
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Genet., 30 May 2023
Sec. RNA
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1201934
This article is part of the Research TopicApplications of RNA-Seq in Cancer and Tumor ResearchView all 12 articles
 Zheyu Niu
Zheyu Niu Xin Gao
Xin Gao Zhaozhi Xia
Zhaozhi Xia Shuchao Zhao
Shuchao Zhao Hongrui Sun
Hongrui Sun Heng Wang
Heng Wang Meng Liu
Meng Liu Xiaohan Kong
Xiaohan Kong Chaoqun Ma
Chaoqun Ma Huaqiang Zhu
Huaqiang Zhu Hengjun Gao
Hengjun Gao Qinggong Liu
Qinggong Liu Faji Yang
Faji Yang Xie Song
Xie Song Jun Lu
Jun Lu Xu Zhou*
Xu Zhou*MicroRNAs (miRNAs) play a crucial role in various biological processes and human diseases, and are considered as therapeutic targets for small molecules (SMs). Due to the time-consuming and expensive biological experiments required to validate SM-miRNA associations, there is an urgent need to develop new computational models to predict novel SM-miRNA associations. The rapid development of end-to-end deep learning models and the introduction of ensemble learning ideas provide us with new solutions. Based on the idea of ensemble learning, we integrate graph neural networks (GNNs) and convolutional neural networks (CNNs) to propose a miRNA and small molecule association prediction model (GCNNMMA). Firstly, we use GNNs to effectively learn the molecular structure graph data of small molecule drugs, while using CNNs to learn the sequence data of miRNAs. Secondly, since the black-box effect of deep learning models makes them difficult to analyze and interpret, we introduce attention mechanisms to address this issue. Finally, the neural attention mechanism allows the CNNs model to learn the sequence data of miRNAs to determine the weight of sub-sequences in miRNAs, and then predict the association between miRNAs and small molecule drugs. To evaluate the effectiveness of GCNNMMA, we implement two different cross-validation (CV) methods based on two different datasets. Experimental results show that the cross-validation results of GCNNMMA on both datasets are better than those of other comparison models. In a case study, Fluorouracil was found to be associated with five different miRNAs in the top 10 predicted associations, and published experimental literature confirmed that Fluorouracil is a metabolic inhibitor used to treat liver cancer, breast cancer, and other tumors. Therefore, GCNNMMA is an effective tool for mining the relationship between small molecule drugs and miRNAs relevant to diseases.
With the development of sequencing technology, the biomedical field has accumulated a large amount of medical data, which provides more convenience for researchers to study the relationship between diseases and drugs using these data. The prediction of the relationship between small molecule (SM) drugs and microRNAs (miRNAs) has become an important and rapidly developing area in pharmacology and pharmacogenomics research (Bartel, 2004; Beermann et al., 2016; Kozomara et al., 2019; Liu et al., 2022). miRNAs are small non-coding RNA molecules that regulate gene expression and play a key role in various biological processes, including the development of diseases (Cai et al., 2021; Peng et al., 2023). On the other hand, small molecule drugs have been widely used to treat diseases, but their impact on miRNA expression is not clear. However, there are still blind issues in using traditional biological experiments to identify small molecule drug-related miRNAs, which require a lot of experimental time and cost. With the increasing availability of large datasets, it is possible to predict the relationship between small molecule drugs and miRNAs and use this information to improve the efficacy and safety of drugs (Wang et al., 2019; Chen et al., 2020). This field has tremendous potential in discovering new therapeutic targets and developing personalized drugs (Chen et al., 2021; Liu et al., 2023; Xu et al., 2023).
Computational methods have played a crucial role in predicting the association between small molecule drugs and miRNAs (Xu et al., 2020; Zhang et al., 2023). As the available data on drugs and miRNAs continues to increase, various computational methods have been proposed to identify and predict their interactions. Lv et al. (2015) constructed a complete network by combining small molecule similarity networks, miRNA similarity networks, and known small molecule-miRNA association networks. They calculated the similarity of small molecules and miRNAs using a weighted combination strategy, and then used the RWR (Random Walk With Restart) algorithm to predict the potential associations between small molecule drugs and miRNAs. BNNRSMMA first defined a new matrix to represent the small molecule-miRNA heterogenous network using miRNA-miRNA similarity, small molecule-small molecule similarity, and known small molecule-miRNA associations. They then completed this matrix by minimizing its kernel parameter count and used alternating direction multiplication to further minimize the kernel parameter count and obtain prediction scores. They introduced a regularization term to tolerate noise in the integrated similarity. Wang et al. (2022a) proposed a novel dual-network collaborative matrix factorization (DCMF) method for predicting potential SM-miRNA associations. They first preprocessed the missing values in the SM-miRNA association matrix using the WKNKN method, and then constructed a matrix factorization model for the dual network to obtain feature matrices containing potential features of small molecules and miRNAs, respectively. Finally, the predicted SM-miRNA association score matrix was obtained by calculating the inner product of the two feature matrices. Li et al. (2016) proposed a network-based inference model for small molecule-miRNA networks (SMiR-NBI), which relies solely on known SM-miRNA associations. For a given SM, the initial resources are evenly allocated to its associated miRNAs. Then, the resources of each miRNA are allocated to all its associated SMs, and the resources are then redistributed from SMs to their associated miRNAs. The final resources obtained by the miRNAs reflect the likelihood of associations between the given SM and miRNAs. Guan et al. (2018) developed a new graphlet interaction-based inference model for predicting small molecule-miRNA associations (GISMMA). The complex relationships among SMs or miRNAs are described by graphlet interactions, which consist of 28 isomers. The association score for an SM-miRNA pair is calculated by counting the number of graphlet interactions. However, if neither the SM nor the miRNA has a known association, the model cannot predict the SM-miRNA association. Wang et al. (2022b) proposed an ensemble method for predicting small molecule-miRNA associations based on kernel ridge regression (EKRRSMMA). This method combines feature dimension reduction and ensemble learning to reveal potential SM-miRNA associations. Firstly, the authors constructed different feature subsets for SMs and miRNAs. Then, homogeneous base learners were trained on different feature subsets, and the average scores obtained from these base learners were used as the association scores for SM-miRNA pairs. Peng et al. (2022) proposed a new computational method based on deep autoencoder and scalable tree boosting model (DAESTB) to predict the associations between small molecules and miRNAs. Firstly, a high-dimensional feature matrix was constructed by integrating small molecule-small molecule similarity, miRNA-miRNA similarity, and known small molecule-miRNA associations. Secondly, the feature dimension of the integrated matrix was reduced using a deep autoencoder to obtain potential feature representations for each small molecule-miRNA pair. Finally, a scalable tree boosting model was used to predict potential associations between small molecules and miRNAs. Although these models have achieved promising results and played important roles in the development of computational methods for small molecule-miRNA association identification, they have certain issues or limitations: the experimental validation of small molecule-miRNA associations is very limited, and there are many negative associations. When performed on this noisy and sparse small molecule-miRNA association network, the predictors often detect many false negative associations.
Therefore, we propose a miRNA-molecule association prediction model (GCNNMMA) by integrating graph convolutional networks (GCNs) (Scarselli et al., 2008) and convolutional neural networks (CNNs) (Chen, 2015) (Figure 1). Firstly, GCNs are used to effectively learn the molecular structural graph data of small molecule drugs, and CNNs are used to learn the sequence data of miRNAs. Due to the black-box nature of deep learning models, it is difficult to analyze and interpret them. Therefore, GCNNMMA introduces a neural attention mechanism (Bahdanau et al., 2014) to address this issue. The neural attention mechanism enables CNNs to learn the weights of sub-sequences in miRNAs, thus predicting the associations between miRNAs and small molecule drugs.
For dataset 1, we obtained a total of 664 known small molecule-miRNA associations from SM2miR database (version 1.0) (Liu et al., 2013). Then a total of 831 small molecules were extracted and integrated from SM2miR, DrugBank (Wishart et al., 2018), and PubChem (Kim et al., 2019). 541 miRNAs were collected from SM2miR, HMDD, miR2Disease, and PhenomiR (Ruepp et al., 2010). To evaluate our model performance more comprehensively, we constructed dataset 2, which contains 680 small molecules, 2,460 miRNAs, and 60,212 known small molecule-miRNA associations. Additionally, we downloaded corresponding small molecule drug SMILES data from DrugBank. The SMILES format data was used to describe the spatial structural information of small molecule drugs. Furthermore, we obtained corresponding miRNA sequence data from the miRbase database (Table 1).
End-to-end learning model GNNs has been shown to achieve good performance in many scenarios. Therefore, we first use two functions [the transformation function
Subgraph embedding with radius
Where,
Vertex transformation function: In the molecular structure graph G, subgraph embedding can start from any vertex.
Where
Were,
The edge transformation function: The process of updating edge embeddings are similar to the process of updating vertex embeddings. Here,
The formula describes
Small molecule output function: To obtain the final output
First, CNNs use filter functions to compute a hidden vector
To apply CNNs to miRNA sequence data, First, miRNA sequences are defined as “words” consisting of
Using
Where
Multiple hidden vectors form a hidden vector set
miRNA sequence output function. In order to obtain the final output
GCNNMMA employs a neural attention mechanism to infer interactions between small molecules and subsequences in miRNA sequences. In the collection of hidden vector sequences
Where
Finally, the model obtains the final classification output vector
Where
In this work, we compared the performance of the latest five models [SMiR-NBI (Li et al., 2016), GISMMA (Guan et al., 2018), SLHGISMMA (Yin et al., 2019), SNMFSMMA (Zhao et al., 2020), EKRRSMMA (Wang et al., 2022b)] with GCNNMMA, and conducted 5-fold cross-validation (CV) on both dataset 1 and dataset 2 to evaluate the predictive performance of GCNNMMA. All predicted small molecule miRNA pairs were ranked according to the obtained scores. Based on the rankings, we used receiver operating characteristic (ROC) curves to illustrate the performance of our models in the cross-validation runs. As shown in Figure 4, we found that GCNNMMA achieved the best predictive performance on both dataset 1 (AUC = 0.9812) and dataset 2 (AUC = 0.9384). This suggests that GCNNMMA performed the best in predicting the correlation between small molecule drugs and miRNAs.
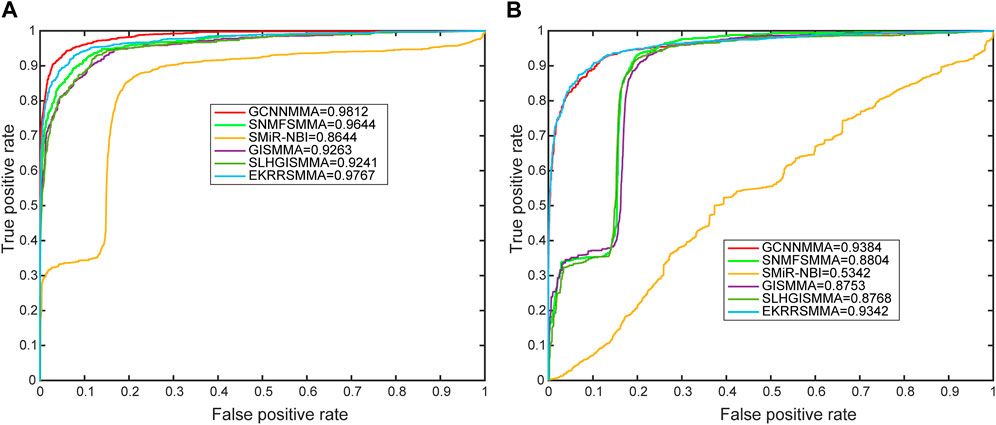
FIGURE 4. The ROC curves for GCNNMMA and benchmark algorithms for 5-fold CV on the (A) dataset 1 and (B) dataset 2.
It is important to examine the performance of the above method in predicting new miRNAs related to small molecule drugs, in addition to testing the performance of global prediction of small molecule drug-miRNA relationships. A leave-one-out experiment is used to evaluate the ability of the algorithm to predict miRNAs related to new small molecule drugs. To compare the fairness of the test, we still use ROC as the indicator of predictive performance. The local LOOCV experiment was carried on the dataset 1 and dataset 2 (see Figure 5). GCNNMMA showed a higher performance over other approaches in terms of AUC on the dataset 2. Specifically, GCNNMMA obtained AUC value of 0.9367, outperforming that of SMiR-NBI (AUC = 0.6754), GISMMA (AUC = 0.8473), SLHGISMMA (AUC = 0.8532), SNMFSMMA (AUC = 0.9254), EKRRSMMA (AUC = 0.8751). In addition, we can find that the performance of GCNNMMA is also second only to SNMFSMMA on the dataset 1. This also sufficient GCNNMMA is also the best way to predict m miRNAs related to new small molecule drugs.
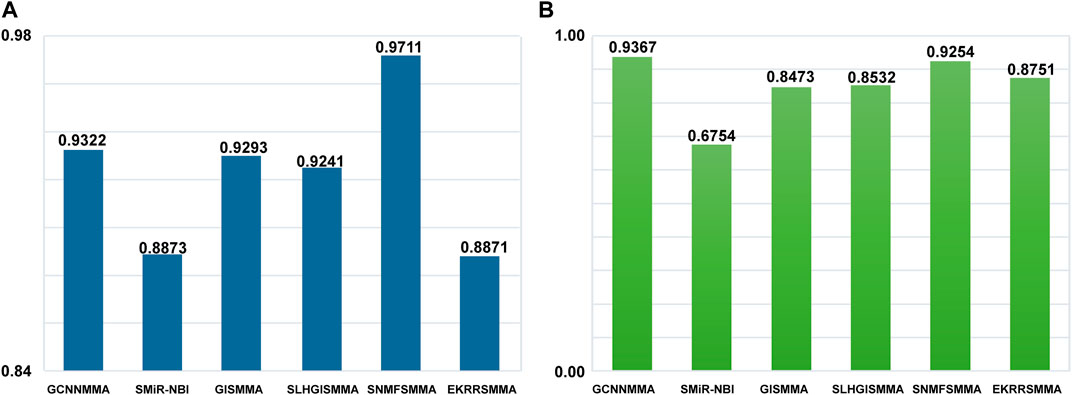
FIGURE 5. The ROC curves for GCNNMMA and benchmark algorithms for local LOOCV on the (A) dataset 1 and (B) dataset 2.
To further verify the reliability capability of GCNNMMA, we take all known miRNAs-small molecule drug associations in the SM2miR dataset 1 as the training set, and regard the missing miRNAs-small molecule drug associations as candidate sets. After GCNNMMA predicted the interaction probabilities of all candidate miRNAs-small molecule drug associations, we then ranked them according to the predicted probabilities so that the top-ranked associations were most likely to interact. We also validated these top 30 associations by searching for corresponding PubMed literature, as shown in Table 2. Among the top 10, 20, and 30 predicted associations, we were able to validate 6, 12, and 20 associations, respectively through literature search. In the top 10 predicted associations, we found that 5 different miRNAs were associated with Fluorouracil (CID: 3385), a small molecule drug that belongs to the class of pyrimidine analogs and is an anti-metabolic drug used to treat tumors. It interferes with DNA synthesis by blocking the conversion of deoxyuridine monophosphate to thymidine monophosphate (Ellison, 1961). Currently, Fluorouracil is used to treat diseases such as actinic keratosis, breast cancer, colon cancer, pancreatic cancer, gastric cancer, liver cancer, and superficial basal cell carcinoma (Lecluse and Spuls, 2015; Guo et al., 2020). Among the top 20 predicted associations, we discovered novel small molecule drugs associated with miRNAs and Estradiol (CID:5757), Testosterone (CID: 6013), and Dihydrotestosterone (CID: 10635). These three hormones have high bioavailability and can enhance cellular metabolism. These three hormones have high bioavailability and can enhance cellular metabolism (Pentikäinen et al., 2000). Among the top 30 predicted associations, we found that the small molecule drugs Etoposide (CID: 36462) (Wang et al., 2003) and Gemcitabine (CID: 60750) are used for cancer treatment. Etoposide is a semi-synthetic derivative with anti-tumor activity. It inhibits DNA synthesis by forming a complex with topoisomerase II and DNA, inducing double-stranded DNA breaks and preventing repair by blocking the binding of topoisomerase II. Accumulation of DNA breaks prevents cells from entering mitosis, leading to cell death (Uesaka et al., 2007). Gemcitabine (CID: 60750) is a nucleoside analog used in chemotherapy that, like fluorouracil and other pyrimidine analogs, replaces a structural group of nucleic acids in DNA replication to form cytidine in this case. The formation of cytidine stops tumor growth as new nucleosides cannot attach to the “defective” nucleosides, leading to cell apoptosis (cell “suicide”) (Hastak et al., 2010; Vogl et al., 2010). Currently, Gemcitabine is used to treat cancers such as non-small cell lung cancer, pancreatic cancer, bladder cancer, and breast cancer.
The development of deep learning provides new approaches for predicting the association between small molecule drugs and miRNAs. We developed a prediction model called GCNNMMA based on graph neural networks (GNNs) and convolutional neural networks (CNNs), and validated its performance on two datasets. Experimental results show that GCNNMMA exhibited the best performance in the datasets. Compared with previous similarity-based models, our model extracts the characteristic information of small molecule drugs and miRNAs through GNN and CNN networks, avoiding the dependence on known association information. Furthermore, when predicting the top 30 associations in the dataset, GCNNMMA identified Gemcitabine (CID: 60750) related to hsa-mir-139 and Fluorouracil (CID: 3385) related to hsa-mir-149, both of which are used in cancer treatment by targeting the relevant miRNAs to inhibit cell division and induce cancer cell death. While GCNNMMA achieved good performance, there is still room for improvement, such as integrating multi-source data which remains a challenging problem. In the future, incorporating more data sources, such as miRNA spatial structure data and miRNA precursor data, could improve GCNNMMA. In addition, three-dimensional structural information can better reflect spatial information. One of the future research directions is to utilize the three-dimensional structural information of miRNAs and small molecule drugs to improve prediction accuracy.
The program and data used in this study are publicly available at: https://github.com/niuzheyu123/GCNNMMA.git.
XZ conceived the study. ZN, XG, ZX, SZ, and HS performed experiments and data analysis. HW, ML, KX, and CM interpreted the data analysis. ZN, HZ HG, QL FY, XS, JL, and XZ drafted the manuscript and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
This study is supported by Natural Science Foundation of Shandong Province, China (Grant Nos ZR2020MH259, ZR2016HB40), Natural Science Foundation of China Youth Project (Grant No. 81802379).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bahdanau, D., Cho, K., and Bengio, Y. (2014). "Neural machine translation by jointly learning to align and translate.", arXiv preprint arXiv:1409.0473.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 1162, 281–297. doi:10.1016/s0092-8674(04)00045-5
Beermann, J., Piccoli, M. T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325. doi:10.1152/physrev.00041.2015
Cai, L., Lu, C., Xu, J., Meng, Y., Wang, P., Fu, X., et al. (2021). Drug repositioning based on the heterogeneous information fusion graph convolutional network. Briefings Bioinforma. 22, bbab319. doi:10.1093/bib/bbab319
Chen, X., Zhou, C., Wang, C. C., and Zhao, Y. (2021). Predicting potential small molecule–miRNA associations based on bounded nuclear norm regularization. Briefings Bioinforma. 22, bbab328. doi:10.1093/bib/bbab328
Chen, X., Guan, N. N., Sun, Y. Z., Li, J. Q., and Qu, J. (2020). MicroRNA-small molecule association identification: From experimental results to computational models. Briefings Bioinforma. 21, 47–61. doi:10.1093/bib/bby098
Chen, Y. (2015). “Convolutional neural network for sentence classification,”. MS thesis (University of Waterloo) Computer Science.
Costa, F., and De Grave, K. (2010). “Fast neighborhood subgraph pairwise distance kernel,” in Proceedings of the 26th International Conference on Machine Learning (Madison, WI, United States: Omnipress), 255–262.
Dong, Q.-W., Wang, X., and Lin, L. (2006). Application of latent semantic analysis to protein remote homology detection. Bioinformatics 22.3, 285–290. doi:10.1093/bioinformatics/bti801
Ellison, R. R. (1961). Clinical applications of the fluorinated pyrimidines. Med. Clin. N. Am. 45.3, 677–688. doi:10.1016/s0025-7125(16)33880-9
Guan, N.-N., Sun, Y. Z., Ming, Z., Li, J. Q., and Chen, X. (2018). Prediction of potential small molecule-associated microRNAs using graphlet interaction. Front. Pharmacol. 9, 1152. doi:10.3389/fphar.2018.01152
Guo, P., Pi, C., Zhao, S., Fu, S., Yang, H., Zheng, X., et al. (2020). Oral co-delivery nanoemulsion of 5-fluorouracil and curcumin for synergistic effects against liver cancer. Expert Opin. Drug Deliv. 17.10, 1473–1484. doi:10.1080/17425247.2020.1796629
Hastak, K., Alli, E., and JamesFord, M. (2010). Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res. 70.20, 7970–7980. doi:10.1158/0008-5472.CAN-09-4521
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic acids Res. 47, D1102–D1109. doi:10.1093/nar/gky1033
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). "miRBase: from microRNA sequences to function." Nucleic acids Res. 47. D155–D162. doi:10.1093/nar/gky1141
Lecluse, L. L. A., and Spuls, P. I. (2015). Photodynamic therapy versus topical imiquimod versus topical fluorouracil for treatment of superficial basal-cell carcinoma: A single blind, non-inferiority, randomised controlled trial: A critical appraisal. Br. J. Dermatology 172.1, 8–10. doi:10.1111/bjd.13460
Li, J., Lei, K., Wu, Z., Li, W., Liu, G., Liu, J., et al. (2016). Network-based identification of microRNAs as potential pharmacogenomic biomarkers for anticancer drugs. Oncotarget 7.29, 45584–45596. doi:10.18632/oncotarget.10052
Liu, W., Hui, L., and Li, H. (2022). Identification of miRNA–disease associations via deep forest ensemble learning based on autoencoder. Briefings Bioinforma. 23, 3. doi:10.1093/bib/bbac104
Liu, W., Yang, Y., Lu, X., Fu, X., Sun, R., Yang, L., et al. (2023). Nsrgrn: A network structure refinement method for gene regulatory network inference. Briefings Bioinforma., bbad129. doi:10.1093/bib/bbad129
Liu, X., Wang, S., Meng, F., Wang, J., Zhang, Y., Dai, E., et al. (2013). SM2miR: A database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics 29.3, 409–411. doi:10.1093/bioinformatics/bts698
Lv, Y., Wang, S., Meng, F., Yang, L., Wang, Z., Wang, J., et al. (2015). Identifying novel associations between small molecules and miRNAs based on integrated molecular networks. Bioinformatics 31.22, 3638–3644. doi:10.1093/bioinformatics/btv417
Peng, L., Cheng, Y., Yifan, C., and Wei, L. (2023). Predicting CircRNA-Disease associations via feature convolution learning with heterogeneous graph attention network. IEEE J. Biomed. Health Inf., 1–11. doi:10.1109/JBHI.2023.3260863
Peng, L., Tu, Y., Huang, L., Li, Y., Fu, X., and Chen, X. (2022). Daestb: Inferring associations of small molecule–miRNA via a scalable tree boosting model based on deep autoencoder. Briefings Bioinforma. 23.6, bbac478. doi:10.1093/bib/bbac478
Pentikäinen, V., Erkkilä, K., Suomalainen, L., Parvinen, M., and Dunkel, L. (2000). Estradiol acts as a germ cell survival factor in the human testis in vitro. J. Clin. Endocrinol. Metabolism 85.5, 2057–2067. doi:10.1210/jcem.85.5.6600
Ruepp, A., Kowarsch, A., Schmidl, D., Buggenthin, F., Brauner, B., Dunger, I., et al. (2010). PhenomiR: A knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 11, R6–R11. doi:10.1186/gb-2010-11-1-r6
Scarselli, F., Gori, M., Tsoi, A. C., Hagenbuchner, M., and Monfardini, G. (2008). The graph neural network model. IEEE Trans. neural Netw. 20.1, 61–80. doi:10.1109/TNN.2008.2005605
Uesaka, T., Shono, T., Kuga, D., Suzuki, S. O., Niiro, H., Miyamoto, K., et al. (2007). Enhanced expression of DNA topoisomerase II genes in human medulloblastoma and its possible association with etoposide sensitivity. J. neuro-oncology 84, 119–129. doi:10.1007/s11060-007-9360-0
Vogl, T. J., Naguib, N. N. N., Nour-Eldin, N. E. A., Eichler, K., Zangos, S., and Gruber-Rouh, T. (2010). Transarterial chemoembolization (TACE) with mitomycin C and gemcitabine for liver metastases in breast cancer. Eur. Radiol. 20, 173–180. doi:10.1007/s00330-009-1525-0
Wang, C.-C., Chen, X., Qu, J., Sun, Y. Z., and Li, J. Q. (2019). Rfsmma: A new computational model to identify and prioritize potential small molecule–mirna associations. J. Chem. Inf. Model. 59, 1668–1679. doi:10.1021/acs.jcim.9b00129
Wang, C.-C., Zhu, C.-C., and Chen, X. (2022). Ensemble of kernel ridge regression-based small molecule–miRNA association prediction in human disease. Briefings Bioinforma. 23, bbab431. doi:10.1093/bib/bbab431
Wang, S.-H., Wang, C. C., Huang, L., Miao, L. Y., and Chen, X. (2022). Dual-network collaborative matrix factorization for predicting small molecule-miRNA associations. Briefings Bioinforma. 23, bbab500. bbab500. doi:10.1093/bib/bbab500
Wang, X., Furukawa, T., Nitanda, T., Okamoto, M., Sugimoto, Y., Akiyama, S. I., et al. (2003). Breast cancer resistance protein (BCRP/ABCG2) induces cellular resistance to HIV-1 nucleoside reverse transcriptase inhibitors. Mol. Pharmacol. 63.1, 65–72. doi:10.1124/mol.63.1.65
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic acids Res. 46.D1, D1074–D1082. doi:10.1093/nar/gkx1037
Xu, J., Cai, L., Liao, B., Zhu, W., and Yang, J. (2020). CMF-impute: An accurate imputation tool for single-cell RNA-seq data. Bioinformatics 36.10, 3139–3147. doi:10.1093/bioinformatics/btaa109
Xu, J., Meng, Y., Lu, C., Cai, L., Zeng, X., et al. (2023). Graph embedding and Gaussian mixture variational autoencoder network for end-to-end analysis of single-cell RNA sequencing data. Cell Rep. Methods 3, 100382. doi:10.1016/j.crmeth.2022.100382
Yin, J., Chen, X., Wang, C. C., Zhao, Y., and Sun, Y. Z. (2019). Prediction of small molecule–microRNA associations by sparse learning and heterogeneous graph inference. Mol. Pharm. 16.7, 3157–3166. doi:10.1021/acs.molpharmaceut.9b00384
Zhang, Z., Xu, J., Wu, Y., Liu, N., Wang, Y., and Liang, Y. (2023). CapsNet-LDA: Predicting lncRNA-disease associations using attention mechanism and capsule network based on multi-view data. Briefings Bioinforma. 24, bbac531. doi:10.1093/bib/bbac531
Keywords: small molecule drug, miRNAs, graph neural networks, convolutional neural networks, CNN, liver cancer
Citation: Niu Z, Gao X, Xia Z, Zhao S, Sun H, Wang H, Liu M, Kong X, Ma C, Zhu H, Gao H, Liu Q, Yang F, Song X, Lu J and Zhou X (2023) Prediction of small molecule drug-miRNA associations based on GNNs and CNNs. Front. Genet. 14:1201934. doi: 10.3389/fgene.2023.1201934
Received: 07 April 2023; Accepted: 17 May 2023;
Published: 30 May 2023.
Edited by:
Junlin Xu, Hunan University, ChinaCopyright © 2023 Niu, Gao, Xia, Zhao, Sun, Wang, Liu, Kong, Ma, Zhu, Gao, Liu, Yang, Song, Lu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Zhou, emhvdXh1MjAwOEBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.