2776–2792. doi:10.1080/15384101.2020.1820697
Zhou, Y., and Zhang, Y. (2020). Inhibition of LncRNAH19 has the effect of anti-tumour and enhancing sensitivity to Gefitinib and Chemotherapy in Non-small-cell lung cancer in vivo. J. Cell Mol. Med. 24 (10), 5811–5816. doi:10.1111/jcmm.15245
Zhou, Y., Zheng, X., Xu, B., Chen, L., Wang, Q., Deng, H., et al. (2019). Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 508 (2), 527–535. doi:10.1016/j.bbrc.2018.11.157
Keywords: long non-coding RNAs (lncRNA), epidermal growth factor receptor (EGFR), tyrosine kinase inhibitors (TKIs), drug resistance, non-small cell lung cancer (NSCLC)
Citation: Liu D, Lu X, Huang W and Zhuang W (2023) Long non-coding RNAs in non-small cell lung cancer: implications for EGFR-TKI resistance. Front. Genet. 14:1222059. doi: 10.3389/fgene.2023.1222059
Received: 13 May 2023; Accepted: 07 June 2023;
Published: 30 June 2023.
Edited by:
Mario Acunzo, Virginia Commonwealth University, United StatesReviewed by:
Daniel Del Valle Morales, Virginia Commonwealth University, United StatesHoward Li, United States Department of Veterans Affairs, United States
Copyright © 2023 Liu, Lu, Huang and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhuang, zhuangwei@csu.edu.cn
 Detian Liu1
Detian Liu1 Xiaolin Lu
Xiaolin Lu Wentao Huang
Wentao Huang Wei Zhuang
Wei Zhuang