Identification of extremely GC-rich micro RNAs for RT-qPCR data normalization in human plasma
- 1Genomics Core Facility, VetCore, University of Veterinary Medicine, Vienna, Austria
- 2Garvan Institute of Medical Research, Sydney, NSW, Australia
- 3Lowy Cancer Research Centre, School of Biomedical Sciences, University of New South Wales, Sydney, NSW, Australia
- 4Institute of Psychology, Karl-Franzens-University Graz, Graz, Austria
- 5Department of Behavioral and Cognitive Biology, University of Vienna, Vienna, Austria
- 6Department of Microbiology, Immunobiology and Genetics, University of Vienna, Vienna, Austria
A Corrigendum on
Identification of extremely GC-rich micro RNAs for RT-qPCR data normalization in human plasma
by Baumann V, Athanasiou A-T, Faridani OR, Schwerdtfeger AR, Wallner B and Steinborn R (2023). Front. Genet. 13:1058668. doi: 10.3389/fgene.2022.1058668
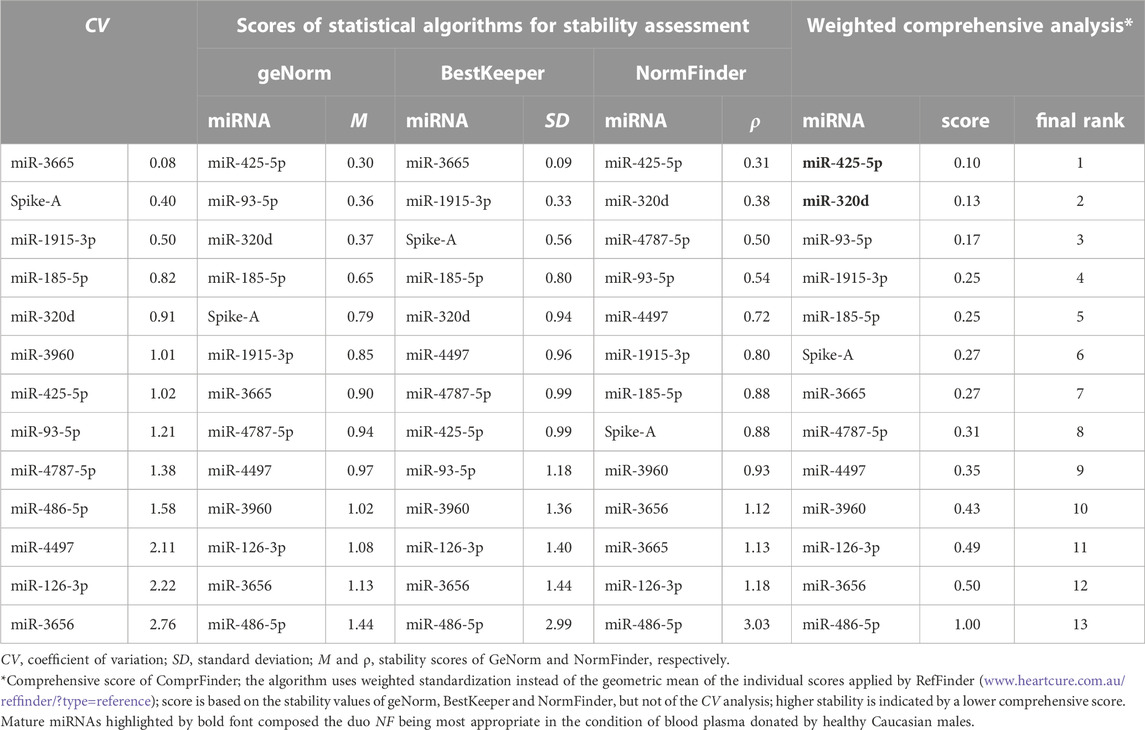
In the original article, the coefficient of variation (CV, ratio between standard deviation and mean) was calculated from Cq values that results in a profound difference compared to the use of real numbers (Kralik and Ricchi, 2017). We now corrected the calculation by converting the Cq values into relative quantities (Hellemans et al., 2007), also termed linear-scale transformation or just linearisation of Cq values (2−Cq) (Marabita et al., 2016; Sundaram et al., 2019). This was achieved by converting the efficiency-adjusted Cq values into n-fold quantities relatively to the lowest expressing sample according to the term
Therefore, we had to correct the CV values contained in Figure 3, Table 3 and the Supplementary Tables S12, S17 as well as the Extended Methods contained in Supplementary Material S1.
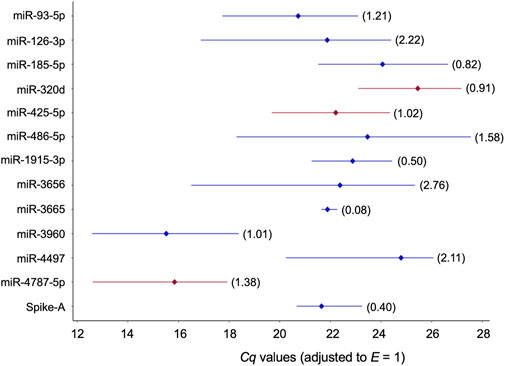
FIGURE 3. Profiles of Cq values of 13 candidate reference miRNAs across 32 blood plasmas derived from probands with different modes of cognitive stress coping. Cq values were corrected with the amplification efficiency value determined for the respective qPCR assay (Table 2). The median value is depicted by the diamond. The percent coefficient of variation (CV) is given in parentheses. miRNAs composing the two-gene NFs either recommended for plasma or the two stress-coping dimensions are highlighted in red.
Based on the adjusted CV values, a correction has been made in the Abstract (single paragraph). The incorrect statement was “In general, inter-individual variance of miRNA abundance was low or very low as indicated by coefficient of variation (CV) values of 0.6%–8.2%. miR-3665 and miR-1915-3p outperformed in this analysis (CVs: 0.6% and 2.4%, respectively).” It was corrected to:
“The lowest inter-individual variance of miRNA abundance was determined for miR-3665 and miR-1915-3p [coefficient of variation (CV) values: 0.08 and 0.50, respectively].”
A correction has been made to the Results section, sub-section Stability assessment of the selected miRNA reference candidates (paragraph 1). The sentence previously stated “They showed high to extraordinary uniformity of abundance as indicated by CV values of 13.9%–0.6% (Table 3; Supplementary Table S12) and mostly low inter- and intragroup variability assessed with the NormFinder algorithm (Figure 4).” The corrected text on the CV range appears below:
“They differed considerably in abundance stability ranging from poor to high or extraordinarily high (CV range: 2.76 down to 0.50 and 0.08; Figure 3, Table 3, and Supplementary Table S12) and showed mostly low inter- and intragroup variability assessed with the NormFinder algorithm (Figure 4).”
Another correction has been made in this sub-section (paragraph 3). The previous sentence “However, MFE of folding was not related to the CV of miRNA’s plasma abundance (Pearson’s correlation coefficient r = 0.17, p = 0.60, n = 10).” was adjusted as follows:
“However, MFE of folding was not related to the CV of miRNA’s plasma abundance (Pearson’s correlation coefficient r = 0.03, p = 0.93, n = 12).”
A correction was made in the Results section, sub-section Folding of spike-in miRNA controls (paragraph 1). Previously we stated: “Its uniformity of recovery was concluded to be good based on a low CV value of 3.1% and was only outperformed by miR-3665 and miR-1915-3p (Table 3).” The corrected sentence appears below:
“Its uniformity of recovery was good based on low CV (0.40) and was only outperformed by miR-3665 (Table 3).”
Based on the corrected CV analysis, an update on the recommended set of miRNA reference genes was made in the Discussion section (paragraph 2). The previous text was: “Our strategy resulted in an extended miRNA repertoire for context-optimised RT-qPCR normalization (n = 6). The normalizers include tumour suppressors (miR-3665, miR-4497 and miR-4787-5p), oncogenic (miR-425-5p), and Janus-faced tumour molecules (miR-320d and miR-1915-3p) that fulfil either tumour-suppressive or oncogenic functions depending on the cellular context and the downstream targets they affect (Han et al., 2020).” Here is the adjusted sentence:
“Our strategy resulted in an extended miRNA repertoire for context-optimised RT-qPCR normalization (n = 8). The set of normalizers included miR-3960-5p, the tumour suppressors miR-185-5p, miR-3665 and miR-4787-5p, an oncogenic miRNA, miR-425-5p, and the Janus-faced tumour molecules miR-93-5p, miR-320d and miR-1915-3p that fulfil either tumour-suppressive or oncogenic functions depending on the cellular context and the downstream targets they affect (Han et al., 2020).”
A correction has been made to the Discussion section (paragraph 3). The previous sentence was: “Second, stability evaluation using CV analysis identified two miRNAs, that showed extraordinary even abundances across the plasma samples of the 32 human males, namely, miR-3665 and miR-1915-3p (CVs: 0.6% and 2.4%, respectively).” The corrected statement appears below:
“Second, stability evaluation using CV analysis identified two miRNAs that showed extraordinary or high uniformity of abundance across the plasma samples of the 32 human males, namely, miR-3665 and miR-1915-3p (CVs: 0.08 and 0.50, respectively).”
Finally, a correction has been made to the Conclusion section (paragraph 1). The previous information on the set of recommended miRNA normalizers was; “Here, we expanded the panel of putative miRNA normalizers for the context of human (and possibly also animal) plasma by adding miR-3665, miR-1915-3p, miR-320d, miR-4497, miR-425-5p, and miR-4787-5p.” The corrected statement appears below:
“Here, we expanded the panel of putative miRNA normalizers for the context of human (and possibly also animal) plasma by adding miR-3665, miR-1915-3p, miR-185-5p, miR-320d, miR-3960-5p, miR-425-5p, miR-93-5p, and miR-4787-5p.”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., and Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19. doi:10.1186/gb-2007-8-2-r19
Kralik, P., and Ricchi, M. (2017). A basic Guide to real time PCR in microbial diagnostics: Definitions, Parameters, and everything. Front. Microbiol. 8, 108. doi:10.3389/fmicb.2017.00108
Marabita, F., de Candia, P., Torri, A., Tegner, J., Abrignani, S., and Rossi, R. L. (2016). Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 17, 204–212. doi:10.1093/bib/bbv056
Keywords: miRNA expression microarray, small-RNA sequencing, stem-loop reverse-transcription quantitative PCR, human plasma miRNAs, miRNA reference genes, cognitive stress-coping, qPCR normalization
Citation: Baumann V, Athanasiou A-T, Faridani OR, Schwerdtfeger AR, Wallner B and Steinborn R (2023) Corrigendum: Identification of extremely GC-rich micro RNAs for RT-qPCR data normalization in human plasma. Front. Genet. 14:1216150. doi: 10.3389/fgene.2023.1216150
Received: 03 May 2023; Accepted: 17 May 2023;
Published: 31 May 2023.
Edited by:
Amaresh Chandra Panda, Institute of Life Sciences (ILS), IndiaReviewed by:
Gopal Pandi, Madurai Kamaraj University, IndiaKotb Abdelmohsen, National Institute on Aging (NIH), United States
Copyright © 2023 Baumann, Athanasiou, Faridani, Schwerdtfeger, Wallner and Steinborn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Steinborn, ralf.steinborn@gmx.at
†ORCID: Ralf Steinborn, orcid.org/0000-0001-9287-5039
 Volker Baumann
Volker Baumann Angelos-Theodoros Athanasiou
Angelos-Theodoros Athanasiou Omid R. Faridani
Omid R. Faridani Andreas R. Schwerdtfeger
Andreas R. Schwerdtfeger Bernard Wallner
Bernard Wallner Ralf Steinborn
Ralf Steinborn