
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 25 July 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1197206
This article is part of the Research TopicSolanaceae VIII: Biodiversity, Climate Change and BreedingView all 7 articles
Potato, tomato, pepper, and eggplant are worldwide important crop and vegetable species of the Solanaceae family. Molecular linkage maps of these plants have been constructed and used to map qualitative and quantitative traits of agronomic importance. This research has been undertaken with the vision to identify the molecular basis of agronomic characters on the one hand, and on the other hand, to assist the selection of improved varieties in breeding programs by providing DNA-based markers that are diagnostic for specific agronomic characters. Since 2011, whole genome sequences of tomato and potato became available in public databases. They were used to combine the results of several hundred mapping and map-based cloning studies of phenotypic characters between 1988 and 2022 in physical maps of the twelve tomato and potato chromosomes. The traits evaluated were qualitative and quantitative resistance to pathogenic oomycetes, fungi, bacteria, viruses, nematodes, and insects. Furthermore, quantitative trait loci for yield and sugar content of tomato fruits and potato tubers and maturity or earliness were physically mapped. Cloned genes for pathogen resistance, a few genes underlying quantitative trait loci for yield, sugar content, and maturity, and several hundred candidate genes for these traits were included in the physical maps. The comparison between the physical chromosome maps revealed, in addition to known intrachromosomal inversions, several additional inversions and translocations between the otherwise highly collinear tomato and potato genomes. The integration of the positional information from independent mapping studies revealed the colocalization of qualitative and quantitative loci for resistance to different types of pathogens, called resistance hotspots, suggesting a similar molecular basis. Synteny between potato and tomato with respect to genomic positions of quantitative trait loci was frequently observed, indicating eventual similarity between the underlying genes.
The Solanaceae are a highly diverse family of land plants. It comprises 3,000 to 4,000 species that are organized into approximately 90 genera, the largest of which is the genus Solanum (Knapp et al., 2004). Some species of the Solanaceae had important roles in the history of mankind as providers of edible fruits and tubers such as tomato, potato, pepper, and eggplant, drugs such as tobacco, or ornamentals such as Petunia. Members of this family have been domesticated thousands of years ago in the Americas by pre-Columbian civilizations (potato, tomato, and pepper) and in South-East Asia (eggplant) (Daunay and Laterrot, 2008). Today, crop or vegetable species of worldwide importance are the potato (Solanum tuberosum Group tuberosum), tomato (Solanum lycopersicum, former Lycopersicon esculentum), eggplant (Solanum melongena), and pepper (Capsicum species). Besides being a crop, the tomato and its wild relatives played an important role as plant models in classical genetic research, whereas potato, pepper, and eggplant were mainly subject to breeding research aimed at crop improvement (Gebhardt, 2016).
The structure of plant genomes became accessible at the molecular level approximately 40 years ago with the advent of DNA-based markers, which allowed for the first time the construction of detailed molecular linkage maps (Helentjaris et al., 1985; Helentjaris et al., 1986; Bernatzky and Tanksley, 1986). The first type of DNA-based genetic marker was restriction fragment length polymorphism (RFLP), which detects DNA variation between individuals of the same or closely related species. The individual’s genomic DNA is digested with a restriction endonuclease; the fragments are size separated by gel electrophoresis and hybridized to a labelled DNA probe (Southern, 1975). The first molecular linkage maps of tomato, potato, pepper, and eggplant were constructed based on the segregation patterns of RFLP markers (Bonierbale et al., 1988; Gebhardt et al., 1989; Gebhardt et al., 1991; Tanksley et al., 1992; Gebhardt et al., 1994; Livingstone et al., 1999; Doganlar et al., 2002; Gebhardt et al., 2003). Based on a common set of tomato RFLP markers used for linkage mapping in tomato, potato, pepper, and eggplant, it was shown that the tomato and potato genomes are highly collinear or syntenic. The 12 chromosomes of both species corresponded to 12 linkage groups. The order of the markers was preserved between the potato and tomato maps, except for a few intrachromosomal inversions (Bonierbale et al., 1988; Tanksley et al., 1992; The Tomato Genome Consortium, 2012). A higher fragmentation in syntenic blocks (genome segments smaller than whole chromosomes sharing the same marker order in different species) and chromosome rearrangements (inversions, intra-, and inter-chromosomal translocations) was observed when comparing the maps of tomato, eggplant, and pepper (Livingstone et al., 1999; Doganlar et al., 2002).
RFLP linkage maps were the starting point for the mapping of qualitative and quantitative traits of agronomic importance, first and predominantly, in potato and tomato. The aim was, on the one hand, the development of molecular diagnostic tools for marker-assisted cultivar selection in breeding programs, and, on the other hand, preparing the ground for the map-based cloning of genes controlling agronomic traits such as pathogen resistance. After the invention of the polymerase chain reaction (PCR) (Saiki et al., 1988), RFLP markers were supplemented and eventually replaced by PCR-based markers, such as microsatellites or simple sequence repeats (SSR’s) (Broun and Tanksley, 1996; Milbourne et al., 1998), amplified fragment length polymorphism (AFLP) (Meksem et al., 1995; Vos et al., 1995), and random amplified polymorphic DNA (RAPD) (Williams et al., 1990; Klein-Lankhorst et al., 1991). For practical reasons, RFLP, AFLP, and RAPD markers linked with traits of interest were often converted into locus-specific PCR-based markers. They allowed the reliable detection of specific DNA polymorphisms in large numbers of plants, such as breeding populations (for example, Kasai et al., 2000). Subsequently, automatization and cost reduction of DNA sequencing facilitated the direct detection of point mutations (single nucleotide polymorphism, SNP) and small insertion/deletion polymorphisms by comparative sequencing of individuals of the same or closely related species (Rickert et al., 2003). The next milestones were whole genome sequences of potato (The Potato Genome Sequencing Consortium, 2011; Sharma et al., 2013; Kyriakidou et al., 2020; Freire et al., 2021; Sun et al., 2022), tomato (The Tomato Genome Consortium, 2012; The 100 Tomato Genome Sequencing Consortium, 2014) and subsequently other members of the Solanaceae (Kim et al., 2014; Kim et al., 2017; Bombarely et al., 2016; Edwards et al., 2017).
Hundreds of linkage mapping studies of qualitative and quantitative traits using DNA-based markers have been performed over the last 35 years on potato and tomato. It started with monogenic traits, such as single dominant genes for pathogen resistance (R genes) (Young et al., 1988; Barone et al., 1990), and was soon followed by the mapping of polygenic or quantitative traits (Paterson et al., 1988; Leonards-Schippers et al., 1994). A molecular linkage map was instrumental in the positional cloning of the tomato Pto gene for resistance to the bacterial pathogen Pseudomonas syringae (Martin et al., 1993a; Martin et al., 1993b). Pto was one of the first plant genes isolated for pathogen resistance and the first one of the Solanaceae family. Quantitative trait locus (QTL) mapping led to the identification of the first plant gene underlying a tomato QTL for fruit size (Grandillo et al., 1999; Frary et al., 2000).
With the increasing number of linkage mapping studies of R genes and quantitative resistance loci (QRL) using tomato and potato RFLP and microsatellite markers of known chromosomal position, it became possible to combine the positional information generated in independent mapping experiments into a single function map for pathogen resistance (Grube et al., 2000; Gebhardt and Valkonen, 2001; Danan et al., 2011). However, positions and distances between loci based on recombination frequencies have lower precision compared with physical positions and distances measured in DNA base pairs. This is due to the large variability of linkage map resolution with the type and size of mapping populations, marker density, and suppressed recombination in centromeric regions. When sequence information is available for DNA-based markers used in linkage mapping experiments, it is now possible to use whole genome reference sequences available in databases (http://spuddb.uga.edu/, https://solgenomics.net/) to construct physical chromosome maps, which combine the positional information of phenotypic characters from independent linkage mapping experiments. In addition, several hundred candidate genes for the traits were available in the literature that could be included in the physical maps. Such maps should facilitate comparisons of the genomic positions of phenotypic characters across species borders, eventually pointing to a common molecular basis of traits mapping to syntenic positions and genomic regions of particular interest.
This paper provides a survey and database of more than four hundred publications on linkage and association mapping of qualitative and quantitative traits with DNA-based markers, and map-based cloning of genes underlying such traits, primarily in potato and tomato and to a lesser extent in pepper and eggplant. The information provided in the literature was used to construct the first physical map of qualitative and quantitative traits on the 12 chromosomes of tomato and potato, including some resistance traits of pepper and eggplant. The purpose of this paper is to summarize 40 years of research in this specific field and to provoke new thoughts and hypotheses about the organization and identity of genes controlling important agronomic characters in crop plants.
Sequences of potato RFLP markers derived from randomly selected genomic (GP***) and cDNA clones (CP***, S****, and P****) (Gebhardt et al., 1989; 1994) were retrieved from the GenBank NCBI (accession numbers in Supplementary Tables S1–S12). Tomato RFLP markers were randomly selected genomic (TG***) or cDNA clones (CT*** or CD***) (Tanksley et al., 1992). Most tomato RFLP marker sequences were available in the Sol Genomics Network (SGN) database (https://solgenomics.net/search/markers). Tomato marker sequences not available in SGN were eventually retrieved from NCBI (Ganal et al., 1998). Primer sequences for PCR-based markers were taken from the literature or the SGN database (tomato microsatellites). AFLP and RAPD markers were not used due to insufficient sequence information. Sequences flanking tomato SNPs (solcap_snp_sl_*****) were obtained from the SGN database. Sequences flanking potato SNPs (solcap_snp_c*_*****) were obtained from http://solcap.msu.edu/potato and the SPUD database (http://spuddb.uga.edu/). Sequences flanking other SNPs were taken from the corresponding reference paper. Gene sequences were retrieved via accession number from NCBI, SPUD, and SGN databases and in some cases directly from the corresponding article. Gene sequences of Arabidopsis thaliana were retrieved from the Arabidopsis information resource (TAIR) database (https://www.arabidopsis.org/). The source of each sequence is provided in Supplementary Tables S1–S12.
DNA sequences or translated polypeptide sequences, for example, A. thaliana sequences, were mapped in silico to the genome sequences of potato and tomato (potato genome versions DM v4.03 and DM_v6.1; tomato genome version SL4.0) using the BLAST tool in the SPUD and SGN databases and default parameters. For short sequences, e.g., PCR primers, the parameter “expect threshold” was increased to 1. The physical position of markers and genes was determined in most cases unambiguously based on high sequence similarity (>70% identity). In cases of multiple sequence matches, the chromosomal position of a marker or gene known from genetic mapping was used to identify the most likely physical position.
Qualitative and quantitative traits were mapped based on the information content of articles published from 1988 to 2022. Articles were selected using the following criteria: articles that established the first molecular linkage maps of tomato and potato with RFLP and microsatellite markers, which were subsequently used for trait mapping; articles on linkage and association mapping of pathogen resistance and QTL for sugar content, yield, and maturity; articles on the cloning of resistance genes and QTL in the Solanaceae. Excluded were articles on mapping exclusively with AFLP and RAPD markers, redundant articles not adding further positional information to a specific locus, a few articles with very unclearly presented results, and articles in which the reported map position of the trait contradicted the physical map position. Traits with known molecular basis (cloned resistance genes and QTL) were physically mapped using the NCBI accessions. The position and size of genomic segments harbouring the genes controlling traits with unknown molecular basis were estimated based on the physical positions of linked, associated, and, whenever possible, flanking markers. The information from multiple mapping experiments performed with different mapping populations and different markers was combined. The physical borders of QTL could not always be precisely determined based on flanking markers and are approximations. For example, when only one linked marker from one experiment was available, it was assumed that the linked QTL was located within 2 Mbp upstream and downstream of the marker. This might have led to an overestimation of the size of a physical genome segment toward chromosome ends and an underestimation in centromeric regions. QTL resulting from epistatic interactions were not considered.
The plant material used for the linkage mapping of phenotypic characters in the self-compatible, inbred tomato was segregating F2 and backcross populations, near-isogenic lines (NILs) and recombinant inbred lines (RILs). Progeny was derived from the hybridization of cultivars with wild tomato species (Supplementary Table S13). In pepper, inbred cultivars and a few wild species (C. chinense, C. frutescens, C. baccatum, and C. chacoense) were hybridized to generate inter- and intra-specific F2 or doubled haploid (DH) populations. The cultivated potato is tetraploid with tetrasomic inheritance, which complicates the linkage analysis. Varieties and cultivars are heterozygous due to breeding schemes based on outcrossing. Linkage mapping in potato was mostly performed in F1 progeny of diploid, self-incompatible, and, therefore, heterozygous parents. Most of them contained in their pedigree one or more introgressions from wild tuber-bearing Solanum species (Supplementary Table S13). The genetics of this material is equivalent to the genetics of human families and has the advantage that up to four alleles at each locus segregate in the progeny. The discovery of the Sli (S-locus inhibitor) gene, which confers self-compatibility to diploid potato plants (Hosaka and Hanneman, 1998a; Hosaka and Hanneman, 1998b) marks the beginning of a new approach in potato genetics and breeding based on inbred lines, similar to tomato (Lindhout et al., 2011; Kaiser et al., 2020). However, such material has no significant role yet in the literature on the genetic mapping of agronomic traits. At the beginning of the 21st century, association or linkage disequilibrium mapping (Zhu et al., 2008; Burghardt et al., 2017) of quantitative trait loci (QTL) in populations of cultivars started to supplement linkage mapping based on experimental populations (Gebhardt et al., 2004; Simko et al., 2004; Mazzucato et al., 2008).
Based on publicly available DNA sequence information, I constructed physical maps of the 12 potato and tomato chromosomes, first with genetically mapped RFLP and microsatellite markers (Bonierbale et al., 1988; Gebhardt et al., 1991; Tanksley et al., 1992; Milbourne et al., 1998; Gebhardt et al., 2003; Feingold et al., 2005; Ghislain et al., 2009) and second with DNA-based markers of various types (RFLP, SSR, SCAR (Sequence Characterized Amplified Region), CAPS (Cleaved Amplified Polymorphic Sequence), KASP (Kompetitive Allele Specific PCR), and SNP (Single Nucleotide Polymorphism)) that were linked or associated with genetic factors for pathogen resistance, the yield and the sugar content of tomato fruits and potato tubers, and maturity. Furthermore, candidate genes, cloned genes for pathogen resistance, and some other cloned genes encoding QTL, self-incompatibility (potato), and colour traits (Supplementary Tables S1–S12) were included in the maps
Matching pairs of physical positions both in the potato and tomato genome were found for 2,741 sequence-based loci (Supplementary Tables S1–S12). In the case of physical clusters of highly similar sequences, for example, NBS-LRR (nucleotide-binding site–leucine-rich repeat)-type gene families, the matching pairs within the clustered gene family were sometimes ambiguous. This was without consequences for the higher-order structural comparisons. No matching pair of physical positions in both potato and tomato were found for 121 genetically mapped loci. The reasons were insufficient or lack of sequence similarity, high sequence redundancy, or possible sequencing errors or errors in genetic mapping or genome assembly. For similar reasons, no matching pair of physical positions either in potato or tomato was found for 321 loci (Supplementary Tables S1–S12). Some of these cases might indicate small genomic rearrangements between potato and tomato, which were not reliably resolved with the density of physical loci, especially in centromeric regions. Inconsistencies between genetic and physical marker order were also observed (Supplementary Tables S1–S12), most likely due to imprecision and errors in genetic mapping or, alternatively, due to genome assembly errors.
Physical locus density was much higher on the chromosome arms comprising approximately 50% of the whole genome as compared with central regions (Figures 1–12; Supplementary Tables S1–S12). This was similar to the density distribution of annotated genes in the genome sequences (genome browsers in SGN and Spud DB). Consequently, the majority of genetically mapped traits were also located on the chromosome arms.
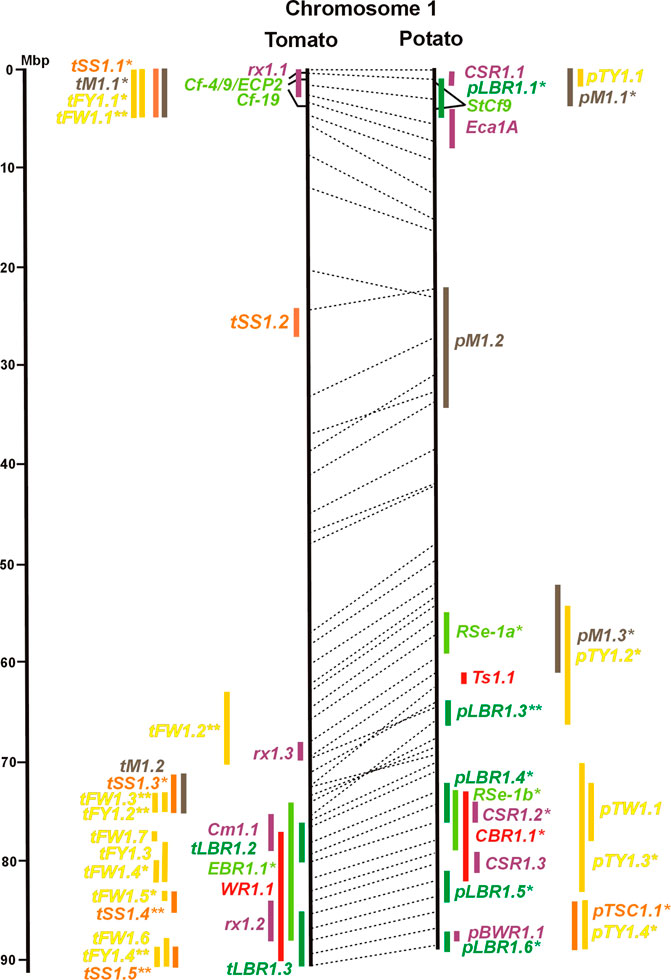
FIGURE 1. Physical maps in megabase pair scale (Mbp) of the 12 tomato/potato chromosomes. Genome version SL 4.0 and DM v6.1 of tomato and potato, respectively, are the basis of the physical chromosome maps represented by vertical black lines. The genome sequences of the chromosomes have the same orientation, except chromosome 12, where the tomato sequence is inversely oriented versus the potato sequence. Each tomato chromosome map is linked to the corresponding potato chromosome map by dotted lines representing the positions of a subset of sequence-based markers (indicated by * in column A of Tables S1 to S12). Clear intrachromosomal translocations are indicated by red dotted lines. The positions of sequenced genes for pathogen resistance and QTL are shown by a single black line connecting the chromosome map with the gene’s colour-coded name to the left (tomato) or right (potato) of the map. Intervals including mapped but not sequence-characterized qualitative resistance genes are shown as two black lines representing the borders that connect the chromosome map with the gene’s colour-coded name. Positions of pepper and eggplant resistance genes are shown at syntenic positions on both maps. QRL for pathogen resistance and QTL for sugar content, yield, and maturity are shown as colour-coded bars to the left (tomato) or right (potato) of the chromosome map. QRL and QTL names (for nomenclature see text and Tables 1 and 3) are shown in the same colour next to the coloured bar. Colour codes: Dark green–resistance to the oomycete P. infestans; light green–resistance to fungi; purple–resistance to bacteria; blue–virus resistance; magenta–nematode resistance; red–insect resistance; orange–QTL for fruit and tuber sugar content; yellow–QTL for fruit and tuber weight and yield; maroon–QTL for maturity. QRL and QTL names extended with * were supported by two or three mapping studies; QRL and QTL names ending with ** were supported by more than three mapping studies. QRL and QTL names without an extension were supported by a single mapping study. Coordinates in base pairs for all loci are provided in Tables S1 to S12.
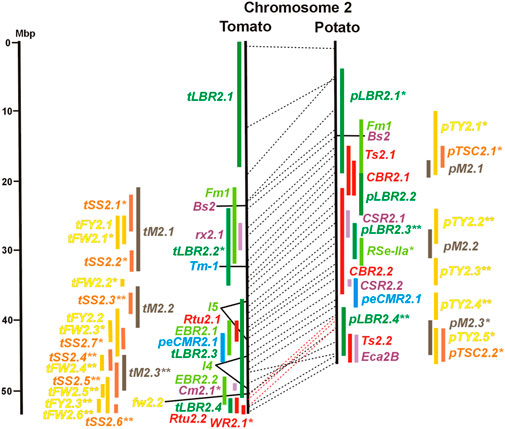
FIGURE 2. See caption for Figure 1 for detailed description.
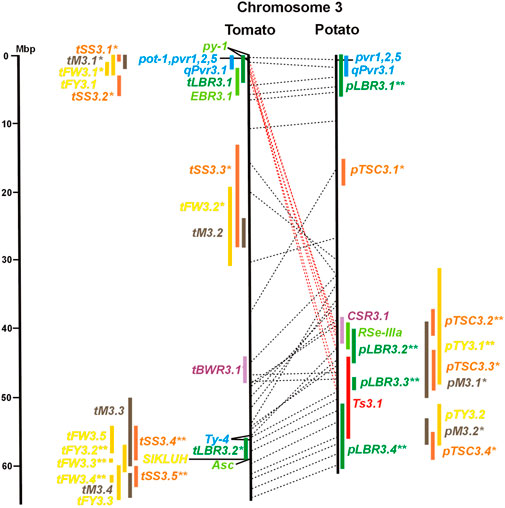
FIGURE 3. See caption for Figure 1 for detailed description.
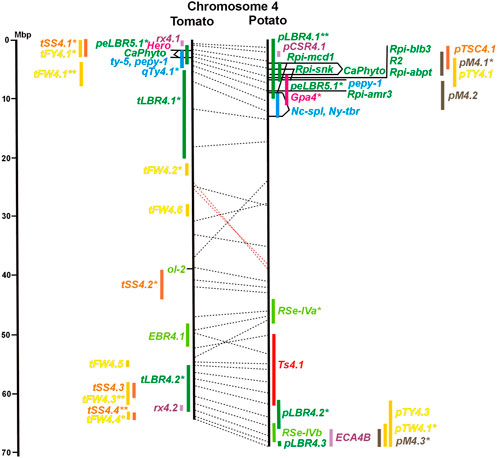
FIGURE 4. See caption for Figure 1 for detailed description.
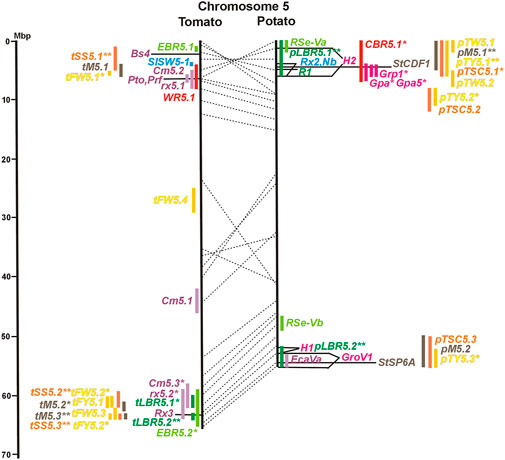
FIGURE 5. See caption for Figure 1 for detailed description.
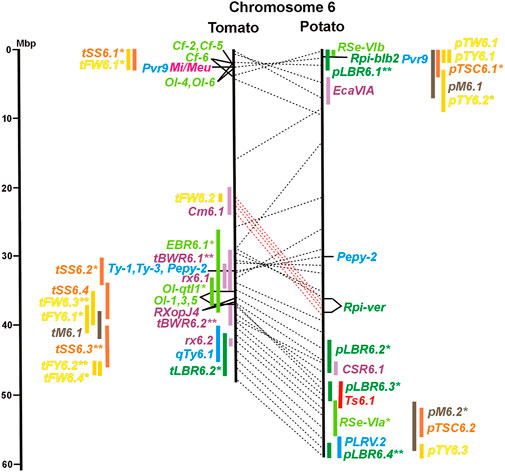
FIGURE 6. See caption for Figure 1 for detailed description.
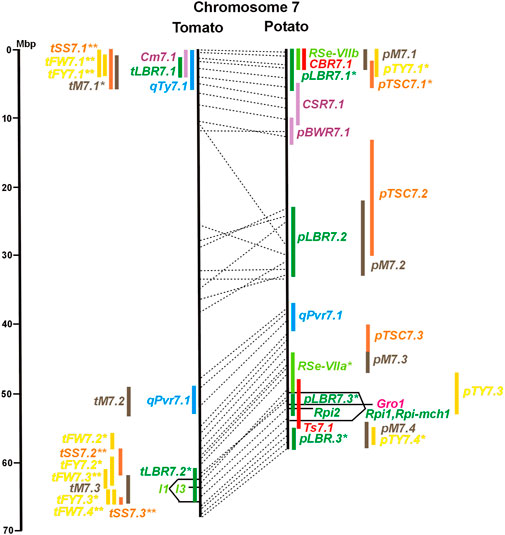
FIGURE 7. See caption for Figure 1 for detailed description.
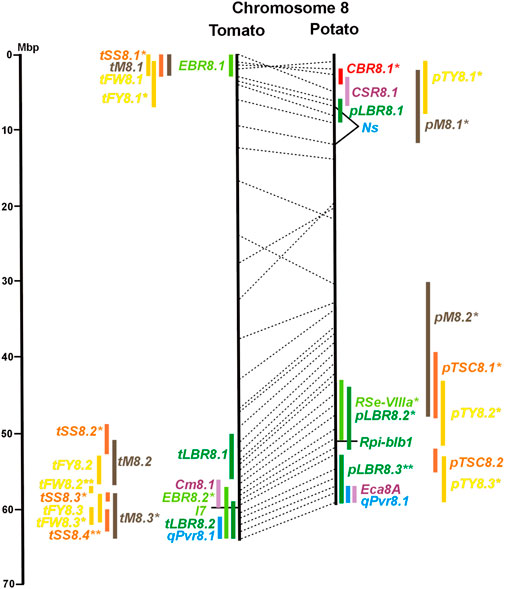
FIGURE 8. See caption for Figure 1 for detailed description.
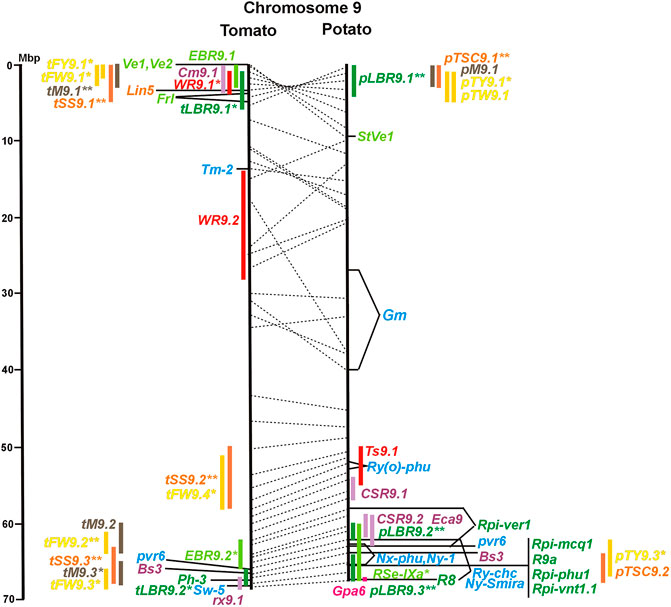
FIGURE 9. See caption for Figure 1 for detailed description.
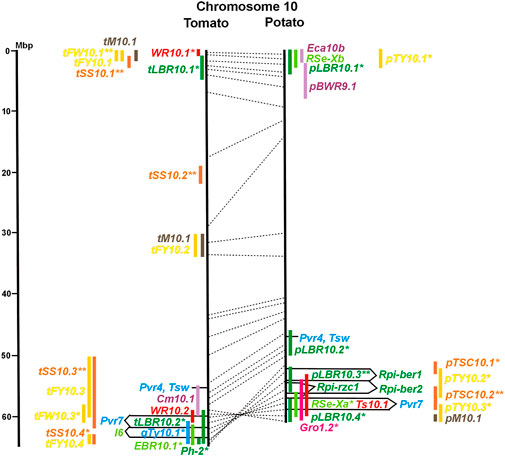
FIGURE 10. See caption for Figure 1 for detailed description.
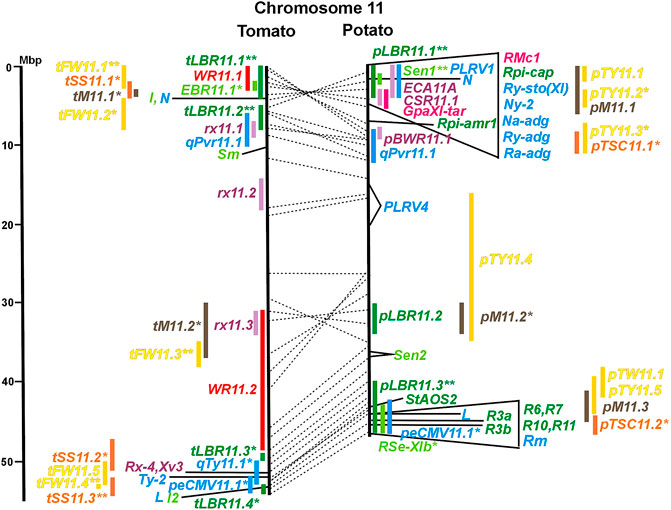
FIGURE 11. See caption for Figure 1 for detailed description.
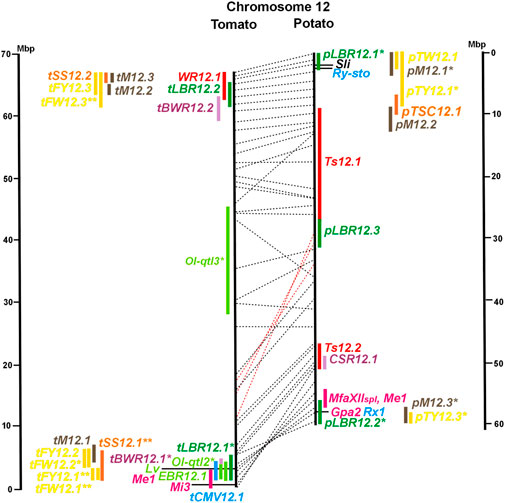
FIGURE 12. See caption for Figure 1 for detailed description.
Comparative molecular linkage maps of tomato and potato have been constructed (Bonierbale et al., 1988; Tanksley et al., 1992; The Tomato Genome Consortium, 2012). They have low resolution compared with physical maps. Comparison based on genomic sequences has the highest resolution. However, the single comparison of this type (The Tomato Genome Consortium, 2012) used primary versions of both the tomato and potato genome sequences, where uncertainty about the orientation of physical contigs still existed. The comparison presented in this paper is based on the physical mapping of 2,741 short sequences to consolidated reference genome sequences of tomato and potato (tomato version SL4.0; potato version DM v6.1). It has an intermediate resolution. Besides the eight genetically or cytogenetically resolved intra-chromosomal inversions between tomato and potato chromosomes 2, 5, 6, 9, 10, 11, and 12 (Tanksley et al., 1992; The Tomato Genome Consortium, 2012), 25 additional inversions and 12 intra-chromosomal translocations with a size range from 0,2–22 megabase pairs (Mbp) were detected (Figures 1–12; Supplementary Tables S1–S12). The translocation and inversion on chromosome 2, the inversion on the short arm of chromosome 6, and the inversion on the long arm of chromosome 10 are consistent with cytogenetic FISH analyses (Peters et al., 2012). Twelve inversions and six translocations were located in central chromosomal regions with low genetic and physical resolution. Some of those might be artefacts of the genome assembly in either tomato or potato. Chromosomes 3, 6, and 11 carried the most rearrangements. Traces of ancient intrachromosomal duplications predating the speciation of tomato and potato were detected on chromosomes 2, 6, and 8 based on the duplication of linked loci in both species, which mapped to two different physical sections of the same chromosome (Gebhardt et al., 2003).
Three hundred and six articles were analysed, which describe the molecular mapping, map-based cloning, and characterization of loci for qualitative (monogenic) and quantitative (polygenic) resistance (quantitative resistance locus QRL) to plant pathogenic oomycetes, fungi, viruses, bacteria, nematodes, and insects in potato (150 articles), tomato (130), pepper (23), eggplant (2), and tobacco (1) (Supplementary Data Sheet S14 and Supplementary Table S15). The phenotypic distinction between qualitative and quantitative resistance was not always clear-cut. The analysis of an observed phenotypic distribution with quantitative statistics may result in one QRL with a major effect, indicating that the resistance is based on a single locus. Vice versa, the distinction between resistant and susceptible phenotypic classes might be blurred by variation within the classes. The literature exhibited large diversity with respect to size and type of mapping populations and genome coverage with DNA-based markers from less than a hundred RFLPs to thousands of SNPs, environments, and methods to assess resistance phenotypes. Nevertheless, in a number of cases, QRL genetically mapped in independent studies were allocated to the same genome segment, suggesting that they eventually tag the same gene(s). The nomenclature used in different studies for QRL to the same pathogen was also diverse and inconsistent. To integrate the information from various sources and make it more accessible, a uniform nomenclature was adopted for some quantitative resistance traits (Table 1). The same name was assigned to a QRL when it was allocated to a similar genome segment in different studies. The originally published locus names, as far as they were assigned, are included in Supplementary Tables S1–S12.
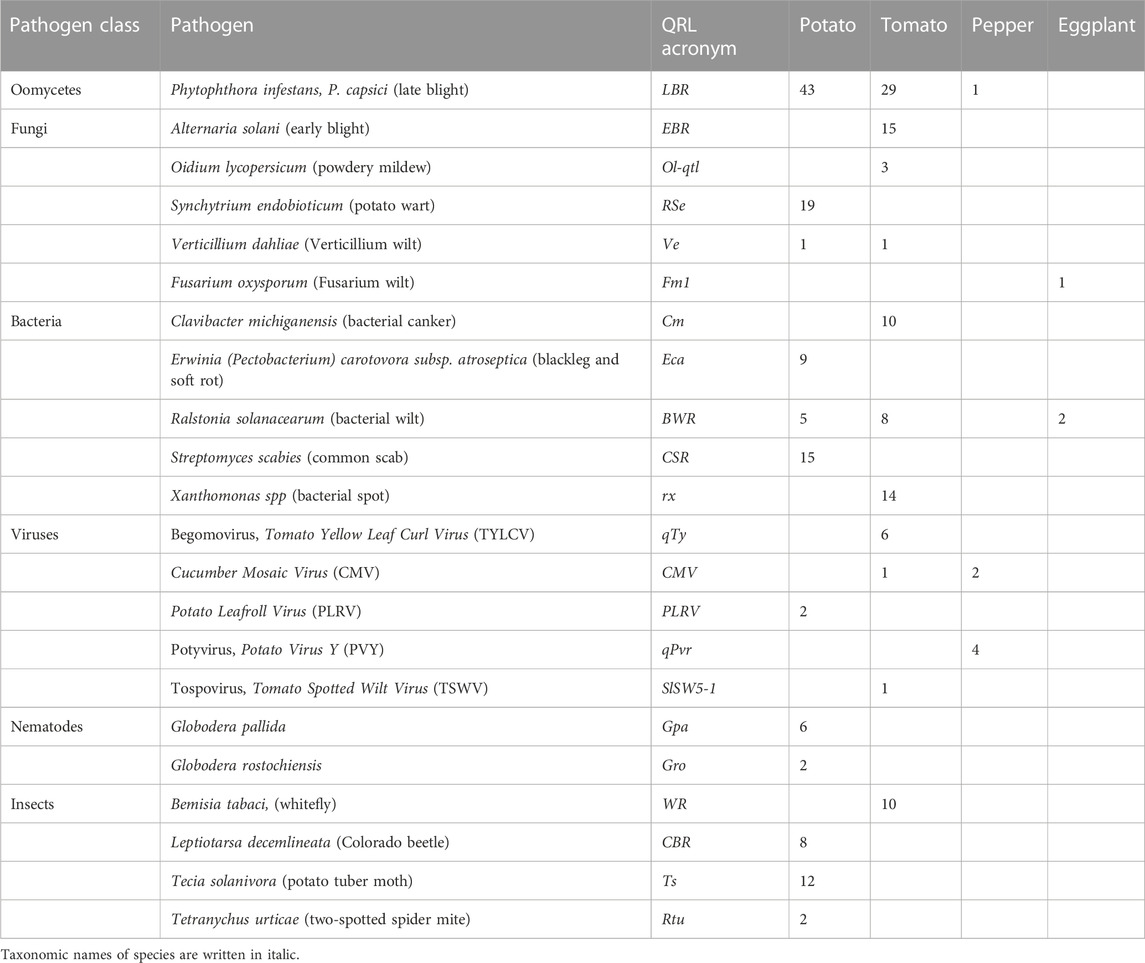
TABLE 1. Number of physically mapped QTL for pathogen resistance in potato, tomato, pepper, and eggplant.
The frequency of genetic studies dealing with plant resistance to a specific pathogen corresponds to some extent with the importance of this pathogen for crop cultivation. On the other hand, studies on important resistance traits might be underrepresented due to the difficulties in analysing the resistance phenotype, for example, insect resistance and resistance to potato wart. The most studied pathogen resistance in potato was resistance to the oomycete Phytophthora infestans, followed by resistance to the root cyst nematodes G. rostochiensis and Globodera pallida, and viruses, particularly, Potato Virus Y (PVY) (Supplementary Data Sheet S14). In tomato, resistance to various fungi and bacteria was important besides resistance to P. infestans, followed by virus resistance, particularly, to Tomato Yellow Leaf Curl Virus (TYLCV), and resistance to the nematode M. incognita. Despite the close phylogenetic relationship between potato and tomato, only six of 33 host-pathogen systems were analysed in both species (P. infestans, Verticillium dahliae, Ralstonia solanacearum, Potyvirus, Globodera rostochiensis, and Meloidogyne incognita) (Supplementary Data Sheet S14). Resistance to Phytophthora species, Potato Virus Y (PVY), and root-knot nematodes (Meloidogyne sp.) was analysed in potato, tomato, and pepper. Resistance to R. solanacearum was analysed in tomato, potato, and eggplant, and resistance to Tobacco Mosaic Virus (TMV) in tomato, pepper, and tobacco. Resistance to Xanthomonas species and the viruses Tomato Yellow Leaf Curl Virus (TYLCV) and Cucumber Mosaic Virus (CMV) was analysed in both tomato and pepper (Supplementary Data Sheet S14). This shows the relevance of these host-pathogen systems across species borders in the Solanaceae family. Compared with potato and tomato, fewer genetic studies on pathogen resistance have been published in pepper and eggplant. Only some of these results could be integrated into the physical maps of potato and tomato due to the more fragmented synteny of pepper and eggplant compared with tomato and potato. Mainly, single pepper genes for virus resistance and some major QRL could be placed on the physical potato/tomato maps. The sequence of the tobacco N gene for resistance to the Tobacco Mosaic Virus (TMV) was also available. N is a prototype of the major class of plant genes for pathogen resistance characterized by a nucleotide-binding site (NBS) and a leucine-rich repeat (LRR) domain (Whitham et al., 1994).
DNA sequences of 57 resistance genes were available in the literature, mostly but not exclusively dominant R genes. They were introgressed into cultivars from wild Solanum species. Twenty-seven resistance genes were from tomato, twenty from potato, nine from pepper, and one from tobacco. With two exceptions, all resistance genes were placed on syntenic physical positions in the tomato and potato genome (Figures 1–12; Supplementary Tables S1–S12). No sequence homology was found on chromosome 5 of potato and tomato for the potato Rx2 gene for resistance to Potato Virus X (PVX). Rx2 originated from S. acaule, and it was mapped on potato chromosome 5 (Ritter et al., 1991) and cloned (Bendahmane et al., 2000) (Figure 5; Supplementary Table S5). It seems unlikely that both the potato and tomato genome assemblies should have sequence gaps at the same position. Therefore, orthologous genes of Rx2 on chromosome 5 are obviously absent in the genotypes of S. phureja and S. lycopersicum, from which the reference genomes were constructed. Furthermore, the potato homologs of the tomato resistance gene Sm on chromosome 11 mapped to non-syntenic positions on the same chromosome (Supplementary Table S11).
Two-thirds of the sequence-characterized resistance genes belong to the major class of NBS-LRR type plant genes for pathogen recognition (reviewed in Andersen et al., 2018). The remaining third includes six recessive resistance genes, three of tomato (pot-1, ty-5, ol-2) and three of pepper (pvr1,2,5, pepy-1, and pvr6). The ‘atypical’ resistance genes represent diverse structures and functions, among others a protein kinase (Pto on chromosome 5) (Martin et al., 1993a; Martin et al., 1993b), the eukaryotic translation initiation factor 4E (pvr2 and pot-1 on chromosome 3) (Ruffel et al., 2002; Ruffel et al., 2005), and RNA-dependent RNA polymerase (Ty-1, Ty-3, and Pepy-2 on chromosome 6) (Verlaan et al., 2013; Koeda et al., 2022).
Fifty-six mapped resistance genes with yet unknown sequences were located on the physical potato/tomato maps by means of linked and flanking markers, thirty-six of potato, seventeen of tomato, and three of pepper (Figures 1–12; Supplementary Tables S1–S12). Fifty-five genes mapped to chromosome arms in physical intervals between 40 Kbp and 6 Mbp with an average of 2,5 Mbp. Only one resistance gene (Gm on potato chromosome 9) mapped to a physical interval >10 Mbp in the centromeric region.
Many qualitative resistance loci colocalized with quantitative resistance loci for the same or other pathogens (see below).
Based on the sequences of linked, associated, and flanking DNA markers, the position of 230 quantitative resistance loci (QRL) (120 in potato, 100 in tomato, 7 in pepper, and 3 in eggplant) on the 12 physical tomato/potato chromosome maps was estimated (Table 1). Sixteen QRL (nine in tomato and seven in potato) were mapped to physical intervals larger than 15 Mbp and were not further considered in comparisons within and between the species. The remaining 214 QRL mapped to intervals between 0,5 and 15 Mbp with an average of 4,2 Mbp in both tomato and potato. Physical intervals of this size contain hundreds of genes. One or more genes may be responsible for any mapped QRL. The number of genes underlying quantitative resistance is therefore certainly higher than the actual number of mapped QRL.
The oomycete P. infestans causes late blight disease in potato and tomato, which can lead to complete crop loss. To reduce the necessity of chemical control, late blight-resistant cultivars are important breeding goals. Qualitative and quantitative resistance to late blight was most extensively studied in potato (Supplementary Data Sheet S14). Twenty-eight R genes were mapped on eight chromosomes, fifteen of which were cloned and sequenced (Figures 1–12; Supplementary Tables S1–S12, and Supplementary Table S15). Besides R genes, quantitative or field resistance to late blight was extensively analysed in potato (33 articles in potato, 12 in tomato, and 2 in pepper (Supplementary Data Sheet S14). Resistance levels were quantified based on different P. infestans races and various evaluation methods, such as detached leaf assays (Leonards-Schippers et al., 1994) or whole plant evaluations in a greenhouse, a growth chamber, and a field (Brouwer et al., 2004). Mostly leaves but also stems and tubers were evaluated for resistance (Bradshaw et al., 2006; Danan et al., 2009). High levels of field resistance to late blight are correlated with late plant maturity, which is a negative agronomic character (see the section on maturity). Therefore, in some studies, the QTL effect on resistance was corrected for the effect on maturity, resulting in QTL for maturity-corrected resistance to late blight (MCR) (Bormann et al., 2004). In total, 43 potato QTL for late blight resistance (pLBR loci) on all 12 chromosomes were distinguished (Table 1) between two and six per chromosome. Twenty-six of these QRL were also detected as QRL for maturity-corrected resistance to late blight. Six late blight QRL were supported by one study, another six by two studies, and thirty-one by three to fourteen studies (Figures 1–12; Supplementary Tables S1–S12). This shows a certain degree of saturation achieved in mapping potato late blight QRL. The most reproducible QRL was pLBR5.1 on the short arm of chromosome 5 in a 6 Mbp interval, which was detected after infection with various Phytophthora races in different genetic backgrounds and geographical regions by linkage as well as association mapping (Figure 5 and Supplementary Table S5). Part but not all of the QTL effect on resistance could be explained by the QTL effect on plant maturity mapping to the same genome section (Visker et al., 2003; Bormann et al., 2004). This genome section includes the potato R1 gene for race-specific resistance to late blight and the tomato Bs4 gene for resistance to the bacterium Xanthomonas campestris. R1 and Bs4 have been cloned and functionally characterized. Both encode typical NBS-LRR genes (Ballvora et al., 2002; Schornack et al., 2004). The same region contains R genes for a virus (Rx2 and Nb) and a nematode (H2) resistance, major QTL for nematode resistance (Gpa, Gpa5, and Grp1), a QTL for insect resistance (Colorado potato beetle = QRL CBR5.1), and a QTL for resistance to the fungus Synchytrium endobioticum (RSe-Vb). The syntenic tomato genome segment harbours a QTL for resistance to the fungus Alternaria solani (Figure 5 and Supplementary Table S5). The early observation of genetic colocalization of R1 with a late blight QRL in potato gave rise to the hypothesis that R genes and QRL might have a common molecular basis before sequences of any R gene and whole genome sequences were known (Leonards-Schippers et al., 1994; Gebhardt and Valkonen, 2001). According to this hypothesis, the best candidate genes for the potato QRL in this 6 Mbp region are the 30 annotated disease resistance genes organized in several clusters, the largest cluster including R1 (http://spuddb.uga.edu/jbrowse). In the syntenic tomato region, there are only 17 annotated disease resistance genes, among them is Bs4 (https://solgenomics.net/jbrowse). This might be the reason why compared with potato, few tomato QRL map to this region.
Colocalization of R genes and late blight QRL was also observed on the short arm of chromosomes 4, 6, and 11 and the long arm of chromosomes 7, 8, 9.10, and 11 (Figures 4–11; Supplementary Tables S4–S11 and Supplementary Data Sheet S14). In agreement with this observation is the finding that the level of field resistance to late blight of potato varieties increased with the number of R genes for late blight present in these varieties (Stewart et al., 2003). Direct experimental evidence for the effect of an R gene on quantitative resistance to late blight was provided in the case of the sequence-characterized potato late blight resistance gene R8 on chromosome 9, which is an NBS-LRR gene (Jiang et al., 2018). On the other hand, the number and physical size of the QRL and the fact that the potato genome contains more than 400 annotated NBS-LRR genes (The Potato Genome Sequencing Consortium, 2011) suggest that colocalizations of QRL and NBS-LRR genes may occur by chance and do not necessarily indicate a functional relationship. Moreover, there are QRL, which do not include genes annotated as disease resistance genes (NBS-LRR genes), at least not in the genome sequences used here. Examples are pLBR1.5 and pLBR1.6 on chromosome 1 (Figure 1 and Supplementary Table S1) or pLBR3.3 on chromosome 3 (Figure 3 and Supplementary Table S3). The StAOS2 locus on chromosome 11 encoded an allene oxide synthase functional in jasmonate biosynthesis and was strongly associated with MCR (Pajerowska-Mukhtar et al., 2009). When knocked down by antisense constructs in transgenic potato plants, late blight resistance was reduced, showing that this locus is functional in quantitative resistance (Pajerowska-Mukhtar et al., 2008). Therefore, genes other than R genes are also functional in quantitative resistance. Based on various criteria, 153 candidate genes for quantitative resistance were included in the physical chromosome maps (Supplementary Tables S1–S12), which might help in the future to identify the molecular basis of additional QRL.
In tomato, 29 late blight QRL (tLBR loci) were mapped on all 12 chromosomes between one and four per chromosome (Table 1; Figures 1–12, and Supplementary Tables S1–S12). Twelve QRL were supported by one study, ten by two, and seven by three to five studies (Supplementary Tables S1–S12). Most reproducible with four studies each were QRL tLBR5.2 and tLBR11.2 on chromosome 5 and 11, respectively, and tLBR11.1 with five studies on chromosome 11 (Figures 5, 11 and Supplementary Tables S5, S11). QRL tLBR9.2 and tLBR10.2 colocalized with the tomato late blight resistance genes Ph-3 and Ph-2, respectively (Table 2). Ph-3 has been cloned and encodes an NBS-LRR gene (Zhang et al., 2014), suggesting that part of the tomato quantitative resistance to late blight is controlled by NBS-LRR genes.
Approximately half of the potato and tomato late blight QRL mapped to syntenic genomic segments (Supplementary Data Sheet S14). Colocalization by chance is to be expected, considering the number and physical size of the QRL. Nevertheless, some of these syntenic QRL might indicate that orthologous genes or gene families of tomato and potato underlie the mapped QRL.
The singular mapped pepper QTL for resistance to P. capsici (Ca-qPca) was located on the short arm of chromosome 4 and colocalized with a large cluster of potato R genes and QRL for late blight (Figure 4; Supplementary Table S4).
A variety of fungi threatens tomato and potato cultivation. To reduce the control by fungicide applications, genetic resistance is highly desirable and therefore part of breeding programs. Fourteen articles analysed and mapped QTL for resistance to five different pathogenic fungi, seven in potato, six in tomato, and one in eggplant (Supplementary Data Sheet S14 and Supplementary Table S15). Most important in potato is potato wart caused by the soil-born fungus S. endobioticum, which is a quarantine disease and difficult to control. Nineteen QRL for S. endobioticum (RSe loci) were physically mapped, one to two per chromosome (Table 1; Figures 1–12, and Supplementary Tables S1–S12). Sen1 (RSe-XIa) on the short arm of chromosome 11 was located between 1,2 and 2.5 Mbp. It was the major quantitative wart resistance locus, which was detected in six studies. The 1,3 Mbp potato genome segment containing Sen1 is syntenic with the inverted 0,6 Mbp tomato segment between 4,8 and 4,2 Mbp (Figure 11 and Supplementary Table S11). This segment in potato contains nineteen annotated disease resistance genes (http://spuddb.uga.edu/jbrowse), among those homologs of the tobacco N gene for resistance to Tobacco Mosaic Virus (TMV), a prototype for NBS-LRR genes (Whitham et al., 1994). Only three annotated disease resistance genes are located in the tomato syntenic region (https://solgenomics.net/jbrowse) plus the I gene for resistance to Fusarium wilt (Fusarium oxysporum). This resistance gene encodes an atypical leucine-rich-repeat receptor-like protein (LRR-RLP) (Catanzariti et al., 2017). The most distal 5 Mbp segment of the short arm of potato chromosome 11 is syntenic with the inverted tomato segment from approximately 2–5,5 Mbp. It harbours besides Sen1 seven R genes (RMc1, Rpi-cap, Ry-sto (XI), Ny-2, Na-adg, Ry-adg, and Ra-adg) plus several QRL for resistance against different pathogens (Figure 11 and Supplementary Table S11). This region constitutes one of several ‘hotspots’ for disease resistance (Gebhardt and Valkonen, 2001) (Table 2). With two exceptions (Soltu.DM11G003310 and Soltu. DM.11G003490) all annotated disease resistance genes are located within the 1,3 Mbp region including Sen1 and N homologs (http://spuddb.uga.edu/jbrowse). The R genes and eventually some QRL might be encoded by alleles of one or more of the nineteen NBS-LRR genes located between 1,2 and 2.5 Mbp.
A single association mapping study located a potato QTL for resistance to V. dahliae (StVe1) on chromosome 9, using as markers homologs of the cloned tomato Ve1 gene for resistance to the same pathogen (Simko et al., 2004). Tomato Ve1 and Ve2 are duplicated genes with syntenic positions on tomato chromosome 9 and encode cell surface-like receptors (Kawchuk et al., 2001), which correspond to three clustered homologs on potato chromosome 9. This is another example of the possible functional relationship between R genes and QRL (Figure 9 and Supplementary Table S9).
The majority of articles on resistance to pathogenic fungi in tomato deal with the mapping and cloning of R genes for resistance to Cladosporium fulvum and F. oxysporum (Supplementary Data Sheet S14). QTL mapping was performed for resistance to early blight (EBR loci) caused by A. solani and powdery mildew (Ol-qtl loci) caused by Oidium lycopersici. Fifteen EBR QRL (one to two per chromosome) were mapped to all chromosomes except chromosome 7 (Figures 1–12 and Supplementary Tables S1–S12). Eight EBR QRL were detected in one, six in two, and one (EBR5.2) in three of three studies. Resistance to powdery mildew (O. lycopersici) appears oligogenic as only three QRL were mapped based on three studies. Ol-qtl1 on chromosome 6 was the only QRL detected in all three studies and colocalized with the Ol-1 locus and EBR6.1 (Figure 6 and Supplementary Table S6). Ol-qtl2 and Ol-qtl3 were both located on chromosome 12. Ol-qtl2 colocalized with EBR12.1 and the Lv gene for resistance to powdery mildew caused by Leveillula taurica (Figure 12 and Supplementary Table S12).
Clear synteny between potato and tomato QTL for resistance to pathogenic fungi was not observed except between StVe1 and EBR9.1 on chromosome 9 and between the Sen1 locus and EBR11.1 on chromosome 11.
Thirty-eight articles report mapping and cloning of genes for resistance to bacterial pathogens (Supplementary Table S15). Twenty-four analysed and mapped QTL for resistance to five species, sixteen in tomato, seven in potato, and one in eggplant (Supplementary Data Sheet S14). Chromosome 1 carried nine QRL, the highest number of QRL for bacteria, followed by chromosomes 6 and 11 with seven QRL each.
Genetics of resistance to bacterial wilt caused by R. solanacearum was studied mainly in tomato (nine articles) but also in potato and eggplant (one article each). Eight QRL (BWR loci) were mapped in tomato, five in potato, and two in eggplant (Table 1). Most reproducible were tomato QRL tBWR6.1 and tBWR6.2 on chromosome 6, each detected in five studies (Figure 6 and Supplementary Table S6). The two eggplant QRL colocalized with tomato BWR6.1 and BWR3.1 on chromosomes 6 and 3, respectively. One of the five potato QRL for R. solanacearum, pBWR11.1 on chromosome 11, could be limited to a 1,7 Mbp interval, which does not contain annotated disease resistance genes in the sequenced potato/tomato genomes (Figure 11 and Supplementary Tables S11).
Three articles located fourteen tomato QTL for resistance to Xanthomonas species (rx loci) on seven chromosomes (Table 1). The only QRL supported by all three studies was QRL rx5.2 on chromosome 5, which colocalized with QRL Cm5.3 and the R gene Rx3 for Xanthomonas resistance (Figure 5; Supplementary Table S5 and Supplementary Data Sheet S14).
Ten tomato QTL for resistance to Clavibacter michiganensis (Cm loci) were mapped to eight chromosomes in four studies (Table 1). Two QRL, Cm2.1 and Cm5.3 on chromosomes 2 and 5 (Figures 2, 5; Supplementary Tables S2, S5), respectively, were supported by two of the four studies.
Specific for potato was a single QTL mapping experiment for resistance to Erwinia carotovora ssp. atroseptica (Eca loci) and five studies on QTL for resistance to Streptomyces scabies (common scab, CSR loci) (Supplementary Data Sheet S14). Nine Eca QRL on nine chromosomes and fifteen CSR QRL on ten chromosomes were distinguished (Table 1; Figures 1–12, and Supplementary Tables S1–S12). There was little consistency between the five mapping experiments on resistance to S. scabies. Only one locus, CSR1.2 on chromosome 1, was supported by two studies (Figure 1 and Supplementary Table S1).
Colocalization and synteny between QTL for resistance to different pathogenic bacteria were observed in eight potato/tomato genome segments (Supplementary Data Sheet S14), which might indicate similarities between the underlying genes. Interestingly, the tomato QRL rx5.1 and Cm5.2 colocalized with the Pto and Prf genes for resistance to P. syringae (Figure 5; Supplementary Table S5 and Supplementary Data Sheet S14).
A multitude of viruses infects Solanaceous crops leading to losses of crop yield and quality. In the vegetatively propagated potato, viruses are transmitted to the next tuber generation leading to progressive yield reduction, which is the reason for seed tuber production under virus-free conditions. The 70 articles on mapping and cloning of virus resistance loci in potato and tomato (Supplementary Table S15) deal mainly with monogenic resistance. Eighteen of 37 mapped virus resistance genes (20 in potato, 8 each in tomato and pepper, and 1 in tobacco) have been identified at the molecular level (7 each in tomato and pepper and 4 in potato). The highest number of virus resistance loci maps to chromosome 11 in two clusters on the short and the long arm (Figure 11 and Supplementary Table S11). Only nine articles dealt with quantitative resistance to five viruses, five in tomato, three in pepper, and one in potato (Supplementary Data Sheet S14 and Supplementary Table S15).
Six QTL for resistance to Tomato Yellow Leaf Curl Virus (qTy loci) mapped to six tomato chromosomes (Table 1; Figures 1–12, and Supplementary Tables S1–S12), three of which were supported by two studies (qTY4.1, qTY10.1, and qTy11.1). Two qTy QRL colocalized with monogenic TYLCV resistance: qTy4.1 on chromosome 4 colocalized with the recessive resistance genes ty-5 of tomato and pepy-1 of pepper, both encoding the messenger RNA surveillance factor Pelota (Lapidot et al., 2015; Koeda et al., 2021). qTy11.1 on chromosome 11 colocalized with the dominant resistance gene Ty-2, an NBS-LRR gene (Yamaguchi et al., 2018).
One QTL for resistance to Cucumber Mosaic Virus (CMV loci) was mapped in tomato and two in pepper (Table 1). The pepper QRL peCMV11.1 colocalized with the pepper NBS-LRR gene L for tobamovirus resistance (Tomita et al., 2011) on the long arm of chromosome 11 (Figure 11 and Supplementary Table S11).
A major tomato QTL for resistance to Tomato Spotted Wilt Virus (TSWV), probably an NBS-LRR gene (Qi et al., 2022), mapped on the short arm of chromosome 5 (Figure 5 and Supplementary Table S5).
Four pepper QTL for resistance to potyviruses were localized on potato/tomato chromosomes 3, 7, 8, and 11 (Figures 3, 7, 8, 11; Supplementary Table S3, S7, S8, S11). qPvr3.1 on chromosome 3 colocalized with the recessive potyvirus resistance genes pvr1,2,5 from pepper and pot-1 from tomato. Both encode the eukaryotic translation initiation factor 4E (Ruffel et al., 2002; 2005), suggesting that this QRL might be an effect of this gene.
One QTL mapping experiment in potato resistance to Potato Leafroll Virus (PLRV) detected a major QRL (PLRV.1) on chromosome 11 and a minor QRL (PLRV.2) on chromosome 6. PLRV.1 was one component of a cluster of potato genes for resistance to different viruses between 0 and 5 Mbp on the short arm of chromosome 11 (Figures 6, 11; Table 2, and Supplementary Tables S6, S11).
Soil parasitic nematodes infest and damage plant roots leading to yield losses. Nematode infestations are difficult to control and therefore quarantined. Twelve R genes for resistance to soil parasitic nematodes of the genus Meloidogyne and Globodera were mapped in potato (seven genes), tomato (four genes), and pepper (one gene), four of which were characterized at the molecular level (Mi, Hero, Gro1-4, and Gpa2). Twelve articles analysed quantitative resistance to nematodes in potato, mainly to the root cyst nematode G. pallida, which is a serious problem in potato cultivation (Supplementary Data Sheet S14 and Supplementary Table S15). Resistance to G. pallida was oligogenic. Only six QRL on chromosomes 4, 5, 9, and 11 were identified in ten mapping studies (Figures 4, 5, 9, 11; Supplementary Tables S4, S5, S9, S11). Three major QTL for nematode resistance were mapped reproducibly to the same potato genome segment between 4 and 7 Mbp on chromosome 5 and colocalized with the H2 gene for resistance to G. pallida. One of these QRL (Grp1) was effective not only against G. pallida but also against the closely related species G. rostochiensis (van der Voort et al., 1998). This major nematode resistance locus constitutes part of the resistance hotspot on the short arm of potato chromosome 5 (Figure 5; Table 2, and Supplementary Table S5).
Regarding the genetics of resistance to insect pests, the information available in the literature was limited to 11 articles, nine dealing with quantitative resistance, four in potato and five in tomato (Supplementary Data Sheet S14 and Supplementary Table S15). The most damaging insect pest in potato is the Colorado potato beetle (Leptiotarsa decemlineata). Based on three studies, eight QTL for resistance to Colorado beetle (CBR loci) were mapped to seven chromosomes (Table 1; Figures 1–12, and Supplementary Tables S1–S12). CBR1.1, CBR5.1, CBR8.1, and CBR10.1 were detected in two studies based on the same mapping population. They did not overlap with the major QRL CBR2.2 on chromosome 2 reported in the third study.
The larvae of the potato tuber moth Tecia solanivora are a serious threat to potato cultivation in South America. One study reported twelve QTL for resistance to T. solanivora (Ts loci) which were located on nine chromosomes (Table 1; Figures 1–12, and Supplementary Tables S1–S12).
Whiteflies transmit viruses and are controlled by insecticides, especially in greenhouses. They are particularly relevant for tomato, where greenhouse cultivation is common. Genetics of quantitative resistance to whitefly in tomato was the subject of four articles (Supplementary Data Sheet S14 and Supplementary Table S15). Ten QTL for whitefly resistance (WR loci) were mapped to seven chromosomes (Table 1), three of which were detected in two studies, WR2.1, WR9.2, and WR10.1 on chromosomes 2, 9, and 11, respectively (Figures 2, 9, 10; Supplementary Table S2, S9, S10). A single study identified two QTL for resistance to two-spotted spider mites (Tetranychus urticae) on chromosome 2 (Rtu2.1 and Rtu2.2).
Chromosome 2 harboured with six the most QTL for resistance to insects. Colocalization was observed between potato QRL Ts2.1 and CBR2.1 and between tomato QRL Rtu2.2 and WR2.1. The latter two tomato QTL for insect resistance mapped syntenic with potato QRL Ts2.2 and might be caused by the type and density of trichomes and the quantitative variation of metabolites, particularly acyl sugars (Vosman et al., 2019). Tomato QRL WR1.1 and potato QRL CBR1.1 also mapped to syntenic segments on chromosome 1. Tomato Rtu2.1 and potato CBR2.2 were syntenic on chromosome 2 and tomato WR10.2 and potato Ts10.1 on chromosome 10 either by chance or by identity or similarity of the underlying genes (Figures 1, 2, 10; Supplementary Tables S1, S2, S10).
Pathogen resistance loci are located on all chromosomes (Figures 1–12). However, some genome segments are outstanding in harbouring numerous single genes for resistance as well as QRL for different types of pathogens. They are summarized in Table 2 and are considered hotspots for pathogen resistance in the genomes of tomato, potato, and their wild relatives, maybe also in syntenic genome segments of other Solanaceous species. Particularly striking are the clusters of R genes and QRL on chromosomes 9 and 11. The clustering of resistance factors of different origins and different pathogen specificity can be explained when orthologous genes from different species confer resistance either to the same pathogen or to different types of pathogens. Alternatively, clustered members of the same gene family may confer resistance to diverse pathogens. Direct experimental evidence for this concept has been provided in some cases. For example, it was shown that the tomato gene Mi for resistance to the nematode M. incognita on chromosome 6 is identical to the gene Meu for resistance to the potato aphid (Rossi et al., 1998; Vos et al., 1998). Mi/Meu is one member of a family of annotated resistance genes located between 2,3 and 2.8 Mbp on tomato chromosome 6, which is syntenic with 700 Kbp between 1,1 and 1.8 Mbp in potato. Furthermore, the sequences of the tomato gene Mi/Meu, the potato gene Rpi-blb2 for late blight resistance, and the pepper gene Pvr9 for resistance to Pepper Mottle Virus matched different members of the same NBS-LRR gene family (Table 2 and Supplementary Table S6). The NBS-LRR gene family in this syntenic genome segment seems to be a source of resistance factors against multiple pathogens in the Solanaceae. Other examples are the potato genes Rx and Gpa2 for resistance to Potato Virus X (PVX) and the nematode G. pallida, respectively, which were shown to be members of the same clustered family of NBS-LRR genes on chromosome 12 (Van der Vossen et al., 2000). Additional examples of cloned R genes from different Solanaceous species matching the same gene or gene family are shown in Table 2 and Supplementary Tables S2, S4, S9, S11.
Besides pathogen resistance, three further quantitative traits were selected for physical mapping and comparisons of QTL positions between potato and tomato, namely, the sugar content of fruits and tubers, fruit and tuber yield, and fruit or plant maturity. The sugar content of tubers and fruits is based on the same biochemical processes, that is carbohydrate metabolism. Yield and maturity of tomato fruits and potato tuber yield and plant maturity might be controlled to some extent by homologous genes. This homology might extend to other species of the Solanaceae. Colocalization and synteny of QTL might point eventually to a common molecular basis. Each mapped QTL comprised several mega base pairs of sequence including hundreds of annotated genes. More than one gene in a particular genome segment might be responsible for the observed QTL effect. It should also be kept in mind that colocalization and synteny of QTL may be by chance, and the underlying genes and mechanisms are functionally independent.
Sixty-nine publications, 45 in tomato and 24 in potato, were evaluated, which describe the genetic mapping and in very few cases the cloning of these QTL (Supplementary Table S15). The nomenclature used for the QTL varied between articles or was altogether absent. A uniform nomenclature (Table 3) was therefore adopted for QTL mapping to the same genome segment to facilitate comparisons.

TABLE 3. Number of physically mapped QTL for sugar content, yield, and maturity in tomato and potato.
The sugar content of tomato fruits and potato tubers is an important quality criterion in breeding programs of both species. The high sugar content of tomato fruits is valued for culinary quality but is negatively correlated with yield (Tanksley et al., 1996). On the contrary, tuber sugar content is considered a negative character as the accumulation of fructose and glucose during tuber storage, particularly at low temperatures, negatively affects the quality of processed potato products, such as chips and French fries. Sucrose, fructose, and glucose are products of carbohydrate metabolism, e.g., starch degradation. Many genes encoding the proteins functional in carbohydrate metabolism and transport have been biochemically and molecularly characterized in many species. They are obvious functional candidates for QTL for sugar content. One hundred and sixteen such candidate genes were included in the physical chromosome maps (Supplementary Tables S1–S12). The sugar contents of fruits and tubers of the individuals of a mapping population were measured directly in aqueous tissue extracts by enzymatic assays or indirectly: in tomato as soluble solids content based on the Brix index and in potato based on the colour of fried potato chips.
Thirty-two articles reported the QTL of tomato fruit sugar content (Supplementary Tables S15 and Supplementary Data Sheet S14). Physical mapping distinguished 47 QTL (tSS loci, Table 3) between two (chromosome 12) and seven (chromosome 2) QTL per chromosome (Figures 1–12 and Supplementary Tables S1–S12). The size range of the QTL was between 1 and 15 Mbp with an average of 3,6 Mbp. Thirty-five tSS loci were supported by three or more studies. Most reproducible were thirteen QTL (tSS1.4, tSS1.5, tSS2.4, tSS2.5, tSS3.4, tSS4.4, tSS5.3, tSS6.3, tSS9.1, tSS9.3, tSS10.1, tSS10.3, and tSS12.1), each supported by six to ten studies (Supplementary Tables S1–S12). Two-thirds of the tSS QTL colocalized with yield QTL, corroborating the correlation between fruit sugar content and yield. Colocalizing QTL for fruit yield and sugar content could be the result of the pleiotropic effects of the same gene(s). So far, only one tomato gene underlying a QTL for fruit sugar content was identified at the molecular level. This turned out to be one (lin5) of a duplicated pair of invertase genes (lin5/lin7) on chromosome 9, to which high-resolution linkage mapping limited the sugar QTL Brix9-2-5 (Fridman et al., 2000). The highly reproducible QTL tSS9.1 (seven studies) includes the invertase locus lin5/lin7. The orthologous potato invertase locus INV-GE/GF is a strong candidate for the highly reproducible QTL pTSC9.1 for tuber sugar content (Figure 9 and Supplementary Tables S9). Recent approaches based on genome-wide association analysis (GWAS) with large numbers of SNP markers identified further candidate genes for fruit sugar QTL, without a priori assumptions on gene function (Tripodi et al., 2021; Zhao et al., 2022), fifteen of which were included in Supplementary Tables S1–S12.
The evaluation of eight QTL studies in potato (Supplementary Table S15 and Supplementary Data Sheet S14) resulted in 27 QTL for tuber sugar content, chip quality, or chip colour (pTSC loci, Table 3) in the size range from 2 to 9 Mbp with an average of 4,5 Mbp (excluding three QTL ≥15 Mbp). As in tomato, chromosome 2 carried with four QTL the highest number of sugar QTL. Due to the limited number of experiments, the reproducibility of QTL for tuber sugar content was rather low. Only four QTL were supported by three or more studies (pTSC3.2, pTSC5.1, pTSC9.1, and pTSC10.2). Among those was the most reproducible QTL pTSC9.1 (six studies), which includes the candidate invertase locus INV_GE/GF (see above). Association between tuber sugar content and DNA variants within the invertase locus INV_GE/GF was observed besides associations of some other genes functional in carbohydrate metabolism (Li et al., 2008; Schreiber et al., 2014).
Synteny between potato pTSC and tomato tSS sugar QTL was observed for 16 of 27 pTSC loci (Figures 1–12; Supplementary Tables S1–S12 and Supplementary Data Sheet S14), suggesting that fruit and tuber sugar content might be partially controlled by homologous genes, for example, invertase genes.
Tomato fruit and potato tuber yield are essential characters in breeding programs. The yield was quantified in various ways as average fruit or tuber weight per plant, per plot, or area unit. Quantitative genetics of tomato average fruit weight and total yield were most extensively studied. The integration of 42 QTL mapping experiments (Supplementary Table S15 and Supplementary Data Sheet S14) into the tomato physical map resulted in 55 QTL for fruit weight (tFW loci) and 30 QTL for fruit yield (tFY loci) (Table 3), demonstrating the complexity of this character. Excluding three QTL ≥15 Mbp, QTL intervals ranged from 0,1 to 10 Mbp with an average of 3,5 Mbp. Fruit weight and total yield are highly correlated traits (Fulton et al., 1997). The majority of fruit yield QTL colocalized with fruit weight QTL (Figures 1–12 and Supplementary Tables S1–S12), suggesting that they are largely based on the same set of genes. The highest density of tomato yield QTL was observed on chromosomes 1, 2, and 3. Forty-six QTL were supported by three or more studies. Most reproducible were QTL tFW2.4, tFW2.5, tFW3.3, and tFW11.4 on chromosomes 2, 3, and 11, respectively, each of which was mapped in 12–16 studies. Yield QTL mapping to the same genome segment in independent experiments was analysed in the progeny of interspecific hybrids between cultivars and wild tomato species, with the exception of the cherry tomato S. lycopersicum var. cerasiforme. Wild tomatoes and S. lycopersicum var. cerasiforme have generally smaller fruits and lower yields compared with cultivated tomatoes. Most tomato yield QTL were therefore species unspecific. They represent rather the genomic landscape of domestication, during which fruit size and yield were increased by selection (Lin et al., 2014). Two genes underlying tomato yield QTL have been identified at the molecular level. The highly reproducible QTL tFW2.5 on chromosome 2 (13 studies) colocalized with tFY2.3 (Figure 2 and Supplementary Table S2) and includes fw2.2, the first gene identified at the molecular level underlying a yield QTL (Frary et al., 2000). Colocalizing QTL tFW3.3 (14 studies) and tFY3.2 on chromosome 3 include a cytochrome P450 gene that controls fruit weight (Chakrabarti et al., 2013). The potato QTL pTY3.2 maps to the syntenic region (Figure 3; Supplementary Tables S3 and Supplementary Data Sheet S14). Natural variation in hundreds of genes in most functional categories has probably a direct or indirect effect on yield. Among them are genes functional in central metabolism such as glycolysis, Calvin cycle, tricarboxylic acid (TCA) cycle, and other pathways. Seventy-six characterized genes of this type were included in the physical maps (Supplementary Tables S1–S12). GWAS identified 37 novel candidate genes for fruit weight QTL (Sacco et al., 2015; Tripodi et al., 2021; Zhao et al., 2022), which are also included in Supplementary Tables S1–S12.
Compared with tomato, few QTL mapping experiments addressed potato tuber yield. The evaluation of 10 articles (Supplementary Table S15 and Supplementary Data Sheet S14) resulted in 41 QTL for tuber yield (pTY loci) and eight for tuber weight (pTW loci) (Table 3). Excluding six QTL ≥15 Mbp, QTL intervals ranged from 1,6 to 13 Mbp with an average of 4,7 Mbp. Like in tomato, most tuber weight QTL colocalized with tuber yield QTL. Ten yield QTL (pTY2.2, pTY2.3, pTY2.4, pTY3.1, pTY5.1, pTY5.2, pTY6.2, pTY7.2, pTY9.3, and pTY12.1) were supported by three or more studies (Figures 1–12; Supplementary Tables S1–S12). The QTL pTY2.2 on chromosome 2 and pTY5.1 on chromosome 5 were most reproducible with five and seven supporting studies, respectively (Figures 2, 5; Supplementary Tables S2, S5). Genes controlling tuber yield QTL have not been formally cloned and characterized. However, candidate genes for yield QTL are available and were included in the physical maps. They resulted from the pairwise comparison of potato genotype pools with differential tuber yield with the 8,3 k SolCap SNP array. SNPs with differential allele frequency between the genotype pools were identified, some of which resulted in amino acid changes in annotated genes (Schönhals et al., 2017). One hundred and ninety-four genes of this type were included as candidate genes in the physical maps (Supplementary Tables S1–S12). None of them were identical to the novel candidate genes for tomato fruit weight identified by GWAS. As for tomato fruit yield, the 67 genes functional in central metabolism are also candidate genes for potato tuber yield. The large and certainly incomplete number of candidate genes combined with the number and genome coverage of yield QTL resulted in multiple colocalizations of QTL and candidate genes. Further functional analysis is required to verify a possible causal relationship between a QTL and natural allelic variation at candidate gene loci.
Twenty-eight yield QTL (tFW, tFY, pTY, and pTW loci) appeared syntenic in tomato and potato (Figures 1–12; Supplementary Tables S1–S12 and Supplementary Data Sheet S14). Due to the presence of yield QTL on every chromosome arm in both tomato and potato, the assumption that synteny is observed because orthologous genes control a syntenic QTL might not be justified in many cases. Nevertheless, the observation that chromosomes 1 and 2 of both tomato and potato are particularly rich in yield QTL suggests that the molecular basis of some yield QTL might be related.
The maturity or earliness of fruits or plants is an agronomic character in annual crops that is relevant for agricultural practice, where early maturity is preferred. In tomato, the trait describes the status of fruit ripening, and in potato, the trait describes the vegetative period from plant emergence, growth, flowering, and tuberization to senescence. Early maturity is correlated with lower yields in tomato and potato (Maris, 1969; Bernacchi et al., 1998). Moreover, earlier maturity in potato is correlated with higher susceptibility to late blight (Collins et al., 1999; Visker et al., 2003; Bormann et al., 2004). High-yielding varieties with acceptable earliness and high field resistance to late blight are highly desirable breeding goals. Maturity or earliness was visually scored in various ways and recorded in numerical scales from 1 to 5 or 1 to 9. The trait was the subject of twelve QTL mapping experiments in potato and eight in tomato (Supplementary Table S15 and Supplementary Data Sheet S14).
Maturity or earliness was evaluated together with late blight resistance in nine of 12 potato QTL studies. In total, 30 QTL were mapped (pM loci) (Table 3), of which 23 overlapped with late blight QRL. Excluding two QTL ≥15 Mbp, QTL size ranged from 1 to 12 Mbp with an average of 4,5 Mbp. Only two maturity QTL were reported in three or more studies (pM3.1 and pM5.1). The most reproducible QTL was pM5.1 on the short arm of chromosome 5 (eight studies) (Figure 5 and Supplementary Table S5). Plant maturity in potato depends on the adaptation of tuberization to the daylength in northern latitudes. Here, the crop is grown under long-day conditions, whereas in its original habitats in central South America, the potato’s life cycle is adapted to short days. Short-day adapted genotypes initiate tubers and mature very late under long-day conditions. The gene StCDF1 encoding a DOF (DNA-binding with one finger) transcription factor has been identified, which controls all or part of the effect of the major QTL pM5.1 (Kloosterman et al., 2013). The tuberization-control gene StSP6A, a paralogue of the Arabidopsis flowering locus T (FT) (Navarro et al., 2011), is located on the long arm of chromosome 5 within the range of maturity QTL pM5.2. It is a functional candidate gene for this QTL.
Twenty-nine QTL for fruit maturity (tM loci) were distinguished in tomato (Table 3), in a size range from 1 to 12 Mbp with an average of 3,6 Mbp. Six maturity QTL were supported by three or more studies (tM1.1, tM2.3, tM5.2, tM5.3, tM9.1, and tM11.2). Most reproducible with five studies each were tM2.3 and tM5.3. The tomato orthologue of StSP6A might be a candidate gene for QTL tM5.3 (Supplementary Data Sheet S14). Three-quarters of the maturity QTL overlapped with yield QTL in accordance with the correlation between these traits, suggesting that genes controlling fruit maturity may have pleiotropic effects on yield and vice versa (Figures 1–12; Supplementary Tables S1–S12 and Supplementary Data Sheet S14). Synteny between tomato and potato maturity QTL was observed in 13 genome segments (Supplementary Data Sheet S14).
The structural comparison between the tomato and potato genomes based on the physical mapping of 2,741 sequences to updated reference genome sequences detected novel intrachromosomal inversions and translocations between the otherwise collinear tomato and potato genomes.
One hundred and twelve single loci for pathogen resistance of potato, tomato, and pepper were integrated into physical maps of the 12 potato/tomato chromosomes. They have been located previously on molecular linkage maps. Linkage mapping with DNA-based markers has led to the map-based cloning and characterization of 56 resistance genes, thus making a significant contribution to the knowledge of the structure and function of plant genes for pathogen resistance. Based on the sequences of cloned resistance genes or markers tightly linked with major effect resistance loci, diagnostic markers have been developed, which have enriched the breeder’s toolbox for the selection of resistant cultivars (Tiwari et al., 2022; Marhadour and Prodhomme, 2023).
Two hundred and thirty QTL for pathogen resistance were positioned on the physical tomato/potato maps, part of which colocalized with R genes for resistance to the same or different pathogens. This and in very few cases the identification of the underlying gene suggest that qualitative and quantitative resistance is at least in part based on the same genes, namely, NBS-LRR type gene families. On the other hand, not all QRL can be explained by the action of R genes. Certain genome segments exhibited a high density of qualitative and quantitative resistance loci, so-called hotspots for pathogen resistance.
Two hundred and sixty-seven QTL for tomato fruit and potato tuber sugar content, yield, and maturity were distinguished on the physical tomato/potato chromosome maps. This is certainly an underestimation of the number of genes involved in these complex traits. Numerous colocalizations of QTL corroborate the phenotypic correlations and suggest that some genes underlying QTL for sugar content or maturity have pleiotropic effects on yield or pathogen resistance or vice versa.
Map-based cloning of genes underlying QTL in the Solanaceae was rarely undertaken. Only about half a dozen cases were reported in the literature. The molecular basis of QTL in the Solanaceae is still largely unknown. The main objective of QTL mapping in the past has been the identification of diagnostic markers to be used for the marker-assisted selection of superior cultivars in breeding programs. This has not materialized in applied breeding programs. Instead of genetic dissection of individual QTL, the method of choice is now genomic selection (GS), which does not require knowledge of individual QTL map positions (Meuwissen et al., 2001; Duangjit et al., 2016; Sverrisdóttir et al., 2018; Budhlakoti et al., 2022). GS is made possible by technical advances in DNA sequencing and data analysis, which make genotyping by sequencing large populations of cultivars manageable and affordable.
The author confirms being the sole contributor of this work and has approved it for publication.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1197206/full#supplementary-material
Andersen, E. J., Ali, S., Byamukama, E., Yen, Y., and Nepal, M. P. (2018). Disease resistance mechanisms in plants. Genes 9, 339. doi:10.3390/genes9070339
Ballvora, A., Ercolano, M. R., Weiß, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi:10.1046/j.1365-313x.2001.01292.x
Barone, A., Ritter, E., Schachtschabel, U., Debener, T., Salamini, F., and Gebhardt, C. (1990). Localization by restriction fragment length polymorphism mapping in potato of a major dominant gene conferring resistance to the potato cyst nematode Globodera rostochiensis. Mol. Gen. Genet. 224, 177–182. doi:10.1007/BF00271550
Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D. C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21, 73–81. doi:10.1046/j.1365-313x.2000.00654.x
Bernacchi, D., Beck-Bunn, T., Eshed, Y., Lopez, J., Petiard, V., Uhlig, J., et al. (1998). Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor. Appl. Genet. 97, 381–397. doi:10.1007/s001220050908
Bernatzky, R., and Tanksley, S. D. (1986). Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112, 887–898. doi:10.1093/genetics/112.4.887
Bombarely, A., Moser, M., Amrad, A., Bao, M., Bapaume, L., Barry, C. S., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2, 16074. doi:10.1038/nplants.2016.74
Bonierbale, M. W., Plaisted, R. L., and Tanksley, S. D. (1988). RFLP Maps Based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120, 1095–1103. doi:10.1093/genetics/120.4.1095
Bormann, C. A., Rickert, A. M., Castillo Ruiz, R. A., Paal, J., Lübeck, J., Strahwald, J., et al. (2004). Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol. Plant Microbe Interact. 17, 1126–1138. doi:10.1094/MPMI.2004.17.10.1126
Bradshaw, J. E., Hackett, C. A., Lowe, R., McLean, K., Stewart, H. E., Tierney, I., et al. (2006). Detection of a quantitative trait locus for both foliage and tuber resistance to late blight [Phytophthora infestans (Mont) de Bary] on chromosome 4 of a dihaploid potato clone (Solanum tuberosum subsp. tuberosum). Theor. Appl. Genet. 113, 943–951. doi:10.1007/s00122-006-0353-8
Broun, P., and Tanksley, S. D. (1996). Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol. Gen. Genet. 250, 39–49. doi:10.1007/BF02191823
Brouwer, D. J., Jones, E. S., and St Clair, D. A. (2004). QTL analysis of quantitative resistance to Phytophthora infestans (late blight) in tomato and comparisons with potato. Genome 47, 475–492. doi:10.1139/g04-001
Budhlakoti, N., Kushwaha, A. K., Rai, A., Chaturvedi, K. K., Kumar, A., Pradhan, A. K., et al. (2022). Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 13, 832153. doi:10.3389/fgene.2022.832153
Burghardt, L. T., Young, N. D., and Tiffin, P. (2017). A guide to genome-wide association mapping in plants. Curr. Protoc. Plant Biol. 2, 22–38. doi:10.1002/cppb.20041
Catanzariti, A.-M., Do, H. T. T., Bru, P., de Sain, M., Thatcher, L. F., Rep, M., et al. (2017). The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 89, 1195–1209. doi:10.1111/tpj.13458
Chakrabarti, M., Zhang, N., Sauvage, C., Muños, S., Blanca, J., Cañizares, J., et al. (2013). A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. U. S. A. 110, 17125–17130. doi:10.1073/pnas.1307313110
Collins, A., Milbourne, D., Ramsay, L., Meyer, R., Chatot-Balandras, C., Oberhagemann, P., et al. (1999). QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol. Breed. 5, 387–398. doi:10.1023/a:1009601427062
Danan, S., Chauvin, J. E., Caromel, B., Moal, J. D., Pellé, R., and Lefebvre, V. (2009). Major-effect QTLs for stem and foliage resistance to late blight in the wild potato relatives Solanum sparsipilum and S. spegazzinii are mapped to chromosome X. Theor. Appl. Genet. 119, 705–719. doi:10.1007/s00122-009-1081-7
Danan, S., Veyrieras, J. B., and Lefebvre, V. (2011). Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biol. 11, 16. doi:10.1186/1471-2229-11-16
Daunay, M.-C., and Laterrot, H. (2008). “Iconography and history of Solanaceae. Antiquity to the 17th century,” in Horticultural reviews 34. Editor J. Janick (John Wiley & Sons), 1–112.
Doganlar, S., Frary, A., Daunay, M.-C., Lester, R. N., and Tanksley, S. D. (2002). A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161, 1697–1711. doi:10.1093/genetics/161.4.1697
Duangjit, J., Causse, M., and Sauvage, C. (2016). Efficiency of genomic selection for tomato fruit quality. Mol. Breed. 36, 29. doi:10.1007/S11032-016-0453-3
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18, 448. doi:10.1186/s12864-017-3791-6
Feingold, S., Lloyd, J., Norero, N., Bonierbale, M., and Lorenzen, J. (2005). Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L). Theor. Appl. Genet. 111, 456–466. doi:10.1007/s00122-005-2028-2
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E., Cong, B., Liu, J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi:10.1126/science.289.5476.85
Freire, R., Weisweiler, M., Guerreiro, R., Baig, N., Hüttel, B., Obeng-Hinneh, E., et al. (2021). Chromosome-scale reference genome assembly of a diploid potato clone derived from an elite variety. G3 (Bethesda) 11, jkab330. doi:10.1093/g3journal/jkab330
Fridman, E., Pleban, T., and Zamir, D. (2000). A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. (USA) 97, 4718–4723. doi:10.1073/pnas.97.9.4718
Fulton, T., Beck-Bunn, T., Emmatty, D., Eshed, Y., Lopez, J., Petiard, V., et al. (1997). QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor. Appl. Genet. 95, 881–894. doi:10.1007/s001220050639
Ganal, M. W., Czihal, R., Hannappel, U., Kloos, D.-U., Polley, A., and Ling, H.-Q. (1998). Sequencing of cDNA clones from the genetic map of tomato (Lycopersicon esculentum). Genome Res. 8, 842–847. doi:10.1101/gr.8.8.842
Gebhardt, C., and Valkonen, J. P. T. (2001). Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 39, 79–102. doi:10.1146/annurev.phyto.39.1.79
Gebhardt, C., Ritter, E., Debener, T., Schachtschabel, U., Walkemeier, B., Uhrig, H., et al. (1989). RFLP analysis and linkage mapping in Solanum tuberosum. Theor. Appl. Genet. 78, 65–75. doi:10.1007/BF00299755
Gebhardt, C., Ritter, E., Barone, A., Debener, T., Walkemeier, B., Schachtschabel, U., et al. (1991). RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83, 49–57. doi:10.1007/BF00229225
Gebhardt, C., Ritter, E., and Salamini, F. (1994). “RFLP map of the potato,” in DNA-Based markers in plants. Advances in cellular and molecular biology of plants. Editors R. L. Phillips, and I. K. Vasil (Kluwer Academic Publishers), 271–285.
Gebhardt, C., Walkemeier, B., Henselewski, H., Barakat, A., Delseny, M., and Stüber, K. (2003). Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J. 34, 529–541. doi:10.1046/j.1365-313x.2003.01747.x
Gebhardt, C., Ballvora, A., Walkemeier, B., Oberhagemann, P., and Schüler, K. (2004). Assessing genetic potential in germ plasm collections of crop plants by marker-trait association: A case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol. Breed. 13, 93–102. doi:10.1023/B:MOLB.0000012878.89855.df
Gebhardt, C. (2016). The historical role of species from the Solanaceae plant family in genetic research. Theor. Appl. Genet. 129, 2281–2294. doi:10.1007/s00122-016-2804-1
Ghislain, M., Núñez, J., del Rosario Herrera, M., Pignataro, J., Guzman, F., Bonierbale, M., et al. (2009). Robust and highly informative microsatellite-based genetic identity kit for potato. Mol. Breed. 23, 377–388. doi:10.1007/s11032-008-9240-0
Grandillo, S., Ku, H., and Tanksley, S. (1999). Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 99, 978–987. doi:10.1007/s001220051405
Grube, R. C., Radwanski, E. R., and Jahn, M. (2000). Comparative genetics of disease resistance within the Solanaceae. Genetics 155, 873–887. doi:10.1093/genetics/155.2.873
Helentjaris, T., King, G., Slocum, M., Siedenstrang, C., and Wegman, S. (1985). Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol. Biol. 5, 109–118. doi:10.1007/BF00020093
Helentjaris, T., Slocum, M., Wright, S., Schaefer, A., and Nienhuis, J. (1986). Construction of genetic linkage maps in maize and tomato using restriction fragment length polymorphisms. Theor. Appl. Genet. 72, 761–769. doi:10.1007/BF00266542
Hosaka, K., and Hanneman, R. E. (1998a). Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 1. Detection of an S locus inhibitor (Sli) gene. Euphytica 99, 191–197. doi:10.1023/a:1018353613431
Hosaka, K., and Hanneman, R. E. (1998b). Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 2. Localization of an S locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica 103, 265–271. doi:10.1023/a:1018380725160
Jiang, R., Li, J., Tian, Z., Du, J., Armstrong, M., Baker, K., et al. (2018). Potato late blight field resistance from QTL dPI09c is conferred by the NB-LRR gene R8. J. Exp. Bot. 69, 1545–1555. doi:10.1093/jxb/ery021
Kaiser, N., Manrique-Carpintero, N. C., DiFonzo, C., Coombs, J., and Douches, D. (2020). Mapping Solanum chacoense mediated Colorado potato beetle (Leptinotarsa decemlineata) resistance in a self-compatible F2 diploid population. Theor. Appl. Genet. 133, 2583–2603. doi:10.1007/s00122-020-03619-8
Kasai, K., Morikawa, Y., Sorri, V. A., Valkonen, J. P., Gebhardt, C., and Watanabe, K. N. (2000). Development of SCAR markers to the PVY resistance gene Ryadg based on a common feature of plant disease resistance genes. Genome 43, 1–8. doi:10.1139/g99-092
Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease-resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U. S. A. 98, 6511–6515. doi:10.1073/pnas.091114198
Kim, S., Park, J., Yeom, S.-I., Kim, Y.-M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi:10.1038/ng.2877
Kim, S., Park, J., Yeom, S.-I., Kim, Y.-M., Lee, J. M., Lee, H. A., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18, 210. doi:10.1186/s13059-017-1341-9
Klein-Lankhorst, R. M., Vermunt, A., Weide, R., Liharska, T., and Zabel, P. (1991). Isolation of molecular markers for tomato (L. esculentum) using random amplified polymorphic DNA (RAPD). Theor. Appl. Genet. 83, 108–114. doi:10.1007/BF00229232
Kloosterman, B., Abelenda, J., Gomez, M., Oortwijn, M., de Boer, J. M., Kowitwanich, K., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi:10.1038/nature11912
Knapp, S., Bohs, L., Nee, M., and Spooner, D. M. (2004). Solanaceae-a model for linking genomics with biodiversity. Comp. Funct. Genomics 5, 285–291. doi:10.1002/cfg.393
Koeda, S., Onouchi, M., Mori, N., Pohan, N. S., Nagano, A. J., and Kesumawati, E. (2021). A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum. Theor. Appl. Genet. 134, 2947–2964. doi:10.1007/s00122-021-03870-7
Koeda, S., Mori, N., Horiuchi, R., Watanabe, C., Nagano, A. J., and Shiragane, H. (2022). PepYLCIV and PepYLCAV resistance gene Pepy-2 encodes DFDGD-Class RNA-dependent RNA polymerase in Capsicum. Theor. Appl. Genet. 135, 2437–2452. doi:10.1007/s00122-022-04125-9
Kyriakidou, M., Achakkagari, S. R., Gálvez López, J. H., Zhu, X., Tang, C. Y., Tai, H. H., et al. (2020). Structural genome analysis in cultivated potato taxa. Theor. Appl. Genet. 133, 951–966. doi:10.1007/s00122-019-03519-6
Lapidot, M., Karniel, U., Gelbart, D., Fogel, D., Evenor, D., Kutsher, Y., et al. (2015). A novel route controlling begomovirus resistance by the messenger RNA surveillance factor Pelota. PLOS Genet. 11, e1005538. doi:10.1371/journal.pgen.1005538
Leonards-Schippers, C., Gieffers, W., Schäfer-Pregl, R., Ritter, E., Knapp, S. J., Salamini, F., et al. (1994). Quantitative resistance to Phytophthora infestans in potato: A case study for QTL mapping in an allogamous plant species. Genetics 137, 67–77. doi:10.1093/genetics/137.1.67
Li, L., Paulo, M. J., Strahwald, J., Lübeck, J., Hofferbert, H.-R., Tacke, E., et al. (2008). Natural DNA variation at candidate loci is associated with potato chip color, tuber starch content, yield and starch yield. Theor. Appl. Genet. 116, 1167–1181. doi:10.1007/s00122-008-0746-y
Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., Zheng, Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. doi:10.1038/ng.3117
Lindhout, P., Meijer, D., Schotte, T., Hutten, R. C. B., Visser, R. G. F., and van Eck, H. J. (2011). Towards F1 hybrid seed potato breeding. Potato Res. 54, 301–312. doi:10.1007/s11540-011-9196-z
Livingstone, K. D., Lackney, V. K., Blauth, J. R., van Wijk., R., and Kyle-Jahn, M. (1999). Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152, 1183–1202. doi:10.1093/genetics/152.3.1183
Marhadour, S., and Prodhomme, C. (2023). “Recent trends in genetics studies and molecular breeding of potato,” in Potato production worldwide. Editors M. E. Çalişkan, A. Bakhsh, and K. Jabran (Academic Press), 273–301. doi:10.1016/B978-0-12-822925-5.00021-9
Maris, B. (1969). Studies on maturity, yield, under water weight and some other characters of potato progenies. Euphytica 18, 297–319. doi:10.1007/BF00397777
Martin, G. B., Brommonschenkel, S. H., Chungwongse, J., Frary, A., Ganal, M. W., Spivey, R., et al. (1993a). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. doi:10.1126/science.7902614
Martin, G. B., de Vicente, M. C., and Tanksley, S. D. (1993b). High-resolution linkage analysis and physical characterization of the Pto bacterial resistance locus in tomato. Mol. Plant Microbe Interact. 6, 26–34. doi:10.1094/MPMI-6-026
Mazzucato, A., Papa, R., Bitocchi, E., Mosconi, P., Nanni, L., Negri, V., et al. (2008). Genetic diversity, structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L) landraces. Theor. Appl. Genet. 116, 657–669. doi:10.1007/s00122-007-0699-6
Meksem, K., Leister, D., Peleman, J., Zabeau, M., Salamini, F., and Gebhardt, C. (1995). A high-resolution map of the vicinity of the R1 locus on chromosome V of potato based on RFLP and AFLP markers. Mol. Gen. Genet. 249, 74–81. doi:10.1007/BF00290238
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi:10.1093/genetics/157.4.1819
Milbourne, D., Meyer, R., Collins, A., Ramsay, L. D., Gebhardt, C., and Waugh, R. (1998). Isolation, characterisation and mapping of simple sequence repeat loci in potato. Mol. Gen. Genet. 259, 233–245. doi:10.1007/s004380050809
Navarro, C., Abelenda, J., Cruz-Oró, E., Cuellar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi:10.1038/nature10431
Pajerowska-Mukhtar, K. M., Mukhtar, M. S., Guex, N., Halim, V. A., Rosahl, S., Somssich, I. E., et al. (2008). Natural variation of potato allene oxide synthase 2 causes differential levels of jasmonates and pathogen resistance in Arabidopsis. Planta 228, 293–306. doi:10.1007/s00425-008-0737-x
Pajerowska-Mukhtar, K., Stich, B., Achenbach, U., Ballvora, A., Lübeck, J., Strahwald, J., et al. (2009). Single nucleotide polymorphisms in the allene oxide synthase 2 gene are associated with field resistance to late blight in populations of tetraploid potato cultivars. Genetics 181, 1115–1127. doi:10.1534/genetics.108.094268
Paterson, A., Lander, E., Hewitt, J., Peterson, S., Lincoln, S. E., and Tanksley, S. D. (1988). Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335, 721–726. doi:10.1038/335721a0
Peters, S. A., Bargsten, J. W., Szinay, D., van de Belt, J., Visser, R. G., Bai, Y., et al. (2012). Structural homology in the Solanaceae: Analysis of genomic regions in support of synteny studies in tomato, potato and pepper. Plant J. 71, 602–614. doi:10.1111/j.1365-313X.2012.05012.x
Qi, S., Shen, Y., Wang, X., Zhang, S., Li, Y., Islam, M.M., et al. (2022). A new NLR gene for resistance to Tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 135, 1493–1509. doi:10.1007/s00122-022-04049-4
Rickert, A. M., Kim, J. H., Meyer, S., Nagel, A., Ballvora, A., Oefner, P. J., et al. (2003). First-generation SNP/InDel markers tagging loci for pathogen resistance in the potato genome. Plant Biotech. J. 1, 399–410. doi:10.1046/j.1467-7652.2003.00036.x
Ritter, E., Debener, T., Barone, A., Salamini, F., and Gebhardt, C. (1991). RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX). Mol. Gen. Genet. 227, 81–85. doi:10.1007/BF00260710
Rossi, M., Goggin, F. L., Milligan, S. B., Kaloshian, I., Ullman, D. E., and Williamson, V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U. S. A. 95, 9750–9754. doi:10.1073/pnas.95.17.9750
Ruffel, S., Dussault, M.-H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., et al. (2002). A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. doi:10.1046/j.1365-313x.2002.01499.x
Ruffel, S., Gallois, J. L., Lesage, M. L., and Caranta, C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274, 346–353. doi:10.1007/s00438-005-0003-x
Sacco, A., Ruggieri, V., Parisi, M., Festa, G., Rigano, M. M., Picarella, M. E., et al. (2015). Exploring a tomato landraces collection for fruit-related traits by the aid of a high-throughput genomic platform. PLOS ONE 10, e0137139. doi:10.1371/journal.pone.0137139
Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., et al. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491. doi:10.1126/science.2448875
Schönhals, E. M., Ding, J., Ritter, E., Cara, N., Tacke, E., Hofferbert, H.-R., et al. (2017). Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array. BMC Genomics 18, 642. doi:10.1186/s12864-017-3979-9
Schornack, S., Ballvora, A., Gürlebeck, D., Peart, J., Ganal, M., Baker, B., et al. (2004). The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37, 46–60. doi:10.1046/j.1365-313x.2003.01937.x
Schreiber, L., Nader-Nieto, A. C., Schönhals, E. M., Walkemeier, B., and Gebhardt, C. (2014). SNPs in genes functional in starch-sugar interconversion associate with natural variation of tuber starch and sugar content of potato (Solanum tuberosum L). G3 Genes|Genomes|Genetics 4, 1797–1811. doi:10.1534/g3.114.012377
Sharma, S. K., Bolser, D., de Boer, J., Sønderkær, M., Amoros, W., Carboni, M. F., et al. (2013). Construction of reference chromosome-scale pseudomolecules for potato: Integrating the potato genome with genetic and physical maps. G3 Genes|Genomes|Genetics 3, 2031–2047. doi:10.1534/g3.113.007153
Simko, I., Costanzo, S., Haynes, K. G., Christ, B. J., and Jones, R. W. (2004). Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. doi:10.1007/s00122-003-1431-9
Southern, E. M. (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517. doi:10.1016/S0022-2836(75)80083-0
Stewart, H. E., Bradshaw, J. E., and Pande, B. (2003). The effect of the presence of R-genes for resistance to late blight (Phytophthora infestans) of potato (Solanum tuberosum) on the underlying level of field resistance. Plant Pathol. 52, 193–198. doi:10.1046/j.1365-3059.2003.00811.x
Sun, H., Jiao, W.-B., Campoy, J. A., Krause, K., Goel, M., Folz-Donahue, K., et al. (2022). Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 54, 342–348. doi:10.1038/s41588-022-01015-0
Sverrisdóttir, E., Sundmark, E. H. R., Johnsen, H. Ø., Kirk, H. G., Asp, T., Janss, L., et al. (2018). The value of expanding the training population to improve genomic selection models in tetraploid potato. Front. Plant Sci. 9, 1118. doi:10.3389/fpls.2018.01118
Tanksley, S. D., Ganal, M. W., Prince, J. P., de Vicente, M. C., Bonierbale, M. W., Broun, P., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. doi:10.1093/genetics/132.4.1141
Tanksley, S. D., Grandillo, S., Fulton, T. M., Zamir, D., Eshed, Y., Petiard, V., et al. (1996). Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92, 213–224. doi:10.1007/BF00223378
The 100 Tomato Genome Sequencing Consortium Aflitos, S., Schijlen, E., De Jong, H., De Ridder, D., Smit, S., et al. (2014). Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80, 136–148. doi:10.1111/tpj.12616
The Potato Genome Sequencing Consortium Xu, X., Pan, S., Cheng, S., Zhang, B., Mu, D., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi:10.1038/nature10158
The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi:10.1038/nature11119
Tiwari, J. K., Yerasu, S. R., Rai, N., Singh, D. P., Singh, A. K., Karkute, S. G., et al. (2022). Progress in marker-assisted selection to genomics-assisted breeding in tomato. Crit. Rev. Plant Sci. 41, 321–350. doi:10.1080/07352689.2022.2130361
Tomita, R., Sekine, K.-T., Mizumoto, H., Sakamoto, M., Murai, J., Kiba, A., et al. (2011). Genetic basis for the hierarchical interaction between Tobamovirus spp. and L resistance gene alleles from different pepper species. Mol. Plant. Microbe Interact. 24, 108–117. doi:10.1094/MPMI-06-10-0127
Tripodi, P., Soler, S., Campanelli, G., Díez, M. J., Esposito, S., Sestili, S., et al. (2021). Genome wide association mapping for agronomic, fruit quality, and root architectural traits in tomato under organic farming conditions. BMC Plant Biol. 21, 481. doi:10.1186/s12870-021-03271-4
van der Voort, J., Lindeman, W., Folkertsma, R., Hutten, R., Overmars, H., van der Vossen, E., et al. (1998). A QTL for broad-spectrum resistance to cyst nematode species (Globodera spp) maps to a resistance gene cluster in potato. Theor. Appl. Genet. 96, 654–661. doi:10.1007/s001220050785
Van der Vossen, E. A. G., van der Voort, J. N. A. M. R., Kanyuka, K., Bendahmane, A., Sandbrink, H., Baulcombe, D. C., et al. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 23, 567–576. doi:10.1046/j.1365-313x.2000.00814.x
Verlaan, M. G., Hutton, S. F., Ibrahem, R. M., Kormelink, R., Visser, R. G. F., Scott, J. W., et al. (2013). The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLOS Genet. 9, e1003399. doi:10.1371/journal.pgen.1003399
Visker, M. H., Keizer, L. C., Van Eck, H. J., Jacobsen, E., Colon, L. T., and Struik, P. C. (2003). Can the QTL for late blight resistance on potato chromosome 5 be attributed to foliage maturity type? Theor. Appl. Genet. 106, 317–325. doi:10.1007/s00122-002-1021-2
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). Aflp: A new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi:10.1093/nar/23.21.4407
Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotech. 16, 1365–1369. doi:10.1038/4350
Vosman, B., Kashaninia, A., van’t Westende, W., Meijer-Dekens, F., van Eekelen, H., Visser, R. G. F., et al. (2019). QTL mapping of insect resistance components of Solanum galapagense. Theor. Appl. Genet. 132, 531–541. doi:10.1007/s00122-018-3239-7
Whitham, S., Dinesh-Kumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. doi:10.1016/0092-8674(94)90283-6
Williams, J. G., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18, 6531–6535. doi:10.1093/nar/18.22.6531
Yamaguchi, H., Ohnishi, J., Saito, A., Ohyama, A., Nunome, T., Miyatak, e. K., et al. (2018). An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 131, 1345–1362. doi:10.1007/s00122-018-3082-x
Young, N. D., Zamir, D., Ganal, M. W., and Tanksley, S. D. (1988). Use of isogenic lines and simultaneous probing to identify DNA markers tightly linked to the Tm-2a gene in tomato. Genetics 120, 579–585. doi:10.1093/genetics/120.2.579
Zhang, C., Liu, L., Wang, X., Vossen, J., Li, G., Li, T., et al. (2014). The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 127, 1353–1364. doi:10.1007/s00122-014-2303-1
Zhao, J., Sauvage, C., Bitton, F., and Causse, M. (2022). Multiple haplotype-based analyses provide genetic and evolutionary insights into tomato fruit weight and composition. Hortic. Res. 9, uhab009. doi:10.1093/hr/uhab009
Keywords: potato, tomato, pepper, physical map, pathogen resistance, yield, sugar content, maturity
Citation: Gebhardt C (2023) A physical map of traits of agronomic importance based on potato and tomato genome sequences. Front. Genet. 14:1197206. doi: 10.3389/fgene.2023.1197206
Received: 30 March 2023; Accepted: 30 May 2023;
Published: 25 July 2023.
Edited by:
Joanna Jankowicz-Cieslak, International Atomic Energy Agency, AustriaReviewed by:
Bodo Trognitz, The International Maize and Wheat Improvement Center (CIMMYT), IndiaCopyright © 2023 Gebhardt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christiane Gebhardt, Z2ViaGFyZHRAbXBpcHoubXBnLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.