- 1Genomics Division, National Institute of Agricultural Sciences, Rural Development Administration, Jeonju, Republic of Korea
- 2Department of Crop Science and Biotechnology, Jeonbuk National University, Jeonju, Republic of Korea
Background: In the Sesamum species complex, the lack of wild species genomic resources hinders the evolutionary comprehension of phylogenetic relationships.
Results: In the present study, we generated complete chloroplast genomes of six wild relatives (Sesamum alatum, Sesamum angolense, Sesamum pedaloides, Ceratotheca sesamoides (syn. Sesamum sesamoides), Ceratotheca triloba (syn. Sesamum trilobum), and Sesamum radiatum) and a Korean cultivar, Sesamum indicum cv. Goenbaek. A typical quadripartite chloroplast structure, including two inverted repeats (IR), a large single copy (LSC), and a small single copy (SSC), was observed. A total of 114 unique genes encompassing 80 coding genes, four ribosomal RNAs, and 30 transfer RNAs were counted. The chloroplast genomes (152, 863–153, 338 bp) exhibited the IR contraction/expansion phenomenon and were quite conserved in both coding and non-coding regions. However, high values of the nucleotide diversity index were found in several genes, including ndhA, ndhE, ndhF, ycf1, and psaC–ndhD. Concordant tree topologies suggest ndhF as a useful marker for taxon discrimination. The phylogenetic inference and time divergence dating indicate that S. radiatum (2n = 64) occurred concomitantly with the sister species C. sesamoides (2n = 32) approximately 0.05 million years ago (Mya). In addition, S. alatum was clearly discriminated by forming a single clade, showing its long genetic distance and potential early speciation event in regards to the others.
Conclusion: Altogether, we propose to rename C. sesamoides and C. triloba as S. sesamoides and S. trilobum, respectively, as suggested previously based on the morphological description. This study provides the first insight into the phylogenetic relationships among the cultivated and wild African native relatives. The chloroplast genome data lay a foundation for speciation genomics in the Sesamum species complex.
Introduction
The Sesamum L. genus belongs to the Pedaliaceae family with approximately 80 species grouped in 17 genera (Cronquist, 1981). Its leaves are alternate or opposite, and the inflorescence appears generally as a solitary type in leaf axils with the presence of extra-floral nectaries (Bedigian, 2015). The number of species in this genus is under constant revision, since the classification criteria were quite diverse according to the authors. The index Kewensis listed 34 species (Nayar and Mehra, 1970). Later on, Kobayashi (1991) reported 38 species. Through the construction of the Sesamum spp. descriptor, the number was revised to 20 species (IPGRI and NBPGR, 2004; Bedigian, 2015). Based on the Plants of the World Online database, a total of 31 species have been accepted, including 22 wild species native from Africa (POWO, 2022).
The wild relatives are mainly distributed across tropical Africa (from Senegal to Somalia), central and southern Africa, and in drought-prone Indian subcontinent areas (Bedigian, 2015). Both dietary habits and traditional medicine practices are marked by the usage of cultivated and wild relatives (Ntwenya et al., 2017; Aworh, 2018; Bedigian, 2018). Among the therapeutic virtues of sesame, lowering cholesterol is one of the important functions that were reported for preventing high blood pressure disease (Hsu and Parthasarathy, 2017). This function was imputed to the presence of singular lignans known as sesamolin and sesamin (Visavadiya and Narasimhacharya, 2008).
The progenitor and the domestication history underpinning the cultivated sesame have been a subject of debate. Despite the high number of wild relatives in Africa, the investigations based on the interspecific hybridization ability (Bedigian, 2014), the presence or absence of sesamolin (Bedigian et al., 1985; Bedigian, 2003), and external transcribed spacer-based phylogeny reconstruction (Gormley et al., 2015) suggested the Indian native wild Sesamum malabaricum as the probable progenitor. In addition, the scientific controversy relative to the center of origin of the cultivated sesame opposed Africa and the Indian subcontinent (Bedigian, 2003).
Moreover, several species belonging to the Josephinia, Dicerocaryum, and Ceratotheca genus were reported to be closely related to Sesamum species based on phenotypic data and limited number of plastid markers (trnL–trnF and ndhF) and external transcribed spacer sequences (Gormley et al., 2015). More specifically, these genera form a species complex with Sesamum, making it difficult to clearly delineate species boundaries (Manning and Magee, 2018).
The chloroplast organelle is referred to as a chemical factory of plant cells involved in the crucial metabolism of green plants known as photosynthesis (Kirchhoff, 2019). Due to its uniparental inheritance and non-recombination intrinsic characteristics, the chloroplast is widely used to infer the phylogenetic relationships at inter- and intra-taxon levels (Biju et al., 2019; Köhler et al., 2020; Zhou et al., 2020). To the best of our knowledge, only the Sesamum indicum taxon chloroplast genome has been assembled (Yi and Kim, 2012; Zhang et al., 2013). Due to the lack of wild relatives’ chloroplast genome, a comprehensive study on the phylogenetic relationship between Sesamum and Ceratotheca has not been elucidated yet. Interestingly, a total of three types of chromosome numbers (2n = 26, 2n = 32, and 2n = 64) have been reported in the Sesamum genus, and only 2n = 32 for Ceratotheca suggests a potential polyploidy or hybridization event occurrence (Raghavan and Krishnamurthy, 1947; Kobayashi, 1991; Patil and Hiremath, 2002; Patil and Hiremath, 2004). Therefore, a complete chloroplast genome offers a relevant opportunity to investigate the evolutionary relationship between the two sister species.
This study was designed to address two main questions: 1) what is the chloroplast genome organization variation between Sesamum and Ceratotheca representatives? 2) How phylogenetically related are the sister species? We took advantage of the whole-genome sequencing data 1) to assemble and annotate the first complete chloroplast genome of Sesamum alatum, Sesamum pedaloides, Sesamum angolense, Sesamum radiatum, Ceratotheca sesamoides, and Ceratotheca triloba; 2) to investigate their sequence polymorphism and divergence; and 3) to infer the phylogenetic relationships and time divergence among the species.
Materials and methods
Taxon sampling and DNA extraction
A total of six wild sesame species, namely, S. alatum (2n = 26), S. angolense (2n = 32), S. radiatum (2n = 64), S. pedaloides (2n = indeterminate), C. sesamoides, and C. triloba were provided by the Korean genebank (Supplementary Table S1). The natural distribution of wild relatives is summarized in Figure 1. In addition, the Korean elite cultivar Goenbaek (2n = 26) was also employed for de novo chloroplast genome assembly. The seeds were grown under field conditions during the 2020 summer season (May–September) at the National Institute of Agricultural Sciences in Jeonju (35°49′50.37″N latitude, 127°3′52.79″E longitude).
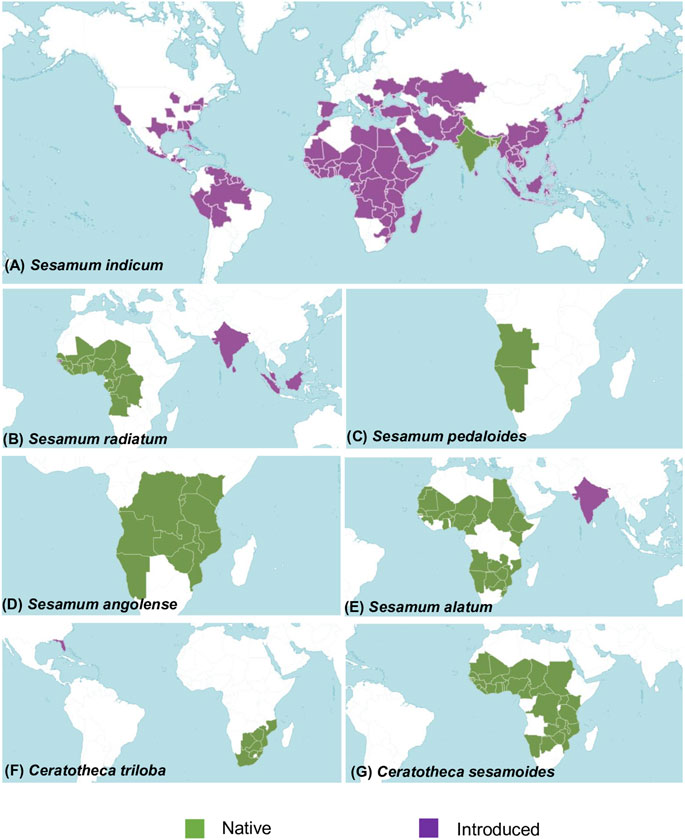
FIGURE 1. Map showing the geographical distribution of the studied species in their native (in green) and host habitat (in purple). (A) Sesamum indicum distribution. The native region covers Assam, Bangladesh, India, and western Himalaya. It has a worldwide distribution. (B) Sesamum radiatum is native to Angola, Benin, Burkina, Cameroon, Cape Verde, the Central African Republic, Chad, Congo, Gabon, the Gambia, Ghana, Guinea, the Gulf of Guinea Is., Ivory Coast, Liberia, Mali, Niger, Nigeria, Senegal, Sierra Leone, Togo, and the Democratic Republic of Congo. It has been introduced in Borneo, Guinea-Bissau, India, Malaya, Sri Lanka, and Sumatera. (C) Sesamum pedaloides is originated from Angola and Namibia. (D) Sesamum angolense came from the southern part of Africa covering Angola, Burundi, Kenya, Malawi, Mozambique, Namibia, Rwanda, Tanzania, Uganda, Zambia, the Democratic Republic of Congo, and Zimbabwe. (E) Sesamum alatum was introduced in India. Its native distribution covers Angola, Benin, Botswana, Burkina Faso, Cameroon, Caprivi Strip, Chad, Egypt, Eritrea, Ethiopia, Ghana, Guinea, Kenya, KwaZulu-Natal, Mali, Mauritania, Mozambique, Namibia, Niger, Nigeria, Senegal, Sudan, Swaziland, Western Sahara, Zambia, and Zimbabwe. (F) Ceratotheca triloba originated from Botswana, Cape Provinces, the Free State, KwaZulu-Natal, Mozambique, Swaziland, and Zimbabwe. It has been introduced in Florida (US). (G) Ceratotheca sesamoides is native to Benin, Botswana, Burkina, Cameroon, the Caprivi Strip, the Central African Republic, Chad, Gambia, Ghana, Guinea, Guinea-Bissau, Ivory Coast, Kenya, Liberia, Malawi, Mali, Mauritania, Mozambique, Namibia, Niger, Nigeria, Senegal, Sierra Leone, Sudan, Tanzania, Togo, Uganda, Zambia, the Democratic Republic of Congo, and Zimbabwe. Distribution data of each species were collected from POWO (2022).
Young leaves of each species were sampled, and their DNA was extracted following a modified CTAB protocol (Allen et al., 2006). Afterward, DNA purity was checked in 1% agarose gel (1× TAE) using the NanoDrop® ND-1000 UV-vis spectrophotometer (Thermo Fisher Scientific, United States). The extracted DNA samples were stored at −20°C prior to further usage.
Library preparation and sequencing
The TruSeq DNA Nano Library Preparation Kit (Illumina, San Diego, United States) was used to construct the library by fragmenting 1 µg high-quality genomic DNA of each sample followed by 5′ and 3′ adapter ligation. The NovaSeq 6000 machine (Illumina, San Diego, United States) served as a platform for short-read sequencing.
Assembly and annotation
For de novo chloroplast assemblies, we used GetOrganelle with default parameters (Jin et al., 2020). The Rubisco subunit gene (GenBank accession: HQ384882.1) of the reference chloroplast genome (GenBank accession: NC_016433.2) from S. indicum cv. Ansanggae was provided as a seed.
All chloroplast assemblies were annotated with GeSeq (Tillich et al., 2017). The setting parameters were defined as follows: HMMER profile search for coding genes and ribosomal RNA annotation, ARAGORN v1.2.38 (Laslett and Canback, 2004) and tRNAscan-SE v2.0.5 (Chan and Lowe, 2019) for transfer RNA gene detection, and the S. indicum L. cv. Ansanggae chloroplast as a reference for the homology-based annotation purpose. Chloë v1.1, a stand-alone chloroplast annotator (https://chloe.plantenergy.edu.au/annotate.html), served as an additional third party annotator for comparison. Using the reference chloroplast annotation, pseudo-genes and trans-spliced genes were manually inspected. The chloroplast genome map was rendered using OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al., 2019).
Wet-lab validation of plastome junction sites
To check the quality of each assembly, an alignment to the reference chloroplast genome (GenBank accession: NC_016433.2) was conducted using the BLASTN tool (Altschul et al., 1990). Subsequently, the position of each chloroplast genome junction was detected. Thus, primers flanking the junction sites were designed using primer3 v2.3.6 (Untergasser et al., 2012), and a PCR-based validation was carried out under the conditions described as follows: the total volume of 20 μL encompassed 15 ng of DNA, 10 pmol of each primer, and the dried SafeDry Taq LTP-480 Premix (CellSafe Co., Ltd., Gyeonggi-do, Korea). PCR experiments were conducted in eight strip tubes in a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States). PCR cycles were 95°C (3 min) and 35 cycles at 95°C (30 s), 55°C (30 s), and 72°C (30 s), followed by the extension step for 5 min at 72°C. The amplified products were separated in 1% agarose gel (1× TAE) and visualized using a UVP GelSolo M-26XV imager (Analytik Jena, CA, United States).
Comparative chloroplast genome analysis
The annotated chloroplast genomes were compared using the mVISTA web server (http://genome.lbl.gov/vista/mvista/submit.shtml) (Frazer et al., 2004), with the cultivar Ansanggae chloroplast genome as a reference. Shuffle-LAGAN was selected as the alignment mode. In order to identify putative gene rearrangements or synteny patterns within the chloroplast genomes, a whole-genome alignment was executed in AliTV (Ankenbrand et al., 2017) and Mauve v.2.4.0 with the progressiveMauve algorithm option (Darling et al., 2004), respectively.
The IR/LSC and IR/SSC boundaries of the chloroplast genomes were visualized using the IRscope R Shiny web app (https://irscope.shinyapps.io/irapp/) (Amiryousefi et al., 2018). The Arabidopsis thaliana chloroplast genome (GenBank accession: NC_000932.1) was included as an outgroup. The nucleotide diversity (π) among the assembled chloroplast genomes was calculated using DnaSP v.6.12.3 (Rozas et al., 2017).
Repeat analysis
Palindrome, complement, forward, and reverse sequence identification was carried out using the REPuter web server (https://bibiserv.cebitec.uni-bielefeld.de/reputer) (Kurtz et al., 2001). The minimal repeat size and hamming distance were set to 30 bp and 3, respectively. Simple sequence repeats (SSRs) were identified using the MISA-web program (https://webblast.ipk-gatersleben.de/misa/) with default parameters (Beier et al., 2017).
Phylogenetic inference
To infer the phylogeny among Sesamum, Ceratotheca, and close members from the Lamiales order, chloroplast genome datasets (Supplementary Table S2) were retrieved from the NCBI. Vitis vinifera was used as an outgroup. A total of 75 common protein-coding genes (Supplementary Table S3) served as the phylogeny inference. A multiple sequence alignment was performed using MAFFT v7.471-0 (Katoh and Standley, 2013). The multiple sequence alignment files were trimmed using trimAl (Capella-Gutierrez et al., 2009) to remove poorly aligned regions. Afterward, the maximum-likelihood tree was constructed using IQ-TREE v2.0.3 (Nguyen et al., 2015), following the automatically selected best-fit model. The ultrafast bootstrap method (Minh et al., 2013) with 1,000 iterations was applied. Bayesian inference was also employed to infer the phylogenetic relationship using MrBayes v3.2.6 (Huelsenbeck and Ronquist, 2001).
Divergence time estimation
To estimate the divergence time among the studied species, the RelTime method and the general time reversible model were performed in MEGA X, following the procedure defined by Mello (2018). Based on the TimeTree database (Kumar et al., 2017), two calibration constraints were set: 1) Pedaliaceae versus Acanthaceae: 34–70 Mya and 2) Linderniaceae versus Pedaliaceae: 41–66 Mya.
Selection pressure analysis
The non-synonymous-to-synonymous substitution ratio (Ka/Ks), for each orthologous pair (with S. indicum as reference), was computed using the codeml package from the PAML tool (Yang, 1997). Prior to the calculation, a codon-based nucleic acid alignment was obtained from the initial protein-coding multiple sequence alignment file using PAL2NAL (Suyama et al., 2006). For a reliable interpretation, the ratios with Ks <0.01 or Ks >2 were filtered out.
Codon usage bias and RNA editing site analyses
Relative synonymous codon usage (RSCU) bias analysis was conducted using CodonW v1.4.4 (http://codonw.sourceforge.net/, accessed 12 February 2021). The PREP-Cp package from the PREP suite (Mower, 2009) was used to predict RNA editing sites for each species, with a cut-off value of 0.8.
Results
General features of the assembled chloroplast genomes
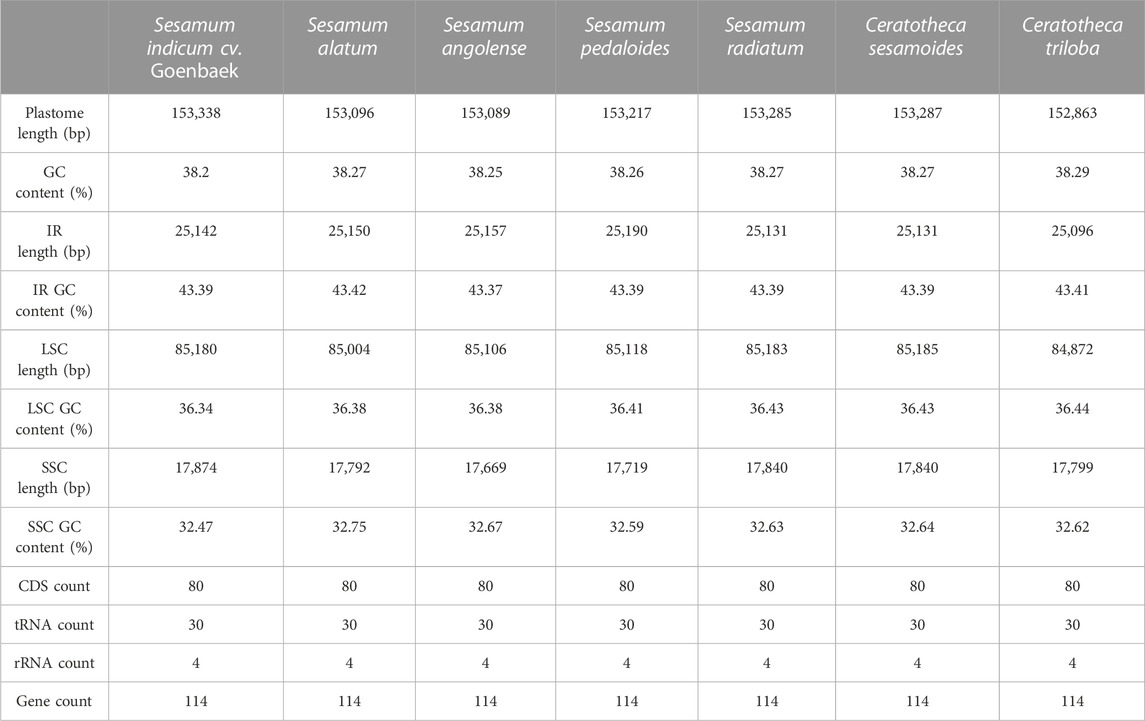
The assembled chloroplast genomes resulted in a single circular quadripartite genome with two typical IRs separated by LSC and SSC (Figure 2). The plastome sizes were 153,089 bp, 153,096 bp, 153,217 bp, 153,285 bp, 153,338 bp, 153,287 bp, and 152,863 bp for S. angolense, S. alatum, S. pedaloides, S. radiatum, S. indicum cv. Goenbaek, C. sesamoides, and C. triloba, respectively (Table 1). The seven genomes contained 114 unique genes, including 80 coding proteins, 30 transfer RNAs, and four ribosomal RNA genes, as previously found by Yi and Kim (2012) and Zhang et al. (2013) using S. indicum cv. Ansanggae and S. indicum cv. Yuzhi 11 as plant models, respectively. Among the assembled chloroplast genomes, 10 coding sequence genes (atpF, rpoC1, rps12, petB, petD, rps16, rpl2, rpl16, ndhA, and ndhB) harbor a single intron, while two coding genes (clpP and ycf3) contain two introns. The GC contents were similar in IR (43.39% ± 0.01%), LSC (36.40% ± 0.03%), SSC (32.62% ± 0.08%), and whole-chloroplast genome scales (38.26% ± 0.03%) (Table 1).
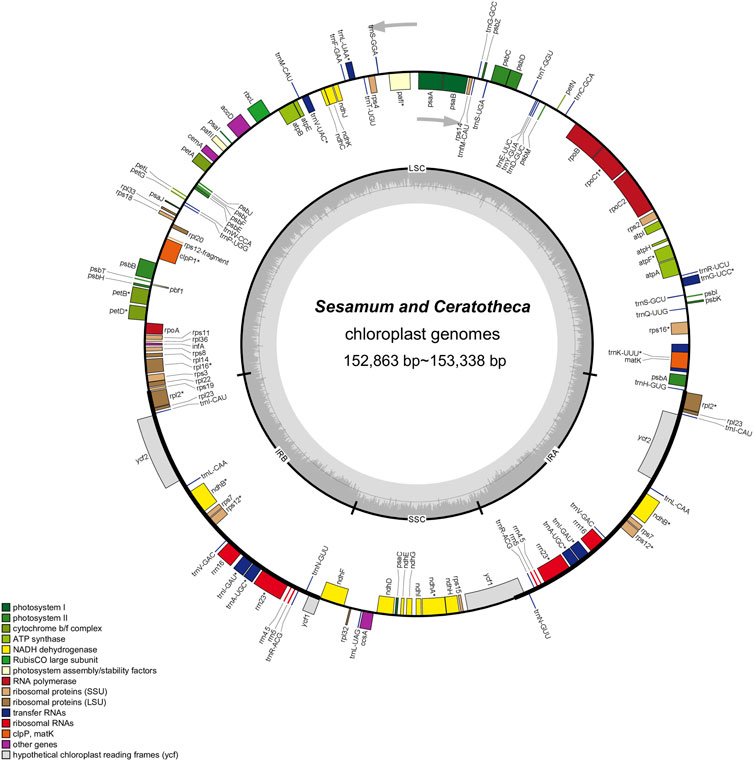
FIGURE 2. Chloroplast genome map of Sesamum and Ceratotheca species. Intron-containing genes are marked with the asterisk symbol (*). The dashed gray area in the inner circular map indicates the GC content. Different functional gene categories are colored as indicated in the lower part of the genome map.
Phylogenetic relationships between Sesamum and Ceratotheca species
To figure out the evolutionary relationship among Sesamum, Ceratotheca, and the closest related species belonging to Acanthaceae and Linderniaceae, a tree (Figure 3) was constructed using 75 shared protein sequences. Vitis vinifera was employed as an outgroup. As expected, Sesamum and Ceratotheca representatives clustered in a single clade, resolving the sister species relationship. A close view of the Pedaliaceae clade highlighted four major sub-clades. The first sub-clade represents a mixture of two ploidy levels with S. radiatum (2n = 64), S. angolense (2n = 32), and C. sesamoides (2n = 32). Interestingly, the second sub-clade encompassed S. pedaloides (2n = unknown) and C. triloba (2n = 32), suggesting that the ploidy of S. pedaloides might also be 2n = 32. The cultivated species S. indicum (2n = 26) is grouped into one sub-clade, while the wild relative S. alatum (2n = 26) constitutes a single sub-clade as being genetically distant from the cultivars and other wild relatives.
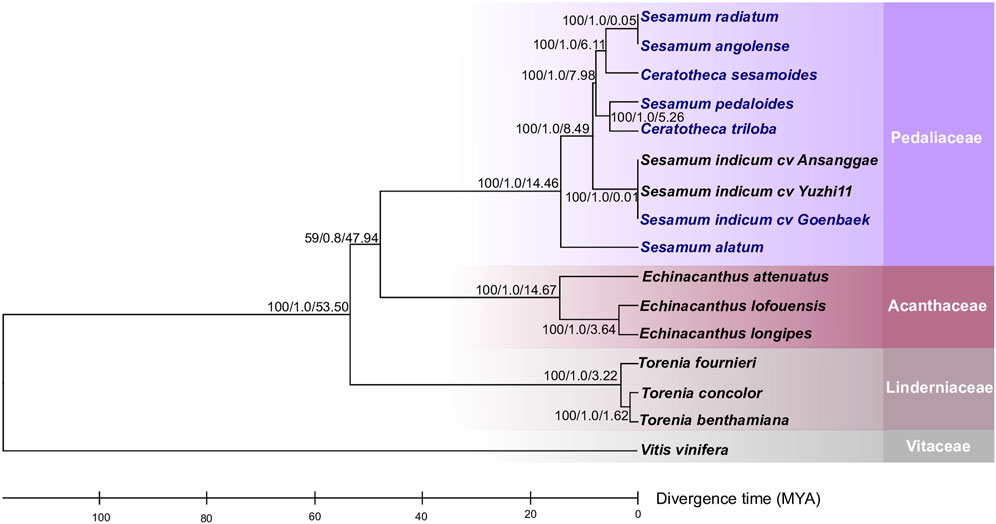
FIGURE 3. Phylogeny placement of newly constructed chloroplast genomes within the Lamiales members. The phylogenetic tree was inferred from the maximum-likelihood and Bayesian analyses of 75 conserved coding genes. The closest Lamiales species cover Acanthaceae and Linderniaceae species. Vitis vinifera was set as an outgroup. The numbers above the nodes are support values with maximum-likelihood bootstrap values on the left, Bayesian bootstrap values in the middle, and divergence times (in millions of years) estimated using the RelTime approach implemented using MEGA X software. The newly assembled chloroplast genomes are indicated in blue.
Divergence time estimation
In order to understand the speciation occurrence time among Sesamum and Ceratotheca species, a time divergence analysis was performed (Figure 3). First, S. alatum was estimated to have occurred 14.46 million years ago (Mya). Second, S. pedaloides and C. triloba split concomitantly approximately 5.26 Mya. Third, C. sesamoides was inferred to have occurred 6.11 Mya, a little bit earlier than S. pedaloides and C. triloba. As expected, the cultivated type (S. indicum) occurred lastly at 0.01 Mya. Interestingly, the time divergence inference revealed that S. radiatum (2n = 64) and C. sesamoides (2n = 32) have recently concomitantly occurred at approximately 0.05 Mya.
Comparative plastome analysis
The chloroplast genome structure is highly conserved (Figure 4A), although the morphological features of the species are distinct (Figure 4B). When comparing the plastid genome structure using mVISTA (Shuffle-LAGAN) (Supplementary Figure S1), Mauve (progressiveMauve alignment) (Supplementary Figure S2), and AliTV (LASTZ all-vs-all alignment) (Figure 4A), the conservative genome structure in coding and non-coding regions was also confirmed. However, IR contraction and expansion has been revealed with a length ranging from 25,096 bp to 25,190 bp, while LSC varied from 84,872 bp to 85,185 bp. SSC was sized from 17,719 bp to 17,874 bp.
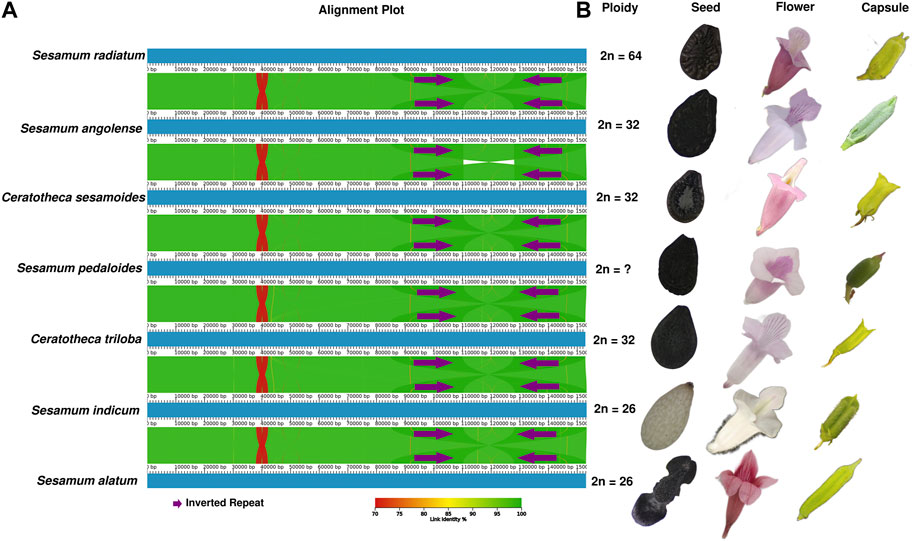
FIGURE 4. Morphological and plastome-based comparative analyses between Sesamum and Ceratotheca species. The alignment view of chloroplast genomes using the all-vs-all alignment approach implemented in AliTV software (A). The purple arrows represent the IR region. Each genome is colored following the link identity percentage as shown in the right corner. The ploidy, seed capsule, and flower photos were added to the figure (B).
By comparing chloroplast genome boundaries of Sesamum and Ceratotheca species, we noted that the IRb/LSC junction is sited between rpl2 and rsp19 genes (Figure 5). The pseudogene ycf1 is located exclusively in IRb for S. indicum cv. Ansanggae, S. indicum cv. Yuzhi 11, and at the border of the IRb/SSC of other species. The ndhF gene is mainly found in the SSC region for all species, except for S. angolense. The ycf1 gene of all species was located at SSC/IRa junctions with a gene size ranging from 5,312 to 5,369 bp. The trnH gene was localized in the LSC region, 1–16 bp away from the IRa–LSC border. Overall, the chloroplast genome structure at different junctions was highly conserved among Sesamum and Ceratotheca species.

FIGURE 5. Chloroplast junction site view showing gene distribution alongside the boundaries and IR contraction and expansion within the Sesamum and Ceratotheca species. Arabidopsis thaliana was set as the outgroup.
Variation hotspots within chloroplast genomes of Sesamum and Ceratotheca species
Despite the high collinearity of the chloroplast genome within Sesamum and Ceratotheca species, substantial variations were noted mainly in SSC regions (Figure 6). The nucleotide diversity calculation revealed a peak value located in ycf1, followed by ndhA, ndhE, psaC–ndhD, and ndhF regions. To estimate their discriminatory power, we inferred the phylogenetic tree using each gene (Figures 7A–E). As a result, only ndhF clearly distinguished the taxa (Figure 7E), as depicted previously with a set of 75 (Figure 3) protein-coding genes. Therefore, ndhF could be used as a marker to delineate Sesamum and Ceratotheca species, since several species are not yet well characterized at both morphological and cytogenetic levels.
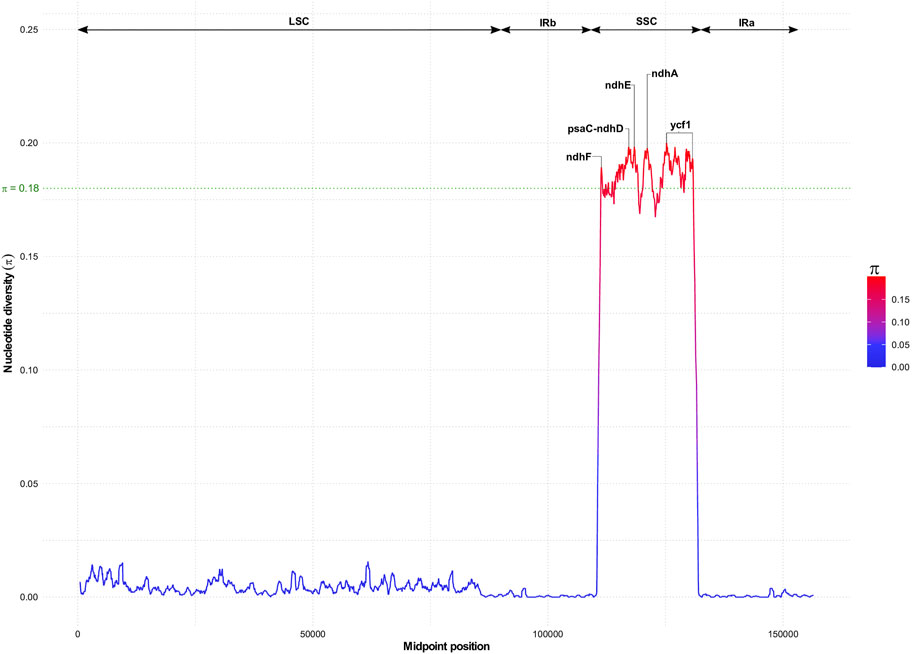
FIGURE 6. Nucleotide diversity variation with chloroplast genomes of Sesamum and Ceratotheca species. Highest values above 0.18 (in green) indicate candidate genes for the population genetics purpose.
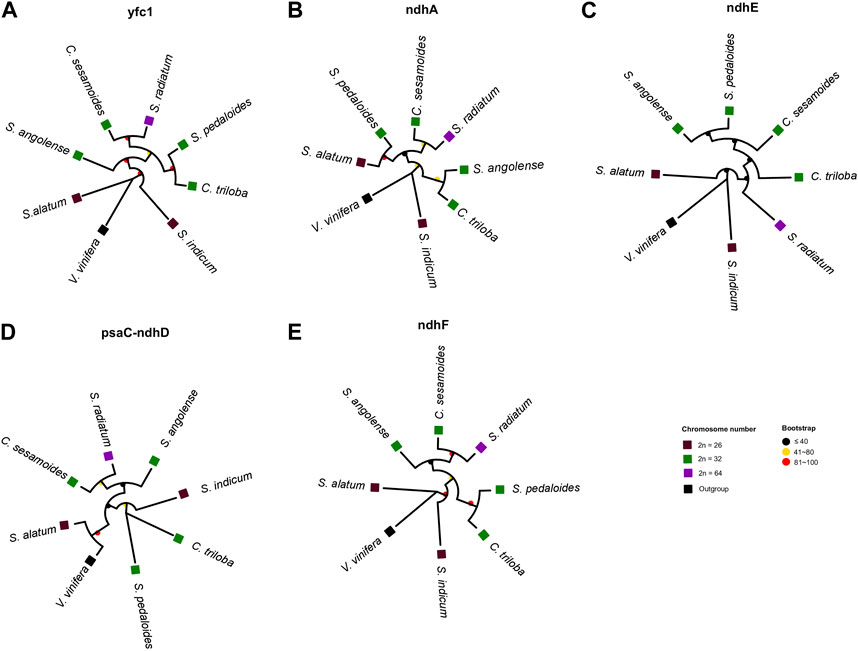
FIGURE 7. Evaluation of the discriminatory power of candidate regions, namely ycf1 (A), ndhA (B), ndhE (C), psaC-ndhD (D), and ndhF (E). The tree topology of ndhF (E) was consistent with the maximum-likelihood phylogenetic tree based on 75 coding genes. Discrepancies were noted for other gene tree inferences. Vitis vinifera was set as an outgroup. Squares and circles were colored following the ploidy and bootstrap values, respectively.
Codon usage bias analysis
Codon usage bias was examined by computing the RSCU (Sharp and Cowe, 1991). The RSCU represents the observed frequency of a codon divided by the expected frequency. A lack of bias referred to codons with RSCU values close to 1. Globally, a slight variation in the RSCU was found within Sesamum and Ceratotheca species (Supplementary Figure S3) values among Sesamum and Ceratotheca species. A total of 27 core codons exhibited RSCU >1, of which 24 were adenine/thymine-ending codons, one guanine-ending codon, and two cytosine-ending codons. In contrast, guanine- or cytosine-ending codons mostly exhibited RSCU <1. The most biased codon was found for the stop codon TAA (RSCU = 1.55 ± 0.01), while a less biased codon was detected for the stop codon TAG (RSCU = 0.69 ± 0.01). A similar trend of A–T bias in codon usage has been observed in other plant species (Wang et al., 2017; Biju et al., 2019).
Interestingly, by examining the species cluster from the heat map, the phylogenetic tree topology is concordant with the coding sequence-based tree inference, indicating a robust estimation of the phylogenetic relationship based on codon usage, as observed in a wide range of families (Gao et al., 2019; Chi et al., 2020; Wu et al., 2021).
Long and simple sequence repeats
Long repeats constitute a driving force for chloroplast genome rearrangement and have been used for phylogenetic inferences between species (Park et al., 2017; Luo et al., 2021). They induce genetic diversity by promoting intermolecular recombination in the chloroplast genome (Park et al., 2018). Long repeats encompass forward, reverse, palindrome, and complement types. In the present study, the mean count of long repeats was 25.71 ± 3.24 bp. The number of long repeats ranged from 21 to 31, among which palindromic (12–18) and forward (9–13) types were the most abundant. Moreover, the size of the repeats was mainly within the range of 30–39 bp. Only S. indicum exhibited repeats in the range of 60–69 bp (Supplementary Figure S4).
Microsatellites are referred to short tandem repeat sequences of one to six nucleotide repeats (Fan and Chu, 2007). SSRs are widely present in the chloroplast genome and have been extensively used as molecular markers for population genetics, phylogenetic relationship inferences, and species identification (Powell et al., 1995; Huang et al., 2018; Lee et al., 2019; Li et al., 2020). We detected 21–32 chloroplast SSRs within the assembled chloroplast genomes (Supplementary Figure S5A; Supplementary Table S4), of which most are monomeric (>87%) (Supplementary Figure S5B). The majority (13–25) of SSRs are located in LSC sequences (Supplementary Figure S5C) and mainly in intergenic regions (>76%) for all species (Supplementary Figure S5D). The most dominant motif (A) count ranged from 19 to 27 and spanned 208–277 bp (Supplementary Figure S6). Trinucleotide repeats (AAT) were only detected for S. alatum occupying 30 bp of the chloroplast genome length (Supplementary Figure S6).
Selection pressure analysis
The pairwise ratio of non-synonymous substitutions (Ka) to the rate of synonymous substitutions (Ks) analysis is presented in Supplementary Figure S7. Ka/Ks ratios with Ks <0.1 or K >0.2 were changed to zero for a reliable estimation of the selection pressure. The results revealed that the NAD(P)H-quinone oxidoreductase subunit I (ndhI) has undergone strong positive selection in S. angolense and S. radiatum. Similar trends were observed for the photosystem I gene ycf4, mainly in S. radiatum, S. alatum, and C. sesamoides. The rpl20 gene also exhibited positive selection only in S. alatum. However, matK, ndhF, and ycf1 Ka/Ks values were approximately equal to 1, implying neutral selection pressure.
RNA editing analysis
RNA editing is an important biological phenomenon that plays a crucial role in the regulation of gene expression, the diversification of proteins, and adaptation to environmental changes (Mohammed et al., 2022). From the seven chloroplast genomes, we predicted 140 ± 5 RNA editing sites, which were found in 27 ± 1 genes (Supplementary Tables S5–S11). The psaB gene contained the highest number (16) of editing sites within the species. Meanwhile, the rpoB gene was predicted to have an average of 14 editing sites, while ndhD and ndhF genes exhibited 10 editing sites, respectively. The abundant editing site conversion within species was C-to-U transition, which is primarily found in a majority of plant chloroplast genomes (Hao et al., 2021). Globally, the change from proline to leucine was most frequent followed by thymine to inosine, proline to serine, and serine to leucine (Supplementary Tables S5–S11).
Discussion
Plastome evolution between the Sesamum and Ceratotheca genus
We reported for the first time, whole-chloroplast genomes of six African native wild sesame species: S. alatum, S. angolense, S. pedaloides, S. radiatum, C. sesamoides, and C. triloba. The information from the generated plastome sequences served as the basis for comparative analyses. A typical quadripartite chloroplast structure, including LSC, SSC, and two IRs, was observed. In-depth comparative analyses revealed contraction and expansion events within all species. IR expansion and contraction is a common phenomenon observed in land plants resulting in the variation of chloroplast length at both intra- and inter-species levels (Asaf et al., 2020; Guo et al., 2021). As expected, the chloroplast genome was highly conserved among Sesamum and Ceratotheca, despite the morphological differences. The conserved structure is consistent with the two previously published S. indicum chloroplast genomes (Yi and Kim, 2012; Zhang et al., 2013).
The variation of microsatellite copy numbers in the chloroplast genome is helpful for population genetics and polymorphism assessment. For instance, microsatellites with one nucleotide motif dataset provided in the current study constitute a useful resource for further population polymorphism assessment within Sesamum, Ceratotheca, and potentially close relative species in Pedaliaceae. Palindromic repeats are known to constitute mutational hotspots contributing to plastome expansion (Smith, 2020). By mining the chloroplast genomes, we detected that palindromic repeats are prominent in all chloroplast genomes. Therefore, they represent a suitable resource for marker development in regard to genetic diversity investigation in the Sesamum species continuum.
Chloroplast genes generally evolved under purifying selection, mainly to maintain the functional continuity of genes over a long period of time (Matsuoka et al., 2002; Jiang et al., 2018). However, previous comparative plastome studies identified some genes that underwent positive selection, including the photosynthetic genes rbcL (Ivanova et al., 2017) and ycf2 (Jiang et al., 2018), among others.
In our study, mainly photosynthesis-related genes including ndhA, ndhI, and ycf4 exhibited strong positive selection. Subsequently, owing to the specific distribution patterns of the studied samples in tropical Africa (See Figure 1) and the drought-prone habitat preference in nature, we postulate that the selection of this category of genes might be related to adaptation to environmental changes including the photosynthetic rate, drought, temperature, carbon dioxide level, or ecological niche (Piot et al., 2018). Moreover, the capability of photosynthetic-oriented gene selection may contribute to the drought tolerance strength of the cultivated sesame S. indicum, as revealed by previous extensive functional genomic studies (Dossa et al., 2019; Yu et al., 2019).
A high-resolution view for the delineation of Pedaliaceae species in the sesame speciation continuum
Delineating species is a challenging aspect in taxonomy, since the methodology is quite heterogeneous depending not only on the taxa but also on the scientists. The Pedaliaceae s.l. family encompasses several tribes, including Sesamothamneae Ihlenf., Sesameae (Endl.) Meisn., and Pedaliae Dumort. Using plastid and nuclear markers, Gormley et al. (2015) provided evidence of the monophyletic pattern of these tribes. However, the authors highlighted that Sesamum is paraphyletic in regards with the Ceratotheca, Josephinia, and Dicerocaryum genus. This latter observation is in contrast with our results that showed that Sesamum and Ceratotheca formed a complex. The low number of markers and the used taxa in the previous study might explain the divergence of the tree topology. In fact, there were the absence of 2n = 64 chromosome set representatives. Therefore, our study provided the first insight regarding the chromosome number variation criterion.
The chloroplast genome is generally well conserved in land plants (Trösch et al., 2018). Despite the highly conserved chloroplast genome arrangement within both genera, remarkable sequence divergence was noted in several genes, including ndhF gene. The ndhF-based tree topology was consistent with the protein-coding gene tree, confirming that ndhF is a powerful candidate gene with promising potential for the DNA barcoding purpose.
Interestingly, a relatively long branch of S. alatum was observed implying its long evolutionary occurrence compared to that of the other species, which is consistent with the tree topology from Gormley et al. (2015), biogeographical and morphological data (Bedigian, 2018). It is noteworthy that S. alatum seeds exhibit a singular characteristic with winged-seeds, which is absent in the other species. The presence of wings is one of the key ecological adaptive trait of ancient wild crops that ensures seed dispersal by wind (Willson and Traveset, 2000). To the best of our knowledge, this trait seems to have been lost during evolution in the Sesamum genus, since no other member (described so far) of the genus harbors it. Moreover, the time divergence estimation revealed that Sesamum alatum diverged earlier (14.46 Mya) than other species, supporting the previous findings.
Implications of the systematic placement of Ceratotheca in the Sesamum tribe (Sesameae)
In the Sesamum tribe, the classification of species has evolved following the descriptor. From the chloroplast genome analyses, we postulate that the nomenclature C. sesamoides and C. triloba might change into Sesamum sesamoides and Sesamum trilobum as suggested by Bruce (1953) and POWO (2022). Consequently, the section Ceratotheca (Endl.) J.C. Manning and Magee might be merged into the section Sesamum.
Conclusion
The chloroplast genomes were highly conserved with respect to gene orientation, GC content, and gene content. However, the divergent sequences within species were detected in ycf1, ndhA, ndhE, and ndhF coding genes. The tree topology showed that Sesamum and Ceratotheca species were confidently resolved as sister species. Ultimately, the chloroplast sequence data from this study lay the foundation for the development of DNA barcoding markers and species-centered genomic research.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YZ and KL designed this study. YZ carried out the laboratory and bioinformatics steps. YZ wrote the initial manuscript. ST, YM, B-OA, J-GK, KL, and YZ edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The present research was supported by the Rural Development Administration under project no. PJ013470, Rural Development Administration, South Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1207306/full#supplementary-material
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi:10.1038/nprot.2006.384
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi:10.1093/bioinformatics/bty220
Ankenbrand, M. J., Hohlfeld, S., Hackl, T., and Förster, F. (2017). AliTV—Interactive visualization of whole genome comparisons. PeerJ Comput. Sci. 3, e116. doi:10.7717/peerj-cs.116
Asaf, S., Khan, A. L., Lubna, , , Khan, A., Khan, A., Khan, G., et al. (2020). Expanded inverted repeat region with large scale inversion in the first complete plastid genome sequence of Plantago ovata. Sci. Rep. 10, 3881. doi:10.1038/s41598-020-60803-y
Aworh, O. C. (2018). From lesser-known to super vegetables: The growing profile of african traditional leafy vegetables in promoting food security and wellness. J. Sci. Food Agric. 98, 3609–3613. doi:10.1002/jsfa.8902
Bedigian, D., Seigler, D. S., and Harlan, J. R. (1985). Sesamin, sesamolin and the origin of sesame. Biochem. Syst. Ecol. 13, 133–139. doi:10.1016/0305-1978(85)90071-7
Bedigian, D. (2003). Evolution of sesame revisited: Domestication, diversity and prospects. Genet. Resour. Crop Evol. 50, 779–787. doi:10.1023/A:1025029903549
Bedigian, D. (2014). A new combination for the Indian progenitor of sesame, Sesamum indicum (Pedaliaceae). Novon 23, 5–13. doi:10.3417/2012062
Bedigian, D. (2015). Systematics and evolution in Sesamum L. (Pedaliaceae), part 1: Evidence regarding the origin of sesame and its closest relatives. Webbia J. Plant Taxon. Geogr. 70, 1–42. doi:10.1080/00837792.2014.968457
Bedigian, D. (2018). Feeding the forgotten: Wild and cultivated Ceratotheca and Sesamum (Pedaliaceae) that nourish and provide remedies in Africa. Econ. Bot. 72, 496–542. doi:10.1007/s12231-018-9437-z
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-Web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi:10.1093/bioinformatics/btx198
Biju, V. C., Shidhi, P. R. S., Vijayan, S., Rajan, V. S., Sasi, A., Janardhanan, A., et al. (2019). The complete chloroplast genome of Trichopus zeylanicus, and phylogenetic analysis with dioscoreales. Plant Genome 12, 190032. doi:10.3835/plantgenome2019.04.0032
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi:10.1093/bioinformatics/btp348
Chan, P. P., and Lowe, T. M. (2019). “tRNAscan-SE: Searching for tRNA genes in genomic sequences,” in Gene prediction: Methods and protocols, methods in molecular biology. Editor K. Martin, 1–14. doi:10.1007/978-1-4939-9173-0_1
Chi, X., Zhang, F., Dong, Q., and Chen, S. (2020). Insights into comparative genomics, codon usage bias, and phylogenetic relationship of species from biebersteiniaceae and nitrariaceae based on complete chloroplast genomes. Plants 9, 1605. doi:10.3390/plants9111605
Cronquist, A. (1981). An integrated system of classification of flowering plants. New York: Colombia University Press.
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi:10.1101/gr.2289704
Dossa, K., Li, D., Zhou, R., Yu, J., Wang, L., Zhang, Y., et al. (2019). The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotechnol. J. 17, 1788–1803. doi:10.1111/pbi.13100
Fan, H., and Chu, J. Y. (2007). A brief review of short tandem repeat mutation. Genomics, Proteomics Bioinforma. 5, 7–14. doi:10.1016/S1672-0229(07)60009-6
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). Vista: Computational tools for comparative genomics. Nucleic Acids Res. 32, 273–279. doi:10.1093/nar/gkh458
Gao, B., Yuan, L., Tang, T., Hou, J., Pan, K., and Wei, N. (2019). The complete chloroplast genome sequence of Alpinia oxyphylla Miq. and comparison analysis within the Zingiberaceae family. PLoS One 14, e0218817. doi:10.1371/journal.pone.0218817
Gormley, I. C., Bedigian, D., and Olmstead, R. G. (2015). Phylogeny of Pedaliaceae and martyniaceae and the placement of trapella in plantaginaceae s. L. Syst. Bot. 40, 259–268. doi:10.1600/036364415x686558
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi:10.1093/nar/gkz238
Guo, Y.-Y., Yang, J.-X., Bai, M.-Z., Zhang, G.-Q., and Liu, Z.-J. (2021). The chloroplast genome evolution of venus slipper (paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 21, 248. doi:10.1186/s12870-021-03053-y
Hao, W., Liu, G., Wang, W., Shen, W., Zhao, Y., Sun, J., et al. (2021). RNA editing and its roles in plant organelles. Front. Genet. 12, 757109. doi:10.3389/fgene.2021.757109
Hsu, E., and Parthasarathy, S. (2017). Anti-inflammatory and antioxidant effects of sesame oil on atherosclerosis: A descriptive literature review. Cureus 9, e1438. doi:10.7759/cureus.1438
Huang, L. S., Sun, Y. Q., Jin, Y., Gao, Q., Hu, X. G., Gao, F. L., et al. (2018). Development of high transferability cpSSR markers for individual identification and genetic investigation in Cupressaceae species. Ecol. Evol. 8, 4967–4977. doi:10.1002/ece3.4053
Huelsenbeck, J. P., and Ronquist, F. (2001). Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi:10.1093/bioinformatics/17.8.754
IPGRI, and NBPGR (2004). Descriptors of sesame (Sesamum spp). Rome, Italy: International Plant Genetic Resources Institute, and National Bureau of Plant Genetic Resources, New Delhi, India.
Ivanova, Z., Sablok, G., Daskalova, E., Zahmanova, G., Apostolova, E., Yahubyan, G., et al. (2017). Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 8, 204. doi:10.3389/fpls.2017.00204
Jiang, P., Shi, F.-X., Li, M.-R., Liu, B., Wen, J., Xiao, H.-X., et al. (2018). Positive selection driving cytoplasmic genome evolution of the medicinally important ginseng plant genus panax. Front. Plant Sci. 9, 359. doi:10.3389/fpls.2018.00359
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., DePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi:10.1186/s13059-020-02154-5
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi:10.1093/molbev/mst010
Kirchhoff, H. (2019). Chloroplast ultrastructure in plants. New Phytol. 223, 565–574. doi:10.1111/nph.15730
Kobayashi, T. (1991). “Cytogenetics of sesame (Sesamum indicum),” in Chromosome engineering in plants genetics, breeding, evolution, Part B. Editors T. Tsuchiya, and P. K. Gupta (Elsevier B.V.), 581–592. doi:10.1016/B978-0-444-88260-8.50036-7
Köhler, M., Reginato, M., Souza-Chies, T. T., and Majure, L. C. (2020). Insights into chloroplast genome evolution across opuntioideae (cactaceae) reveals robust yet sometimes conflicting phylogenetic topologies. Front. Plant Sci. 11, 729. doi:10.3389/fpls.2020.00729
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi:10.1093/molbev/msx116
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi:10.1093/nar/29.22.4633
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi:10.1093/nar/gkh152
Lee, K. J., Raveendar, S., Choi, J. S., Gil, J., Lee, J. H., So, Y. S., et al. (2019). Development of chloroplast microsatellite markers for identification of Glycyrrhiza species. Plant Genet. Resour. Characterisation Util. 17, 95–98. doi:10.1017/S1479262118000308
Li, C., Zheng, Y., and Huang, P. (2020). Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci. Rep. 10, 12188. doi:10.1038/s41598-020-68092-1
Luo, C., Huang, W., Sun, H., Yer, H., Li, X., Li, Y., et al. (2021). Comparative chloroplast genome analysis of Impatiens species (Balsaminaceae) in the karst area of China: Insights into genome evolution and phylogenomic implications. BMC Genomics 22, 571. doi:10.1186/s12864-021-07807-8
Manning, J. C., and Magee, A. R. (2018). Additional new combinations in Sesamum L. (Pedaliaceae: Sesameae). Bothalia 48, 1–2. doi:10.4102/abc.v48i1.2363
Matsuoka, Y., Yamazaki, Y., Ogihara, Y., and Tsunewaki, K. (2002). Whole chloroplast genome comparison of rice, maize, and wheat: Implications for chloroplast gene diversification and phylogeny of cereals. Mol. Biol. Evol. 19, 2084–2091. doi:10.1093/oxfordjournals.molbev.a004033
Mello, B. (2018). Estimating TimeTrees with MEGA and the TimeTree resource. Mol. Biol. Evol. 35, 2334–2342. doi:10.1093/molbev/msy133
Minh, B. Q., Nguyen, M. A. T., and Von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi:10.1093/molbev/mst024
Mohammed, T., Firoz, A., and Ramadan, A. M. (2022). RNA editing in chloroplast: Advancements and opportunities. Curr. Issues Mol. Biol. 44, 5593–5604. doi:10.3390/cimb44110379
Mower, J. P. (2009). The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 37, 253–259. doi:10.1093/nar/gkp337
Nayar, N. M., and Mehra, K. L. (1970). Sesame: Its uses, botany, cytogenetics, and origin. Econ. Bot. 24, 20–31. doi:10.1007/BF02860629
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Ntwenya, J. E., Kinabo, J., Msuya, J., Mamiro, P., Mamiro, D., Njoghomi, E., et al. (2017). Rich food biodiversity amid low consumption of food items in kilosa district, Tanzania. Food Nutr. Bull. 38, 501–511. doi:10.1177/0379572117708647
Park, I., Yang, S., Choi, G., Jin Kim, W., and Moon, B. C. (2017). The complete chloroplast genome sequences of Aconitum pseudolaeve and Aconitum longecassidatum, and development of molecular markers for distinguishing species in the Aconitum subgenus lycoctonum. Molecules 22, 2012. doi:10.3390/molecules22112012
Park, I., Yang, S., Kim, W. J., Noh, P., Lee, H. O., and Moon, B. C. (2018). The complete chloroplast genomes of six Ipomoea species and indel marker development for the discrimination of authentic Pharbitidis semen (Seeds of I. nil or I. Purpurea). Front. Plant Sci. 9, 965. doi:10.3389/fpls.2018.00965
Patil, C. G., and Hiremath, S. C. (2002). Genome relations among octaploid species of Sesamum L. (Pedaliaceae). Cytol. (Tokyo) 67, 403–409. doi:10.1508/cytologia.67.403
Patil, C. G., and Hiremath, S. C. (2004). Karyotypic studies in octaploid species of Sesamum L. J. Cytol. Genet. 5, 73–76.
Piot, A., Hackel, J., Christin, P.-A., and Besnard, G. (2018). One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 247, 255–266. doi:10.1007/s00425-017-2781-x
Powell, W., Morgante, M., McDevitt, R., Vendramin, G. G., and Rafalski, J. A. (1995). Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. U. S. A. 92, 7759–7763. doi:10.1073/pnas.92.17.7759
POWO (2022). Plants of the World online. Facilitated by the royal botanic gardens, kew. POWO. Published on the Internet: http://www.plantsoftheworldonline.org/ (Retrieved January 1, 2022).
Raghavan, T. S., and Krishnamurthy, K. V. (1947). Cytogenetical studies in Sesamum. Part I. Cytology of the parents, Sesamum orientale Linn, and Sesamum prostratum Retz. and the cytology of the sterile hybrid between them and of the fertile amphidiploid. Proc. Indian Acad. Sci. - Sect. B 26, 236–275. doi:10.1007/BF03051810
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Sharp, P. M., and Cowe, E. (1991). Synonymous codon usage in Saccharomyces cerevisiae. Yeast 7, 657–678. doi:10.1002/yea.320070702
Smith, D. R. (2020). Can green algal plastid genome size Be explained by DNA repair mechanisms? Genome Biol. Evol. 12, 3797–3802. doi:10.1093/gbe/evaa012
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi:10.1093/nar/gkl315
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi:10.1093/nar/gkx391
Trösch, R., Barahimipour, R., Gao, Y., Badillo-Corona, J. A., Gotsmann, V. L., Zimmer, D., et al. (2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants 4, 564–575. doi:10.1038/s41477-018-0211-0
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—New capabilities and interfaces. Nucleic Acids Res. 40, e115. doi:10.1093/nar/gks596
Visavadiya, N. P., and Narasimhacharya, A. V. R. L. (2008). Sesame as a hypocholesteraemic and antioxidant dietary component. Food Chem. Toxicol. 46, 1889–1895. doi:10.1016/j.fct.2008.01.012
Wang, W., Yu, H., Wang, J., Lei, W., Gao, J., Qiu, X., et al. (2017). The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (oleaceae). Int. J. Mol. Sci. 18, 2288. doi:10.3390/ijms18112288
Willson, M. F., and Traveset, A. (2000). “The ecology of seed dispersal,” in Seeds. The ecology of regeneration in plant communities. Editor M. Fenner (Wallingford, UK: CABI Publishing), 410. doi:10.1079/SSR2003142
Wu, L., Nie, L., Wang, Q., Xu, Z., Wang, Y., He, C., et al. (2021). Comparative and phylogenetic analyses of the chloroplast genomes of species of Paeoniaceae. Sci. Rep. 11, 14643. doi:10.1038/s41598-021-94137-0
Yang, Z. (1997). Paml: A program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13, 555–556. doi:10.1093/bioinformatics/13.5.555
Yi, D. K., and Kim, K. J. (2012). Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One 7, e35872. doi:10.1371/journal.pone.0035872
Yu, J., Golicz, A. A., Lu, K., Dossa, K., Zhang, Y., Chen, J., et al. (2019). Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol. J. 17, 881–892. doi:10.1111/pbi.13022
Zhang, H., Li, C., Miao, H., and Xiong, S. (2013). Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PLoS One 8, e80508. doi:10.1371/journal.pone.0080508
Keywords: phylogeny, chloroplast genome, Sesamum, Ceratotheca, wild relatives, species complex
Citation: Zoclanclounon YAB, Thamilarasan SK, Mo Y, Ahn B-O, Kim J-G and Lee K (2023) Insights into chloroplast genome structure and phylogenetic relationships within the Sesamum species complex (Pedaliaceae). Front. Genet. 14:1207306. doi: 10.3389/fgene.2023.1207306
Received: 17 April 2023; Accepted: 15 May 2023;
Published: 30 May 2023.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Senouwa Segla Koffi Dossou, Chinese Academy of Agricultural Sciences, ChinaDiaga Diouf, Cheikh Anta Diop University, Senegal
Copyright © 2023 Zoclanclounon, Thamilarasan, Mo, Ahn, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong-Gu Kim, amtpbTVhdWdAa29yZWEua3I=; Keunpyo Lee, a3BsZWVAa29yZWEua3I=
 Yedomon Ange Bovys Zoclanclounon
Yedomon Ange Bovys Zoclanclounon Senthil Kumar Thamilarasan
Senthil Kumar Thamilarasan Youngjun Mo2
Youngjun Mo2 Jeong-Gu Kim
Jeong-Gu Kim Keunpyo Lee
Keunpyo Lee