- 1Engineering Research Center of Chestnut Industry Technology, Ministry of Education, Hebei Normal University of Science and Technology, Qinhuangdao, Hebei, China
- 2Hebei Collaborative Innovation Center of Chestnut Industry, Qinhuangdao, Hebei, China
- 3Hebei Key Laboratory of Horticultural Germplasm Excavation and Innovative Utilization, Qinhuangdao, Hebei, China
The basic helix-loop-helix (bHLH) transcription factors (TFs) gene family is an important gene family in plants, and participates in regulation of plant apical meristem growth, metabolic regulation and stress resistance. However, its characteristics and potential functions have not been studied in chestnut (Castanea mollissima), an important nut with high ecological and economic value. In the present study, 94 CmbHLHs were identified in chestnut genome, of which 88 were unevenly distributed on chromosomes, and other six were located on five unanchored scaffolds. Almost all CmbHLH proteins were predicted in the nucleus, and subcellular localization demonstrated the correctness of the above predictions. Based on the phylogenetic analysis, all of the CmbHLH genes were divided into 19 subgroups with distinct features. Abundant cis-acting regulatory elements related to endosperm expression, meristem expression, and responses to gibberellin (GA) and auxin were identified in the upstream sequences of CmbHLH genes. This indicates that these genes may have potential functions in the morphogenesis of chestnut. Comparative genome analysis showed that dispersed duplication was the main driving force for the expansion of the CmbHLH gene family inferred to have evolved through purifying selection. Transcriptome analysis and qRT-PCR experiments showed that the expression patterns of CmbHLHs were different in different chestnut tissues, and revealed some members may have potential functions in chestnut buds, nuts, fertile/abortive ovules development. The results from this study will be helpful to understand the characteristics and potential functions of the bHLH gene family in chestnut.
Introduction
Transcription factors (TFs) are a class of functional protein factors widely existing in eukaryotes, and they regulate the expression of downstream genes through specific binding with cis-acting elements of DNA, and participate in many biological processes (Yu et al., 2023). Basic helix-loop-helix protein (bHLH) transcription factors widely exist in eukaryotes and their member number next only to the MYB family, whose name was derived from their specific protein structure contained basic amino acid regions and helix-loop-helix regions (Ledent and Vervoort, 2001; Baudry et al., 2006). The bHLH genes mostly exist in the form of gene family in plant genome, and have important functions in plant signal transduction, regulation of plant apical meristem growth, and stress resistance (Bailey et al., 2003; Komatsu et al., 2003). The bHLH gene family has been analyzed in multiple plants, such as Arabidopsis thaliana (Bailey et al., 2003), rice (Oryza sativa) (Li et al., 2006) and tomato (Solanum lycopersicum) (Sun et al., 2015; Wang et al., 2015), and the bHLH genes in this three model plant species were divided into subgroups 21, 22 and 24, respectively, which means that members of the bHLH gene family have different subgroups in other plants (Pires and Dolan, 2010b).
The functions of the bHLH genes in model plants have been widely characterized, participating in multiple biological processes such as morphogenesis, stress resistance, signal transduction and secondary metabolism (Li et al., 2006). For example, PIF4 plays a role in the elongation of the hypocotyl of A. thaliana by participating in phyB-regulated photomorphogenesis (Huq and Quail, 2002). During the development of A. thaliana embryo, the expression of RGE1 gene in endosperm can regulate the growth of embryo (Kondou et al., 2008). AtbHLH112 has a positive regulatory effect on salt resistance, drought resistance and osmotic resistance, but inhibits root development (Yujia et al., 2015). OsbHLH148, a bHLH gene in rice, constitutes the initial response of the OsbHLH148-OsJAZ-OsCOI1 signal module in rice to the expression of drought resistance genes regulated by jasmonic acid (JAs) (Seo et al., 2010). SlbHLH59 is directly bound to the PMM, GMP2 and GMP3 gene promoters in the D-mannose/L-galactose biosynthesis pathway, one of the ascorbic acid biosynthesis pathways, to promote the accumulation of ascorbic acid in tomato (Ye et al., 2019). Overexpression of ZmPTF1 in maize (Zea mays) can promote root cap development and increase biomass under low phosphorus stress (Li et al., 2011). However, the potential functions of bHLH genes in chestnut are still unclear.
Chestnut is one of the earliest domesticated plants in ancient times, with a cultivation history of more than 6,000 years in China, which is widely distributed in the northern hemisphere due to its good adaptability (Hu et al., 2022; Yu et al., 2023). Chestnut is not only an ecological tree species, but also has nutritional value. The worldwide chestnut production is about 2.27 million tons (Food and Agriculture Organization of the United Nations), and this number has been increasing continuously for more than 10 years, which indicates that consumers have great confidence in the health of chestnut nut (Massantini et al., 2021). In addition to being a good source of starch, chestnut nut is also rich in a variety of sugars, vitamins, and other biological active substances and minerals required by human body (Hu et al., 2022; Yu et al., 2023). Notably, more and more attention has been paid to the medicinal value of chestnut nut. The proper amylose/amylopectin ratio in chestnut nut has attracted attention as a preventive factor against diabetes and allergy (Choupina, 2019). Some terpenoids in chestnut nut have effects on inhibiting the expansion of HeLa tumor cells (Gao et al., 2010). Chestnut nut is rich in antioxidant substances such as ascorbic acid, phenols and flavonoids, which are very beneficial to human antibacterial and anti-cancer (Abe-Matsumoto et al., 2010; Dinis et al., 2012; Chang et al., 2020; Xu et al., 2020). In fact, the research on the antiviral effects, common disease prevention and anti-aging of chestnut nut has been carried out, which undoubtedly further increases the potential utilization value of chestnut nut (Kawasaki et al., 2009; Lupini et al., 2009; Noh et al., 2010). However, as a monoecious plant, the proportion of female and male flowers in chestnut can reach 1:2,400–4,000, and the low number of female flowers and excessive consumption of tree nutrition by abundant male flowers are significant causes for limiting the yield of chestnut (Chen et al., 2019; Yu et al., 2023). One of the important functions of the ovule is to promote the normal growth and development of flowers, and the ovule is the female reproductive organ of flower and an important part of sexual reproduction (Gómez et al., 2019). Furthermore, excavating genes that regulate the accumulation of bioactive substances in chestnut nut can be used as the first step in cultivating new high-quality chestnut varieties (Hu et al., 2022). Notably, the functions of bHLH genes in plants are diversified, such as GA signal transduction regulation, meristem growth and fruit development, and these are all related to factors that limit the development of the chestnut industry, such as imbalanced male and female flowers, poor nut quality (Sun et al., 2020; Yu et al., 2023). Thus, it is very significant to excavate the genes that regulate the fertility of chestnut ovules and nut development. The publication of the whole genome sequence of chestnut has enabled us to excavate the bHLH genes which potentially regulate growth and development by comprehensively studying the gene structure and evolutionary characteristics of bHLH gene family (Wang et al., 2020).
Chestnut is an excellent tree species with nutritional, medicinal and ecological values, and the extremely small ratio of female to male flowers (1:2,400–4,000) is an important reason for limiting chestnut yield (Chen et al., 2019; Yu et al., 2023). Studies have confirmed that bHLH factors play an important role in the development of ovule, which is an important part of sexual reproduction and promote the normal development of flowers (Heisler et al., 2001; Huq and Quail, 2002). Whether bHLH genes play a role in chestnut development has not been reported. Here, the bHLH gene family in chestnut has been identified and characterized first time, with analyzing the physicochemical properties, phylogenetic comparison, gene structure, cis-acting elements and duplication model. Moreover, we also compared the expression patterns of all identified bHLH genes in chestnut buds, nuts, fertile/sterile ovules at different development stages. To verify the above analysis, several bHLH genes were validated using qRT-PCR and subcellular localization assays. This study provided a foundation for further analysis of the potential mechanism of bHLH genes in chestnut growth and development, especially for aiming to identify candidate genes that may be involved in the development of chestnut buds, nuts, fertile/sterile ovules.
Materials and methods
Gene identification, enrichment and subcellular location analysis
The published 158 bHLH genes in A. thaliana were used as a query sequence to run the BLAST (Basic Local Alignment Search Tool) program on the protein dataset of chestnut with default parameters. Furthermore, we used the latest Hidden Markov Model (HMM) file of the bHLH domain (PF00010) to search against the all proteins of chestnut using HMMER 3.0 (Evalue < 1e-5). After determine the existence of the bHLH domain using the NCBI Batch CD Search tool, we obtained 94 bHLH genes in chestnut, which were renamed CmbHLH1 to CmbHLH94 based their order on chromosome. The ExPasy website was used to obtain their physicochemical properties, and we used the eggNOG-mapper (Cantalapiedra et al., 2021) to obtain GO and KEGG annotation. We performed GO and KEGG enrichment analysis and visualization using TBtools (Chen et al., 2020). The subcellular localization of CmbHLH genes were predicted by using the online tool Cell-Ploc (Chou and Shen, 2010), and the full length coding sequence (CDS) without stop codon was cloned into the pAN580 and N-terminal fused with the green fluorescent protein (GFP) to verify the location of CmbHLH9, CmbHLH25 and CmbHLH55. The fusion constructs then transiently expressed into A. thaliana protoplasts.
Phylogenetic and sequence analysis
MEGA 7.0 (Kumar et al., 2016) was used to construct a phylogenetic tree for the bHLH genes in chestnut with the maximum likelihood method, and we used the “Find Best DNA/Protein Models (ML)” function to find the best amino acid substitution model (partial deletion 95%). The final parameters were as follows: Jones–Taylor–Thornton (JTT) model; Gamma Distributed (G); Partial deletion 95%; 1,000 bootstrap replications. A phylogenetic tree of CmbHLH genes was constructed with neighbor-joining method using MEGA 7.0 with parameters: Poisson model, pairwise deletion, and 1,000 bootstrap replications. Additionally, Bayesian method was used to construct a phylogenetic tree of CmbHLHs. Briefly, MAFFT 7.0 (Katoh and Standley, 2013) and ModelFinder (Kalyaanamoorthy et al., 2017) were used to perform multiple sequence alignments and select optimum protein substitution model, respectively. MrBayes 3.2 (Ronquist et al., 2012) was used to construct Bayesian tree based on the best-fit model: JTT + F + I + G4. TBtools was used to visualize the structure of CmbHLH genes. The conserved domain and motif information of the CmbHLH genes were obtained from the Batch-CDD and MEME online programs, respectively. The combined analysis of phylogenetic tree, conserved domain and motif distribution was used to explore the sequence characteristics in different subgroups. The psRNATarget (Dai and Zhao, 2011) and Simple Sequence Repeat Identification Tool (SSRIT) on Gramene website (Tello-Ruiz et al., 2021) were used to predict miRNA targets and SSRs of the CmbHLH genes, respectively. The 3D structure models of all CmbHLH proteins were predicted through Swiss Model.
Collinearity analysis of CmbHLH genes
To further explore the evolution of the CmbHLH genes, the Multiple Collinearity Scan Toolkit (MCScanX) program (Wang et al., 2012) was used to perform a collinearity analysis within chestnut and with A. thaliana, rice, oak and grape genomes. Notably, we used a detailed method as in our previous study to determine the duplication model of the CmbHLH genes (Yu et al., 2022a). Based on the algorithm of the MCScanX program (Wang et al., 2012), the protein-coding genes from chestnut genome was compared against itself, and the duplicate genes were first defined as i) “dispersed duplicates”; ii) If the distance or gene rank between two duplicate genes is less than 20, they are re-labeled as “proximal duplicates”; iii) If the duplicate genes are adjacent (gene rank is 1), they are re-labeled as “tandem duplicates”; iv) The duplicated genes in the collinearity regions are re-labeled as “WGD or segmental ". The MCScanX program was run to directly obtain CmbHLH genes from proximal, dispersed and tandem duplication. Specifically, based on the collinearity results within the chestnut genome, we drawn the homologous collinearity dot-plot within the chestnut genome. Further, non-synonymous (Ka) and synonymous substitution sites (Ks) values of homologous gene pairs on the homologous collinear blocks were calculated by KaKs_Calculator (Wang et al., 2010), and the median Ks values was calculated by writing script (Yu et al., 2023). The median Ks values of homologous collinear blocks containing the CmbHLH genes were labeled on the dot-plots. Finally, based on the distribution of Ks values corresponding to the whole genome duplication (WGD) event that occurred in the chestnut genome in our earlier stage (Yu et al., 2023), combined with the complementarity of homologous collinear blocks, we finally determined the CmbHLH genes formed by WGD event.
Cis-acting elements analysis
Based on the structure annotation of the chestnut genome, the 2000 bp upstream sequences of 94 CmbHLH genes were extracted from the chestnut genome sequence. The upstream sequences of CmbHLH genes were submitted to PlantCARE software for identification of cis-acting elements and function categorization. We used Tbtools to visualize the predicted cis-acting elements, except for those members without specific function.
Expression analysis of CmbHLH genes
In order to explore the potential function of the CmbHLH genes in different tissues of chestnut, public database resources were utilized. RNA-seq data of chestnut buds at different stages (20, 25 and 30 days post-anthesis), chestnut nuts at different stages (70, 82 and 94 days post-anthesis), chestnut fertile/abortive ovules at different stages (15-July, 20-July and 25-July) were obtained from NCBI, and corresponding accession numbers were in Supplementary Table S1. The above transcriptome data from chestnut buds, nuts and fertile/abortive ovules were all three biological replicates. All RNA-seq reads were mapped on the reference genome (N11-1 from Castanea Genome Database) and the Kallisto software was used to quantify transcriptome data into Transcripts Per Kilobase of exon model per Million mapped reads (TPM). Tbtools was used to map the heatmaps for exhibiting the expression level of CmbHLH genes. The “Normalized” (scale method) in TBtools was used to normalize the expression, and the heatmaps were created by TBtools based on the transformed data of log2 (TPM + 1) values.
Plant material and qRT-PCR analysis
The samples used for real-time quantitative PCR experiments were the nuts of “Yan Long” chestnut planted by Hebei Normal University of Science and Technology at 70, 82, and 94 days post-anthesis with three biological replicates. All samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. The HiPure Total RNA kit (Magen, R4111, China) was used to extracted and isolated total RNA. PrimeScript RT Master Mix (Takara Biotechnology Co., Dalian, China) was used to reverse transcribe RNA into single stranded cDNA. Real-time quantitative PCR experiments were performed on ABI 7500 Real-Time PCR system (Applied Biosystems Inc., Foster City, CA, United States) with TB Green Premix Ex Taq (Takara). The relative gene expression values were calculated using the comparative CT (2-△△CT) method (Livak and Schmittgen, 2001), and 18S gene of chestnut was used an internal control.
Results
Identification, physicochemical properties, classification and function annotation
We identified 94 bHLH genes in chestnut genome using the BLASTP and HMMER 3.0 programs, and they were designated as CmbHLH1 to CmbHLH94 based on their relative position on chromosome and scaffold (Figure 1A; Supplementary Table S2). Eighty-eight CmbHLH genes were randomly distributed on all 12 chromosomes and other six CmbHLH genes were located on five unanchored scaffolds. Chromosome 8 had the highest number of CmbHLH members (13), while chromosome 6 had the fewest CmbHLH members (3) (Figure 1A). The analysis of all CmbHLH protein sequences showed that they had extensive variation in physicochemical properties (Supplementary Table S2). The length of CmbHLH proteins varied from 159 to 711 aa, molecular weight ranged from18.0 to 79.2 KDa. Most CmbHLH proteins were weakly acidic, because their average theoretical isoelectric point (pI) was 6.48. Only the instability index of CmbHLH42 protein was less than 40, and only the grand average of hydropathicity (GRAVY) value of CmbHLH49 protein was greater than zero, which meant that most CmbHLH proteins were unstable and hydrophilic. The aliphatic index of CmbHLH proteins varied from 49.33 to 109.53. A total of 6,831 microRNA (miRNA) targets were identified in CmbHLHs, indicating that corresponding miRNAs can activate or inhibit gene expression (Supplementary Table S3). In addition, 180 simple sequence repeats (SSRs) were predicted in the CmbHLHs, and the 3D structure models of CmbHLH proteins were predicted (Supplementary Tables S4, S5; Supplementary Figure S1). All these results provided valuable information reference for further research on the CmbHLH genes.
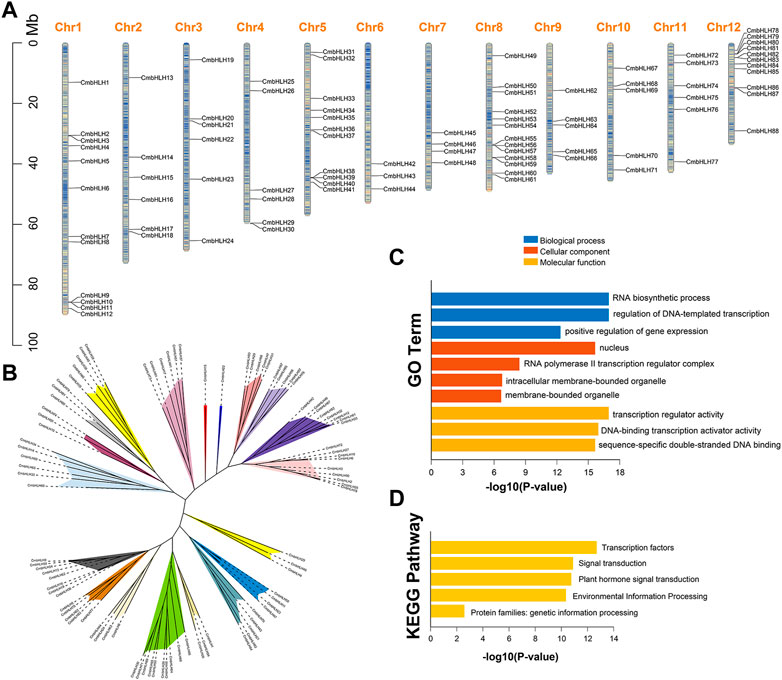
FIGURE 1. Chromosome distribution, phylogenetic analysis and functional enrichment analysis of the CmbHLH genes. (A) Chromosome distribution of CmbHLH genes. The color of segments in the chromosomes shows the gene density of the corresponding region. (B) Unrooted phylogenetic tree of CmbHLH proteins. MEGA 7.0 was used to construct the phylogenetic tree based on the protein sequences with maximum likelihood method. The proteins were clustered into 19 groups. Different background colors indicate the different group of the CmbHLH proteins. (C) GO function enrichment analysis of CmbHLH genes. (D) KEGG function enrichment analysis of CmbHLH genes.
The classification of plant bHLH proteins had always been vague, but it was generally considered to be divided into 15–32 subgroups (Pires and Dolan, 2010b; Carretero-Paulet et al., 2010). We used the maximum likelihood method to construct an unrooted phylogenetic tree to explore the evolutionary relationships of CmbHLH genes (Figure 1B). The 94 CmbHLH proteins were classified into 19 subgroups based on the unrooted tree, which were named A to S (Figure 1B). Interestingly, subgroup J and K contained a single CmbHLH protein (CmbHLH16 and CmbHLH22), respectively. Except subgroup J and K, the number of CmbHLH genes in each subgroup varies greatly from 3 (subgroup D, H, Q and R) to 10 (subgroup E). In addition, we also constructed two phylogenetic tree of CmbHLH proteins with neighbor-joining and Bayesian method, respectively (Supplementary Figures S2, S3). The results showed that the phylogenetic trees of CmbHLHs constructed by the three methods were almost identical, with 94 CmbHLHs divided into 19 subgroups. Gene Ontology (GO) and The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to characterize the potential functions of the identified CmbHLH genes. As shown in Figures 1C, D, CmbHLH genes were mainly involved in RNA biosynthetic process, positive regulation of gene expression, and transcription regulator activity, and these functions and processes were closely related to transcription factors. KEGG enrichment results indicated that they were mainly enriched in transcription factors, signal transduction, plant hormone signal transduction, environmental information processing, which were the main mechanisms by which bHLH family transcription factors regulate the expression of downstream genes.
Motif distribution, gene structure and promoter cis-elements analysis
The motif diversity and gene structure of CmbHLH genes were exhibited to reveal their evolution (Figure 2). The online MEME program was performed to identify motif patterns, and CmbHLH genes contained varying numbers of conserved motifs (Figure 2A). CmbHLH members in the same subgroup usually had similar motif arrangement patterns, while members in different subgroups had obvious differences. Although motifs 1 and 2 were present in almost all CmbHLH genes, some of the other motifs were detected in some specific subgroups. For example, motif 8 only can be detected in subgroup A, and motif 10 only can be detected in subgroup N. Interestingly, it was found that some CmbHLH genes in M subgroup had the same motif composition as the members in L subgroup, but some members of the M subgroup had completely different motif arrangements. This result implied that some CmbHLH genes had undergone dramatic changes in their non-motif sequences during evolution and formed the new subgroups without motif composition changing. Although the gene structure analysis indicated that there was extensive variation in the number of introns (0–11 introns) and gene structure of the CmbHLH genes, the members in the same subgroup had similar intron-exon structure (Figure 2B). For example, members in subgroup A had 0 or 1 intron, while members in subgroup Q had 3 introns. Interestingly, some CmbHLH members had exactly motif arrangement and extreme similar gene structure, such as CmbHLH39 and CmbHLH40, and subsequent analysis showed that they were formed from tandem duplication. These results indicated that the members in the same subgroup had similar motifs arrangement pattern and gene structure, which suggested that they may have similar functions.
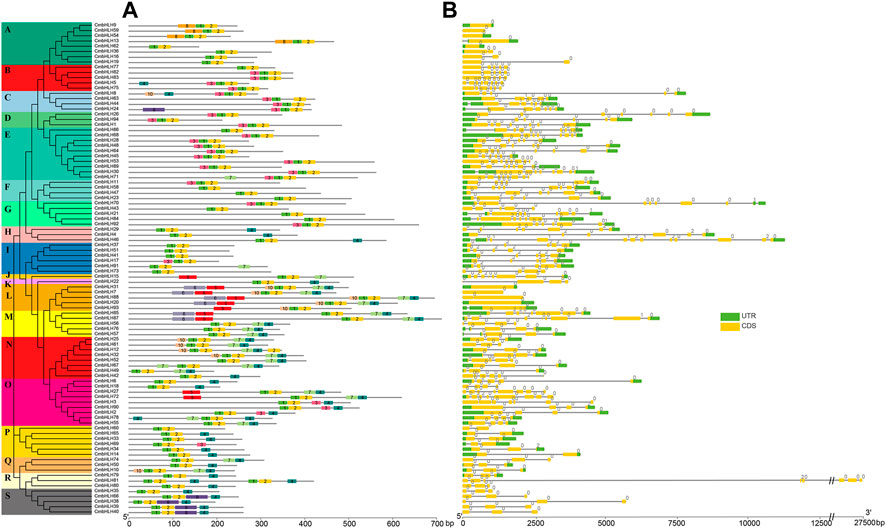
FIGURE 2. The conserved motifs distribution and gene structure of CmbHLH genes. (A) Distribution of conserved motifs of CmbHLH genes. The left side dendrogram shows the phylogenetic tree of CmbHLH genes. (B) Gene structure of CmbHLH genes.
The upstream 2,000 bp sequences of the CmbHLH genes were extracted to predict cis-acting elements in promoter regions, which can further explore the potential function and regulatory mechanism of the CmbHLHs (Figure 3; Supplementary Table S6). A total of 1,562 cis-acting elements were detected in promoter regions of CmbHLHs by the PlantCare software. Abundant cis-acting regulatory elements related to endosperm expression, meristem expression, and responses to gibberellin (GA) and auxin were identified in the upstream sequences of CmbHLH genes. This indicated that these genes may have potential functions in the morphogenesis of chestnut. Additionally, numerous cis-acting elements related to environment and stress were detected, such as cis-acting element involved in defense and stress responsiveness, cis-acting element involved in low-temperature responsiveness, cis-acting regulatory element essential for the anaerobic induction, and wound-responsive element. Notably, the cis-acting elements commonly considered important in relation to hormone response were detected, such as auxin-responsive element, gibberellin-responsive element, and salicylic acid responsive element, which were believed to be widely involved in all aspects of plant growth and development (Woodward and Bartel, 2005; Yamaguchi, 2008; Peng et al., 2021). The cis-acting elements involved in many functions were identified, which suggested that CmbHLH genes played diverse roles in chestnut.
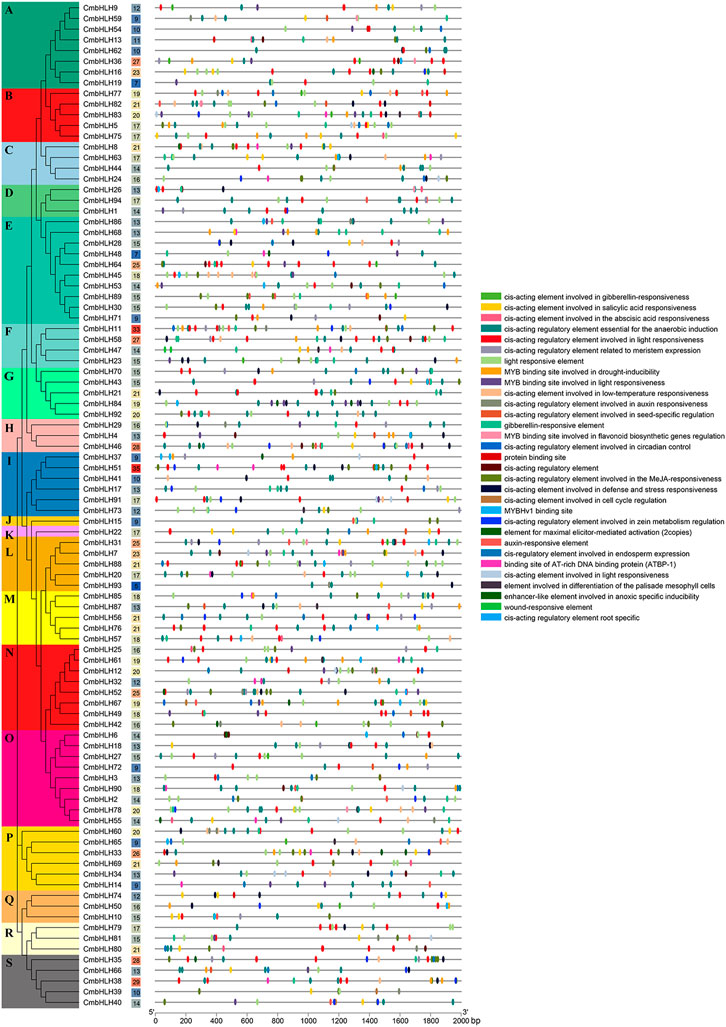
FIGURE 3. The cis-acting elements detected in the promoter regions of CmbHLH genes. The left side dendrogram shows the phylogenetic tree of CmbHLH genes. The number of cis-acting elements in the promoter region of each CmbHLH gene was shown in the middle of the figure. The distributions of cis-acting elements in the 2,000 bp upstream promoter are shown. The different functions of cis-acting elements are represented by different colors, as shown on the right.
Collinearity analysis of CmbHLHs
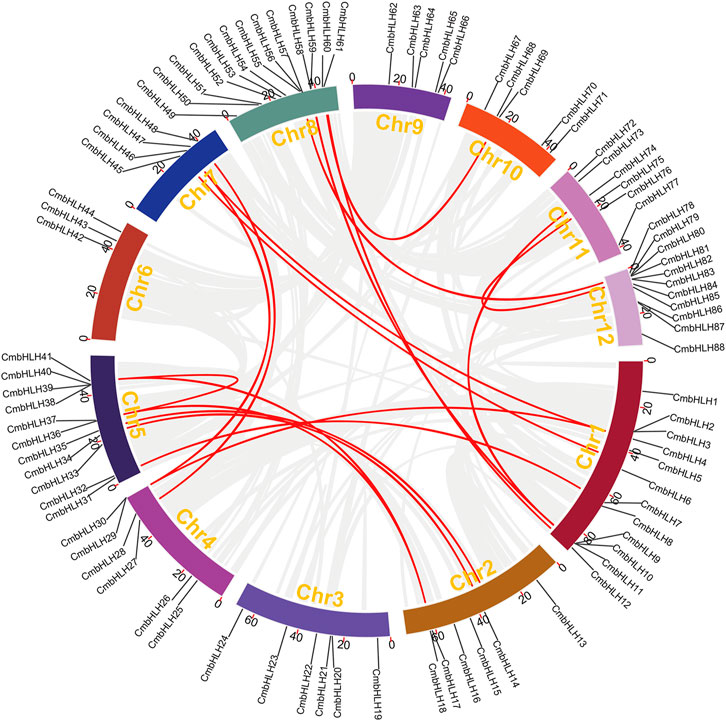
The collinearity analysis among chestnut, grape, A. thaliana, oak, and rice genomes was performed to further investigate the evolution of the CmbHLH genes (Figure 4). Interestingly, we detected 68 CmbHLHs in the collinearity regions of the chestnut and grape genomes, and there were only 51, 50 and 25 CmbHLHs in the collinear regions between chestnut and A. thaliana, chestnut and oak, chestnut and rice, respectively (Supplementary Tables S7–S10). Additionally, there were 88 orthologous gene pairs contained CmbHLHs between chestnut and grape genome, and only 74, 57 and 44 orthologous gene pairs were found between chestnut and A. thaliana, chestnut and oak, chestnut and rice, respectively (Supplementary Tables S7–S10). As we all know, grape had a relatively stable genome structure, which was often used as a good reference for understanding the genome or related gene evolution of other eudicot plants (Consortium et al., 2012; Yu et al., 2022a). Therefore, the chestnut and grape genomes had not experienced additional whole genome duplication (WGD) events after the eudicot common hexaploidization event (ECH) (Yu et al., 2022b; Hu et al., 2022), and the grape genome had a relatively stable genomic structure, which resulted in the above statistical results about chestnut and grape. The distant genetic relationship between chestnut and rice, and rice genome had experienced additional WGD after ECH, may result in the least CmbHLHs in their collinear regions, and the least orthologous gene pairs contained CmbHLHs between them.
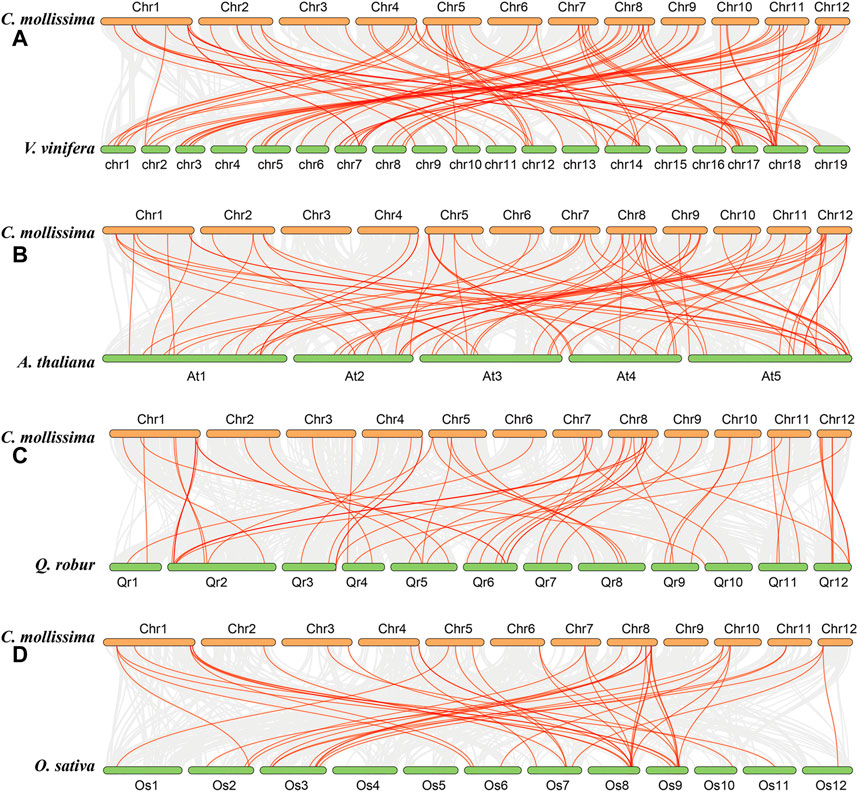
FIGURE 4. The collinearity among chestnut, grape, Arabidopsis thaliana, oak, and rice genomes obtained through MCScanX. (A) Collinearity relationship between chestnut and grape genomes. (B) Collinearity relationship between chestnut and Arabidopsis thaliana genomes. (C) Collinearity relationship between chestnut and oak genomes. (D) Collinearity relationship between chestnut and rice genomes.
Significantly, we can detect some CmbHLHs existed in three or more collinear gene pairs, such as CmbHLH1, which suggesting that more copies of their orthologous genes in A. thaliana, rice, oak and grape genomes had been retained (Figure 4). According to the gene balance hypothesis, the products of these CmbHLHs may participate in macromolecular complexes or signaling networks (Birchler and Veitia, 2007). The subsequent transcriptome analysis revealed that they may have important functions in the bud development and ovule fertility of chestnut. Additionally, the collinear blocks contained CmbHLHs generally including more gene pairs in chestnut and grape (average of 49 gene pairs) (Supplementary Table S11). Contrasty, there were only 19, 27 and 11 (average value) gene pairs per block contained CmbHLHs between chestnut and A. thaliana, between chestnut and oak, between chestnut and rice, respectively (Supplementary Tables S12–S14). We speculated that the differences in WGD experience and genetic relationships of these species may be the cause of these results.
Gene duplication event was an important motivation for the expansion of gene families and the occurrence of new functions (Liu et al., 2018; Quan et al., 2019; Yu et al., 2021). The Multiple Collinearity Scan toolkit (MCScanX) program (Wang et al., 2012) and writing script were used to conduct collinearity analysis of chestnut genome to determine the duplication model of CmbHLHs (Yu et al., 2021). MCScanX could identify most duplication types except WGD and segmental duplication, and the reason was that the plant genome undergone a large number of chromosome recombination after WGD, which made them difficult to distinguish (Figure 5) (Wang et al., 2018; Yu et al., 2022a; Wang et al., 2022; Yu et al., 2023). As in our previous study (Yu et al., 2023), by drawing the homologous collinearity dot-plot within chestnut genome labeled with the median Ks of the homologous blocks, combining their length and complementarity, WGD and segmental duplications were finally distinguished (Figure 6). The results indicated that most of the CmbHLHs formed from dispersed duplication, followed by WGD, and no CmbHLH member was considered as singletons (Supplementary Table S15). These results showed that dispersed duplication was the main reason for the increase in the member of the bHLH gene family in chestnut. Additionally, Ka/Ks values of homologous gene pairs were calculated, which indicated that all of CmbHLHs were experienced purifying selection pressure during evolution (Ka/Ks < 1) (Supplementary Table S16).
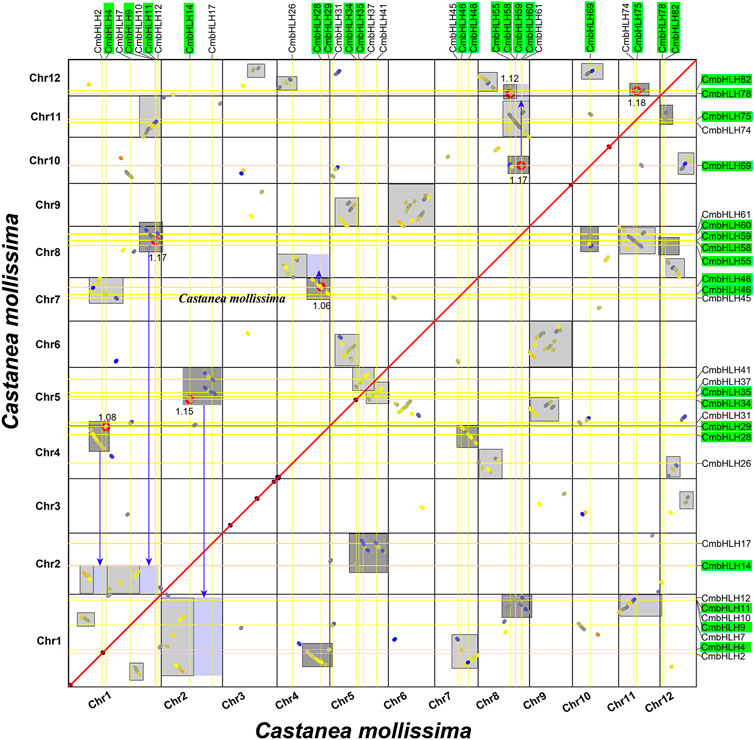
FIGURE 6. Homologous collinearity dot-plot within chestnut genome. The collinear blocks from WGD containing CmbHLH genes are marked in the gray boxes of the figure, and the median Ks of the collinear blocks are marked.
Expression analysis of CmbHLHs
The transcriptome data of chestnut buds at different stages from NCBI were investigated to explore their potential function in chestnut bud development (Figure 7A). Some CmbHLH genes had high expression level at the all stages of chestnut bud development, suggesting their potential function in the chestnut bud development process. For example, the average TPM values of CmbHLH88 were 138.16, 165.03 and 155.20 at three stages, respectively. Notably, the expression level of some CmbHLH genes decreased (CmbHLH25) or increased (CmGRAS51) sharply as the buds continue to growth. The TPM value of CmbHLH25 in chestnut buds at 20 days post-anthesis was 107.91, while the TPM values at 25 and 30 days post-anthesis were 27.07 and 3.44, respectively. The TPM value of CmbHLH51 in chestnut buds at 20 days post-anthesis was 34.37, while the TPM values at 25 and 30 days post-anthesis were 71.61 and 109.33, respectively. These results indicated that CmbHLHs play an important role in chestnut bud development.
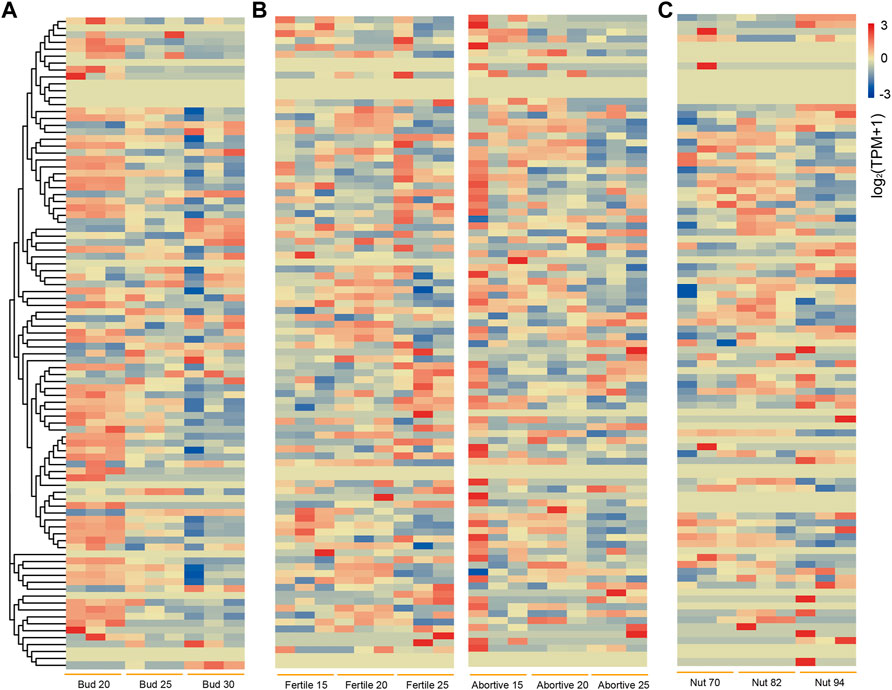
FIGURE 7. Heatmaps of bHLH genes expression in different stages of chestnut buds, nuts, fertile/abortive ovules. (A) Heatmap of bHLH genes expression in chestnut buds at 20, 25 and 30 days post-anthesis. The left side dendrogram shows the phylogenetic tree of CmbHLH genes. (B) Heatmap of bHLH genes expression in fertile and abortive ovules of chestnut on 15-July, 20-July and 25-July. (C) Heatmap of bHLH genes expression in chestnut nuts at 70, 82 and 94 days post-anthesis.
In addition, we analyzed transcriptome data from fertile/abortive ovules of chestnut at different developmental stages to characterize the possible role of the CmbHLH genes in ovule fertility (Figure 7B) (Du et al., 2021). We found that the expression levels of a few CmbHLHs in fertile/abortive ovules showed significant differences, such as CmbHLH55 and CmbHLH69. The expression level (TPM of all samples was more than 200) of CmbHLH55 in fertile ovules was significantly higher than that in abortive ovules at all three stages, while the expression of CmbHLH69 of fertile ovule (TPM of all samples were less than 50) was significantly lower than that of abortive ovules at all three stages. Additionally, the expression levels of some CmbHLH members had significantly changed during the growth of fertile ovules. For example, the TPM value of CmbHLH69 on 15-July was 86.24, while the TPM values on 20-July and 25-July reached 151.44 and 144.10, respectively. Notably, multitudinous of gibberellin (GA) responsive elements had been detected in their promoter regions, and GA was considered to be a plant hormone that was extremely important for plant reproductive organs (Figure 3) (Plackett et al., 2014). Cis-regulatory element involved in endosperm expression had also been identified in the promoter regions of these CmbHLHs, which indicated that they may have impact on the fertility or development of chestnut ovules.
Similarly, transcriptome data from chestnut nuts at different developmental stages were analyzed (Figure 7C) (Li et al., 2021). Obviously, the expression of some CmbHLH genes at 94 days post-anthesis of chestnut nut development had reduced dramatically, such as CmbHLH25 and CmbHLH78. For example, the average TPM values of CmbHLH25 were 274.67 at 70 days post-anthesis, and this values were 164.14 and 1.29 at 82 and 94 days post-anthesis, respectively. Considering that some bHLH genes had been verified to be involved in the regulation of secondary metabolite formation (Li et al., 2006) and that some cis-acting regulatory elements involved in the regulation of zein metabolism and flavonoid biosynthesis had been discovered (Figure 3; Supplementary Table S6), we believed that these CmbHLH members may be related to chestnut nuts ripening and related quality. To verify the expression patterns of CmbHLHs, six CmbHLH genes were analyzed using qRT-PCR, and the results were shown in Figure 8A. Overall, the qRT-PCR experiments analysis results were consistent with the RNA-seq analysis. For example, CmbHLH25 was significantly highly expressed in nuts at 70 days post-anthesis than in nuts at 82 and 94 days post-anthesis. The expression level of CmbHLH2 was highest at 70 days post-anthesis, and decreased to the lowest at 82 days post-anthesis, and increased at 94 days post-anthesis. The expression trends of other four CmbHLH genes analyzed using qRT-PCR during chestnut nut development were also consistent with the results of RNA-seq analysis. In addition, almost all CmbHLH proteins (85/94) were predicted in the nucleus, based on subcellular localization analysis (Supplementary Table S2). We instantaneously expressed GFP-CmbHLH9, GFP-CmbHLH25, and GFP-CmbHLH55 fusion proteins in A. thaliana protoplasts for subcellular localization. As shown in Figure 8B, CmbHLH25 and CmbHLH55 were co-localized to both the nucleus and cytoplasm, and CmbHLH9 was localized to the nucleus.
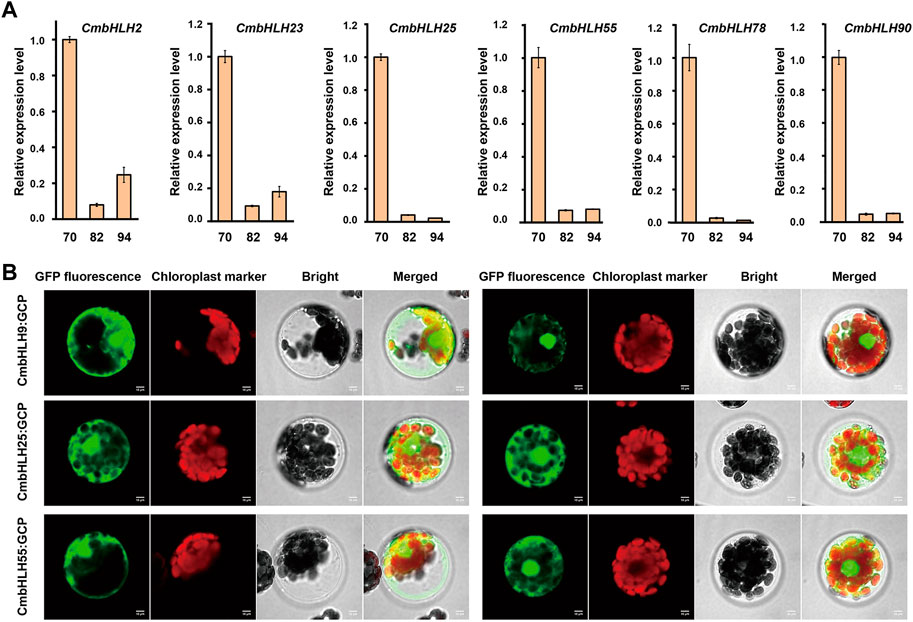
FIGURE 8. qRT-PCR of six CmbHLHs (CmbHLH2, CmbHLH23, CmbHLH25, CmbHLH55, CmbHLH78 and CmbHLH90) in chestnut nuts at different developmental stages and subcellular location of three CmbHLHs (CmbHLH9, CmbHLH25 and CmbHLH55) proteins in Arabidopsis thaliana protoplasts. (A) qRT-PCR of CmbHLH2, CmbHLH23, CmbHLH25, CmbHLH55, CmbHLH78 and CmbHLH90 in chestnut nuts at different developmental stages. (B) Subcellular location of CmbHLH9, CmbHLH25, CmbHLH55 proteins in Arabidopsis thaliana protoplasts.
Discussion
Numerous studies have shown that the bHLH gene family had been formed in algae, but their number was relatively small, and the bHLH domain had not undergone significant differentiation (Robinson and Lopes, 2000; Pires and Dolan, 2010a; Fan et al., 2021; Song et al., 2021). Accordingly, the function of the bHLH genes in algae may be limited, and the number of downstream genes that can be regulated was also small (Robinson and Lopes, 2000; Pires and Dolan, 2010a). In the process of algae evolving into various complex plants, the number of members in bHLH gene family had greatly expanded, indicating the important role of the bHLH gene function in plant evolution (Kurbidaeva et al., 2014). The current functional characterization of the bHLH genes in many higher plants indicates that they have important functions in various biological processes such as morphogenesis, stress resistance, signal transduction, and secondary metabolism (Li et al., 2006).
The number and classification of bHLH genes in plants showed great diversity. For example, there were 161, 167, and 152 bHLH genes in A. thaliana, rice, and tomato, and they were divided into 21, 22, and 24 subgroups, respectively (Sun et al., 2015). In this study, we identified 94 bHLH genes in the chestnut genome and divided them into 19 subgroups using three methods (the neighbour-joining, maximum likelihood, and Bayesian methods). Similar gene structures were found in CmbHLHs within the same subgroup, while members belonging to the same subgroup showed differences in motif composition (Figure 2). For example, CmbHLH77 lacked a motif 3 compared to other members in the same subgroup B, and the motif composition of CmbHLH77 was completely consistent with most members of subgroup A. These results suggested that CmbHLH77 may loss motif 3 during evolution, but retained other features of members in subgroup B. Considering that gene duplication was an important driving force for the expansion of many gene families (Yu et al., 2022b; Yu et al., 2023), we in-depth analyzed the duplication models of the CmbHLH genes. Notably, previous studies on gene duplication had rarely distinguished WGD from segmental duplication, due to the large number of gene losses and chromosome fusion that accompany WGD in plant genomes (Yu et al., 2022a). Here, we conducted combined analyses of the collinear blocks of the chestnut genome and the Ks that can characterize the time of gene duplication, as in our previous study (Yu et al., 2022a). Grossly, the collinear regions formed by WGD were usually larger blocks (covering more gene pairs), and the Ks values of gene pairs on that collinear blocks were similar, corresponding to the Ks value at the time of the WGD event (Supplementary Table S16). Based on the above principles, we distinguished the duplication models of all CmbHLH genes, and the results showed that dispersed duplication and WGD were the main reasons for the expansion of the CmbHLH genes, which highlighted the importance of transposable elements and polyploidization in the expansion of chestnut bHLH gene family. WGD is one of the important origins of early plant genes, but the mechanism of dispersed duplication is still unclear (Bai et al., 2022). However, transposable elements (TEs) are considered the most common source of dispersed duplication (Kroon et al., 2016). Transposable elements have the flexibility to move their positions within the genome and play an important role in shaping the fate of gene function (Fueyo et al., 2022). In this study, a large number of dispersed duplication genes showed significant diversity in gene structure and expression patterns (Figures 2, 7). All the above analysis suggested that dispersed duplication may be one of the important reasons for the expansion and functional differentiation of the chestnut bHLH genes.
The bHLH genes played diverse roles in plant, especially in plant meristem and morphological development (Heisler et al., 2001; Bernhardt et al., 2005). LAX PANICLE (LAX), a bHLH gene in rice, was a major regulatory factor controlling the apical meristem of rice, and ectopic LAX expression in rice resulted in dwarfing (Komatsu et al., 2003). The SPATULA gene was first found in A. thaliana to participate in the control of the peripheral region of stem tip meristem and the development of specific tissues in leaves, petals, stigmas, and roots (Heisler et al., 2001). In this study, some CmbHLH genes that may have functions in the development of chestnut buds and fertile/abortive ovules were discovered based on analysis of transcriptome data. CmbHLH88 was highly expressed at all stages of chestnut bud development (TPM >130 in examples of all stages), and cis-regulatory elements related to meristem expression had been detected in the CmbHLH88 promoter region. These results prompted us to speculate that CmbHLH88 may be involved in the chestnut buds development. Furthermore, the bHLH genes had also been found to participate in the formation of secondary metabolism in plants (Zhang et al., 2011; Nims et al., 2015). OsbHLH148 in rice was involved in the jasmonic acid (JAs) signaling pathway, and JAs was an important hormone involved in regulating secondary metabolic biosynthesis in plants (Seo et al., 2011). It was found that the expression of some CmbHLH genes increased sharply during the late ripening stage (94 days post-anthesis) of chestnut nuts, such as CmbHLH9, and cis-acting regulatory elements involved in the regulation of zein metabolism and flavonoid biosynthesis in its promoter region were identified, which suggested that it may have potential functions for chestnut ripening.
Notably, the involvement of the bHLH genes in plant embryos has received widespread attention. RGE1 had been proven to control the normal growth of embryos (Kondou et al., 2008). PIF4 plays a role in the elongation of the hypocotyl of A. thaliana by participating in phyB-regulated photomorphogenesis (Huq and Quail, 2002). The SPATULA gene can ultimately affect the formation of pistils by promoting the growth of carpels (Heisler et al., 2001). Here, interesting phenomena were found in the transcriptome of ovules. The expression level of CmbHLH55 in fertile ovules was significantly higher than that in abortive ovules during all developmental stages. Contrasty, the expression of CmbHLH69 in fertile ovule was significantly lower than that of abortive ovules during all developmental stages. Additionally, multitude researchers had reported that gibberellin has important function in plant reproductive organs, especially in gender determination of flowers (Song et al., 2013; Yu et al., 2023). The large ratio of female and male flowers (1:2,400–1:4,000) severely consumes tree nutrients and limits the number of female flowers, which was the key reason for limiting chestnut production (Chen et al., 2019). Due to the gibberellin responsive elements (GRE) (Figure 3) identified in these CmbHLH genes promoter regions, and the significant differences in the expression of these genes in fertile/abortive ovules, we speculated that they may have a significant potential role in the fertility of chestnut ovules.
The important function of bHLH genes in plant was continuously being proven, such as GA signal transduction regulation, secondary metabolite formation and stress resistance, and these functions all may be used to lift restrictions on chestnut yield and quality (Li et al., 2006; Liu et al., 2015; Chen et al., 2019). Paying attention to the potential function of bHLH genes in plants was of great significance for improving the yield and quality of chestnut nuts, as well as the development of the entire chestnut industry. Here, the physicochemical properties, cis-acting elements, phylogenetic analysis, gene structure and duplication model of 94 CmbHLH genes had been comprehensively characterized in chestnut. Transcriptome data, qRT-PCR and subcellular localization were used to analyze the expression patterns of CmbHLHs in different development stages of chestnut buds, nuts, fertile/sterile ovules, and revealed some members may have potential functions in chestnut tissue development and nut quality. This study provides a reference for elucidating the CmbHLH genes evolution and their potential functions in the development of chestnut buds, nuts, and fertile/abortive ovules.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
LiY, CF, DW, RH, and JZ conceived and designed the experiments and drafted the manuscript. LiY, WX, CG, LJ, WM, LuY, and HZ analyzed the data. All authors read and approved the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Hebei Collaborative Innovation Center of Chestnut Industry (2022) and Hebei Province Key R&D Program Project (21326304D).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1193953/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Prediction of 3D structure of CmbHLH proteins by Swiss model.
SUPPLEMENTARY FIGURE S2 | Phylogenetic tree of CmbHLH genes was constructed by neighbor-joining method.
SUPPLEMENTARY FIGURE S3 | Phylogenetic tree of CmbHLH genes was constructed by Bayesian method.
References
Abe-Matsumoto, L., Lajolo, F., and Genovese, M. (2010). Comparison of phenol content and antioxidant capacity of nuts. Cienc. E Tecnol. De. Aliment. - Cienc. Tecnol. Aliment. 30, 254–259. doi:10.1590/S0101-20612010000500038
Bai, J., Song, M. J., Gao, J., and Li, G. (2022). Whole genome duplication and dispersed duplication characterize the evolution of the plant PINOID gene family across plant species. Gene 829, 146494. doi:10.1016/j.gene.2022.146494
Bailey, P. C., Martin, C., Toledo-Ortiz, G., Quail, P. H., Huq, E., Heim, M. A., et al. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 15 (11), 2497–2502. doi:10.1105/tpc.151140
Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. Cell. Mol. Biol. 46 (5), 768–779. doi:10.1111/j.1365-313x.2006.02733.x
Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 (2), 291–298. doi:10.1242/dev.01565
Birchler, J., and Veitia, R. (2007). The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell. 19, 395–402. doi:10.1105/tpc.106.049338
Cantalapiedra, C., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi:10.1093/molbev/msab293
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153 (3), 1398–1412. doi:10.1104/pp.110.153593
Chang, X., Liu, F., Lin, Z., Qiu, J., Peng, C., Lu, Y., et al. (2020). Phytochemical profiles and cellular antioxidant activities in chestnut (Castanea mollissima BL) kernels of five different cultivars. Mol. (Basel, Switz. 25. doi:10.3390/molecules25010178
Chen, G., Li, J., Liu, Y., Zhang, Q., Gao, Y., Fang, K., et al. (2019). Roles of the GA-mediated spl gene family and miR156 in the floral development of Chinese chestnut (Castanea mollissima). Int. J. Mol. Sci. 20 (7), 1577. doi:10.3390/ijms20071577
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chou, K.-C., and Shen, H. B. (2010). Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 02, 1090–1103. doi:10.4236/ns.2010.210136
Choupina, A. (2019). Nutritional and health potential of European chestnut Potencial nutricional e de saúde da castanha europeia. Rev. Ciencias Agrar. 42, 801–807. doi:10.19084/rca.17701
Consortium, T., Monforte, A., and Fawcett, J. (2012). The tomato genome sequence provides insights intofleshy fruit evolution. Nature 485, 635–641. doi:10.1038/nature11119
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. Web Server issue). doi:10.1093/nar/gkr319
Dinis, L.-T., Oliveira, M. M., Almeida, J., Costa, R., Gomes-Laranjo, J., and Peixoto, F. (2012). Antioxidant activities of chestnut nut of Castanea sativa Mill. (cultivar ‘Judia’) as function of origin ecosystem. Food Chem. 132 (1), 1–8. doi:10.1016/j.foodchem.2011.09.096
Du, B., Zhang, Q., Cao, Q., Xing, Y., Qin, L., and Fang, K. (2021). Morphological observation and protein expression of fertile and abortive ovules in Castanea mollissima</i>. PeerJ9. doi:10.7717/peerj.11756
Fan, Y., Yang, H., Lai, D., He, A., Xue, G., Feng, L., et al. (2021). Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in sorghum [Sorghum bicolor (L) Moench]. BMC Genomics 22 (1), 415. doi:10.1186/s12864-021-07652-9
Fueyo, R., Judd, J., Feschotte, C., and Wysocka, J. (2022). Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell. Biol. 23 (7), 481–497. doi:10.1038/s41580-022-00457-y
Gao, H., Wang, X.-B., Xi, R.-G., Sun, B.-H., Huang, J., and Wu, L. J. (2010). Structure and absolute configuration of a diterpenoid from Castanea mollissima. Nat. Prod. Commun. 5, 13. doi:10.1177/1934578X1000500104
Gómez, M. D., Fuster-Almunia, C., Ocaña-Cuesta, J., Alonso, J. M., and Pérez-Amador, M. A. (2019). RGL2 controls flower development, ovule number and fertility in Arabidopsis. Plant Sci. 281, 82–92. doi:10.1016/j.plantsci.2019.01.014
Heisler, M. G., Atkinson, A., Bylstra, Y. H., Walsh, R., and Smyth, D. R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Dev. Camb. Engl. 128 (7), 1089–1098. doi:10.1242/dev.128.7.1089
Hu, G., Cheng, L., Cheng, Y., Mao, W., Qiao, Y., and Lan, Y. (2022). Pan-genome analysis of three main Chinese chestnut varieties. Front. Plant Sci. 13, 916550. doi:10.3389/fpls.2022.916550
Huq, E., and Quail, P. H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J. 21 (10), 2441–2450. doi:10.1093/emboj/21.10.2441
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi:10.1038/nmeth.4285
Katoh, K., and Standley, D. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi:10.1093/molbev/mst010
Kawasaki, K., Shiromizu, T., and Katsui, M. (2009). Suppressive effect of polyphenols extracted from chestnut skins on lipid absorption in rats. J. Jpn. Soc. Food Sci. Technology-nippon Shokuhin Kagaku Kogaku Kaishi 56, 545–548. doi:10.3136/nskkk.56.545
Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., et al. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. U. S. A. 100 (20), 11765–11770. doi:10.1073/pnas.1932414100
Kondou, Y., Nakazawa, M., Kawashima, M., Ichikawa, T., Yoshizumi, T., Suzuki, K., et al. (2008). RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147 (4), 1924–1935. doi:10.1104/pp.108.118364
Kroon, M., Lameijer, E. W., Lakenberg, N., Hehir-Kwa, J. Y., Thung, D. T., Slagboom, P. E., et al. (2016). Detecting dispersed duplications in high-throughput sequencing data using a database-free approach. Bioinforma. Oxf. Engl. 32 (4), 505–510. doi:10.1093/bioinformatics/btv621
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Kurbidaeva, A., Ezhova, T., and Novokreshchenova, M. (2014). Arabidopsis thaliana ICE2 gene: Phylogeny, structural evolution and functional diversification from ICE1. Plant Sci. 229, 10–22. doi:10.1016/j.plantsci.2014.08.011
Ledent, V., and Vervoort, M. (2001). The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. doi:10.1101/gr.177001
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141 (4), 1167–1184. doi:10.1104/pp.106.080580
Li, Z., Gao, Q., Liu, Y., He, C., Zhang, X., and Zhang, J. (2011). Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta 233 (6), 1129–1143. doi:10.1007/s00425-011-1368-1
Li, S., Shi, Z., Zhu, Q., Tao, L., Liang, W., and Zhao, Z. (2021). Transcriptome sequencing and differential expression analysis of seed starch accumulation in Chinese chestnut Metaxenia. BMC Genomics 22 (1), 617. doi:10.1186/s12864-021-07923-5
Liu, Y., Ji, X., Nie, X., Qu, M., Zheng, L., Tan, Z., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New phytologist 207 (3), 692–709. doi:10.1111/nph.13387
Liu, B., Sun, Y., Xue, J., Jia, X., and Li, R. (2018). Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L). PeerJ 6, e4796. doi:10.7717/peerj.4796
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lupini, C., Cecchinato, M., Scagliarini, A., Graziani, R., and Catelli, E. (2009). In vitro antiviral activity of chestnut and quebracho woods extracts against avian reovirus and metapneumovirus. Res. Veterinary Sci. 87 (3), 482–487. doi:10.1016/j.rvsc.2009.04.007
Massantini, R., Moscetti, R., and Frangipane, M. T. (2021). Evaluating progress of chestnut quality: A review of recent developments. Trends Food Sci. Technol. 113, 245–254. doi:10.1016/j.tifs.2021.04.036
Nims, E., Vongpaseuth, K., Roberts, S. C., and Walker, E. L. (2015). TcJAMYC: A bHLH transcription factor that activates paclitaxel biosynthetic pathway genes in yew. J. Biol. Chem. 290 (33), 20104. doi:10.1074/jbc.A109.026195
Noh, J.-R., Kim, Y.-H., Gang, G.-T., Yang, K.-J., Lee, H.-S., Nguyen, P. H., et al. (2010). Chestnut (Castanea crenata) inner shell extract inhibits development of hepatic steatosis in C57BL/6 mice fed a high-fat diet. Food Chem. 121 (2), 437–442. doi:10.1016/j.foodchem.2009.12.061
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi:10.1146/annurev-arplant-081320-092855
Pires, N., and Dolan, L. (2010a). Early evolution of bHLH proteins in plants. Plant Signal. Behav. 5 (7), 911–912. doi:10.4161/psb.5.7.12100
Pires, N., and Dolan, L. (2010b). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27 (4), 862–874. doi:10.1093/molbev/msp288
Plackett, A., Tidy, A., Powers, s. J., Wanchoo, A., Phillips, A., Wilson, Z., et al. (2014). DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New phytologist 201, 825–836. doi:10.1111/nph.12571
Quan, S., Niu, J., Zhou, L., Xu, H., Ma, L., and Qin, Y. (2019). Genome-wide identification, classification, expression and duplication analysis of GRAS family genes in juglans regia L. Sci. Rep. 9 (1), 11643. doi:10.1038/s41598-019-48287-x
Robinson, K. A., and Lopes, J. M. (2000). Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic acids Res. 28 (7), 1499–1505. doi:10.1093/nar/28.7.1499
Ronquist, F., Teslenko, M., Mark, P., Ayres, D., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi:10.1093/sysbio/sys029
Seo, J.-S., Joo, J., Kim, M.-J., Nahm, B., Song, S., Cheong, J.-J., et al. (2010). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. Cell. Mol. Biol. 65, 907–921. doi:10.1111/j.1365-313X.2010.04477.x
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65 (6), 907–921. doi:10.1111/j.1365-313X.2010.04477.x
Song, S., Qi, T., Huang, H., and Xie, D. (2013). Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 6 (4), 1065–1073. doi:10.1093/mp/sst054
Song, M., Wang, H., Wang, Z., Huang, H., Chen, S., and Ma, H. (2021). Genome-Wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in fig (Ficus carica L). Front. Plant Sci. 12, 730692. doi:10.3389/fpls.2021.730692
Sun, H., Fan, H.-J., and Ling, H.-Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16 (1), 9. doi:10.1186/s12864-014-1209-2
Sun, W., Jin, X., Ma, Z., Chen, H., and Liu, M. (2020). Basic helix–loop–helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 155, 1478–1490. doi:10.1016/j.ijbiomac.2019.11.126
Tello-Ruiz, M. K., Naithani, S., Gupta, P., Olson, A., Wei, S., Preece, J., et al. (2021). Gramene 2021: Harnessing the power of comparative genomics and pathways for plant research. Nucleic acids Res. 49 (D1), D1452–D1463. doi:10.1093/nar/gkaa979
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics, Proteomics Bioinforma. 8 (1), 77–80. doi:10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, J., Hu, Z., Zhao, T., Yang, Y., Chen, T., Yang, M., et al. (2015). Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16 (1), 39. doi:10.1186/s12864-015-1249-2
Wang, J., Sun, P., Li, Y., Liu, Y., Yang, N., Yu, J., et al. (2018). An overlooked paleotetraploidization in cucurbitaceae. Mol. Biol. Evol. 35 (1), 16–26. doi:10.1093/molbev/msx242
Wang, J., Tian, S., Sun, X., Cheng, X., Duan, N., Tao, J., et al. (2020). Construction of pseudomolecules for the Chinese chestnut (Castanea mollissima) genome. G3 (Bethesda, Md.) 10, 3565. doi:10.1534/g3.120.401532
Wang, J., Yuan, M., Feng, Y., Zhang, Y., Bao, S., Hao, Y., et al. (2022). A common whole-genome paleotetraploidization in Cucurbitales. Plant Physiol. 190 (4), 2430–2448. doi:10.1093/plphys/kiac410
Woodward, A. W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. 95 (5), 707–735. doi:10.1093/aob/mci083
Xu, Z., Meenu, M., Chen, P., and Xu, B. (2020). Comparative study on phytochemical profiles and antioxidant capacities of chestnuts produced in different geographic area in China. Antioxidants (Basel, Switz. 9, 190. doi:10.3390/antiox9030190
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi:10.1146/annurev.arplant.59.032607.092804
Ye, J., Li, W., Ai, G., Li, C., Liu, G., Chen, W., et al. (2019). Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLOS Genet. 15 (5), e1008149. doi:10.1371/journal.pgen.1008149
Yu, L., Zhang, G., Lyu, Z., He, C., and Zhang, J. (2021). Genome-wide analysis of the GRAS gene family exhibited expansion model and functional differentiation in sea buckthorn (Hippophae rhamnoides L). Plant Biotechnol. Rep. 15 (4), 513–525. doi:10.1007/s11816-021-00694-1
Yu, L., Diao, S., Zhang, G., Yu, J., Zhang, T., Luo, H., et al. (2022a). Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides. Plant Biotechnol. J. 20 (7), 1257–1273. doi:10.1111/pbi.13802
Yu, L., Lyu, Z., Liu, H., Zhang, G., He, C., and Zhang, J. (2022b). Insights into the evolutionary origin and expansion of the BBX gene family. Plant Biotechnol. Rep. 16 (2), 205–214. doi:10.1007/s11816-022-00745-1
Yu, L., Hui, C., Huang, R., Wang, D., Fei, C., Guo, C., et al. (2023). Genome-wide identification, evolution and transcriptome analysis of GRAS gene family in Chinese chestnut (Castanea mollissima). Front. Genet. 13, 1080759. doi:10.3389/fgene.2022.1080759
Yujia, L., Xiaoyu, J., Xianguang, N., Min, Q., Lei, Z., Zilong, T., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytologist 207 (3), 692–709. doi:10.1111/nph.13387
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pré, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. Cell. Mol. Biol. 67 (1), 61–71. doi:10.1111/j.1365-313x.2011.04575.x
Keywords: Castanea mollissima, bHLH family, duplication model, expression pattern, collinearity analysis
Citation: Yu L, Fei C, Wang D, Huang R, Xuan W, Guo C, Jing L, Meng W, Yi L, Zhang H and Zhang J (2023) Genome-wide identification, evolution and expression profiles analysis of bHLH gene family in Castanea mollissima. Front. Genet. 14:1193953. doi: 10.3389/fgene.2023.1193953
Received: 26 March 2023; Accepted: 05 May 2023;
Published: 12 May 2023.
Edited by:
Madhav P. Nepal, South Dakota State University, United StatesReviewed by:
Anuj Kumar, Dalhousie University, CanadaMehdi Hosseini Mazinani, National Institute for Genetic Engineering and Biotechnology, Iran
Nasreen Bano, University of Pennsylvania, United States
Moyang Liu, Shanghai Jiao Tong University, China
Copyright © 2023 Yu, Fei, Wang, Huang, Xuan, Guo, Jing, Meng, Yi, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingzheng Zhang, emhhbmdqaW5nemhlbmdAMTI2LmNvbQ==
 Liyang Yu
Liyang Yu Cao Fei2,3
Cao Fei2,3 Dongsheng Wang
Dongsheng Wang Wang Xuan
Wang Xuan Haie Zhang
Haie Zhang