- 1Department of Neurology, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan
- 2Genomics and Proteomics Core Laboratory, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan
- 3Medical School, College of Medicine, Chang Gung University, Taoyuan, Taiwan
Introduction: The selection of antiseizure medication usually requires a trial-and-error process. Our goal is to investigate whether genetic markers can predict the outcome of perampanel (PER) use in patients with epilepsy.
Method: The studied participants were selected from our previous epilepsy genetics studies where whole exome sequencing was available. We reviewed the medical records of epilepsy patients older than 20 years old treated with PER. The outcome of PER treatment included the response to PER, the occurrence of any adverse drug reaction (ADR), the presence of behavior ADR, and the ability to adhere to PER for more than 1 year. We investigated the association between the rare variants of the glutamate receptor genes and the outcomes of PER use.
Result: A total of 83 patients were collected. The gene group burden analysis showed that enriched genetic variants of the glutamate receptor gene group were statistically significantly associated with the occurrence of ADR, while the glutamate ionotropic receptor delta type subunit had a nominal association with the occurrence of ADR. The gene collapse analysis found that GRID1 had a nominal association with the occurrence of ADR and GRIN3A had a nominal association with the occurrence of behavior ADR. However, these nominal associations did not remain statistically significant once adjusted for multiple testing.
Discussion: We found that enriched rare genetic variants of the glutamate receptor genes were associated with the occurrence of ADR in patients taking PER. In the future, combining the results of various pharmacogenetic studies may lead to the development of prediction tools for the outcome of antiseizure medications.
Introduction
The treatment of epilepsy requires the use of antiseizure medications (ASMs) to cease or control seizures. To choose the optimal ASMs, the treating physicians often consider the patient’s seizure types, age, gender, co-medications, and comorbidities (Perucca and Tomson, 2011) before offering the ideal drug. However, the response to ASMs is unpredictable and the treating physician can only balance the effect and adverse drug reactions (ADRs) of ASMs in a trial-and-error process, which relies heavily on the physician’s judgment and experience. During this process, the patient would suffer from inadequate seizure control from low ASM dose to prevent ADR or endure unpleasant ADR due to the rapid titrating of ASM to control the seizures. There is an unmet need for biomarkers that can guide or suggest the optimal ASM for individual patients to achieve the goal of “no seizures, no side effects” (Glauser, 2011).
Perampanel (PER) is a non-competitive α-amino-3-hydroxy-5-methyl- 4-isoxazole propionic acid (AMPA) receptor antagonist (Plosker, 2012) that inhibits the excitatory neurotransmission to achieve its antiseizure activity (Rogawski and Hanada, 2013). PER is a new-generation ASM that effectively controls both focal and generalized seizures (Tsai et al., 2018). It had the advantage of few drug-drug interactions (Kwan et al., 2015) and long potency (Villanueva et al., 2016a), making it easier to use for physicians and increasing adherence for patients. One of its drawbacks is behavior disturbance including irritation, aggression, or other psychotic symptoms (Fong et al., 2022). Hence, an objective indicator predicting the outcome of PER use may greatly enhance the ability of treating physicians to achieve seizure control and minimize ADRs.
The advance of massively parallel genetic sequencing has expanded our understanding of the relationship between genetic factors and phenotype. This technology allows the incorporation of pharmacogenomic data into the clinical decision-making process (Balestrini and Sisodiya, 2018). One example was the association of HLA-B*1502 and the development of Stevens-Johnson syndrome/toxic epidermal necrolysis in Taiwanese and Southeast Asian populations (Chung et al., 2004) that led physicians to perform a genetic test before prescribing aromatic ASMs to these ethnic groups to prevent severe skin ADR. The association between ASM responsiveness and genetic variants was not as robust as that for dermatological side effects. The association between the responsiveness of sodium channel-blocking ASMs and sodium channel genes is by far the most extensively studied but the results are inconsistent among different ethnic groups (Abe et al., 2008; Sanchez et al., 2010; Manna et al., 2011; Haerian et al., 2013; Kumari et al., 2013; Ma et al., 2014; Angelopoulou et al., 2017; Lin et al., 2021). A large study conducted by EpiPGX Consortium suggested the resistance of ASM may be related to damaging genetic variants (Wolking et al., 2020a). The accumulation of data from these pharmacogenetic studies may 1 day provide robust genetic data to assist the decision-making of physicians in prescribing ASM.
We hypothesize that genetic variants in the glutamate receptor genes may affect the binding between PER and the glutamate receptors or interfere with the interaction of various subtypes of glutamate receptors. These alternations change the downstream neurotransmission and result in differences in the outcome of PER treatment. Currently, no research has focused on the association between the genetic variants in glutamate receptors and the effect of PER. In this study, we searched the pharmacogenomic relationship between genetic variations of the glutamate receptors and the outcome of PER treatment.
Material and methods
Ascertainment of subjects
The studied participants were selected from our previous epilepsy genetics studies where whole exome sequencing (WES) was available. All participants are Taiwanese aged more than 20 years old and provided written, informed consent before sequencing. The studies were approved by the Chang Gung Medical Foundation Institutional Review Board.
Patients with a history of psychogenic nonepileptic seizures were excluded due to the difficulty in determining reliable seizure frequency. Clinical data including gender, onset age of seizure, types of seizure, etiology of seizure, seizure frequency, other medical diseases, the types and maintenance dose of ASMs, ADRs, electroencephalograms, and brain images were collected.
Outcomes related to perampanel treatment
We defined four separate outcomes regarding the result of using PER, which were the response to PER, the occurrence of ADR, the presence of behavior ADRs, and the 1-year retention of PER. These outcomes represent different aspects of treating with PER. The response to PER was to evaluate its effectiveness in controlling seizures. The ADRs and behavior ADRs are annoying side effects to the patient, caregiver, and healthcare provider, which may result in a decreased quality of life or withdrawal of the medication (Hansen et al., 2018). The 1-year retention denotes a balance between the effects and side effects of PER, patients with acceptable seizure control and tolerable side effects may have better adherence to the medication being used (Im et al., 2021).
The response to PER was defined according to the seizure frequency after its prescription and categorized according to the International League Against Epilepsy (ILAE) consensus (Kwan et al., 2010) and EpiPGX Consortium (Androsova et al., 2017; Wolking et al., 2020b). Responding to PER was defined as seizure-free under ongoing PER treatment for at least 1 year and before initiation of any other treatment. Failure to respond to PER was defined as still having seizures during the use of PER or the need for initiation of any other treatment. The occurrence of ADR was defined as PER-related adverse events (Tsai et al., 2018) both occurring after PER use and deemed to be caused by PER by the treating physician. The PER-related ADRs included dizziness, somnolence, headache, fatigue, behavior problems, and falls (Steinhoff et al., 2013; Leppik et al., 2017). The presence of behavior ADRs was defined as irritation, violent behavior, and/or psychotic symptoms that occurred after PER use and were considered caused by PER by the treating physician. One-year retention was defined as patients who kept PER use for more than 1 year.
Gene group and genotype
We specifically focus on the glutamate receptor genes obtained from the HUGO Gene Nomenclature Committee at the European Bioinformatics Institute (https://www.genenames.org/) (Seal et al., 2023). The glutamate receptor gene list includes 26 genes, GRIA1-4, GRID1-2, GRIK1-5, GRIN1, GRIN2A-D, GRIN3A-B, and GRM1-8, which is further separated into five functional subunit gene groups.
We followed the grouping method of HUGO, including 1. The glutamate ionotropic receptor AMPA type subunit (GRIA1-4), 2. The glutamate ionotropic receptor delta type subunit (GRID1-2), 3. The glutamate ionotropic receptor kainate type subunit (GRIK1-5), 4. The glutamate ionotropic receptor N-methyl-D-aspartate (NMDA) type subunit (GRIN1, GRIN2A-D, and GRIN3A-B), and 5. The glutamate metabotropic receptor (GRM1-8).
Statistical analysis
In our study, we focused on the rare variants of the glutamate receptor genes and defined them as minor allele frequency (MAF) ≦ 0.05 in gnomAD (https://gnomad.broadinstitute.org/) (Karczewski et al., 2020). We excluded all synonymous single nucleotide variants.
First, we conduct a gene group burden analysis to search for the different enrichment of rare variants in the gene group that might be associated with the outcome of PER use. This was done by testing the association between the presence/absence of phenotypes and the number of rare variants in each gene group using the Mann–Whitney U test. Second, a gene collapse analysis was performed to find the association between the presence/absence of phenotypes and the presence/absence of at least one rare variant in each glutamate receptor gene using the Fisher exact test. Significance was defined as p-values of less than 0.05 but to cope with the problem of multiple testing, we adjusted p-values by the Bonferroni correction. The clinical statistics were presented as percentages for categorical data and median plus interquartile range for skewed continuous variables.
Results
Demographic characteristics
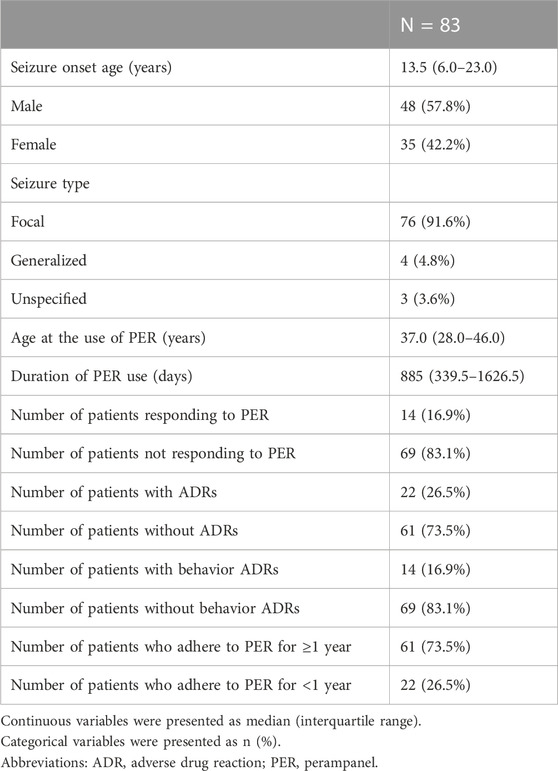
Overall, 83 Taiwanese patients who took PER for seizure control were collected for this study. The demographic data is presented in Table 1. In the study cohort, we included 35 females and 48 males. There were 76 patients (91.6%) with focal seizures, four (4.8%) generalized seizures, and three (3.6%) unclassified. The etiology of seizure included genetic in seven patients (8.4%), immunologic in four (4.8%), infectious in four (4.8%), metabolic in one (1.2%), structural in 33 (39.8%), and unknown in 34 (41.0%). The age of seizure onset was 13.5 (interquartile range (IQR) = 6.0–23.0) years old and the age at PER use was 37.0 (IQR = 28.0–46.0) years old. Ten patients had an underlying intellectual disability and one with an autism spectrum disorder. The interval from seizure onset to the use of PER was 22 (IQR = 8.5–33) years and the duration of PER use was 885 (IQR = 339.5–1626.5) days. The median number of ASMs used before PER was 4 (IQR = 2–6) and three patients underwent non-medical treatment before the use of PER. One had callosotomy, another received deep brain stimulation, and the other received vagus nerve stimulation. In our cohort, 14 patients were responding to PER, and 69 failed; 22 patients had ADRs and 61 did not; 14 patients had behavior ADRs and 69 did not; a total of 61 patients were able to adhere to PER for more than 1 year, and 22 were not. Among patients with ADRs, five had dizziness, three had somnolence, and 14 had behavior ADRs. Behavior ADR occurred in two patients with underlying intellectual disability and one with an autism spectrum disorder. There were no patients with psychiatric histories before PER use in the study cohort. Neither the types of seizure, the etiologies of seizure, nor the interval from seizure onset to the use of PER was associated with the outcome of perampanel use, the result was presented in Supplementary Table S1–S3, respectively.
Burden analysis of glutamate receptor genes and outcomes related to PER
We first analyzed the association between the rare variants of the glutamate receptor gene group, which includes 26 genes, and the outcomes of PER therapy. The result showed that enriched genetic variants of the glutamate receptor gene group were statistically significant associated with the occurrence of ADR (OR = 0.35, CI = 0.14–0.75, p = 0.0093; Figure 1). No statistically significant association between the enrichment of rare variants and the response to PER, behavior ADR, or 1-year retention (Figure 1).
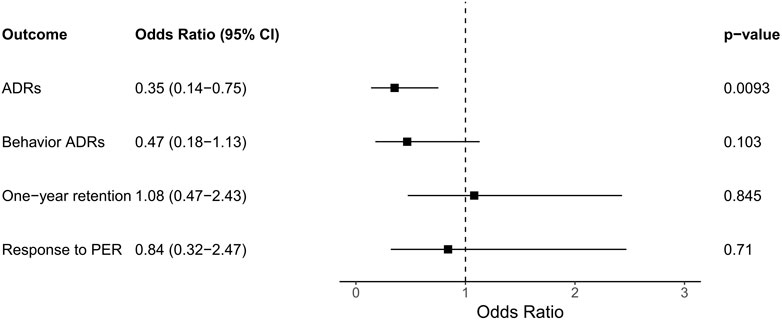
FIGURE 1. The association of the outcomes and the enrichment of rare variants in the glutamate receptor gene group. Abbreviations: ADR, adverse drug reaction; CI, confidence interval; PER, perampanel.
Next, we performed burden analysis to find associations between the rare variants of each of the five function sub-groups (AMPA, NMDA, delta, kainate, and metabotropic) and the outcomes of PER treatment. The occurrence of the ADR had a nominal association with having more rare variants in the glutamate ionotropic receptor delta type subunit compared to those without ADRs (OR = 0.24, CI = 0.03–1.16, p = 0.0354; Figure 2A.). With five function sub-groups in the analysis, Bonferroni correction was applied to correct for multiple testing, defining a p-value threshold for significance of 0.01. Therefore, this nominal difference did not remain statistically significant after the Bonferroni correction. Neither the response to PER, the presence of behavior ADR, nor the 1-year retention of PER was associated with the enrichment of rare variants in any of the five subgroups (Figure 2).
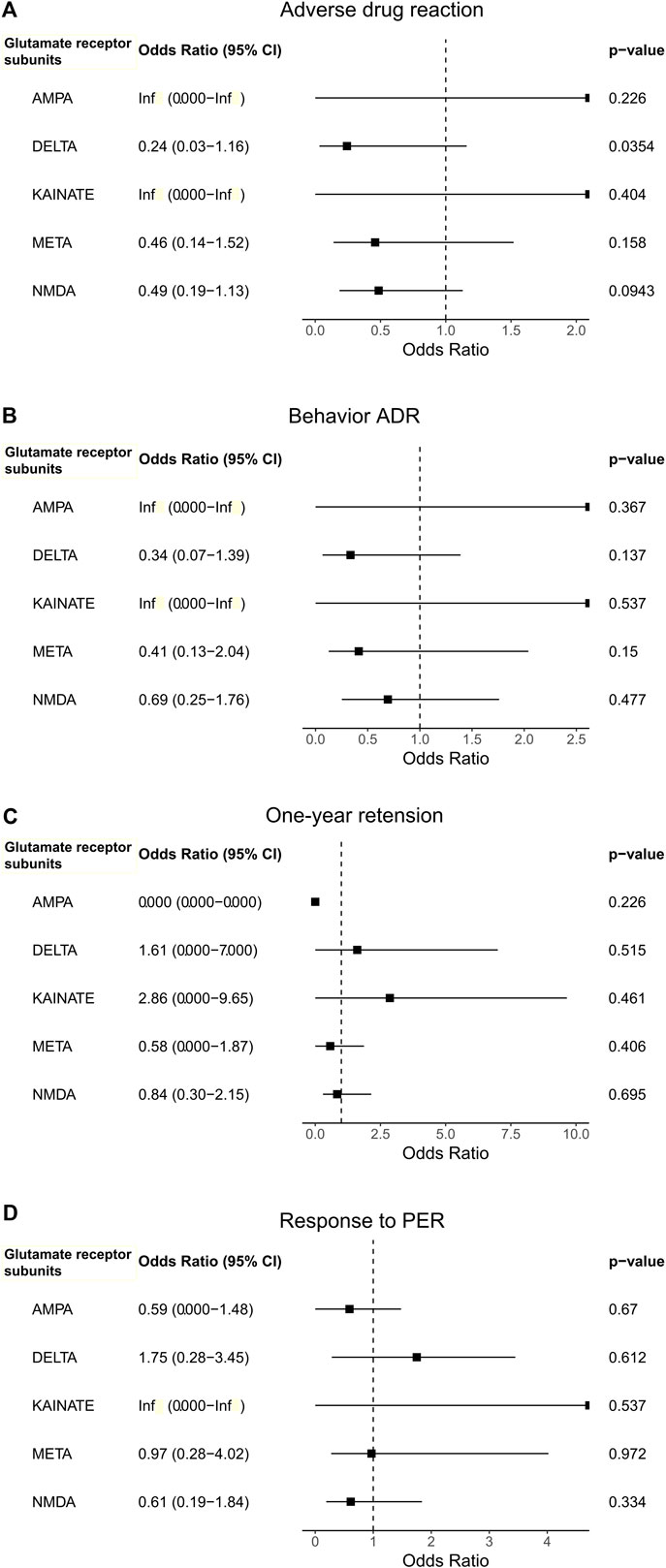
FIGURE 2. The association between outcomes [(A). Adverse drug reaction, (B). Behavior ADR, (C). One-year retension, and (D). Response to PER)] with the use of perampanel and the enrichment of rare variants in the glutamate receptor subunit gene groups.
Gene collapse analysis of glutamate receptor genes and phenotypes related to PER
We performed a gene collapse analysis to find the association between the presence of the rare variants in the glutamate receptor genes and the outcomes of PER use. In terms of ADR, the presence of rare variants in GRID1 had a nominal association with the occurrence of ADR (OR = 5.54, CI = 0.966–39.36, p = 0.0278, Figure 3A) compared to those without. Two variants were observed, GRID1:c.1585G>A and GRID1:c.1287_1288delinsGG. GRID1:c.1585G>A results in changing the amino acid from valine to isoleucine (NM_017551.3:p(Val529Ile)) in the agonist binding domain (ABD) of GluD1. GRID1:c.1287_1288delinsGG results in a non-frameshift substitution of methionine to valine (NM_017551.3:p(Met430Val)) between the N-terminal domain (NTD) and ABD of GluD1.
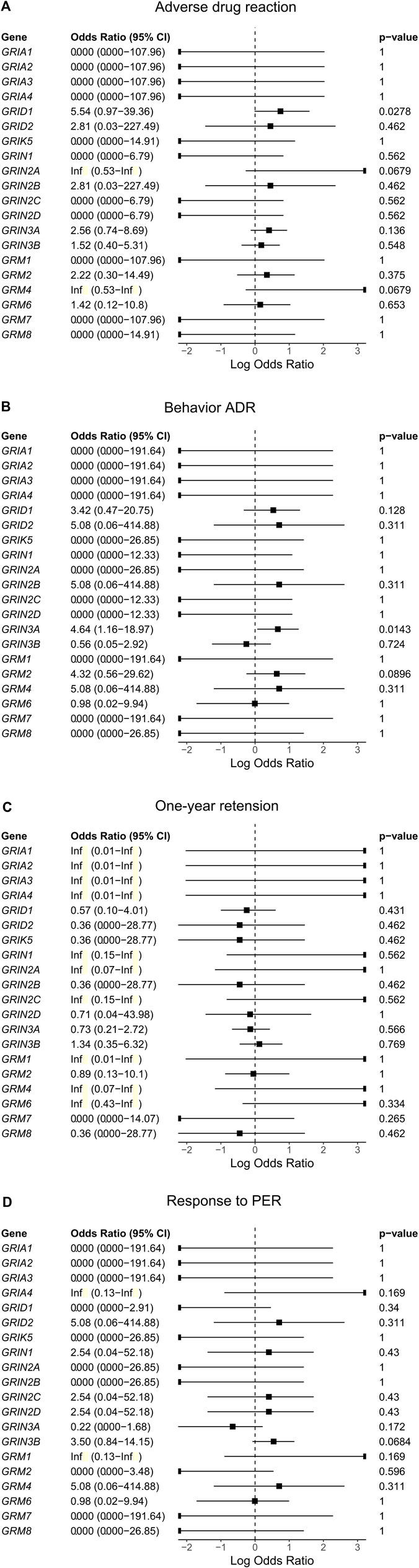
FIGURE 3. The association between rare variants of glutamate receptor genes and the outcomes [(A). Adverse drug reaction, (B). Behavior ADR, (C). One-year retension, and (D). Response to PER)] of using perampanel (PER).
For behavior ADRs, the presence of rare variants in GRIN3A had a nominal association with the occurrence of behavior ADR (OR = 4.64, CI = 1.16–18.97, p = 0.0143, Figure 3B) compared with those without. Five variants were identified, including GRIN3A:c.55C>T, GRIN3A:c.322C>G, GRIN3A:c.1438C>G, GRIN3A:c.1462A>G, and GRIN3A:c.2501_2502delinsCA. GRIN3A:c.55C>T results in changing the amino acid from proline to serine (NM_133445.3:p(Pro19Ser)) in the N-terminal signal peptide of GluN3A. GRIN3A:c.322C>G results in changing the amino acid from proline to alanine (NM_133445.3:p.(Pro108Ala)) in the NTD of GluN3A. GRIN3A:c.1438C>G, and GRIN3A:c.1462A>G result in changing the amino acid from arginine to glycine (NM_133445.3:p.(Arg480Gly)) and lysine to glutamate (NM_133445.3:p.(Lys488Glu)), respectively, in the ABD of GluN3A. GRIN3A:c.2501_2502delinsCA results in a non-frameshift substitution of aspartate to asparagine (NM_133445.3:p.(Asp834Asn)) in the transmembrane domain of GluN3A.
Rare variants of the glutamate receptor genes were neither associated with the response to PER nor 1-year retention (Figure 3).
With 26 genes involved in our gene collapse analysis, Bonferroni correction was again applied to correct for multiple testing, defining a p-value threshold for significance of 1.92*10–3. As a result, none of the above-mentioned nominal associations remain statistically significant after the Bonferroni correction.
Two variants in GRID1 and five in GRIN3A were observed in our cohort. The location of the amino acid changes in their corresponding proteins is demonstrated in Figure 4. The distribution of these variants in our cohort and functional predictions by PolyPhen-2 (Polymorphism Phenotyping v2) (Adzhubei et al., 2010), Mutation Taster (Schwarz et al., 2014), and SIFT (Sorting Intolerant From Tolerant) (Sim et al., 2012) as well as variant classification according to the American College of Medical Genetics and Genomics guideline (Richards et al., 2015) are presented in Table 2.
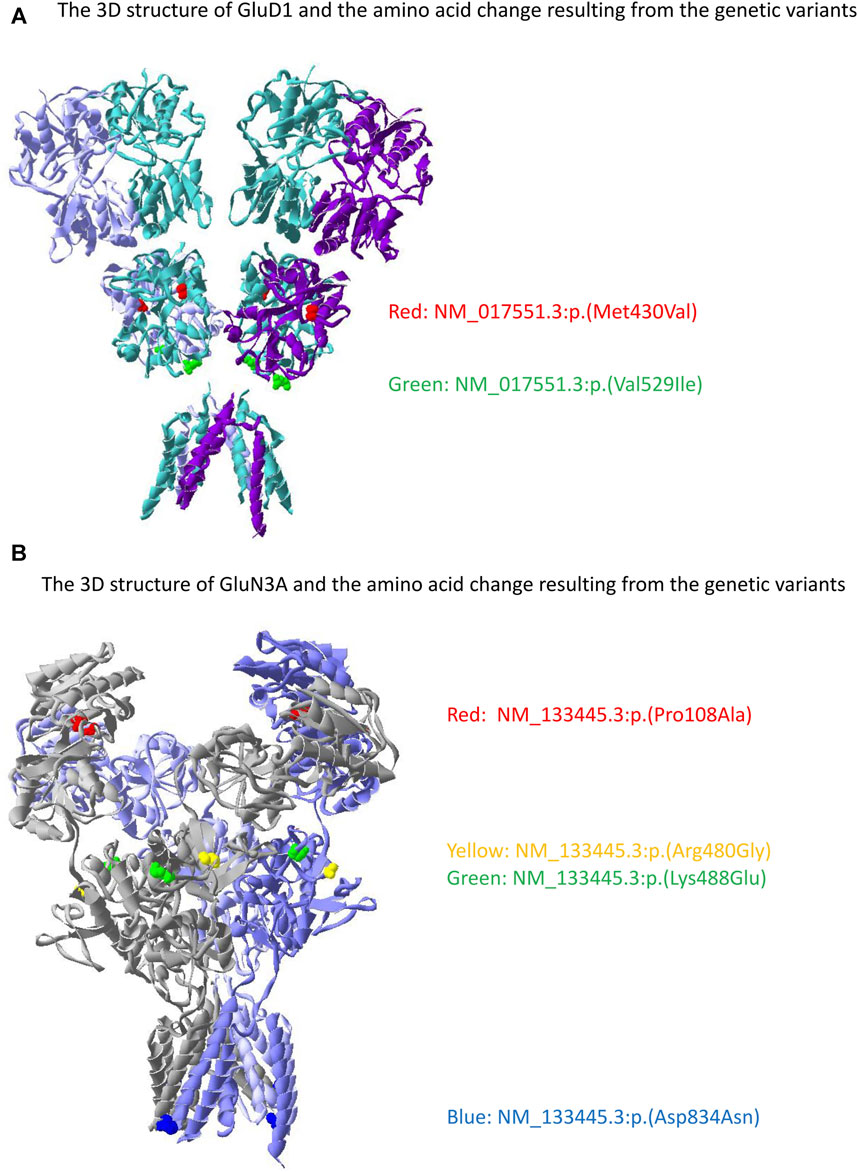
FIGURE 4. The location of the genetic variant in GRID1 and GRIN3A with its corresponding amino acid change on GluD1 and GluN3A proteins. (A) Two variants were identified in GluD1 with the red denotes NM_017551.3:p.(Met430Val) and green denotes NM_017551.3:p.(Val529Ile). (B) Four variants were identified in GluN3A with the red denotes NM_133445.3:p.(Pro108Ala), yellow denotes NM_133445.3:p.(Arg480Gly), green denotes NM_133445.3:p.(Lys488Glu), and blue denotes NM_133445.3:p.(Asp834Asn). NM_133445.3:p.(Pro19Ser) is in the N-terminal signal peptide that is not presented in the mature protein. The 3D structure was generated by the Swiss-Pdb Viewer (Guex and Peitsch 1997).
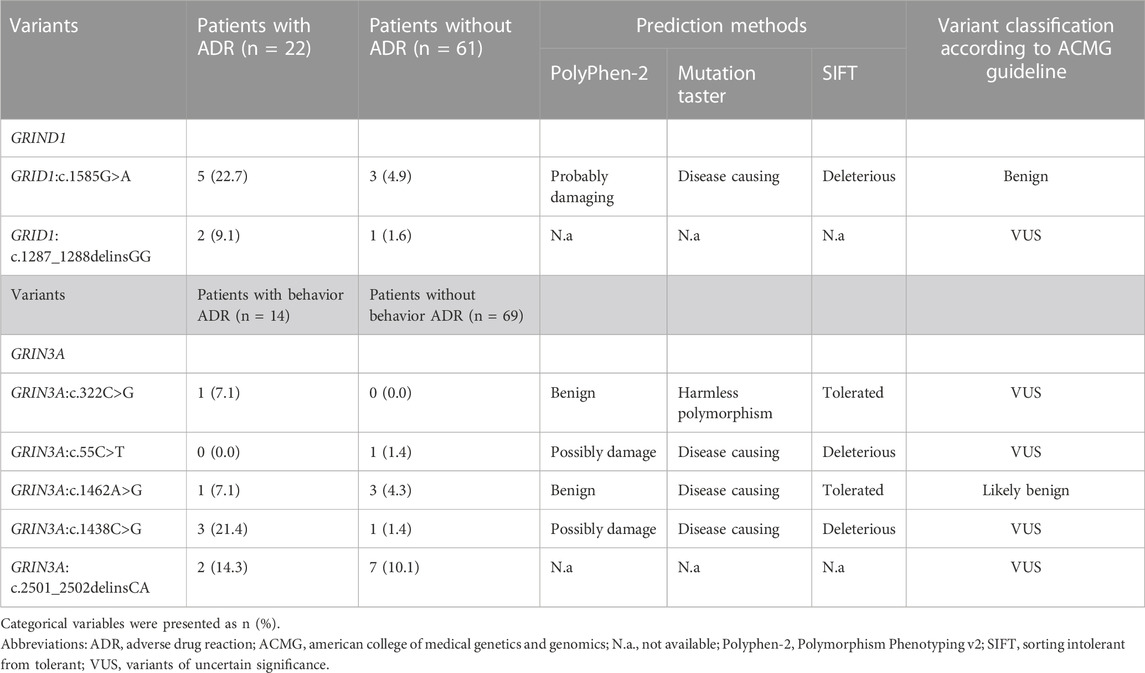
TABLE 2. The genetic variants of GRIND1 and GRIN3A with their distribution and functional prediction.
Discussion
We analyzed the association between the rare variants of the glutamate receptor genes and the outcomes of PER therapy. The gene group burden analysis revealed a statistically significant association between the occurrence of ADR and enriched rare variants of the glutamate gene group. Further analysis revealed the enriched rare variants in the glutamate ionotropic receptor delta subunit had a nominal association with the occurrence of ADR. Furthermore, the gene burden analysis showed that GRID1 had a nominal association with the occurrence of ADR and GRIN3A had a nominal association with the occurrence of behavior ADR. In the end, the nominal association we found did not remain statistically significant after the Bonferroni correction.
Perampanel (PER) is a non-competitive AMPA receptor antagonist (Plosker, 2012), which belongs to one of the glutamate receptors, along with the NMDA receptor, kainite receptor, delta receptor, and metabotropic glutamate receptor (Hansen et al., 2021). These glutamate receptors work in harmony to mediate the excitatory neurotransmission of glutamate (Reiner and Levitz, 2018). The AMPA and NMDA receptors are located in the postsynaptic density at the postsynaptic membrane just opposite the presynaptic vesicle release site and participate in synaptic signal transmission (Scheefhals and MacGillavry, 2018). The metabotropic glutamate receptor was located outside but surrounding the postsynaptic density and served to integrate the function of synapse at high-frequency glutamate release (Rondard and Pin, 2015). The kainite receptor regulates synaptic plasticity with the AMPA and NMDA receptors (Nair et al., 2021). The delta receptors participate in the synaptic organization (Burada et al., 2022) and may alter the number of AMPA receptors in the synapse (Yamasaki et al., 2011). In line with our finding that enriched genetic variants of the glutamate receptor gene group are associated with the presence of ADR. This could be partially explained by the fact that the combined effect of the genetic variants might alter the global interaction of different types of glutamate receptors. These alternations result in changing the overall reaction of the glutamate receptors after PER use and lead to an increase in the risk of ADR in susceptible patients. It is known that PER binds to the AMPA (Yelshanskaya et al. 2016) and kainite receptor (Taniguchi et al. 2022) but our study did not find variants in their corresponding genes, GRIA1-4 and GRIK1-5, respectively, to be associated with PER response through either gene group or gene burden analysis. This could be the result of a relatively small sample size. Genetic variants of glutamate receptors are also linked to epilepsy, such as polymicrogyria is associated with mutations in GRIN1 (Fry et al. 2018), epilepsy-aphasia spectrum disorders with mutations in GRIN2A (Carvill et al. 2013), epilepsy and movement disorders with GRIN2B (Platzer et al. 2017), epileptic encephalopathy with GRIN2D (Li et al. 2016), and developmental epileptic encephalopathy with GRIA2 (Salpietro et al. 2019). We did not find the association of PER responsiveness to be related to variants among these genes.
The nominal association of the presence of ADR after PER use with enriched rare variants in glutamate ionotropic receptor delta type subunit as well as GRID1 suggests the delta receptors could have some role in PER-associated ADRs. Two types of glutamate ionotropic receptor delta-type subunits are identified, glutamate receptor delta 1 (GluD1) and glutamate receptor delta 2 (GluD2) (Burada et al., 2022). Their role is mainly to mediate the synaptic formation and organization, regulate the function of other glutamate ionotropic receptors, and control the trafficking of other glutamate ionotropic receptors (Burada et al., 2022). In the hippocampus, GluD1 enhances the AMPA and NMDA current as knockdown of GluD1 results in the reduction of excitatory synaptic transmission (Tao et al., 2018); in the cerebellar, GluD2 helps the formation of synapses between parallel fibers-Purkinje cells (Yuzaki, 2013). Most interestingly, there was evidence that knock-out of GluD1 decreased the expression of AMPA and NMDA receptors in mice (Yadav et al., 2012; Gupta et al., 2015) and coupling of GluD2 with synaptic organizer complex may trigger the internalization of the AMPA receptor (Elegheert et al., 2016). Therefore, functional alternation from genetic variants of the delta receptor might cause different expression or activity of AMPA and NMDA receptors on the neuronal membrane. This may result in a different response of AMPA receptors when interacting with PER and possibly increase the risk of the presence of ADRs. There is also evidence that GluD is a functional ion channel that interacts with the G protein-coupled glutamate metabotropic receptor (Kumar et al. 2023). They exert a tonic current (Lemoine et al. 2020; Copeland et al. 2023) to maintain the resting membrane potential and activate glutamate metabotropic receptor to promote bursts of action potential (Prisco et al. 2002; Zheng and Johnson 2002). GluD1 is expressed strongly in the cortex, basal ganglia, hippocampus, thalamic, dorsal raphe, and hypothalamic nuclei, while GluD2 in the cerebellum and hypothalamic nuclei (Nakamoto et al. 2020). This wide distribution of GluD1 could explain the association between variants in the GRID1 and the overall occurrence of ADR. In our cohort, GRID1:c.1585G>A was predicted to have functional consequence. There are evidences linking the variants of GRID1 with schizophrenia and bipolar disorder (Treutlein et al., 2009; Greenwood et al., 2011). In vivo study have demonstrated the coupling of glutamate metabotropic receptor 1/5 and GluD1 is essential to the firing of dopamine neurons in the midbrain (Benamer et al., 2018) that are correlated with the physiopathology of schizophrenia (Paladini and Roeper, 2014; Stopper and Floresco, 2015). Although variants of GRID1 are not linked to behavior ADRs in our cohort, our finding may warrant a further investigation of this variant to elucidate its role in the function of glutamate delta receptors.
Our cohort revealed that GRIN3A had a nominal association with behavior ADR. GRIN3A encoded the GluN3A subunit of the NMDA receptor (Eriksson et al., 2002). The GluN3A-containing NMDA receptor had a reduction of the NMDA-induced current (Sasaki et al., 2002). This phenomenon usually occurs at the early stage of development, mostly in utero, when the GluN3A-containing NMDA receptor serves as a negative modulator to limit or eliminate the formation of weakly connected synapses and stabilized the highly specific and durable synaptic connections (Roberts et al., 2009; Kehoe et al., 2014). The effect of GluN3A on synaptic formation implicated its role in memory and cognitive functions. Genetic studies had linked this subunit to other psychiatric diseases, such as schizophrenia (Takata et al., 2013), nicotine dependence (Yang et al., 2015), and delirium (Kazmierski et al., 2014). In our cohort, one of the GRIN3A variants, c.1438C>G, was also found to be associated with schizophrenia (Takata et al., 2013). The roles that GluN3A involved with cognitive function and various psychiatric diseases might make patients harboring variants in GRIN3A more susceptible to behavior ADR. Of note, variants among the GRIN3A are considered to be more relevant to autism spectrum disorders or schizophrenia (Tarabeux et al. 2011), while GRIN1, GRIN2A, GRIN2B, and GRIN2D are more epilepsy-related (Xu and Luo 2018). This may explain why variants of the GRIN3A are associated with behavior ADR but not the overall occurrence of ADR.
To our best knowledge, our study is the first to evaluate the association between the rare variants of the glutamate receptor genes and the outcomes of PER treatment. The development of ADRs after PER might occur in 66.5% of the patients but were usually tolerable (Tsai et al., 2018). Most ADRs are associated with high dose or fast titration while dizziness, falls, and behavior ADRs may be prevented by slow titration (Tsai et al., 2018). Bedtime administration also reduces the occurrence of somnolence and dizziness (Takata et al., 2013). Around 5%–15% of patients taking PER may develop irritation, violent behavior, and/or psychotic symptoms (Gil-Nagel et al., 2018; Tsai et al., 2018). These behavior ADRs could be catastrophic to patient care and decrease their quality of life. Another ASM that might cause behavior ADR was levetiracetam (Chen et al., 2017). A pharmacogenomic study of psychiatric ADR and levetiracetam revealed no association in either common or rare genetic variants (Campbell et al., 2022). However, Campbel et al. utilized a hypothesis-free analysis that included a greater number of genes, and no statistically significant results were found after correction for multiple comparisons. Thus, we proposed a hypothesis based on the pharmacokinetics property of PER that variants in the glutamate receptor genes might alter the response of PER. This approach enabled us to find some associations relevant to the pharmacokinetic property of PER. PER mainly acts on AMPA receptors to exert its antiseizure effect (Ceolin et al., 2012). It is currently unknown whether GluD1 and GluN3A interact with the AMPA receptor (Hansen et al., 2021). Perhaps, a future study allocating function MRI to study patients with and without ADR after PER use may reveal the change in brain function, such as that used in the neuropsychiatric studies in patients with systemic lupus erythematosus (Mikdashi 2016). Therefore, the impact of the GRID1 and GRIN3A variants is still largely unknown based on the available literature. Nevertheless, our findings give a glimpse into the possible underlying genetic variants that may affect the response of the drugs.
Although the variants observed in GRID1 and GRIN3A only had a nominal association with the occurrence of ADR and behavior ADR, respectively, the fact that some of the variants are located in NTD is interesting. NTD is the site that regulates the assembly, trafficking, and function of glutamate receptors (Hansen et al., 2021). Currently, there is no functional evaluation regarding the consequences of the variants we found and the exact interactions of different glutamate receptors remain unknown. However, NTD has the important function of forming a trans-synaptic complex to maintain the integrity of synapses by recruiting neurotransmitter receptors and postsynaptic scaffolding proteins (Sheng and Hoogenraad 2007). It is essential in the formation of excitatory and inhibitory synapses (Fossati et al. 2019) and changes resulting from genetic variants of this region could affect the interaction of various receptors. Since PER acts mainly on AMPA receptors, indirect evidence suggests that non-AMPA glutaminergic receptors, such as glutamate ionotropic receptor delta type subunit and NMDA can affect the function of AMPA receptors. Glutamate ionotropic receptor subunit GluD1 affects the expression of AMPA and NMDA receptors in mice (Yadav et al., 2012; Gupta et al., 2015). The AMPA and NMDA receptors aggregate together in the postsynaptic area to maintain synaptic transmission (Scheefhals and MacGillavry, 2018). As a result, functional changes of non-AMPA glutaminergic receptors may alter the response of AMPA receptors to PER and lead to different outcomes.
In our cohort, 16.9% of patients achieved seizure-free for 1 year after the use of PER. A real-world observational study revealed that 7.2% of patients achieved seizure-free after receiving PER for 1 year (Villanuevaet al. 2016b). With a median of four ASMs before PER use in our cohort and a mean of 7.8 ASMs in the work of Villanueva et al., these patients were having drug resistance epilepsy defined by the International League Against Epilepsy (Kwan et al., 2010). This may explain the reason for the low responder rate in both studies. Various mechanisms contribute to drug resistance epilepsy, including the change of structure or expression of the target of ASMs, alteration of the drug delivery system across the brain, the pharmacodynamic modification that reduces the absorption of ASMs, or formation of the abnormal neural circuits (Loscher et al., 2020). Our study only focused on the association of outcome and the genetic variants in the glutamate receptors. Other genetic variants in the drug-transporting system of the CNS or drug metabolism may also contribute to the responsiveness of PER, which warrants further studies. Besides the change caused by genetic variants, the prolonged seizure may change the expression of the ASM target, such that the expression of sodium channels in human hippocampal tissue is decreased in patients with temporal lobe epilepsy (Remy and Beck, 2006). Currently, no evidence has suggested a similar mechanism existed for AMPA receptors (Lukawski and Czuczwar, 2021) to explain the resistance of PER and may be an interesting topic of future research.
Our study had several limitations. First, our cohort only included Taiwanese patients. Our findings might not be able to apply to patients with different ethnic backgrounds. Just as the association of HLA-B*1502 and Stevens-Johnson syndrome was mainly found in Southeast Asian populations (Chung et al., 2004). Second, we had a relatively low number of patients in our cohort. This limited our ability to find an association between the responsiveness of PER and 1-year retention of PER use. A larger cohort will be needed to validate our findings. Despite the small sample group we had, our research did suggest genetic variants in the glutamate receptor gene group were associated with ADR after PER use. Third, our study only examines the impact rare variants have on the responsiveness of PER, common variants may also have contributions. One study regarding the common variants of the AMPA receptors and the response to antidepressants revealed no association between the two (Chiesa et al. 2012). However, the association between the common variants of the AMPA receptor and the responsiveness of PER has not been studied, which may also provide valuable data for the application of predicting the outcome of PER use. In addition, analysis of the common variants requires a larger patient group to achieve adequate statistical power to find any significant difference. Fourth, we only focused on the glutamate receptors, while auxiliary proteins that interact with glutamate receptors could also affect its response during PER use. Transmembrane AMPA receptor regulatory proteins (TARPs), for example, enhance the conductance level of AMPA receptors (Zhang et al. 2014) and alter their pharmacological properties (Tomita et al. 2006). Variants of the TARPs could result in changing the interplay of TARPs and AMPA receptors leading to different responses to PER use. With ongoing study and the advances in genetic analysis methods, we hope the combination of more genetic association data can reach the ultimate goal of providing patients with the most suitable ASM based on genetic testing.
Data availability statement
The datasets generated and analyzed in the current study are available from the corresponding author upon reasonable request.
Ethics statement
The studies involving humans were approved by Chang Gung Medical Foundation Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization, M-HT; Methodology, C-HL and M-HT; Formal analysis, C-HL, C-JH, and M-HT; Investigation, C-HL, C-JH, Y-TL, and S-YC; Writing–Original Draft Preparation, C-HL; Writing–Review and Editing, M-HT; Supervision, M-HT. All authors contributed to the article and approved the submitted version.
Funding
The publication of this article has been funded by grants from Kaohsiung Chang Gung Memorial Hospital (CMRPG8K1521) to C-HL.
Acknowledgments
The authors would like to thank the Epi25 Collaborative for sequencing part of the patients included in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1215493/full#supplementary-material
References
Abe, T., Seo, T., Ishitsu, T., Nakagawa, T., Hori, M., and Nakagawa, K. (2008). "Association between SCN1A polymorphism and carbamazepine-resistant epilepsy.". Br. J. Clin. Pharmacol. 66 (2), 304–307. doi:10.1111/j.1365-2125.2008.03203.x
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations.". Nat. Methods 7 (4), 248–249. doi:10.1038/nmeth0410-248
Androsova, G., Krause, R., Borghei, M., Wassenaar, M., Auce, P., Avbersek, A., et al. (2017). "Comparative effectiveness of antiepileptic drugs in patients with mesial temporal lobe epilepsy with hippocampal sclerosis.". Epilepsia 58 (10), 1734–1741. doi:10.1111/epi.13871
Angelopoulou, C., Veletza, S., Heliopoulos, I., Vadikolias, K., Tripsianis, G., Stathi, C., et al. (2017). Association of SCN1A gene polymorphism with antiepileptic drug responsiveness in the population of Thrace, Greece. Arch. Med. Sci. 13 (1), 138–147. doi:10.5114/aoms.2016.59737
Balestrini, S., and Sisodiya, S. M. (2018). "Pharmacogenomics in epilepsy.". Neurosci. Lett. 667, 27–39. doi:10.1016/j.neulet.2017.01.014
Benamer, N., Marti, F., Lujan, R., Hepp, R., Aubier, T. G., Dupin, A. A. M., et al. (2018). "GluD1, linked to schizophrenia, controls the burst firing of dopamine neurons.". Mol. Psychiatry 23 (3), 691–700. doi:10.1038/mp.2017.137
Burada, A. P., Vinnakota, R., Bharti, P., Dutta, P., Dubey, N., and Kumar, J. (2022). "Emerging insights into the structure and function of ionotropic glutamate delta receptors.". Br. J. Pharmacol. 179 (14), 3612–3627. doi:10.1111/bph.15313
Campbell, C., McCormack, M., Patel, S., Stapleton, C., Bobbili, D., Krause, R., et al. (2022). A pharmacogenomic assessment of psychiatric adverse drug reactions to levetiracetam.". Epilepsia 63 (6), 1563–1570. doi:10.1111/epi.17228
Carvill, G. L., Regan, B. M., Yendle, S. C., O'Roak, B. J., Lozovaya, N., Bruneau, N., et al. (2013). "GRIN2A mutations cause epilepsy-aphasia spectrum disorders.". Nat. Genet. 45 (9), 1073–1076. doi:10.1038/ng.2727
Ceolin, L., Bortolotto, Z. A., Bannister, N., Collingridge, G. L., Lodge, D., and Volianskis, A. (2012). A novel anti-epileptic agent, perampanel, selectively inhibits AMPA receptor-mediated synaptic transmission in the hippocampus.". Neurochem. Int. 61 (4), 517–522. doi:10.1016/j.neuint.2012.02.035
Chen, B., Choi, H., Hirsch, L. J., Katz, A., Legge, A., Buchsbaum, R., et al. (2017). "Psychiatric and behavioral side effects of antiepileptic drugs in adults with epilepsy.". Epilepsy Behav. 76, 24–31. doi:10.1016/j.yebeh.2017.08.039
Chiesa, A., Crisafulli, C., Porcelli, S., Han, C., Patkar, A. A., Lee, S. J., et al. (2012). "Influence of GRIA1, GRIA2 and GRIA4 polymorphisms on diagnosis and response to treatment in patients with major depressive disorder.". Eur. Arch. Psychiatry Clin. Neurosci. 262 (4), 305–311. doi:10.1007/s00406-011-0270-y
Chung, W. H., Hung, S. I., Hong, H. S., Hsih, M. S., Yang, L. C., Ho, H. C., et al. (2004). Medical genetics: a marker for Stevens-Johnson syndrome. Nature 428 (6982), 486. doi:10.1038/428486aMedical genetics
Copeland, D. S., Gugel, A., and Gantz, S. C. (2023). "Potentiation of neuronal activity by tonic GluD1 current in brain slices.". EMBO Rep. 24 (7), e56801. doi:10.15252/embr.202356801
Elegheert, J., Kakegawa, W., Clay, J. E., Shanks, N. F., Behiels, E., Matsuda, K., et al. (2016). "Structural basis for integration of GluD receptors within synaptic organizer complexes.". Science 353 (6296), 295–299. doi:10.1126/science.aae0104
Eriksson, M., Nilsson, A., Froelich-Fabre, S., Akesson, E., Dunker, J., Seiger, A., et al. (2002). "Cloning and expression of the human N-methyl-D-aspartate receptor subunit NR3A.". Neurosci. Lett. 321 (3), 177–181. doi:10.1016/s0304-3940(01)02524-1
Fong, Y. O., Huang, P., Hsu, C. Y., and Yang, Y. H. (2022). Effects of perampanel on seizure control, cognition, behavior, and psychological status in patients with epilepsy: a systematic review.". J. Clin. Neurol. 18 (6), 653–662. doi:10.3988/jcn.2022.18.6.653
Fossati, M., Assendorp, N., Gemin, O., Colasse, S., Dingli, F., Arras, G., et al. (2019). "Trans-Synaptic signaling through the glutamate receptor delta-1 mediates inhibitory synapse formation in cortical pyramidal neurons.". Neuron 104 (6), 1081–1094. doi:10.1016/j.neuron.2019.09.027
Fry, A. E., Fawcett, K. A., Zelnik, N., Yuan, H., Thompson, B. A. N., Shemer-Meiri, L., et al. (2018). "de novo mutations in GRIN1 cause extensive bilateral polymicrogyria.". Brain 141 (3), 698–712. doi:10.1093/brain/awx358
Gil-Nagel, A., Burd, S., Toledo, M., Sander, J. W., Lebedeva, A., Patten, A., et al. (2018). A retrospective, multicentre study of perampanel given as monotherapy in routine clinical care in people with epilepsy.". Seizure 54, 61–66. doi:10.1016/j.seizure.2017.10.015
Glauser, T. A. (2011). "Biomarkers for antiepileptic drug response.". Biomark. Med. 5 (5), 635–641. doi:10.2217/bmm.11.75
Greenwood, T. A., Lazzeroni, L. C., Murray, S. S., Cadenhead, K. S., Calkins, M. E., Dobie, D. J., et al. (2011). "Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia.". Am. J. Psychiatry 168 (9), 930–946. doi:10.1176/appi.ajp.2011.10050723
Guex, N., and Peitsch, M. C. (1997). "SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling.". Electrophoresis 18 (15), 2714–2723. doi:10.1002/elps.1150181505
Gupta, S. C., Yadav, R., Pavuluri, R., Morley, B. J., Stairs, D. J., and Dravid, S. M. (2015). "Essential role of GluD1 in dendritic spine development and GluN2B to GluN2A NMDAR subunit switch in the cortex and hippocampus reveals ability of GluN2B inhibition in correcting hyperconnectivity.". Neuropharmacology 93, 274–284. doi:10.1016/j.neuropharm.2015.02.013
Haerian, B. S., Baum, L., Kwan, P., Tan, H. J., Raymond, A. A., and Mohamed, Z. (2013). "SCN1A, SCN2A and SCN3A gene polymorphisms and responsiveness to antiepileptic drugs: a multicenter cohort study and meta-analysis.". Pharmacogenomics 14 (10), 1153–1166. doi:10.2217/pgs.13.104
Hansen, C. C., Ljung, H., Brodtkorb, E., and Reimers, A. (2018). Mechanisms underlying aggressive behavior induced by antiepileptic drugs: focus on topiramate, levetiracetam, and perampanel. Behav. Neurol. 2018, 2064027. doi:10.1155/2018/2064027
Hansen, K. B., Wollmuth, L. P., Bowie, D., Furukawa, H., Menniti, F. S., Sobolevsky, A. I., et al. (2021). "Structure, function, and pharmacology of glutamate receptor ion channels.". Pharmacol. Rev. 73 (4), 1469–1658. doi:10.1124/pharmrev.120.000131
Im, K., Lee, S. A., Kim, J. H., Kim, D. W., Lee, S. K., Seo, D. W., et al. (2021). Long-term efficacy and safety of perampanel as a first add-on therapy in patients with focal epilepsy: three-year extension study. Epilepsy Behav. 125, 108407. doi:10.1016/j.yebeh.2021.108407
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alfoldi, J., Wang, Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans.". Nature 581 (7809), 434–443. doi:10.1038/s41586-020-2308-7
Kazmierski, J., Sieruta, M., Banys, A., Jaszewski, R., Sobow, T., Liberski, P., et al. (2014). The assessment of the T102C polymorphism of the 5HT2a receptor gene, 3723G/A polymorphism of the NMDA receptor 3A subunit gene (GRIN3A) and 421C/A polymorphism of the NMDA receptor 2B subunit gene (GRIN2B) among cardiac surgery patients with and without delirium.". Gen. Hosp. Psychiatry 36 (6), 753–756. doi:10.1016/j.genhosppsych.2014.06.002
Kehoe, L. A., Bellone, C., De Roo, M., Zandueta, A., Dey, P. N., Perez-Otano, I., et al. (2014). "GluN3A promotes dendritic spine pruning and destabilization during postnatal development.". J. Neurosci. 34 (28), 9213–9221. doi:10.1523/JNEUROSCI.5183-13.2014
Kumar, J., Popescu, G. K., and Gantz, S. C. (2023). "GluD receptors are functional ion channels.". Biophys. J. 122 (12), 2383–2395. doi:10.1016/j.bpj.2023.05.012
Kumari, R., Lakhan, R., Kumar, S., Garg, R. K., Misra, U. K., Kalita, J., et al. (2013). SCN1AIVS5-91G-->A polymorphism is associated with susceptibility to epilepsy but not with drug responsiveness. Biochimie 95 (6), 1350–1353. doi:10.1016/j.biochi.2013.02.006
Kwan, P., Arzimanoglou, A., Berg, A. T., Brodie, M. J., Allen Hauser, W., Mathern, G., et al. (2010). "Definition of drug resistant epilepsy: consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies.". Epilepsia 51 (6), 1069–1077. doi:10.1111/j.1528-1167.2009.02397.x
Kwan, P., Brodie, M. J., Laurenza, A., FitzGibbon, H., and Gidal, B. E. (2015). Analysis of pooled phase III trials of adjunctive perampanel for epilepsy: impact of mechanism of action and pharmacokinetics on clinical outcomes. Epilepsy Res. 117, 117–124. doi:10.1016/j.eplepsyres.2015.09.002
Lemoine, D., Mondoloni, S., Tange, J., Lambolez, B., Faure, P., Taly, A., et al. (2020). Probing the ionotropic activity of glutamate GluD2 receptor in HEK cells with genetically-engineered photopharmacology. Elife 9, e59026. doi:10.7554/eLife.59026
Leppik, I. E., Yang, H., Williams, B., Zhou, S., Fain, R., Patten, A., et al. (2017). "Analysis of falls in patients with epilepsy enrolled in the perampanel phase III randomized double-blind studies.". Epilepsia 58 (1), 51–59. doi:10.1111/epi.13600
Li, D., Yuan, H., Ortiz-Gonzalez, X. R., Marsh, E. D., Tian, L., McCormick, E. M., et al. (2016). "GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers.". Am. J. Hum. Genet. 99 (4), 802–816. doi:10.1016/j.ajhg.2016.07.013
Lin, C. H., Ho, C. J., Lu, Y. T., and Tsai, M. H. (2021). "Response to Sodium Channel blocking Antiseizure medications and coding polymorphisms of Sodium Channel genes in Taiwanese epilepsy patients.". BMC Neurol. 21 (1), 367. doi:10.1186/s12883-021-02395-2
Loscher, W., Potschka, H., Sisodiya, S. M., and Vezzani, A. (2020). Drug resistance in epilepsy: clinical impact, potential mechanisms, and new innovative treatment options. Pharmacol. Rev. 72 (3), 606–638. doi:10.1124/pr.120.019539
Lukawski, K., and Czuczwar, S. J. (2021). "Understanding mechanisms of drug resistance in epilepsy and strategies for overcoming it.". Expert Opin. Drug Metab. Toxicol. 17 (9), 1075–1090. doi:10.1080/17425255.2021.1959912
Ma, C. L., Wu, X. Y., Zheng, J., Wu, Z. Y., Hong, Z., and Zhong, M. K. (2014). "Association of SCN1A, SCN2A and ABCC2 gene polymorphisms with the response to antiepileptic drugs in Chinese Han patients with epilepsy.". Pharmacogenomics 15 (10), 1323–1336. doi:10.2217/pgs.14.89
Manna, I., Gambardella, A., Bianchi, A., Striano, P., Tozzi, R., Aguglia, U., et al. (2011). A functional polymorphism in the SCN1A gene does not influence antiepileptic drug responsiveness in Italian patients with focal epilepsy.". Epilepsia 52 (5), e40–e44. doi:10.1111/j.1528-1167.2011.03097.x
Mikdashi, J. A. (2016). Altered functional neuronal activity in neuropsychiatric lupus: a systematic review of the fMRI investigations.". Semin. Arthritis Rheum. 45 (4), 455–462. doi:10.1016/j.semarthrit.2015.08.002
Nair, J. D., Wilkinson, K. A., Henley, J. M., and Mellor, J. R. (2021). "Kainate receptors and synaptic plasticity.". Neuropharmacology 196, 108540. doi:10.1016/j.neuropharm.2021.108540
Nakamoto, C., Konno, K., Miyazaki, T., Nakatsukasa, E., Natsume, R., Abe, M., et al. (2020). "Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain.". J. Comp. Neurol. 528 (6), 1003–1027. doi:10.1002/cne.24792
Paladini, C. A., and Roeper, J. (2014). "Generating bursts (and pauses) in the dopamine midbrain neurons.". Neuroscience 282, 109–121. doi:10.1016/j.neuroscience.2014.07.032
Perucca, E., and Tomson, T. (2011). The pharmacological treatment of epilepsy in adults.". Lancet Neurol. 10 (5), 446–456. doi:10.1016/S1474-4422(11)70047-3
Platzer, K., Yuan, H., Schutz, H., Winschel, A., Chen, W., Hu, C., et al. (2017). GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J. Med. Genet. 54 (7), 460–470. doi:10.1136/jmedgenet-2016-104509
Plosker, G. L. (2012). Perampanel: as adjunctive therapy in patients with partial-onset seizures. CNS Drugs 26 (12), 1085–1096. doi:10.1007/s40263-012-0021-2
Prisco, S., Natoli, S., Bernardi, G., and Mercuri, N. B. (2002). "Group I metabotropic glutamate receptors activate burst firing in rat midbrain dopaminergic neurons.". Neuropharmacology 42 (3), 289–296. doi:10.1016/s0028-3908(01)00192-7
Reiner, A., and Levitz, J. (2018). Glutamatergic signaling in the central nervous system: ionotropic and metabotropic receptors in concert. Neuron 98 (6), 1080–1098. doi:10.1016/j.neuron.2018.05.018
Remy, S., and Beck, H. (2006). "Molecular and cellular mechanisms of pharmacoresistance in epilepsy.". Brain 129 (Pt 1), 18–35. doi:10.1093/brain/awh682
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). "Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and Genomics and the association for molecular pathology.". Genet. Med. 17 (5), 405–424. doi:10.1038/gim.2015.30
Roberts, A. C., Diez-Garcia, J., Rodriguiz, R. M., Lopez, I. P., Lujan, R., Martinez-Turrillas, R., et al. (2009). "Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation.". Neuron 63 (3), 342–356. doi:10.1016/j.neuron.2009.06.016
Rogawski, M. A., and Hanada, T. (2013). "Preclinical pharmacology of perampanel, a selective non-competitive AMPA receptor antagonist.". Acta Neurol. Scand. Suppl(197) 127, 19–24. doi:10.1111/ane.12100
Rondard, P., and Pin, J. P. (2015). "Dynamics and modulation of metabotropic glutamate receptors.". Curr. Opin. Pharmacol. 20, 95–101. doi:10.1016/j.coph.2014.12.001
Salpietro, V., Dixon, C. L., Guo, H., Bello, O. D., Vandrovcova, J., Efthymiou, S., et al. (2019). "AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders.". Nat. Commun. 10 (1), 3094. doi:10.1038/s41467-019-10910-w
Sanchez, M. B., Herranz, J. L., Leno, C., Arteaga, R., Oterino, A., Valdizan, E. M., et al. (2010). "Genetic factors associated with drug-resistance of epilepsy: relevance of stratification by patient age and aetiology of epilepsy.". Seizure 19 (2), 93–101. doi:10.1016/j.seizure.2009.12.004
Sasaki, Y. F., Rothe, T., Premkumar, L. S., Das, S., Cui, J., Talantova, M. V., et al. (2002). "Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons.". J. Neurophysiol. 87 (4), 2052–2063. doi:10.1152/jn.00531.2001
Scheefhals, N., and MacGillavry, H. D. (2018). "Functional organization of postsynaptic glutamate receptors.". Mol. Cell Neurosci. 91, 82–94. doi:10.1016/j.mcn.2018.05.002
Schwarz, J. M., Cooper, D. N., Schuelke, M., and Seelow, D. (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11 (4), 361–362. doi:10.1038/nmeth.2890
Seal, R. L., Braschi, B., Gray, K., Jones, T. E. M., Tweedie, S., Haim-Vilmovsky, L., et al. (2023). Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51 (D1), D1003–D1009. doi:10.1093/nar/gkac888
Sheng, M., and Hoogenraad, C. C. (2007). The postsynaptic architecture of excitatory synapses: a more quantitative view.". Annu. Rev. Biochem. 76, 823–847. doi:10.1146/annurev.biochem.76.060805.160029
Sim, N. L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., and Ng, P. C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40 (Web Server issue), W452–W457. doi:10.1093/nar/gks539
Steinhoff, B. J., Ben-Menachem, E., Ryvlin, P., Shorvon, S., Kramer, L., Satlin, A., et al. (2013). "Efficacy and safety of adjunctive perampanel for the treatment of refractory partial seizures: a pooled analysis of three phase III studies.". Epilepsia 54 (8), 1481–1489. doi:10.1111/epi.12212
Stopper, C. M., and Floresco, S. B. (2015). Dopaminergic circuitry and risk/reward decision making: implications for schizophrenia. Schizophr. Bull. 41 (1), 9–14. doi:10.1093/schbul/sbu165
Takata, A., Iwayama, Y., Fukuo, Y., Ikeda, M., Okochi, T., Maekawa, M., et al. (2013). A population-specific uncommon variant in GRIN3A associated with schizophrenia.". Biol. Psychiatry 73 (6), 532–539. doi:10.1016/j.biopsych.2012.10.024
Taniguchi, S., Stolz, J. R., and Swanson, G. T. (2022). The antiseizure drug perampanel is a subunit-selective negative allosteric modulator of kainate receptors.". J. Neurosci. 42 (28), 5499–5509. doi:10.1523/JNEUROSCI.2397-21.2022
Tao, W., Diaz-Alonso, J., Sheng, N., and Nicoll, R. A. (2018). Postsynaptic δ1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc. Natl. Acad. Sci. U. S. A. 115 (23), E5373–E5381. doi:10.1073/pnas.1802737115
Tarabeux, J., Kebir, O., Gauthier, J., Hamdan, F. F., Xiong, L., Piton, A., et al. (2011). Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl. Psychiatry 1 (11), e55. doi:10.1038/tp.2011.52
Tomita, S., Sekiguchi, M., Wada, K., Nicoll, R. A., and Bredt, D. S. (2006). "Stargazin controls the pharmacology of AMPA receptor potentiators.". Proc. Natl. Acad. Sci. U. S. A. 103 (26), 10064–10067. doi:10.1073/pnas.0603128103
Treutlein, J., Muhleisen, T. W., Frank, J., Mattheisen, M., Herms, S., Ludwig, K. U., et al. (2009). "Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter.". Schizophr. Res. 111 (1-3), 123–130. doi:10.1016/j.schres.2009.03.011
Tsai, J. J., Wu, T., Leung, H., Desudchit, T., Tiamkao, S., Lim, K. S., et al. (2018). Perampanel, an AMPA receptor antagonist: from clinical research to practice in clinical settings. Acta Neurol. Scand. 137 (4), 378–391. doi:10.1111/ane.12879
Villanueva, V., Garces, M., Lopez-Gonzalez, F. J., Rodriguez-Osorio, X., Toledo, M., Salas-Puig, J., et al. (2016a). Safety, efficacy and outcome-related factors of perampanel over 12 months in a real-world setting: the FYDATA study. Epilepsy Res. 126, 201–210. doi:10.1016/j.eplepsyres.2016.08.001
Villanueva, V., Majid, O., Nabangchang, C., Yang, H., Laurenza, A., Ferry, J., et al. (2016b). "Pharmacokinetics, exposure-cognition, and exposure-efficacy relationships of perampanel in adolescents with inadequately controlled partial-onset seizures.". Epilepsy Res. 127, 126–134. doi:10.1016/j.eplepsyres.2016.08.025
Wolking, S., Moreau, C., Nies, A. T., Schaeffeler, E., McCormack, M., Auce, P., et al. (2020a). "Testing association of rare genetic variants with resistance to three common antiseizure medications.". Epilepsia 61 (4), 657–666. doi:10.1111/epi.16467
Wolking, S., Moreau, C., Nies, A. T., Schaeffeler, E., McCormack, M., Auce, P., et al. (2020b). Testing association of rare genetic variants with resistance to three common antiseizure medications. Epilepsia 61, 657–666. doi:10.1111/epi.16467
Xu, X. X., and Luo, J. H. (2018). "Mutations of N-Methyl-D-Aspartate receptor subunits in epilepsy.". Neurosci. Bull. 34 (3), 549–565. doi:10.1007/s12264-017-0191-5
Yadav, R., Gupta, S. C., Hillman, B. G., Bhatt, J. M., Stairs, D. J., and Dravid, S. M. (2012). "Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors.". PLoS One 7 (3), e32969. doi:10.1371/journal.pone.0032969
Yamasaki, M., Miyazaki, T., Azechi, H., Abe, M., Natsume, R., Hagiwara, T., et al. (2011). Glutamate receptor δ2 is essential for input pathway-dependent regulation of synaptic AMPAR contents in cerebellar Purkinje cells. J. Neurosci. 31 (9), 3362–3374. doi:10.1523/JNEUROSCI.5601-10.2011
Yang, J., Wang, S., Yang, Z., Hodgkinson, C. A., Iarikova, P., Ma, J. Z., et al. (2015). The contribution of rare and common variants in 30 genes to risk nicotine dependence.". Mol. Psychiatry 20 (11), 1467–1478. doi:10.1038/mp.2014.156
Yelshanskaya, M. V., Singh, A. K., Sampson, J. M., Narangoda, C., Kurnikova, M., and Sobolevsky, A. I. (2016). "Structural bases of noncompetitive inhibition of AMPA-subtype ionotropic glutamate receptors by antiepileptic drugs.". Neuron 91 (6), 1305–1315. doi:10.1016/j.neuron.2016.08.012
Yuzaki, M. (2013). Cerebellar LTD vs. motor learning-lessons learned from studying GluD2. Neural Netw. 47, 36–41. doi:10.1016/j.neunet.2012.07.001
Zhang, W., Devi, S. P., Tomita, S., and Howe, J. R. (2014). "Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors.". Eur. J. Neurosci. 39 (7), 1138–1147. doi:10.1111/ejn.12519
Keywords: perampanel, glutamate receptor gene, rare genetic variants, drug response, adverse drug reaction, 1-year retention
Citation: Lin C-H, Ho C-J, Chen S-Y, Lu Y-T and Tsai M-H (2023) A pharmacogenetic study of perampanel: association between rare variants of glutamate receptor genes and outcomes. Front. Genet. 14:1215493. doi: 10.3389/fgene.2023.1215493
Received: 02 May 2023; Accepted: 07 November 2023;
Published: 24 November 2023.
Edited by:
Zaid Afawi, Erasmus Medical Center, NetherlandsReviewed by:
Alberto Lazarowski, University of Buenos Aires, ArgentinaChristopher P. Ptak, The University of Iowa, United States
Copyright © 2023 Lin, Ho, Chen, Lu and Tsai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng-Han Tsai, bWVuZ2hhbkBjZ21oLm9yZy50dw==
 Chih-Hsiang Lin
Chih-Hsiang Lin Chen-Jui Ho
Chen-Jui Ho Shih-Ying Chen1
Shih-Ying Chen1 Yan-Ting Lu
Yan-Ting Lu Meng-Han Tsai
Meng-Han Tsai