
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 24 August 2023
Sec. Livestock Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1199681
This article is part of the Research Topic Regulation of Fertility in Livestock Species View all 6 articles
 Birgitte Narud1*
Birgitte Narud1* Abdolrahman Khezri1
Abdolrahman Khezri1 Teklu T. Zeremichael1
Teklu T. Zeremichael1 Anne-Lene Eriksen2
Anne-Lene Eriksen2 Inger S. Grevle2
Inger S. Grevle2 Anna Nordborg3
Anna Nordborg3 Geir Klinkenberg3
Geir Klinkenberg3 Robert C. Wilson1
Robert C. Wilson1 Elisabeth Kommisrud1
Elisabeth Kommisrud1Cold storage and freezing/thawing of milt may affect sperm functionality and the subsequent fertilization ability of milt. This study aimed to investigate sperm quality parameters and fertilization potential of Atlantic salmon milt, stored cold and subsequently cryopreserved, using different storage conditions. The objective was also to assess if analysis of milt metabolites and sperm DNA methylation signatures could be applicable to further elucidate sperm quality and fertilization following preservation. Milt samples were collected from eight mature Atlantic salmon males and stored for 4 days at 2°C and 8°C. Samples were taken on day one of storage at 2°C and on day four of storage at 2°C and 8°C. Storage for 4 days at 8°C is expected to be detrimental to sperm quality, and was included to create contrasts. Correspondingly, aliquots of cold-stored milt were prepared for cryopreservation, resulting in a total of six experimental conditions. Samples from all six experimental conditions were used in fertilization trials and analyzed for sperm viability, motility, ATP content, DNA fragmentation index, and High DNA stainability. In addition, milt samples from four of the males were analyzed for targeted metabolites and DNA methylation signatures by reduced representation bisulfite sequencing. The fertilization trials were performed using sperm:egg ratios of 75 × 103 and 500 × 103, respectively. Storage duration, temperature, and cryopreservation of cold-stored milt influenced several sperm quality parameters, metabolites, and DNA methylation signatures. The total motility, progressive motility, ATP, and velocity parameters were the sperm parameters with the strongest correlation to fertilization rates (p < 0.01). Several metabolites were correlated with fertility rates in both cold-stored and cryopreserved samples (p < 0.05). The fertilizing capacity of cold-stored milt was significantly reduced after 4 days of storage at 8°C, while corresponding cryopreserved milt showed reduced fertilization at both storage temperatures (2°C and 8°C) (p < 0.05). The results indicate that cryopreservation of milt stored for 1 day does not compromise either fertilization ability or DNA methylation signatures.
Artificial fertilization (AF) using cryopreserved milt has proven advantageous in aquaculture, as it facilitates cross-fertilizing fish spawning in different geographical locations or at different times (Blaxter, 1953). Furthermore, cryopreservation allows for the improvement of broodstock management at hatcheries and preservation of genetically selected breeds as well as endangered aquatic species (Asturiano et al., 2017). Thus, cryopreservation of fish gametes could be a valuable method for increased breeding progress. The large diversity of fish species, differences in reproductive physiology, and a relatively low focus on fish breeding may have limited the research interest in cryopreservation of milt (Tiersch et al., 2007; Cabrita et al., 2010). Nevertheless, aquaculture is a rapidly developing industry, with a growing focus on cryobanking services for several salmonids and other fish species (Asturiano et al., 2017).
In salmonids, cold storage of milt at 2°C–4°C is commonly used and an alternative to cryopreservation (Trigo et al., 2015; Merino et al., 2017). However, this method limits flexibility compared to long-term storage in liquid nitrogen. It will be beneficial for the aquaculture industry if cold storage of milt can be followed by cryopreservation, allowing for increased flexibility and applicability of cryopreservation (Kommisrud et al., 2020). Correct handling and storage of sperm in fish production are important, as several factors can affect sperm quality and hence the paternal contribution to the offspring (Herráez et al., 2017).
It is well known that both cold storage and freezing/thawing of spermatozoa may affect factors such as sperm plasma membrane integrity, motility, ATP content, and DNA integrity (Cabrita et al., 2005; Balamurugan et al., 2019; Kommisrud et al., 2020). Furthermore, these parameters are recognized as important factors for sperm cells to reach and fertilize the eggs and for proper embryo development (Liu et al., 2007; Cabrita et al., 2008; Pérez-Cerezales et al., 2010; Dzyuba et al., 2017). Thus, in vitro sperm quality assessment is crucial to objectively compare different preservation protocols and to select the samples most suitable for freezing and fertilization. However, different external factors (e.g., temperature, nutrition, toxins) may affect fertility by inducing molecular alterations undetectable by traditional in vitro assays (Riesco and Robles, 2013; Herráez et al., 2017). To broaden current knowledge regarding factors affecting reproductive success in fish, different omics techniques have been introduced (Egea et al., 2014), such as epigenomics and metabolomics (Labbé et al., 2017; Dietrich et al., 2019a).
Metabolites are defined as intermediates or final products of metabolic pathways and play significant roles in sperm physiology (Dietrich et al., 2019a). In mammals, several studies have been conducted to investigate the relationship between metabolites in semen and fertility (Zhao et al., 2018; Engel et al., 2019; Menezes et al., 2019; Narud et al., 2020). Though there are studies in fish investigating the metabolome of different cells, tissues, and biofluids [reviewed by Samuelsson and Larsson (2008); Dietrich et al. (2019a)], few have investigated metabolites in fish milt (Dietrich et al., 2019b; Tsentalovich et al., 2020).
The mammalian sperm epigenome is quite well described and reviewed previously by Champroux et al. (2018), and recently, there has been an increase in epigenetic studies performed in fish gametes and embryos (Labbé et al., 2017; Woods et al., 2018; Depincé et al., 2020). Environmental factors such as temperature and water quality, and reproductive biotechnologies including cryopreservation and AF, are examples of factors that may cause epigenetic modifications in fish gametes and embryos (Labbé et al., 2017).
The main objective of this study was to assess if cold storage of milt can be followed by cryopreservation without compromising post-thaw sperm quality and the fertilizing potential in Atlantic salmon (Salmo salar L.). Another goal was to investigate if analysis of milt metabolites and sperm DNA methylation signatures could be applicable to further elucidate sperm quality and fertilization following preservation. To obtain the aims, milt was stored at two different temperatures, 2°C and 8°C, where 8°C was hypothesized to be detrimental to sperm quality. Furthermore, two different sperm:egg ratios were employed in the fertilization trials.
Milt from eight mature Atlantic salmon males kept at a broodstock facility in Central Norway was collected by gonad extraction, during spawning season. Briefly, males were culled, and the gonads were removed, cleaned, and homogenised using a mechanical grinder. The homogenate was weighed and a 1:1 amount of AquaBoost® SpermCoat (Cryogenetics AS, Hamar, Norway) was added, before filtration and storage in 650 mL cell culture flasks (VWR International, Radnor, PA, United States). The milt samples were shipped overnight to the laboratory (Cryogenetics AS, Hamar, Norway) while kept cold on paper on top of crushed ice to ensure a temperature of about 2°C on arrival. Aliquotes of milt from each male (10 mL) were stored at 2°C and 8°C in cell culture flasks (50 mL) with filter screw caps (VWR International, Radnor, PA, United States) for 4 days. Samples were taken on day one of storage at 2°C (hereafter referred to as D1-2°C) and on day four of storage at 2°C and 8°C (hereafter referred to as D4-2°C, D4-8°C). Correspondingly aliquots of cold-stored milt were prepared for cryopreservation (hereafter referred to as D1-2°C/Cryo, D4-2°C/Cryo, D4-8°C/Cryo), resulting in a total of six experimental conditions. Samples from all six experimental conditions were analyzed for sperm viability, motility, ATP content, DNA fragmentation index (DFI), and High DNA stainability (HDS). In addition, samples from four of the males, selected based on their contrasting performance in the fertilization trials (described in 2.7), were analyzed for sperm metabolites and DNA methylation signatures by reduced representation bisulfite sequencing (RRBS). Cryopreservation was conducted using the following procedure. Sperm concentration was estimated after mixing 10 µL milt with 4 mL 0.9% NaCl using an SDM6 photometer (Minitüb GmbH, Tiefenbach, Germany) at 546 nm using a pre-determined calibration curve for Atlantic salmon sperm. Sperm concentration varied between 10.2 and 12.2 × 109 sperm/mL. Based on the concentration estimate, the milt was diluted using a proprietary fish sperm extender (Cryogenetics AS, Hamar, Norway) to a standardized sperm concentration. Pre-printed 0.5 mL medium straws (IMV technologies, L’Aigle, France) were filled with milt and sealed with ultrasound using an MPP Uno automated filling and sealing machine (Minitüb GmbH, Tiefenbach, Germany). Cryopreservation was performed according to a proprietary protocol (Cryogenetics AS, Hamar, Norway) before transfer and storage in liquid N2. The preprint ensured identification of the male, storage condition, and date of cryopreservation.
Samples from the six different experimental conditions from all individuals were treated identically according to the following procedure. Samples were analyzed in triplicate for viability, ATP content, and DFI, and in duplicate for motility assessment. For the cryopreserved milt, three doses per batch were thawed in a water bath (25°C, 30 s) and pooled. All samples were kept cold on paper on top of crushed ice before in vitro analysis. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, United States) unless otherwise noted.
The analysis of sperm plasma membrane integrity (sperm viability) was assessed using propidium iodide (PI, L-7011, LIVE/DEAD®Sperm Viability Kit, Molecular Probes, ThermoFisher Scientific, Waltham, MA United States). The spermatozoa were stained in dark (4°C, 10 min) in PBS (137 mM NaCl, 2.7 mM KCl, 1.76 mM KH2PO4, 8.1 mM Na2HPO4·2H2O, pH 7.4) staining solution with a final concentration of 1.5 × 106 spermatozoa/mL and 0.48 mM PI. The flow cytometry analysis was performed using a Cell Lab Quanta ™ SC MPL flow cytometer (Beckman Coulter, Fullerton, CA, United States), equipped with a 22 mW argon laser with an excitation wavelength of 488 nm. Data were analyzed using Cell Lab Quanta SC MPL Analysis software program (Software Version 1.0, Beckman Coulter, Fullerton, CA, United States). Side scatter (SS) signals were used for the identification of spermatozoa, and PI fluorescence in SS-gated spermatozoa was detected using a 670 nm long pass (LP) filter (FL3).
Sperm motility analysis was performed using a SCA evolution CASA system (Sperm Class Analyzer® version 6.1, Microptic SL, Barcelona, Spain). The CASA system was equipped with a phase contrast Eclipse Ci-S/Ci-L microscope (Nikon, Japan) with a temperature-controlled stage using a water circulation pump, and a Basler digital camera (Basler Vision Technologies, Ahrensburg, Germany). Chamber slides were kept on the cooled microscopic stage before analysis. Samples were diluted to a working sperm concentration of 87 × 106 cells/mL in AquaBoost® Dilutor (Cryogenetics AS, Hamar, Norway) prior to analysis. Sperm activation was initiated by the addition of AquaBoost® Activator (Cryogenetics AS, Hamar, Norway) simultaneously with loading the sample to the microscope slide. Using a Picus (0.2–10 µL) electronic 1-channel pipette (Sartorius Stedim Biotech GmbH, Göttingen, Germany) in Multi-Aspirating Mode, aspiration was done sequentially in the following order: 1.2 µL diluted milt—air—1.2 µL AquaBoost® Activator. For each CASA measurement, the aspiration volume was dispensed into a single chamber on a Leja 20 µm 4 chamber microscope slide (Leja, Nieuw-Vennep, the Netherlands). Image analysis was performed on four consecutive fields within approximately 8 s after activation, with a total of at least 400 cells analyzed per sample. The parameters recorded were total motility (TM, %), progressive motility (PM, %), straight-line velocity (VSL, mm/sec), average path velocity (VAP, mm/sec), curvilinear velocity (VCL, mm/sec), straightness (STR, ratio VSL/VAP) and linearity (LIN, ratio VSL/VCL). Sperm cells were identified by sperm head area of 10–50 μm2, a 50 Hz frame rate was used, and 45 frames were captured per object. Motile sperm cells were defined based on VAP (µm/sec) 15< Slow < 30 < Medium < 50 < Rapid. Cells were classified as progressive motile if STR >45%.
The sperm ATP content was determined using the CellTiter-Glo® Luminescent Cell Viability Assay (G7571, Promega, Madison, WI, United States), a FLUOstar OPTIMA multiwell plate reader (BMG Labtech GmbH, Offenburg, Germany) and MARS data analysis software (Version 1.10, BMG Labtech GmbH, Offenburg, Germany). In brief, milt samples were diluted to 5 × 106 spermatozoa/mL in AquaBoost® SpermCoat (Cryogenetics AS, Hamar, Norway), and each sample (50 µL) was transferred to wells in an opaque 96-well microtiter plate (NUNC™, Roskilde, Denmark). CellTiter-Glo® Reagent (50 µL) was added to each well and the mixture was gently shaken for 2 min in a rotary shaker (300 rpm) (IKA® MS 3 digital, IKA®-Werke GmbH & Co., Staufen, Germany), followed by 10 min incubation in room temperature (RT). The data recorded was measured in relative luminescence units (RLU), and converted to corresponding ATP values (nM) according to a prepared standard curve.
Sperm chromatin integrity was analyzed using the Sperm Chromatin Structure Assay (SCSA), as previously described for bovine semen (Narud et al., 2020), with minor modifications. In brief, milt samples were diluted to about 1.5 × 107 spermatozoa/mL in TNE buffer (10 mM Tris-HCL, 0.1 M NaCl, 1 mM EDTA, pH 7.4) in a final volume of 200 μL. Next, 400 μL acid detergent solution [0.38 M NaCl, 120 mM HCL, 0.1% (w/v) Triton X-100, pH 1.2] was added, followed by incubation for 30 s in RT. Next, 1.2 mL acridine orange (AO) staining solution [6 mg/mL AO (A3568, Life Technologies, OR, United States)] in a buffer containing 37 mM citric acid, 0.126 M Na2HPO4, 1.1 mM EDTA, and 0.15 M NaCl (pH 6) was added. Data acquisition started after a 3 min setup mode, in which 5000 events were captured for each sample at a rate of ∼200 events/sec. Analysis in FCS Express 6 Flow cytometry Software (Denovo Software, Los Angeles, CA, United States) revealed the percentage of red (ssDNA) and green (dsDNA) fluorescence. Based on a histogram of the fluorescence ratio red/(red + green), the sperm cells with fragmented DNA (DFI, %) were calculated. The percentage of spermatozoa with high DNA stainability (HDS, %) was determined by the bivariate cytogram.
Fertilization trials were performed using milt from all six different experimental conditions (2.1). For each trial, eggs from one female were stripped and shipped overnight on ice from a broodstock facility to Cryogenetics AS in Hamar. Eggs were kept in ovarian fluid and shipped in 650 mL cell culture flasks (filled with approximately 300 mL per flask). In vitro fertilization was carried out using two different sperm:egg ratios, 75 × 103 sperm cells/egg (duplicates) and 500 × 103 sperm cells/egg. Each fertilization unit consisted of approximately 150 eggs estimated by weight (18 g). Cryopreserved straws were thawed for 30 s in a water bath at 25°C. Fertilization was performed in a climate-controlled room at 4°C. The temperature difference between eggs and other media throughout the fertilization trial was at all times below 2.5°C. For each fertilization, sperm and eggs were gently mixed in a plastic cup while adding AquaBoost® Activator sperm activating solution to slightly cover the eggs. After 2 min, eggs were rinsed in physiological saline solution (0.9% NaCl) and transferred to nylon mesh-bottomed incubation rings. Incubation rings were then incubated in fresh water. After incubation for approximately 150–200 h-degrees post-fertilization, eggs were transferred to a plastic cup and AquaBoost® Quattro (Cryogenetics AS, Hamar, Norway) was added. After fixation for 15 min in this solution, each egg was visually inspected and categorized as fertilized or unfertilized, based on the presence or absence of cell division.
Samples from all six experimental conditions were analyzed for targeted metabolites. For the cold-stored samples, 2 mL milt prediluted 1:1 with AquaBoost® SpermCoat was centrifuged (800 × g, 10 min) to separate the supernatant (seminal plasma and extender, hereafter referred to as seminal plasma) from the pellet (sperm cells). For the cryopreserved samples, one straw of frozen-thawed milt was centrifuged (500 × g, 5 min). All pellet and supernatant samples were snap-frozen in liquid N2 and shipped on dry ice to SINTEF (SINTEF Industry, Trondheim, Norway) for analyses of targeted amines and amino acids. Samples of the proprietary extenders used were also analyzed for background levels of the targeted metabolites.
At SINTEF the samples were thawed, and an aliquot was collected for sample preparation and analysis. Sample preparation consisted of protein precipitation by the addition of four volumes of ice-cold methanol. Following centrifugation, a 100 μL aliquot of the supernatant was collected for amine analysis, dried using a speed-vac, and dissolved in an aqueous internal standard mix prior to analysis. A 10 μL aliquot of the supernatant was collected for amino acid analysis. The sample was mixed with the amino acid internal standard mixture and derivatized using an AccQ-Tag Derivatization Kit (Waters, Milford, MA, United States) prior to analysis.
Metabolite analysis of amino acids and amines was performed on an Agilent 1290 Infinity II LC system (Agilent, Santa Clara, United States) coupled to an Agilent 6495 QqQ mass spectrometer, using one method for amino acids and one for amines. The QqQ-MS was equipped with a jet-stream ESI source operated in positive mode. The QqQ-MS was operated in dynamic multiple-reaction monitoring (MRM) mode (delta Rt = 1 min) with a unit mass resolution for both mass filters. The MRM transitions for standards and internal standards and the employed collision energies, gas temperatures, and flows are given in Supplementary Tables S1, S2. For the amines (choline, creatine, and L-carnitine), the chromatographic separation was performed in HILIC mode employing a BEH Amide (2.1 × 150 mm, 2.7 μM) column (Waters, Milford, United States) and gradient elution using an 80:20 mixture of 25 mM Formic acid and 50 mM ammonium acetate as eluent A, and acetonitrile as eluent B, at a flow rate of 0.3 mL/min. The gradient used started at 90% B and was decreased stepwise to 5% B at 6.5 min. The column thermostat was maintained at 35°C and the autosampler at 8°C. The injection volume was 1 μL. Mixed standards at 0, 0.5, 1, 10, 50, 100, 500, 1,000, and 5,000 nM were used for calibration and quantitation. Amino acids (alanine, arginine, asparagine, GABA, glutamate, glutamine, glycine, Histidine, isoleucine, leucine, lysine, methionine, ornithine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine) were analyzed in reversed-phase mode employing an Ascentis Express C18 (2.1 × 150 mm, 2.7 μM) column (Sigma-Aldrich), and gradient elution using 25 mM Formic acid as eluent A, and acetonitrile as eluent B, at a flow rate of 0.3 mL/min. The gradient used started at 5% B and was increased stepwise to 50% B at 15 min. The column thermostat was maintained at 20°C and the autosampler at 6°C. The injection volume was 2 μL. Mixed standards at 0, 0.1, 1, 5, 10, 25, 50, 100, 250, 500, and 1,000 μM were used for calibration and quantitation.
Parallel samples from four of the eight individuals belonging to D1-2°C, D1-2°C-Cryo, D4-2°C, and D4-8°C groups were considered for the construction of RRBS libraries using a gel-free multiplexed technique (Boyle et al., 2012), previously optimized to study sperm DNA methylation in boar (Khezri et al., 2019) and bull (Khezri et al., 2020). In brief, 100 ng genomic DNA isolated from milt samples was digested overnight at 37°C using MspI and Taqα1 enzymes (New England Biolabs). Fragmented DNA was subjected to Gap filling, A-tailing, and size selection (300–500 bp). Size selected DNA samples were adapter-ligated using NEXTflex™ Bisulfite-Seq barcodes (Bio Scientific Corporation) and were bisulfite converted using EpiTect kit (QIAGEN, Germany) following the manufacturer’s protocol. The product was cleaned up according to recommendations in the QIAGEN EpiTect kit (Gu et al., 2011) and PCR amplified. The PCR product was further enriched by adding 1× SPRI AMPure XP beads. The DNA concentration of eluted and cleaned RRBS libraries was measured using the PicoGreen dsDNA absorbance method. The prepared libraries were sent to the Norwegian Sequencing Centre where sequencing was performed by Illumina HiSeq 4000 in paired-end (2 × 150 bp) mode.
The quality of paired-end Illumina reads was checked using fastQC (v 0.11.8 for Linux). Illumina adapters and low-quality sequences (below 20 bp and Phred score of 30) were trimmed using the Trim-galore (v 0.4.4 for Linux) (Martin, 2011).
The Atlantic salmon reference genome (ICSASG_v2) was downloaded from the UCSC database and indexed using bismark_genome_preperation script in Bismark (v 0.19.0 for Linux) (Krueger and Andrews, 2011). Clean reads after trimming were collected and mapped against the indexed genome using bowtie2 aligner, integrated with Bismark using default parameters. Global CpG methylation level in Bismark was internally calculated for all covered cytosines (Cs) using the following formula: % of global methylation = 100 × number of methylated Cs/number of methylated Cs + number of unmethylated Cs.
Differential methylated analysis was performed using the methylKit package (v 1.6.1) (Akalin et al., 2012) in Rstudio (v 1.1.453 for Linux). The analysis was performed for five different experiments (Experiments A to E). For each experiment, test and control groups were defined as described in Table 1.
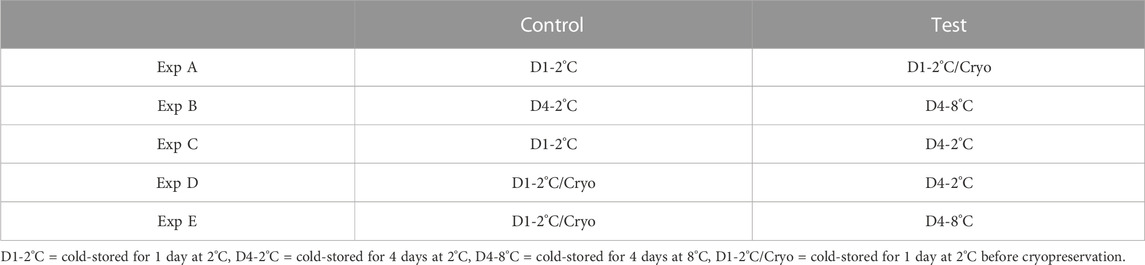
TABLE 1. An overview of the different experiments (Exp A to E) conducted for differential methylation analysis. For each experiment, milt samples from four different salmon were considered. The same milt samples (treated differently) were considered for both the control and test.
For differential methylated analysis, only CpGs ≥ 10 × coverage depth (CpG10x) were considered, and reads containing CpGs with more than 99. Ninth percentile coverage were excluded. After applying logistic regression analysis with a sliding linear model to correct for multiple comparisons, only DMCs with ≥10% methylation difference and q value < 0.05 (filtered DMCs onwards) were considered for downstream analysis. In this study, hypermethylated and hypomethylated Cs are defined as differential methylation over 10% or smaller than −10% in the test group compared to the control group, respectively.
BED12 files containing gene and CpG annotation for the Atlantic salmon assembly (ICSASG_v2) were downloaded from the UCSC table browser (UCSC, 2018). The genomation package (v 1.14.0) in Rstudio was employed to annotate filtered DMCs (contained coordinate) with the nearest transcriptional start site (TSS), gene elements (exons, introns, promoters, and intergenic regions), and CpG features (CpG islands, CpG shores, other). In this study, promoters and CpG shores were defined as ±1,000 bp and ±2,000 bp of the TSS and CpG islands, respectively.
Corresponding transcript stable ID(s) to annotated TSSs were converted to UniProt accession ID using the gene ID conversion tool available from g: Profiler (Raudvere et al., 2019). Obtained UniProt accession IDs were submitted as a list to DAVID Bioinformatics resources for gene ontology functional annotation analysis (Huang da et al., 2009). The list was recognized automatically and DAVID assigned Salmo salar as the target organism. Furthermore, in order to get a better annotation and reduce database bios, multiple databases were selected for enrichment analyses: Interpro, KEGG pathway, Go terms including biological process, cellular components, molecular functions, SMART, and UniprotKB Keywords including biological process, cellular components, molecular functions. Gene enrichment for each identified term or pathway was calculated using Fisher’s exact test and the p-value was Benjamini corrected for multiple testing and set to 0.05.
The statistical analyses of the different in vitro sperm quality traits, metabolites, and fertility performance were performed using SAS Version 9.4 for Microsoft Windows (SAS Institute, Cary, NC, United States). The data were tested for normal distribution by the Shapiro-Wilk test. Parameters that did not show a normal distribution were log-transformed to approach a normal distribution prior to further statistical analysis. The mixed procedure in SAS was used to perform the least square means analyses. For the sperm quality parameters, the effect of the explanatory variables on the outcome variables; motility parameters, viability, DFI, HDS, and ATP, were estimated by the following mixed linear model: Yijk = µ + Ci + Sj + Mk eijk, where: Yijk = observation of in vitro sperm parameter per semen sample; µ = overall mean of the in vitro sperm parameter; Ci = fixed effect of storage condition, i = 1 (day one milt), 2 (stored at two degrees) or 3 (stored at eight degrees); Sj = fixed effect of semen state, j = 0 (fresh) or 1 (frozen-thawed); Mk = random effect of male, k = 1 to 8; eijk = random error. The same model was used to perform a least square means analysis for the metabolites in sperm and seminal plasma, using the different amines and amino acids as outcome variables. For the fertilization outcome, the same model was used, however, the outcome variables were the fertilization outcome either from a sperm:egg ratio of 75 or 500 × 103, respectively. The Tukey test was applied for pairwise comparisons between means. The correlation between in vitro sperm quality parameters or metabolites and fertilization outcome was assessed, where correlation coefficients (Spearman) were calculated. The significance level in all statistical tests was set to 0.05.
The sperm viability of cold-stored milt in D1-2°C was on average 95.1% ± 0.7 and decreased significantly in D4-2°C (p < 0.0001) and D4-8°C (p < 0.0002) (Table 2).
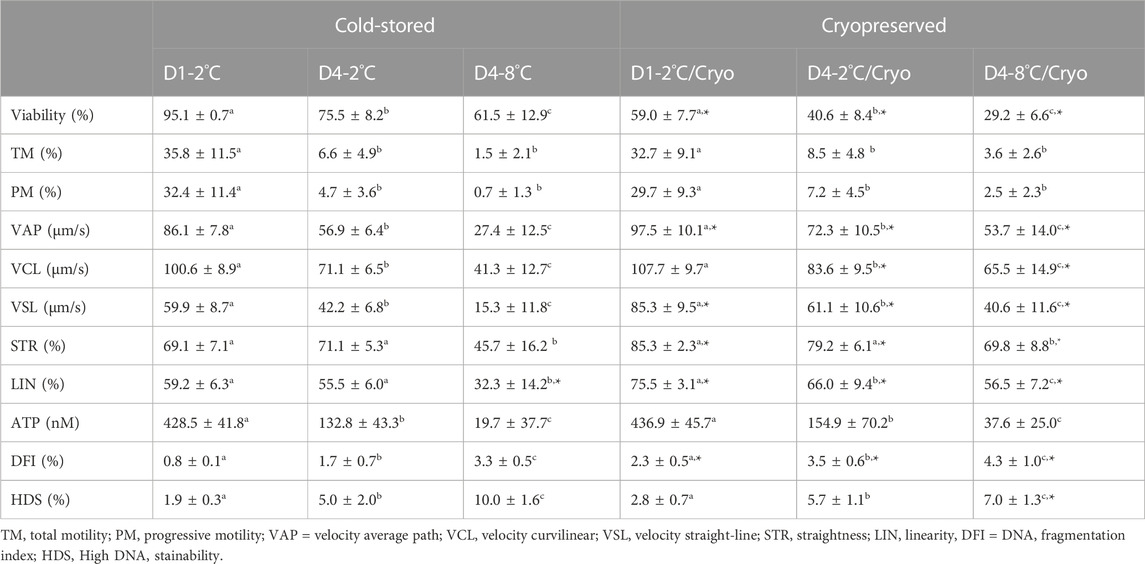
TABLE 2. In vitro sperm parameters of milt collected from eight Atlantic salmon (Salmo salar L.) males are presented as mean ± SD for cold-stored and cryopreserved milt. Milt was stored cold for 1 day at 2°C (D1-2°C) and 4 days at 2°C (D4-2°C) and 8°C (D4-8°C), respectively, with subsequent cryopreservation (D1-2°C/Cryo, D4-2°C/Cryo and D4-8°C/Cryo). Different superscripts denote significant differences (p < 0.05) between storage conditions within cold-stored and cryopreserved samples based on a linear mixed model. Asterix denotes significant differences (p < 0.05) between cold-stored and cryopreserved samples.
Frozen-thawed milt samples showed significantly lower viability than the cold-stored samples (p < 0.0001), for all experimental conditions. There was a significant higher post-thaw sperm viability in D1-2°C/Cryo compared to D4-2°C/Cryo and D4-8°C/Cryo samples (p < 0.002).
Sperm motility of cold-stored milt samples measured as both total (TM) and progressive (PM) motility, were higher in D1-2°C compared to D4-2°C and D4-8°C (p < 0.0001) (Table 2). The TM and PM measured in D4-2°C were not significantly different from D4-8°C. The sperm velocity parameters VAP, VCL, and VSL were higher in D1-2°C compared to D4-2°C and D4-8°C (p < 0.0005). The parameters STR and LIN were not significantly different between D1-2°C and D4-2°C. However, both parameters decreased in D4-8°C (p < 0.0001).
Cryopreservation of milt samples after cold storage did not affect the average sperm motility measured as TM and PM (p > 0.05). The post-thaw TM and PM were higher in D1-2°C/Cryo compared to D4-2°C/Cryo and D4-8°C/Cryo (p < 0.0001). Post-thaw TM and PM did not differ between D4-2°C/Cryo and D4-8°C/Cryo (p > 0.05). Average VAP, VSL, and VCL in frozen-thawed samples increased (p < 0.02) compared to cold-stored samples, except for VCL in D1-2°C/Cryo (p > 0.05). The velocity parameters VAP, VCL, and VSL were higher in D1-2°C/Cryo compared to D4-2°C/Cryo and D4-8°C/Cryo (p < 0.001). Samples cryopreserved after cold storage had higher post-thaw STR and LIN compared to cold-stored samples (p ≤ 0.05). The parameter LIN was lower in D4-2°C/Cryo and D4-8°C/Cryo compared to D1-2°C/Cryo (p < 0.03). Post-thaw STR decreased in D4-8°C/Cryo compared to D1-2°C/Cryo and D4-2°C/Cryo (p < 0.0005).
Sperm ATP content was significantly lower in D4-2°C compared to D1-2°C and was further reduced in D4-8°C (p < 0.0001) (Table 2). The ATP content of frozen-thawed milt samples was not significantly different from the cold-stored samples. Post-thaw ATP content was lower in D4-2°C/Cryo compared to D1-2°C/Cryo and was further reduced in D4-8°C/Cryo (p < 0.0001).
Higher percentages of the sperm chromatin integrity parameters DFI (p < 0.003) and HDS (p < 0.0001) were observed in milt from D4-2°C and D4-8°C compared with D1-2°C. The percentages of DFI and HDS were higher in D4-8°C compared to D4-2°C (p < 0.0001).
Samples cryopreserved after cold storage showed a higher percentage of DFI compared to cold-stored samples (p < 0.05). The post-thaw percentages of DFI and HDS were higher in D4-2°C/Cryo compared to D1-2°C/Cryo (p < 0.0001) and was further increased in D4-8°C (p < 0.05).
There was a significant influence of cold storage duration and temperature using both sperm:egg ratios (Figure 1). For the samples cryopreserved after cold storage, there was a reduction in fertilized eggs from D1-2°C/Cryo to D4-2°C/Cryo and a further reduction in D4-8°C/Cryo (p < 0.05). The cold stored samples in D4-2°C were not significantly different from D1-2°C. However, D4-8°C samples significantly reduced the number of fertilized eggs. Overall, cryopreservation of cold-stored samples resulted in a significant reduction of fertilized eggs compared to cold-stored samples, except for D1-2°C using sperm:egg ratio of 500 × 103.
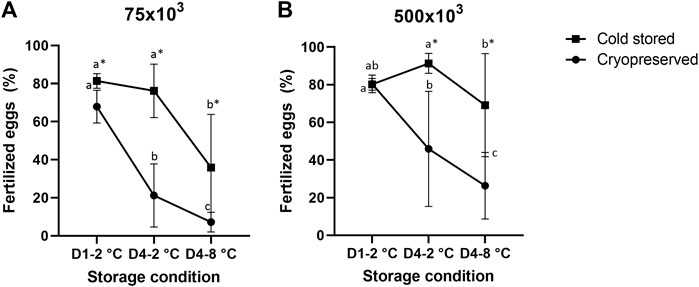
FIGURE 1. Percentage of fertilized eggs (mean ± SD after fertilization with cold stored or frozen-thawed sperm stored cold for 1 day at 2°C (D1-2°C) and 4 days at 2°C and 8°C (D4-2°C and D4-8°C) prior to cryopreservation. Fertilizations were carried out in triplicate with sperm:egg ratios of 75 × 103 (A) or 500 × 103 (B). Different letters denote significant differences (p < 0.05) between storage conditions within cryopreserved or cold-stored samples, respectively. Asterix denotes significant differences (p < 0.05) between cold-stored and cryopreserved samples within the same storage condition.
Spearman correlations between all in vitro sperm parameters and fertilization rate were significantly different from zero (p < 0.05) using a sperm:egg ratio of 75 × 103 for both cold-stored milt and milt cryopreserved after cold storage (Supplementary Table S3). The highest correlations were found for the motility parameters TM (rs = 0.96) and PM (rs = 0.96), ATP (rs = 0.93) content, and the kinematic parameters VAP (rs = 0.91), VCL (rs = 0.91) and VSL (rs = 0.91) of frozen-thawed samples. The post-thaw chromatin integrity parameters DFI (rs = −0.82) and HDS (rs = −0.66) were negatively correlated with the fertilization outcome. No significant correlations were found for cold-stored samples using sperm:egg ratio of 500 × 103. For the frozen-thawed samples, all correlations were weaker for fertilizations with a sperm:egg ratio of 500 × 103 compared with 75 × 103 (Supplementary Table S3).
The effects of storage duration and temperature on amine and amino acid concentrations were assessed in sperm cells (Table 3) and seminal plasma (Supplementary Table S4) from all experimental conditions. For the cold-stored sperm samples, the concentration of the amines choline and creatine increased in D4-2°C and D4-8°C compared with samples in D1-2°C (p < 0.05). The concentration of L-carnitine increased from D1-2°C to D4-2°C (p < 0.0001) but was not further increased in D4-8°C (p > 0.05). There was a significant overall effect of cryopreservation on all amine concentrations (p < 0.002). The concentration of choline was not significantly affected by storage duration and temperature in samples cryopreserved after cold storage. The concentration of carnitine/L-carnitine was significantly reduced from D1-2°C/Cryo to D4-2°C/Cryo but was not further reduced in D4-8 °C/Cryo.
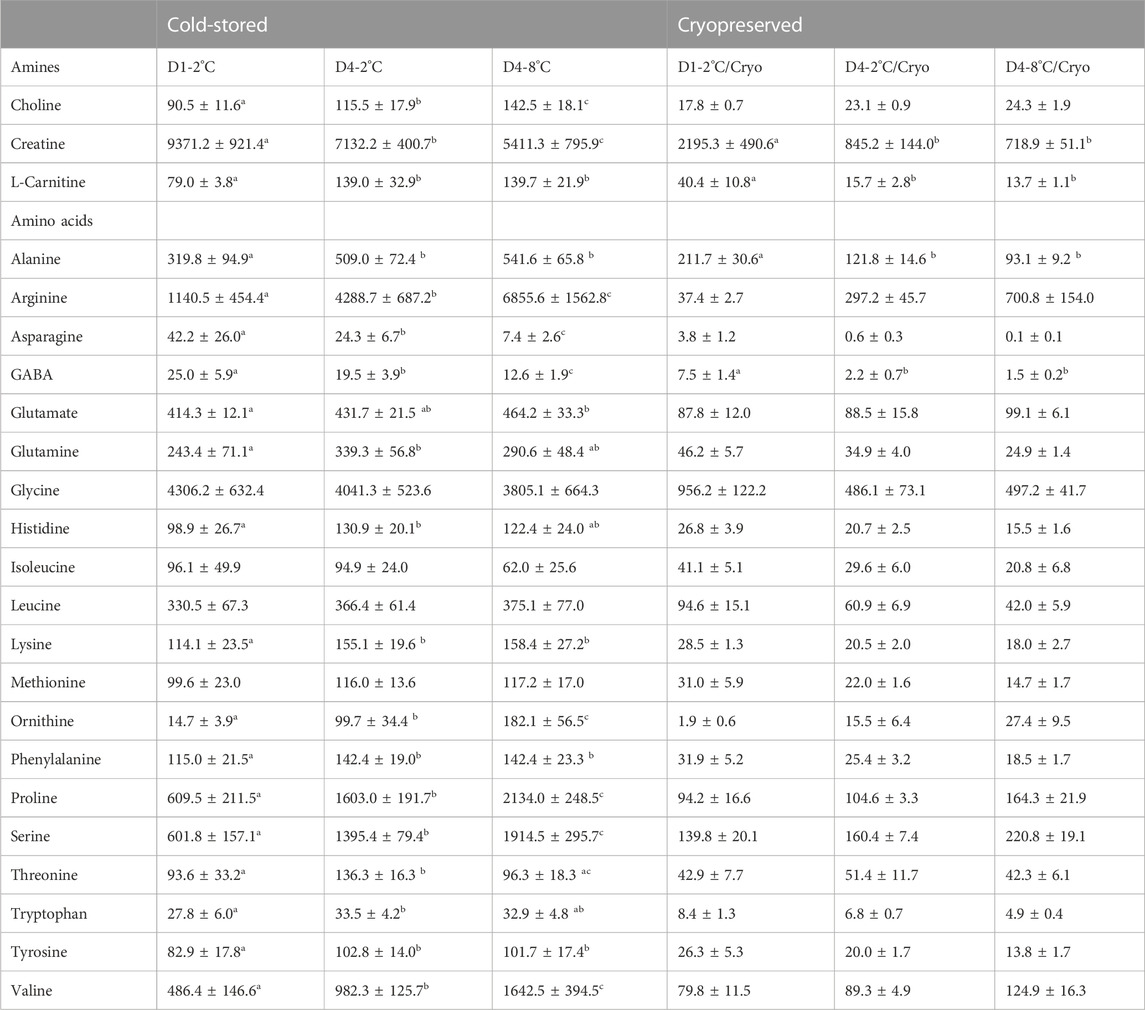
TABLE 3. Mean ± SD for amine and amino acid concentrations (µM) in sperm cells of cold-stored milt and milt cryopreserved after cold storage. Samples were collected from four Atlantic salmon (Salmo salar L.) males. Milt was stored cold for 1 day at 2°C (D1-2°C) and 4 days at 2°C (D4-2°C) and 8°C (D4-8°C), respectively, with subsequent cryopreservation (D1-2°C/Cryo, D4-2°C/Cryo and D4-8°C/Cryo). Different superscripts denote significant differences (p < 0.05) between storage days and temperature within cold-stored and cryopreserved samples based on a linear mixed model.
The concentrations of the amino acids arginine, ornithine, proline, serine, and valine measured in sperm from cold-stored milt samples, significantly increased with both storage time and temperature (p < 0.001). The concentrations of alanine, lysine, phenylalanine, and tyrosine were significantly increased in D4-2°C compared to D1-2°C (p < 0.05). However, there was no significant difference between D4-2°C and D4-8°C samples. The glutamate concentration was significantly higher in D4-8°C, compared to D1-2°C (p < 0.01). For the amino acids glutamine, histidine and tryptophan, the concentration was significantly higher in D4-2°C compared to D1-2°C (p < 0.05). However, there was no significant effect of increasing the storage temperature to eight degrees. Overall, there was a significant effect of cryopreservation for all amino acid concentrations except for asparagine in D4-8°C and ornithine in D1-2°C. For the cryopreserved samples, only the concentrations of alanine (p < 0.05) and GABA (p < 0.02) were significantly reduced during storage. No significant changes were observed after increasing the storage temperature to eight degrees. The results for metabolite concentrations in seminal plasma showed similar patterns to the sperm cell samples, where several metabolites were affected by storage conditions during cold storage (Supplementary Table S4). None of the metabolites in the seminal plasma of frozen-thawed samples were significantly affected by the storage conditions prior to cryopreservation.
Correlations (Spearman correlation) between sperm metabolites and fertilization outcome using a sperm:egg ratio of 75 × 103 is presented in Table 4. For the cold-stored samples, the highest positive correlations were found for asparagine (rs = 0.88) and GABA (rs = 0.86), while choline (rs = −0.87) showed the highest negative correlation. For the cryopreserved samples, all the amines and several amino acids showed significant correlations to the fertilization outcome. The highest positive correlation was observed for GABA (rs = 0.90), creatine (rs = 0.88), and L-carnitine (rs = 0.85). Arginine (rs = −0.81) and choline (rs = −0.78) showed the highest negative correlation.
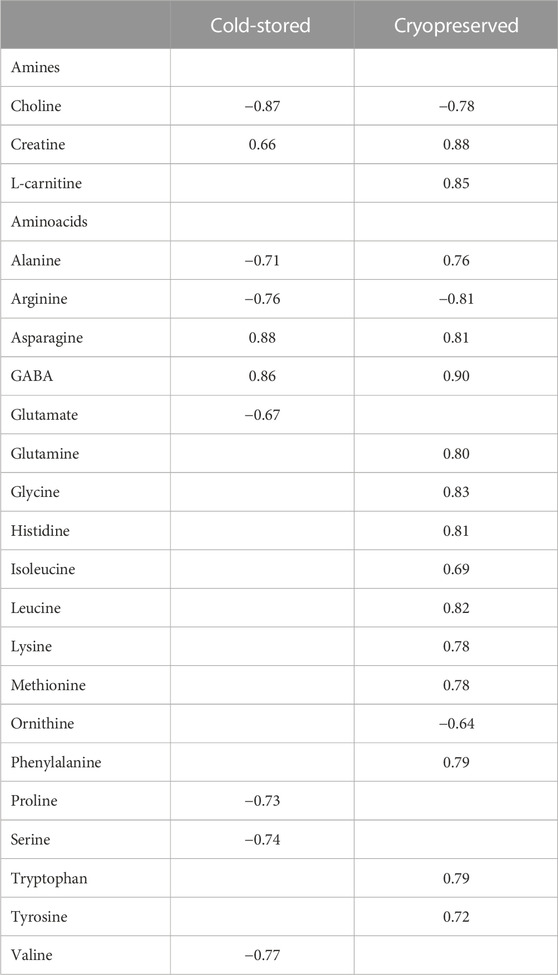
TABLE 4. Spearman correlation coefficient values between metabolite concentrations in sperm cells and percentage of fertilized eggs (sperm:egg ratio of 75 × 103) in Atlantic salmon (Salmo Salar L.) using cold-stored or the corresponding cryopreserved milt after cold storage. Only metabolites significantly (p < 0.05) different from zero are presented.
Overall, minimal variation was observed between libraries regarding retrieved clean data, mapping efficiency, and global CpG methylation levels (Table 5). Using Trim Galore default options, we were able to recover, on average, 99.7% of the reads, corresponding to 83.8% of the total bases in the original reads. Libraries mapped against the Salmo salar reference genome, showed on average 59.3% unique mapping efficiency. Furthermore, the libraries had a high degree of global methylation percentage (largely close to 87.3%). In this study, only CpGs with equal or greater than 10x coverage were considered for methylation analysis. The number of CpG10X varied between samples from 2 million to 4.5 million Cs, corresponding to an average of 23.6% of all covered Cs in the libraries. To compare the methylation profiles, all samples were considered for correlation and cluster analyses based on CpG10X methylation values. Pearson’s correlation revealed a high similarity between samples (Pearson’s correlation coefficient > 0.96) and cluster analysis showed that test and control groups from the same individuals always were clustered together (Supplementary Figure S1; Supplementary Table S5).
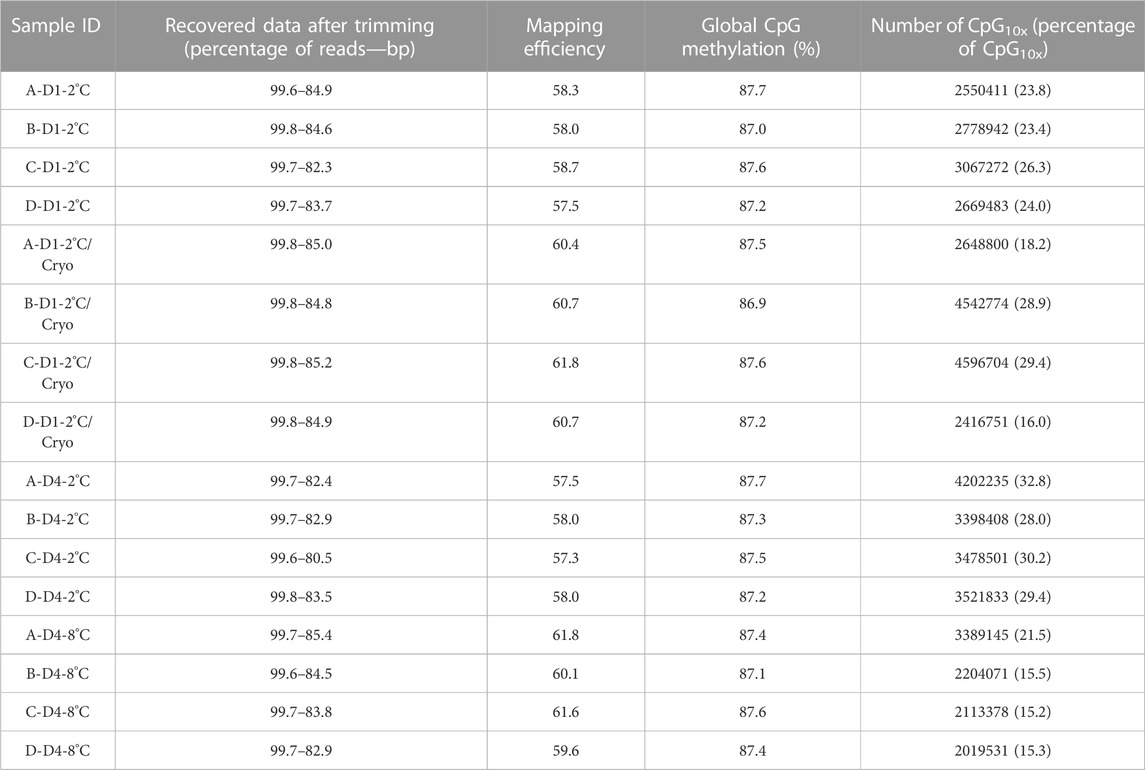
TABLE 5. An overview of basic statistics for RRBS libraries. Sample IDs reflecting Atlantic salmon (Salmo salar L.) individuals (A, B, C, D), days of milt storage (D1 and D4), and temperature of storage (2°C and 8°C), with subsequent cryopreservation (Cryo). Libraries, quality filtered and trimmed using trim-galore, and the number of reads and bp were compared with pre-trimmed data. Mapping efficiency shows the percentage of uniquely mapped clean reads with the reference genome. CpG methylation shows the percentage of global methylation in clean reads. The number of CpG10X quantified in clean reads after trimming and the percentage of CpG10X were calculated relative to all Cs with any coverage in libraries.
Differential methylation analysis was performed using CpG10X after applying a linear regression model and 10% methylation cut-off as well as a q-value < 0.05. This resulted in identifying diverse numbers of DMCs (from 226 to 3483), with different levels of methylation difference ranging from −64% to 59% among different experiments (Table 6). In experiments B and E, test groups were less methylated compared to control groups, while in experiments C and D an opposite trend was observed.
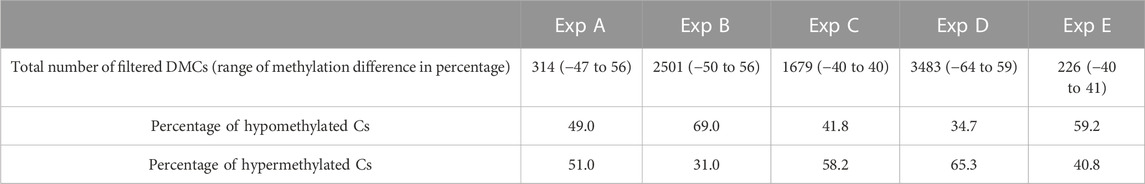
TABLE 6. An overview of the number of filtered DMCs and range of differences in methylation level identified in Atlantic salmon (Salmo salar L.) milt samples from different experiments (Exp A to E). Both hypo- and hyper-methylated Cs percentages refer to differences in test samples compared to control. For instance, in Exp A, 49 percent of mutual Cs in test individuals were less methylated compared to the same Cs in the control group.
Both hypo- and hyper-methylated Cs in all experiments were separately annotated with different genomic and CpG features. Considering gene features, results showed that the majority of DMCs were annotated with intergenic regions followed by introns (Table 7). Furthermore, a higher percentage of DMCs (both in hypo- and hyper-methylated groups) was annotated to promoter regions compared to exons. Regarding CpG features, on average, at least 95% of filtered DMCs in both hypomethylation and hypermethylation groups were annotated within regions outside of CpG islands (CGI)/CpG shores.
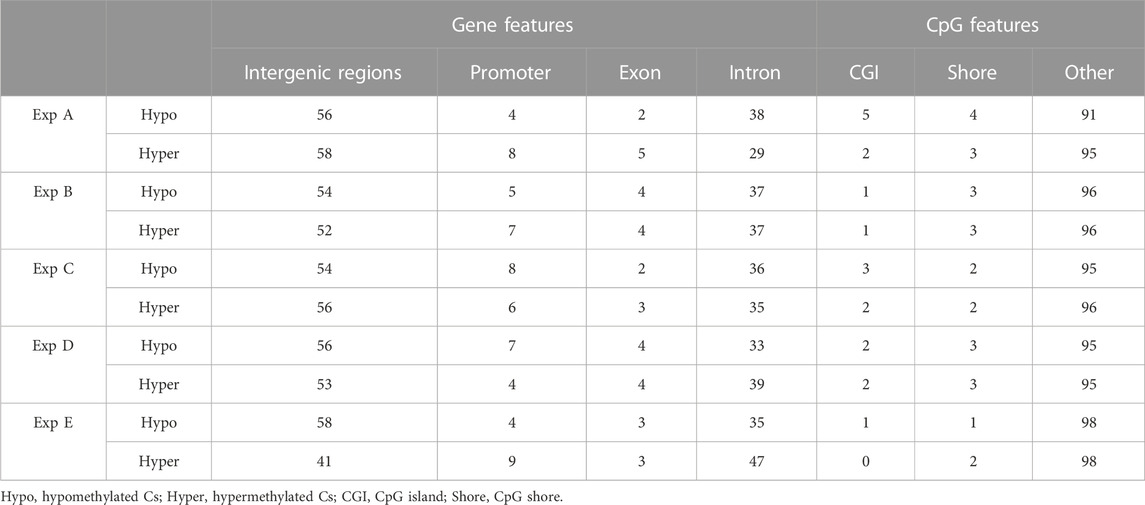
TABLE 7. An overview of the distribution (in percentage) of filtered DMCs identified in different experiments (Exp A to E) across gene and CpG features in the Atlantic salmon (Salmo salar L.) genome. Numbers indicate the percentage of annotated DMCs.
Genes whose TSSs were annotated with DMCs were first identified and subsequently subjected to functional annotation analysis. In the current study, significantly enriched terms were observed for all experiments, except experiment A. For each experiment, the top five significant terms and pathways per group (hypo and hyper) are shown in Table 8. A full list of significant terms and pathways can be found in (Supplementary Table S6). We observed the highest number of significant terms and pathways for experiment C, followed by D, B, and E (Figure 2). The analysis clearly showed that a wide variety of significant terms related to signaling, membrane structure, ion exchange, energy metabolism, cell adhesion, hormone, and embryo development can be linked to genes proximal to DMCs (Supplementary Table S6). The term entitled leucine-rich repeats, was the only mutually identified term in the hypo group of experiments B, C, and D (Figure 2A). No terms or pathways were found mutual among hyper groups between experiments (Figure 2B).
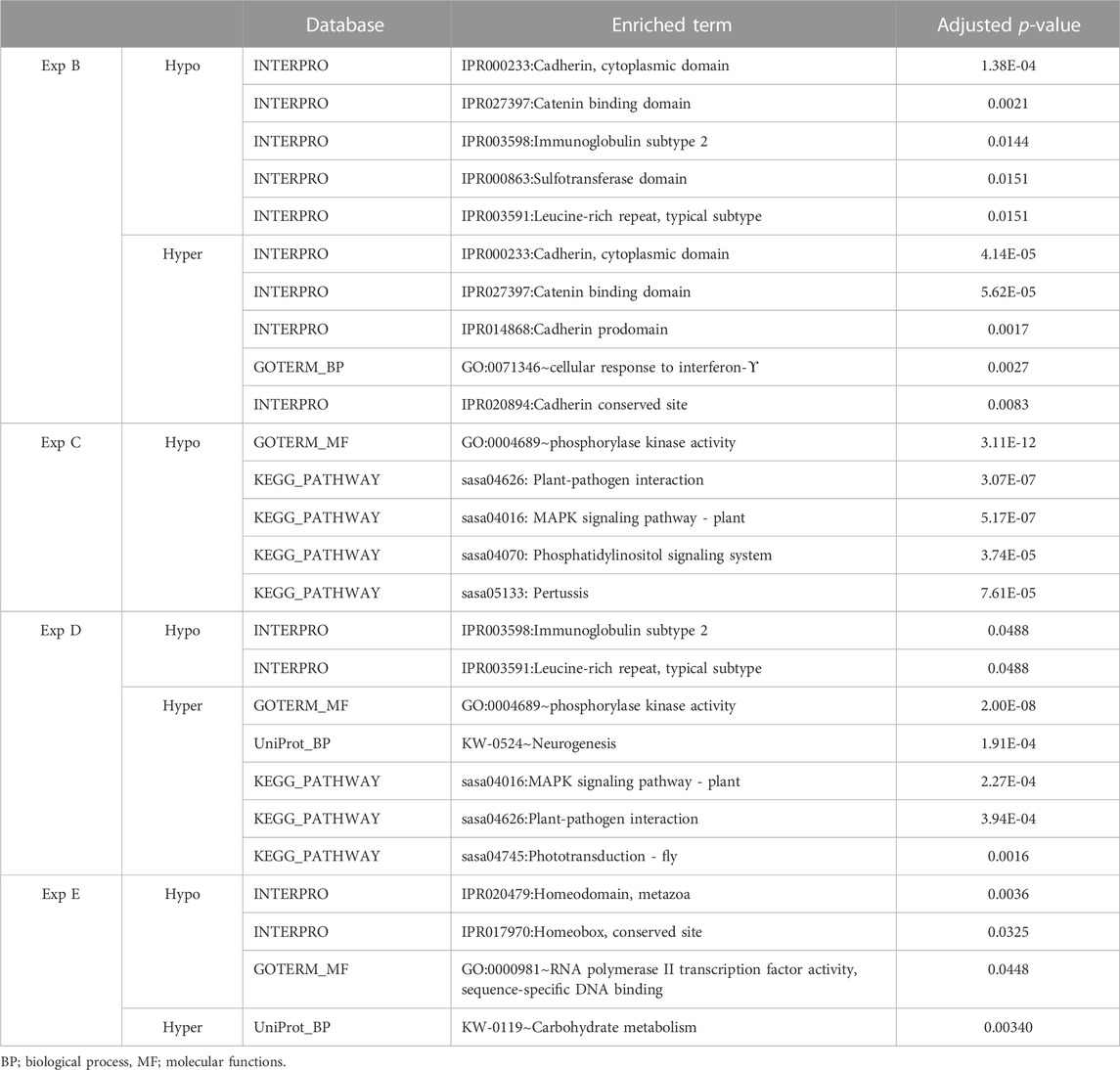
TABLE 8. An overview of functional annotation analysis for annotated genes associated with filtered DMCs in experiments B, C, D and E. No significant terms were enriched for experiment A. The p-value was benjamini corrected to correct for multiple testing.
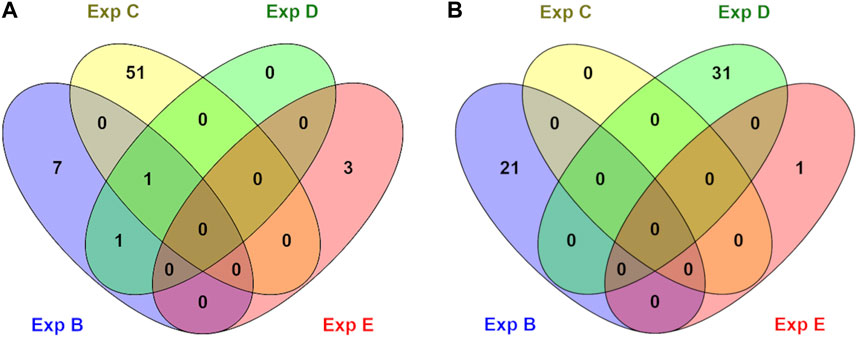
FIGURE 2. An overview of numbers of exclusive and shared identified significant GO terms in hypo (A) and hyper (B) methylated groups in different experiments (B, C, D, and E). No significant terms were identified for experiment A.
The main aim of the present study was to assess if cold storage of milt from Atlantic salmon prior to cryopreservation compromises post-thaw sperm quality and fertilizing potential, and if metabolomics and epigenetic signatures may reveal further aspects of milt quality evaluation. The results show that the fertilizing capacity of milt is preserved when stored for up to 4 days at two degrees. As hypothesized, cold storage for 4 days at eight degrees was detrimental to sperm quality and reduced the fertilization capacity of milt. Further, the results show that cryopreservation should be conducted shortly after milt collection, as milt cryopreserved after cold storage for 4 days exhibited reduced fertilization upon thawing. The negative effect of storage time, temperature, and cryopreservation was more pronounced when the sperm:egg ratio was reduced from 500 × 103 to 75 × 103 sperm per egg. Most of the in vitro sperm quality parameters were negatively influenced by storage time, temperature, and subsequent freezing/thawing. These findings are in agreement with previous studies reporting that cold storage and cryopreservation of Atlantic salmon sperm may affect in vitro sperm quality and fertilizing ability (Díaz et al., 2019; Kommisrud et al., 2020; Erraud et al., 2022).
The sperm viability decreased significantly during cold storage for 4 days. This is in agreement with previous studies in fish (Trigo et al., 2015; Kommisrud et al., 2020), and also reflects the observed decline in fertilizing ability during cold storage for 4 days at eight degrees. Interestingly, Kommisrud et al. (2020) found a small positive effect of storage on fertilization even though the viability was reduced during 6 days of cold storage. The discrepancy with our results may be explained by the lower sperm:egg ratio used in our study. The frozen-thawed samples had significantly lower sperm viability compared to cold-stored samples. This is congruent with previous reports in mammals (Gürler et al., 2016; Narud et al., 2021a) and fish (Díaz et al., 2019; Lee-Estevez et al., 2019).
The presented CASA results on TM and PM are significantly lower for cold-stored milt than otherwise reported for Atlantic salmon (Dziewulska et al., 2010; Parodi et al., 2017). For frozen-thawed milt, our results for TM and PM are higher for day one than previously reported by others (Dziewulska et al., 2011), while comparable to others when cold-stored for 4 days prior to cryopreservation (Kommisrud et al., 2020). There are several factors that may affect motility results, including temperature and sperm activation procedures (Caldeira and Soler, 2018), as well as differences between CASA systems and the settings used for the analysis (Boryshpolets et al., 2013).
For both cold-stored and frozen-thawed samples, there was a significant decline in TM, PM, and ATP levels from day one to day four of storage. Surprisingly, the frozen-thawed sperm cells did not show significantly reduced motility and ATP levels compared to cold-stored samples, in disagreement with previous results reported by others (Dziewulska et al., 2011; Figueroa et al., 2016; Kommisrud et al., 2020). Interestingly, the velocity parameters and LIN and STR were significantly higher in samples cryopreserved after cold storage compared to those stored cold. This is in disagreement with previous studies, reporting that the kinematic parameters decrease after cryopreservation (Dziewulska et al., 2011; Figueroa et al., 2016; Kommisrud et al., 2020). Our results may have been influenced by the activation procedure, which was identical for fresh and cryopreserved samples. A dedicated activation procedure for the fresh milt could have been more optimal and given results more in compliance with other reports. However, the motility values for the different samples are comparable relative to each other within storage conditions. This is also reflected by the correlation analysis, where the post-thaw in vitro motility parameters displayed a greater correlation with fertilization rates than for the cold-stored samples.
Sperm DNA integrity is crucial for successful fertilization and embryo survival (Pérez-Cerezales et al., 2010; Kumaresan et al., 2020). Both cold-stored and frozen-thawed milt showed significantly higher percentages of DFI and HDS in samples stored for 4 days at two and eight degrees, compared to samples analyzed on day one. Furthermore, frozen-thawed sperm had significantly higher levels of fragmented DNA compared with cold-stored sperm. This is congruent with the literature, reporting that both cold-stored and cryopreserved milt are prone to sperm DNA damage (Cabrita et al., 2005; Pérez-Cerezales et al., 2011; Contreras et al., 2020).
All the in vitro post-thaw parameters assessed in this study were correlated with fertilization rates for 75 × 103 sperm per egg. The highest correlations were found for the motility parameters TM and PM, ATP content, and the kinematic parameters VAP, VCL, and VSL, consistent with results obtained by others (Dziewulska et al., 2011; Figueroa et al., 2016; Kommisrud et al., 2020). Also, the DNA integrity parameters DFI and HDS were significantly negatively correlated with fertilization rates. As expected, the correlations were weaker with 500 × 103 sperm per egg. Cryopreservation of cold-stored samples resulted in a significant reduction of fertilized eggs compared to cold-stored samples, except for day one using the sperm:egg ratio 500 × 103. However, the relatively low fertilization rate obtained for day one with cold-stored milt might be explained by poor egg quality observed this day.
Previous studies have investigated the metabolome of fish milt and seminal plasma (Dietrich et al., 2019b; Tsentalovich et al., 2020). However, to our knowledge, this is the first study investigating sperm metabolites in relation to fertilizing potential of cold-stored and cryopreserved salmon milt. Overall, our results showed that the different storage conditions of cold-stored milt significantly affected the level of amines and several amino acids. In frozen-thawed samples, only a few metabolites were affected by storage conditions prior to cryopreservation. The process of freezing and thawing milt significantly reduced the levels of all amines and most of the amino acids studied. Our in vitro sperm quality analysis showed a significant reduction in sperm viability after freezing and thawing. Thus, there was a higher percentage of dead sperm cells in the cryopreserved samples analyzed for targeted metabolites compared to the cold-stored samples. It is possible that the damaged sperm membranes have leaked intracellular metabolites causing the significant reduction in metabolite concentrations observed after cryopreservation. However, the seminal plasma samples did not necessarily show a corresponding increase in metabolite concentration. This may be explained by the metabolite concentrations of seminal plasma samples of cold-stored and cryopreserved milt being not directly comparable. The cryopreserved samples were diluted based on a standardized cell number while the cold-stored samples were diluted 1:1 independent of the sperm number. Thus, due to a higher dilution factor for frozen-thawed samples, the concentrations of metabolites in the seminal plasma samples will be lower. For future studies, the dilution of cold-stored and cryopreserved samples should be standardized such that the metabolite concentrations of cold-stored and frozen-thawed milt samples can be compared directly.
Metabolites such as different amino acids and amines have been shown to act as antioxidants, protecting sperm cells from oxidative stress during freezing and thawing (Davoodian et al., 2017; Dietrich et al., 2019b; Ugur et al., 2019). However, the exact mechanisms of metabolites as cryo-protectants have not been clearly understood and there are few studies documenting the role of metabolites during freezing and thawing of milt. Interestingly, after cryopreservation, the level of L-carnitine was significantly reduced in the samples stored for 4 days compared to day one samples. Furthermore, the level of L-carnitine was positively correlated with fertilization rate of cryopreserved samples. In mammals, L-carnitine has been shown to be highly concentrated in the epididymis and sperm (Jeulin and Lewin, 1996). It is essential for energy metabolism and is involved in sperm motility, sperm maturation and fertilization. Furthermore, L-carnitine is shown to protect sperm cells against oxidative damage (Abdelrazik and Agarwal, 2009; Aliabadi et al., 2012). Thus, one hypothesis for this reduction in L-carnitine is that more L-carnitine has been utilized to protect the sperm cells from reactive oxygen species during the 4 days of cold-storage. For future studies, it would be interesting to further investigate the effect of L-carnitine supplementation on milt quality during storage and cryopreservation. Overall, our results show that storage conditions and cryopreservation of salmon milt affect the concentration of sperm metabolites. Furthermore, several of the metabolites studied were highly correlated with the fertilization ability of salmon milt. Metabolomics of sperm and seminal plasma has thus been shown applicable for Atlantic salmon milt, and a possible tool for broader fertility research. Further studies applying a larger number of individuals are needed to increase knowledge of the fish sperm metabolome, its relationship with fertilizing ability, and the possible role of metabolites as cryo-protectors.
In the current study, the average global sperm DNA methylation level after different milt handling regimes was 87.3%. Similar global hypermethylated status was previously reported for Atlantic salmon milt (Wellband et al., 2021), fertilized salmon eggs (Moghadam et al., 2017) and Rainbow trout milt (Gavery et al., 2018). We previously reported 33%–43% of global methylation levels in boar (Khezri et al., 2019) and bull sperm cells (Khezri et al., 2020; Narud et al., 2021b) using the RRBS technique. This indicates different parental DNA methylation patterns among mammalian species and salmonids. Although, in this study, the methylation profiles of CpG10x were similar between samples, clustering the control and test samples from the same individuals together might also be explained by the low number of samples.
In this study the overall distribution trend of filtered DMCs over gene and CpG features was similar to the trend previously seen in boar (Khezri et al., 2019) and bull (Khezri et al., 2020). Similar distribution and annotation rates previously reported for Atlantic salmon (Moghadam et al., 2017; Robinson et al., 2019). However, the percentages of annotated filtered DMCs with intron in salmon were considerably higher compared to bull and boar reported previously by our group and might be explained by the interspecies differences. Worth mentioning that the differences in percentages of annotated filtered DMCs with introns and CGIs among salmon and bull were smaller than the differences in salmon and boar for the same regions. Several factors could explain this observation. Firstly, the genome size of salmon (2.8 Gb) and bull (2.7 Gb) is bigger than pig (2.5 Gb) and secondly, we sequenced the libraries for both salmon and bull in paired-end mode vs. single-end mode in boar. This potentially provides more coverage for the analyses.
To our knowledge, this is the first study investigating the effects of storage on sperm DNA methylation patterns in Atlantic salmon. A previous study documented the sperm DNA methylation pattern in European carp (Cyprinus carpio) and revealed that similar to our study (Exp A), refrigeration of milt for 24 h significantly increased methylation levels (Cheng et al., 2021). This is similar to the results presented here for Exp A, although we observed a slight hypermethylation. It seems reasonable to consider milt handling and storage time as environmental stress factors. Previous studies have shown that DNA methylation in salmonid sperm cells is highly dynamic and can be influenced by different environmental factors. For instance, sperm cells taken from wild salmon showed very distinct epigenetic signatures compared to sperm cells from hatcher-reared Atlantic salmons (Rodriguez Barreto et al., 2019) and rainbow trout (Nilsson et al., 2021).
Results in this study revealed no significant enriched pathways and Go terms for Exp A. This is comparable with similar and not significant fertilization rates between D1–2°C and D1–2°C/Cryo for both sperm:egg ratios. Therefore, both experiments indicate that cryopreservation within 1 day of milt collection, might be the optimal protocol among tested methods. It is worth mentioning that optimization of the extender formula used for cryopreservation, might have some effects on sperm DNA methylation levels. For instance, it has been shown that cryoprotectant agents decreased the global sperm DNA methylation level in Tambaqui fish, Colossoma macropomum (de Mello et al., 2017).
Results in this study documented that milt handling and storage could significantly change site-specific methylation levels proximal to genes involved in important biological pathways. We observed a significant enrichment of terms related to hormones (renin, oxytocin, insulin, GnRH, aldosterone, estrogen) and signaling, in both hypo group of exp C and hyper group of exp D. Terms reflecting developmental pathways such as melanogenesis, oocyte meiosis (hypo group of exp C), neurogenesis, angiogenesis and nervous system development (hyper group of exp D) were also identified. This is in agreement with previously published results where rearing condition changed sperm DNA methylation status close to genes involved in neuronal development in Atlantic salmons (Rodriguez Barreto et al., 2019). Similar to our study, previously published results in rainbow trout showed that important biological pathways such as development and signaling can be linked with differentially methylated regions in sperm cells taken from wild and hatchery-reared rainbow trout (Nilsson et al., 2021).
Furthermore, we observed a significant overrepresentation of GO terms related to immune response (hypo groups of both exp B and D) and calcium signaling in hypo group of exp C and hyper group of exp D. This is in line with previously published results where authors showed epigenetic modifications induced by hatchery rearing condition in pacific salmon (Le Luyer et al., 2017). Furthermore, our analysis showed that the GO term homeobox protein is significantly enriched in hypo group of Exp E. Different protein members of the Homeobox family also were significantly enriched following changes in sperm DNA methylation in wild and hatchery-reared rainbow trout (Gavery et al., 2018). It has been shown that a homeobox protein expressed in the male reproductive system is involved in developmental events in different species including salmon and cattle (Cánovas et al., 2014; Mobley et al., 2021).
GO terms representing the membrane integrity and cell adhesion process were specifically enriched in Exp B. Leucine-rich repeats (LPR) was the only term significantly enriched in the hypo groups of experiments B, C, and D. The LPR motifs have been reported in a variety of proteins involved in different biological processes such as signaling, cell adhesion, apoptosis, and the immune response (Ng and Xavier, 2011). Interestingly, in this study, leucine was identified as an amino acid whose concentration in sperm cells changed dramatically upon cryopreservation.
The research to date has tended to focus on environmental stressors such as rearing condition effects on epigenetic modification, rather than milt handling effects. The current results highlight the importance of handling and storage of Atlantic salmon milt and their potential to induce epigenetic modifications in sperm cells. The current study showed that milt handling and storage changed DNA methylation patterns close to genes involved mostly in developmental processes. Hence, it could conceivably be hypothesized that developmental pathways are at greater risk following incorrect milt handling and storage. The same conclusion can be drawn based on previous publications where different rearing conditions were tested. Therefore, one could speculate that environmental factors have the potential to alter DNA methylation in Salmonid sperm cells (whether the semen sample is exposed to those factors or the donor) in regions proximal to genes that are potentially involved in the development. More broadly research is needed to determine these effects in the offspring of Atlantic salmon.
In this study, we aimed to create contrasts to explore the potential value of metabolomics and epigenetics on Atlantic salmon sperm and milt. It was successfully shown that storage duration and corresponding cryopreservation influenced metabolites and sperm DNA methylation signatures. Further studies should include shorter preservation intervals to reveal the possibility of applying an extended period of storage before cryopreservation.
In conclusion, the fertilization capacity of cold-stored milt from Atlantic salmon is preserved when stored for up to 4 days at two degrees. Cold storage for 4 days at eight degrees and corresponding cryopreservation was detrimental to sperm quality and influenced levels of milt metabolites and DNA methylation signatures. Cryopreservation of milt stored for 1 day at two degrees do not compromise either fertilization ability or sperm DNA methylation signatures.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB61229.
Ethical review and approval was not required for the animal study because the milt and eggs used for this study was routinely collected at broodstocks. The broodstocks are under supervision/surveillance of an official oppointed veterinarian. Salmon donors of milt and eggs were kept under the Norwegian legislation for aquaculture.
BN: Data curation, formal analysis, visualization, and writing—original draft. AK: Data curation, formal analysis, and writing—original draft. TZ, AN, A-LE, and IG: Investigation, data curation, and writing—review, and editing. GK: Methodology, writing—review, and editing. RW: Validation, Writing—review, and editing. EK: Conceptualization, supervision, writing—review, and editing. All authors contributed to the article and approved the submitted version.
This work was funded by an internal scholarship of Inland Norway University of Applied Sciences and financial support was received from Inland Regional Research Fund, Norway (grant number 313350).
We kindly thank master student Madhuri Angeeru for her contribution in sperm quality analyses. The Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by Oslo University Hospital and the University of Oslo, supported by the Research Council of Norway and the Southeastern Regional Health Authority, provided the sequencing service.
Authors A-LE and IG were employed by Cryogenetics AS.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1199681/full#supplementary-material
Abdelrazik, H., and Agarwal, A. (2009). Invited reviewL-carnitine and assisted reproduction. Archives Med. Sci. Special Issues 2009, 47.
Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., et al. (2012). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87. doi:10.1186/gb-2012-13-10-r87
Aliabadi, E., Soleimani Mehranjani, M., Borzoei, Z., Talaei-Khozani, T., Mirkhani, H., and Tabesh, H. (2012). Effects of L-carnitine and L-acetyl-carnitine on testicular sperm motility and chromatin quality. Iran. J. Reprod. Med. 10, 77–82.
Asturiano, J. F., Cabrita, E., and Horváth, Á. (2017). Progress, challenges and perspectives on fish gamete cryopreservation: A mini-review. Gen. Comp. Endocrinol. 245, 69–76. doi:10.1016/j.ygcen.2016.06.019
Balamurugan, R., Prapaporn, W., and Munuswamy, N. (2019). Sperm activation and effects of cryopreservation on motility, ultrastructure and DNA integrity in Grey mullet Mugil cephalus. Aquac. Rep. 14, 100204. doi:10.1016/j.aqrep.2019.100204
Blaxter, J. H. S. (1953). Sperm storage and cross-fertilization of spring and autumn spawning herring. Nature 172, 1189–1190. doi:10.1038/1721189b0
Boryshpolets, S., Kowalski, R. K., Dietrich, G. J., Dzyuba, B., and Ciereszko, A. (2013). Different computer-assisted sperm analysis (CASA) systems highly influence sperm motility parameters. Theriogenology 80, 758–765. doi:10.1016/j.theriogenology.2013.06.019
Boyle, P., Clement, K., Gu, H., Smith, Z. D., Ziller, M., Fostel, J. L., et al. (2012). Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13, R92. doi:10.1186/gb-2012-13-10-r92
Cabrita, E., Robles, V., and Herraez, P. (2008). Methods in reproductive aquaculture: Marine and freshwater species. Boca Raton, United States: CRC Press.
Cabrita, E., Robles, V., Rebordinos, L., Sarasquete, C., and Herráez, M. P. (2005). Evaluation of DNA damage in rainbow trout (Oncorhynchus mykiss) and gilthead sea bream (Sparus aurata) cryopreserved sperm. Cryobiology 50, 144–153. doi:10.1016/j.cryobiol.2004.12.003
Cabrita, E., Sarasquete, C., Martínez-Páramo, S., Robles, V., Beirão, J., Pérez-Cerezales, S., et al. (2010). Cryopreservation of fish sperm: Applications and perspectives. J. Appl. Ichthyol. 26, 623–635. doi:10.1111/j.1439-0426.2010.01556.x
Caldeira, C., and Soler, C. (2018). Fish sperm assessment using software and cooling devices. J. Vis. Exp. 2018, 56823. doi:10.3791/56823
Cánovas, A., Reverter, A., DeAtley, K. L., Ashley, R. L., Colgrave, M. L., Fortes, M. R. S., et al. (2014). Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLOS ONE 9, e102551. doi:10.1371/journal.pone.0102551
Champroux, A., Cocquet, J., Henry-Berger, J., Drevet, J. R., and Kocer, A. (2018). A decade of exploring the mammalian sperm epigenome: Paternal epigenetic and transgenerational inheritance. Front. Cell Dev. Biol. 6, 50. doi:10.3389/fcell.2018.00050
Cheng, Y., Vechtova, P., Fussy, Z., Sterba, J., Linhartová, Z., Rodina, M., et al. (2021). Changes in phenotypes and DNA methylation of in vitro aging sperm in common carp Cyprinus carpio. Int. J. Mol. Sci. 22, 5925. doi:10.3390/ijms22115925
Contreras, P., Dumorné, K., Ulloa-Rodríguez, P., Merino, O., Figueroa, E., Farías, J. G., et al. (2020). Effects of short-term storage on sperm function in fish semen: A review. Rev. Aquac. 12, 1373–1389.
Dietrich, M. A., Samczuk, P., Ciborowski, M., Nynca, J., Parfieniuk, E., Pietrowska, K., et al. (2019b). Metabolic fingerprinting of carp and rainbow trout seminal plasma. Aquaculture 501, 178–190. doi:10.1016/j.aquaculture.2018.11.017
Davoodian, N., Kadivar, A., Ahmadi, E., and Mohebbi, A. (2017). Effects of two amino acids on motion parameters and enzymatic antioxidant activity of freeze-thawed stallion spermatozoa. J. Equine Vet. Sci. 59, 49–56. doi:10.1016/j.jevs.2017.09.007
de Mello, F., Garcia, J. S., Godoy, L. C., Depincé, A., Labbé, C., and Streit, D. P. (2017). The effect of cryoprotectant agents on DNA methylation patterns and progeny development in the spermatozoa of Colossoma macropomum. General Comp. Endocrinol. 245, 94–101. doi:10.1016/j.ygcen.2016.06.003
Depincé, A., Gabory, A., Dziewulska, K., Le Bail, P. Y., Jammes, H., and Labbé, C. (2020). DNA methylation stability in fish spermatozoa upon external constraint: Impact of fish hormonal stimulation and sperm cryopreservation. Mol. Reprod. Dev. 87, 124–134. doi:10.1002/mrd.23297
Díaz, R., Lee-Estevez, M., Quiñones, J., Dumorné, K., Short, S., Ulloa-Rodríguez, P., et al. (2019). Changes in Atlantic salmon (Salmo salar) sperm morphology and membrane lipid composition related to cold storage and cryopreservation. Anim. Reprod. Sci. 204, 50–59. doi:10.1016/j.anireprosci.2019.03.004
Dietrich, M. A., Nynca, J., and Ciereszko, A. (2019a). Proteomic and metabolomic insights into the functions of the male reproductive system in fishes. Theriogenology 132, 182–200. doi:10.1016/j.theriogenology.2019.04.018
Dziewulska, K., Rzemieniecki, A., Czerniawski, R., and Domagała, J. (2011). Post-thawed motility and fertility from Atlantic salmon (Salmo salar L) sperm frozen with four cryodiluents in straws or pellets. Theriogenology 76, 300–311. doi:10.1016/j.theriogenology.2011.02.007
Dziewulska, K., Rzemieniecki, A., and Domagała, J. (2010). Motility and energetic status of Atlantic salmon (Salmo salar L) sperm after refrigerated storage. J. Appl. Ichthyol. 26, 668–673. doi:10.1111/j.1439-0426.2010.01538.x
Dzyuba, B., Bondarenko, O., Fedorov, P., Gazo, I., Prokopchuk, G., and Cosson, J. (2017). Energetics of fish spermatozoa: The proven and the possible. Aquaculture 472, 60–72. doi:10.1016/j.aquaculture.2016.05.038
Egea, R. R., Puchalt, N. G., Escrivá, M. M., and Varghese, A. C. (2014). OMICS: Current and future perspectives in reproductive medicine and technology. J. Hum. Reprod. Sci. 7, 73–92. doi:10.4103/0974-1208.138857
Engel, K. M., Baumann, S., Rolle-Kampczyk, U., Schiller, J., von Bergen, M., and Grunewald, S. (2019). Metabolomic profiling reveals correlations between spermiogram parameters and the metabolites present in human spermatozoa and seminal plasma. PLoS One 14, e0211679. doi:10.1371/journal.pone.0211679
Erraud, A., Cornet, V., Baekelandt, S., Neus, Y., Antipine, S., Lambert, J., et al. (2022). Optimal sperm–egg ratios for successful fertilization using fresh and cryopreserved sperm in wild anadromous Atlantic salmon (Salmo salar L 1758). Aquaculture 549, 737758. doi:10.1016/j.aquaculture.2021.737758
Figueroa, E., Valdebenito, I., Merino, O., Ubilla, A., Risopatrón, J., and Farias, J. G. (2016). Cryopreservation of atlantic salmon Salmo salar sperm: Effects on sperm physiology. J. Fish. Biol. 89, 1537–1550. doi:10.1111/jfb.13052
Gavery, M. R., Nichols, K. M., Goetz, G. W., Middleton, M. A., and Swanson, P. (2018). Characterization of genetic and epigenetic variation in sperm and red blood cells from adult hatchery and natural-origin steelhead, Oncorhynchus mykiss. G3 Genes|genomes|genetics. 8, 3723–3736. doi:10.1534/g3.118.200458
Gu, H., Smith, Z. D., Bock, C., Boyle, P., Gnirke, A., and Meissner, A. (2011). Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481. doi:10.1038/nprot.2010.190
Gürler, H., Malama, E., Heppelmann, M., Calisici, O., Leiding, C., Kastelic, J. P., et al. (2016). Effects of cryopreservation on sperm viability, synthesis of reactive oxygen species, and DNA damage of bovine sperm. Theriogenology 86, 562–571. doi:10.1016/j.theriogenology.2016.02.007
Herráez, M. P., Ausió, J., Devaux, A., González-Rojo, S., Fernández-Díez, C., Bony, S., et al. (2017). Paternal contribution to development: Sperm genetic damage and repair in fish. Aquaculture 472, 45–59. doi:10.1016/j.aquaculture.2016.03.007
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Jeulin, C., and Lewin, L. M. (1996). Role of free L-carnitine and acetyl-L-carnitine in post-gonadal maturation of mammalian spermatozoa. Hum. Reprod. Update 2, 87–102. doi:10.1093/humupd/2.2.87
Khezri, A., Narud, B., Stenseth, E. B., Johannisson, A., Myromslien, F. D., Gaustad, A. H., et al. (2019). DNA methylation patterns vary in boar sperm cells with different levels of DNA fragmentation. BMC Genomics 20, 897. doi:10.1186/s12864-019-6307-8
Khezri, A., Narud, B., Stenseth, E. B., Zeremichael, T. T., Myromslien, F. D., Wilson, R. C., et al. (2020). Sperm DNA hypomethylation proximal to reproduction pathway genes in maturing elite Norwegian red bulls. Front. Genet. 11, 922. doi:10.3389/fgene.2020.00922
Kommisrud, E., Myromslien, F. D., Stenseth, E.-B., Zeremichael, T. T., Hofman, N., Grevle, I., et al. (2020). Viability, motility, ATP content and fertilizing potential of sperm from Atlantic salmon (Salmo salar L) in milt stored before cryopreservation. Theriogenology 151, 58–65. doi:10.1016/j.theriogenology.2020.04.008
Krueger, F., and Andrews, S. R. (2011). Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinforma. Oxf. Engl. 27, 1571–1572. doi:10.1093/bioinformatics/btr167
Kumaresan, A., Das Gupta, M., Datta, T. K., and Morrell, J. M. (2020). Sperm DNA integrity and male fertility in farm animals: A review. Front. Vet. Sci. 7, 321. doi:10.3389/fvets.2020.00321
Labbé, C., Robles, V., and Herraez, M. P. (2017). Epigenetics in fish gametes and early embryo. Aquaculture 472, 93–106. doi:10.1016/j.aquaculture.2016.07.026
Le Luyer, J., Laporte, M., Beacham, T. D., Kaukinen, K. H., Withler, R. E., Leong, J. S., et al. (2017). Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc. Natl. Acad. Sci. 114, 12964–12969. doi:10.1073/pnas.1711229114
Lee-Estevez, M., Herrera, L., Díaz, R., Beltrán, J., Figueroa, E., Dumorné, K., et al. (2019). Effects of cryopreservation on cAMP-dependent protein kinase and AMP-activated protein kinase in Atlantic salmon (Salmo salar) spermatozoa: Relation with post-thaw motility. Anim. Reprod. Sci. 209, 106133. doi:10.1016/j.anireprosci.2019.106133
Liu, Q. H., Li, J., Xiao, Z. Z., Ding, F. H., Yu, D. D., and Xu, X. Z. (2007). Use of computer-assisted sperm analysis (CASA) to evaluate the quality of cryopreserved sperm in red seabream (Pagrus major). Aquaculture 263, 20–25. doi:10.1016/j.aquaculture.2006.11.017
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10. doi:10.14806/ej.17.1.200
Menezes, E. B., Velho, A. L. C., Santos, F., Dinh, T., Kaya, A., Topper, E., et al. (2019). Uncovering sperm metabolome to discover biomarkers for bull fertility. BMC Genomics 20, 714. doi:10.1186/s12864-019-6074-6
Merino, O., Figueroa, E., Cheuquemán, C., Valdebenito, I., Isachenko, V., Isachenko, E., et al. (2017). Short-term storage of salmonids semen in a sodium alginate-based extender. Andrologia 49, e12661. doi:10.1111/and.12661
Mobley, K. B., Aykanat, T., Czorlich, Y., House, A., Kurko, J., Miettinen, A., et al. (2021). Maturation in atlantic salmon (Salmo salar, salmonidae): A synthesis of ecological, genetic, and molecular processes. Rev. Fish Biol. Fish. 31, 523–571. doi:10.1007/s11160-021-09656-w
Moghadam, H. K., Johnsen, H., Robinson, N., Andersen, Ø., Jørgensen, E. H., Johnsen, H. K., et al. (2017). Impacts of early Life stress on the methylome and transcriptome of atlantic salmon. Sci. Rep. 7, 5023. doi:10.1038/s41598-017-05222-2
Narud, B., Khezri, A., Nordborg, A., Klinkenberg, G., Zeremichael, T. T., Stenseth, E.-B., et al. (2021a). Semen quality parameters including metabolites, sperm production traits and fertility in young Norwegian Red AI bulls. Livest. Sci. 255, 104803. doi:10.1016/j.livsci.2021.104803
Narud, B., Khezri, A., Zeremichael, T. T., Stenseth, E.-B., Heringstad, B., Johannisson, A., et al. (2021b). Sperm chromatin integrity and DNA methylation in Norwegian Red bulls of contrasting fertility. Mol. Reproduction Dev. 88, 187–200. n/a. doi:10.1002/mrd.23461
Narud, B., Klinkenberg, G., Khezri, A., Zeremichael, T. T., Stenseth, E.-B., Nordborg, A., et al. (2020). Differences in sperm functionality and intracellular metabolites in Norwegian Red bulls of contrasting fertility. Theriogenology 157, 24–32. doi:10.1016/j.theriogenology.2020.07.005
Ng, A., and Xavier, R. J. (2011). Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 7, 1082–1084. doi:10.4161/auto.7.9.16464
Nilsson, E., Sadler-Riggleman, I., Beck, D., and Skinner, M. K. (2021). Differential DNA methylation in somatic and sperm cells of hatchery vs wild (natural-origin) steelhead trout populations. Environ. Epigenetics 7, dvab002. doi:10.1093/eep/dvab002
Parodi, J., Guerra, G., Cuevas, M., Ramírez-Reveco, A., and Romero, F. (2017). Effects of storage time on the motility, mortality and calcium levels of Atlantic salmon Salmo salar spermatozoa. J. Fish. Biol. 90, 1506–1516. doi:10.1111/jfb.13245
Pérez-Cerezales, S., Gutiérrez-Adán, A., Martínez-Páramo, S., Beirão, J., and Herráez, M. P. (2011). Altered gene transcription and telomere length in trout embryo and larvae obtained with DNA cryodamaged sperm. Theriogenology 76, 1234–1245. doi:10.1016/j.theriogenology.2011.05.028
Pérez-Cerezales, S., Martínez-Páramo, S., Beirão, J., and Herráez, M. P. (2010). Fertilization capacity with rainbow trout DNA-damaged sperm and embryo developmental success. Reproduction 139, 989–997. doi:10.1530/REP-10-0037
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. doi:10.1093/nar/gkz369
Riesco, M. F., and Robles, V. (2013). Cryopreservation causes genetic and epigenetic changes in zebrafish genital ridges. PLoS One 8, e67614. doi:10.1371/journal.pone.0067614
Robinson, N. A., Johnsen, H., Moghadam, H., Andersen, Ø., and Tveiten, H. (2019). Early developmental stress affects subsequent gene expression response to an acute stress in atlantic salmon: An approach for creating robust fish for aquaculture? G3 Genes|Genomes|Genetics 9, 1597–1611. doi:10.1534/g3.119.400152
Rodriguez Barreto, D., Garcia de Leaniz, C., Verspoor, E., Sobolewska, H., Coulson, M., and Consuegra, S. (2019). DNA methylation changes in the sperm of captive-reared fish: A route to epigenetic introgression in wild populations. Mol. Biol. Evol. 36, 2205–2211. doi:10.1093/molbev/msz135
Samuelsson, L. M., and Larsson, D. G. (2008). Contributions from metabolomics to fish research. Mol. Biosyst. 4, 974–979. doi:10.1039/b804196b
Tiersch, T. R., Yang, H., Jenkins, J. A., and Dong, Q. (2007). Sperm cryopreservation in fish and shellfish. Soc. Reprod. Fertil. Suppl. 65, 493–508.
Trigo, P., Merino, O., Figueroa, E., Valdebenito, I., Sánchez, R., and Risopatrón, J. (2015). Effect of short-term semen storage in salmon (Oncorhynchus mykiss) on sperm functional parameters evaluated by flow cytometry. Andrologia 47, 407–411. doi:10.1111/and.12276
Tsentalovich, Y. P., Zelentsova, E. A., Yanshole, L. V., Yanshole, V. V., and Odud, I. M. (2020). Most abundant metabolites in tissues of freshwater fish pike-perch (Sander lucioperca). Sci. Rep. 10, 17128. doi:10.1038/s41598-020-73895-3
UCSC (2018). Table browser for bosTau 9 assembly. Available at: https: //genome.ucsc.edu/cgi-bin/hgTables.
Ugur, M. R., Dinh, T., Hitit, M., Kaya, A., Topper, E., Didion, B., et al. (2019). Amino acids of seminal plasma associated with freezability of bull sperm. Front. Cell Dev. Biol. 7, 347. doi:10.3389/fcell.2019.00347
Wellband, K., Roth, D., Linnansaari, T., Curry, R. A., and Bernatchez, L. (2021). Environment-driven reprogramming of gamete DNA methylation occurs during maturation and is transmitted intergenerationally in Atlantic Salmon. G3 (Bethesda) 11, jkab353. doi:10.1093/g3journal/jkab353
Woods, L. C., Li, Y., Ding, Y., Liu, J., Reading, B. J., Fuller, S. A., et al. (2018). DNA methylation profiles correlated to striped bass sperm fertility. BMC Genomics 19, 244. doi:10.1186/s12864-018-4548-6
Keywords: Atlantic salmon, spermatozoa, cold storage, cryopreservation, metabolite, RRBS
Citation: Narud B, Khezri A, Zeremichael TT, Eriksen A-L, Grevle IS, Nordborg A, Klinkenberg G, Wilson RC and Kommisrud E (2023) Sperm quality parameters, fertilizing potential, metabolites, and DNA methylation in cold-stored and cryopreserved milt from Atlantic salmon (Salmo salar L.). Front. Genet. 14:1199681. doi: 10.3389/fgene.2023.1199681
Received: 03 April 2023; Accepted: 25 July 2023;
Published: 24 August 2023.
Edited by:
Martino Cassandro, University of Padua, ItalyReviewed by:
Avril Maellen Harder, Auburn University, United StatesCopyright © 2023 Narud, Khezri, Zeremichael, Eriksen, Grevle, Nordborg, Klinkenberg, Wilson and Kommisrud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Birgitte Narud, YmlyZ2l0dGUubmFydWRAaW5uLm5v
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.