- 1Center for Integrative Conservation and Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, Nay Pyi Taw, Myanmar
- 4Key Laboratory of Ecology of Rare and Endangered Species and Environmental Protection (Ministry of Education) and Guangxi Key Laboratory of Landscape Resources Conservation and Sustainable Utilization in Lijiang River Basin, Guangxi Normal University, Guilin, Guangxi, China
- 5Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 6College of Biological Resource and Food Engineering, Qujing Normal University, Qujing, China
Cassytha is the sole genus of hemiparasitic vines (ca. 20 spp.) belonging to the Cassytheae tribe of the Lauraceae family. It is extensively distributed in tropical and subtropical regions. In this study, we determined the complete plastid genome sequences of C. filiformis and C. larsenii, which do not possess the typical quadripartite structure. The length of C. filiformis plastomes ranged from 114,215 to 114,618 bp, whereas that of C. larsenii plastomes ranged from 114,900 to 114,988 bp. Comparative genomic analysis revealed 1,013 mutation sites, four large intragenomic deletions, and five highly variable regions in the eight plastome sequences. Phylogenetic analyses based on 61 complete plastomes of Laurales species, 19 ITS sequences, and trnK barcodes from 91 individuals of Cassytha spp. confirmed a non-basal group comprising individuals of C. filiformis, C. larsenii, and C. pubescens in the family Lauraceae and proposed a sister relationship between C. filiformis and C. larsenii. Further morphological comparisons indicated that the presence or absence of hairs on the haustoria and the shape or size of fruits were useful traits for differentiating C. filiformis and C. larsenii.
1 Introduction
The genus Cassytha L., belonging to the family Lauraceae, encompasses more than 20 hemiparasitic vines found in tropical and subtropical regions worldwide (Weber, 1981). Among these regions, Africa, Asia, and Australia host three, four, and nineteen species, respectively, and Australia stands as the center of species diversity for Cassytha. The dodder laurel (C. filiformis L.), which is the only pantropical species of Cassytha, is used as a medicinal plant in various regions, such as Bahamas, China, Indonesia, Nigeria, and the West Indies. It contains alkaloids, flavonoids, phenol, saponin, terpenoids, and tannin (Burkill, 1995; Tsai et al., 2008; Brophy et al., 2009; Adamu et al., 2017; Nazar et al., 2019). Cassytha, as a member of plants that are adapted to grow in open environments (Jordan et al., 2014; Carpenter et al., 2015), possesses tiny triangular leaves and filiform stems that feature stomata located on any part of both cuticles (Awang et al., 2018). The cuticular characters, along with the glabrous or pubescent stem and petal, have been utilized to distinguish different Cassytha species (Kokubugata et al., 2012). Species identification has often been challenging due to the greatly reduced plant populations and similar habitats. For instance, in 1971, Hatusima described the stems of C. filiformis as thin and reddish. However, later C. filiformis was redefined as a Ryukyu endemic taxon, C. pergracilis, by Hatusima in 1976 (Hatusima, 1971; Hatusima, 1976). Over the last two decades, molecular diagnostic methods for Cassytha species have continuously improved.
In order to distinguish Cassytha species, molecular analyses have utilized a partial sequence of the trnK intron (trnK). Three separate analyses have been conducted, with differing results. Rohwer and Rudolph (2005) found C. ciliolata was sister to C. pubescens, although without bootstrap support. Meanwhile, Wang et al. (2010) identified C. ciliolata as sister to C. filiformis, with C. pubescens as the subsequent sister species, and C. melantha as the most basal species with high support. Kokubugata et al. (2012) conducted a more extensive sampling, which revealed a clade comprising C. pubescens, C. muelleri, and C. rufa as sister to another clade containing C. filiformis, C ciliolata, C. capillaris, and C. pergracilis. C. glabella was identified as the next sister group, followed by C. melantha.
At the genus level, previous molecular phylogenetic analyses based on diverse datasets have revealed incongruent placements of Cassytha species in the family Lauraceae. Rohwer’s original research Rohwer, (2000) employed the plastid marker trnK and sampling 48 species, which estimated that C. ciliolata formed a sub-basal clade within the family Lauraceae. A year later, Chanderbali et al. (2001) utilized plastid sequences, including psbA-trnH, rpl16, trnL-trnF, and trnT-trnL, as well as 26 S nuclear ribosomal DNA (nrDNA), to reconstruct phylogenetic relationships among 77 species in the family Lauraceae. Their analyses suggested that C. filiformis and C. pubescens were most closely related to Neocinnamomum mekongense (Hand.-Mazz.) Kosterm. Subsequently, a third study by Rohwer and Rudolph (2005), based on trnK sequences of 49 species, indicated that the monophyletic Cassytha group was not sub-basal within the family Lauraceae; Wang et al. (2010) used plastid sequences (psbA-trnH and trnK) and nrDNA to reconstruct the phylogenetic relationships of Neocinnamomum and showed a close relationship between the genera Cassytha and Neocinnamomum. However, Li et al. (2016) utilized nuclear gene RPB2 fragment and ITS to reconstruct the phylogenetic relationships of Caryodaphnopsis and indicated an independent clade of C. filiformis.
Plastid genome sequencing has proven to be a valuable tool for elucidating the phylogenetic relationships of Angiosperm plants (Li et al., 2019; Li et al., 2021; Dong et al., 2022b). In order to determine the phylogenetic location of nineteen genera, Song et al. (2017b) conducted comparisons based on 47 Lauraceae plastid genomes and found support for the monophyletic clade of Cassytha within the family Lauraceae. Subsequently, a second study by Song et al. (2020) utilized a higher sampling of plastomes for 97 species and reconstructed a monophyletic Lauraceae clade that included the independent Cassytha subclade. This robust monophyletic Cassytha group was further supported by the analysis of complete nrDNA sequences with a length of 6,281 bp (Liu Z. F. et al., 2021).
In this study, we selected eight individuals of two Cassytha species to obtain their complete plastid genomes and nrDNA sequences. By comparing these sequences, we aim to answer three questions. Firstly, which types of mutation events occurred in the plastid genomes of Cassytha? Secondly, is there any highly variable region in the plastid genomes of Cassytha for DNA barcoding? Finally, what is the phylogenetic placement of C. larsenii? Comparisons were made with the taxonomic character data between C. filiformis and C. larsenii.
2 Materials and methods
2.1 Plant materials
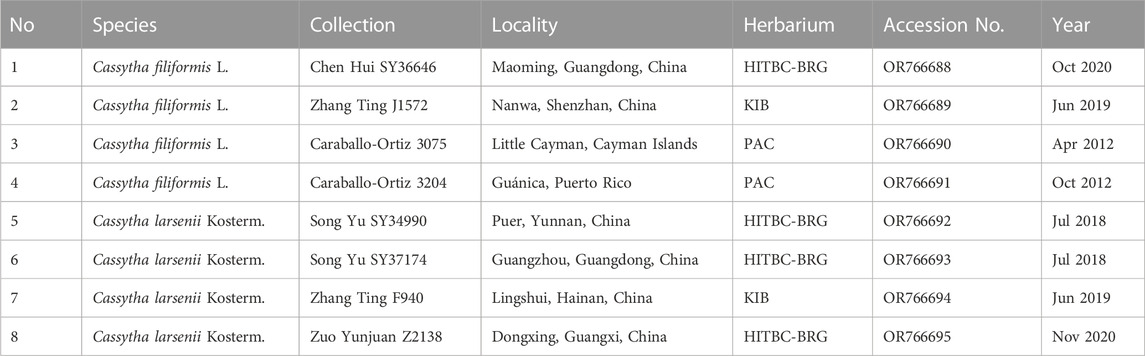
In this study, a total of eight individuals from two species, C. filiformis and C. larsenii, were sampled. Fresh stems of wild vines were collected from China and Puerto Rico, and quickly dried with silica gel (Table 1). Voucher specimens were deposited in the herbarium of Guangxi Normal University. The specimens were identified by Yun-Hong Tan (Xishuangbanna Tropical Botanical Garden, CAS) and Yu Song (Guangxi Normal University). Furthermore, plastid genome sequences of related taxa of Lauraceae were downloaded from Lauraceae Chloroplast Genome Database (LCGDB) (https://lcgdb.wordpress.com) and GenBank of NCBI (https://www.ncbi.nlm.nih.gov) and a total of 61 taxa from 27 genera of Laurales were included. In addition, the partial trnK intron, including matK gene sequences, of eight newly sequenced and 83 individuals of Cassytha obtained from the NCBI database were used to conduct phylogenetic analyses (Figure 1; Supplementary Figure S1).
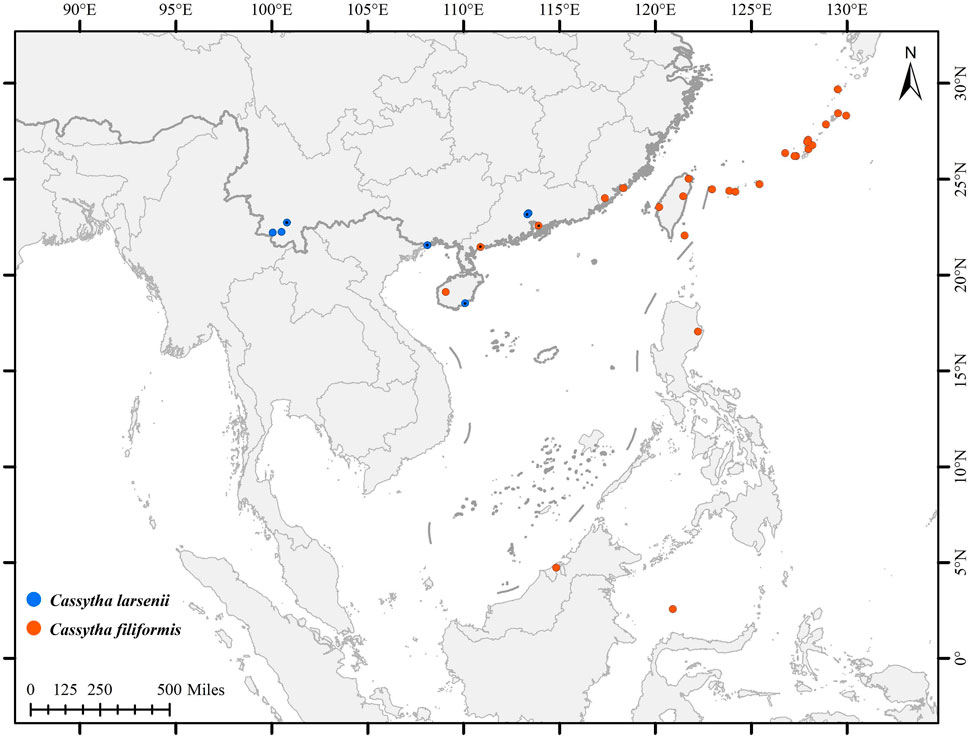
FIGURE 1. Distribution of C. filiformis and C. larsenii. Blue pots indicate C. larsenii and red pots indicate C. filiformis.
2.2 Plastid genome sequencing and assembly
Whole-genomic DNA was extracted from the stem tissues using the CTAB method (Doyle and Doyle, 1987). Libraries were constructed with fragments of approximately 300 bp according to the manufacturer’s protocol (Illumina, San Diego, CA, United States). All samples were sequenced using the Illumina HiSeq 2,500 at Kunming Institute of Botany, Chinese Academy of Sciences (KIB, CAS). The plastid genomes were de novo assembled using the GetOrganelle pipeline under default settings (Jin et al., 2020; Dong et al., 2022a). The assembly quality of all plastid genomes was checked with Bandage software (Wick et al., 2015).
2.3 Genome annotation
The newly assembled plastid genomes were annotated using the GeSeq application, followed by manual verification using the Geneious software (Kearse et al., 2012; Tillich et al., 2017). The annotation sequences and their corresponding information were then submitted to Genbank and assigned accession numbers OR766688 to OR766695. Finally, the physical map of the annotated plastid genomes was drawn using the online Chloroplot program (https://irscope.shinyapps.io/Chloroplot/).
2.4 Plastome sequence divergence and microstructural mutation analysis
In order to compare the divergence within eight newly assembled plastid genomes, the online mVISTA program in Shuffle-LAGAN mode (https://genome.lbl.gov/vista/mvista/submit.shtml) was employed. Additionally, the plastid genome sequence’s nucleotide diversity (Pi) was estimated using DnaSP, with a step size of 200 bp and a window length of 600 bp for sliding window analysis (Rozas et al., 2017). The number and position of Indel and single-nucleotide polymorphism (SNP) events were determined by manual statistics and analyzed in the aligned eight plastid genome sequences of two Cassytha species.
2.5 Phylogenetic analyses
To determine the phylogenetic relationships within Cassytha and its relatives, the eight newly assembled plastid genomes were compared to the other 53 Laurales species, with Illigera celebica (LAU00199) and I. grandiflora (LAU00198) selected as outgroups. The sequences were aligned using MAFFT (Katoh et al., 2019) and manually adjusted in BioEdit (Hall et al., 2011). Maximum likelihood (ML) analysis was conducted using IQ-tree v2, and the best-fit model was determined using ModelFinder (Kalyaanamoorthy et al., 2017), with the GTR + F + I + G4 model and a bootstrap value of 1,000 (Minh et al., 2020). Then, the trnK and ITS sequences of the eight sequenced Cassytha individuals were intercepted and aligned with available trnK and ITS sequences from GenBank. A previous study showed C. melantha was basal to other Cassytha taxa based on extensive sampling (Kokubugata et al., 2012). Consequently, C. melantha was used as an outgroup in phylogenetic analyses. The best-fit DNA substitution models were chosen as TPM3uf + I + G (trnK data matrix) and TIM3+I (ITS data matrix) in jmodeltest v.2.1 (Darriba et al., 2012). Bayesian inference (BI) was performed for ten million generations, sampling every 1,000 generations in MrBayes v.3.2, with independent Markov chain Monte Carlo (MCMC) chains (Ronquist et al., 2012). The first 25% of the trees were discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. The MCMC output was examined and the effective sample size (ESS) values were above 200. Finally, the generated trees were visualized and adjusted using FigTree software (https://tree.bio.ed.ac.uk).
2.6 Morphological analyses
The pan-tropical C. filiformis species are the most widely studied compared with other Cassytha species, but the morphology of C. larsenii has not been fully reported to date. To this end, the morphological characters of two Cassytha species were observed by stereoscopic microscope (SM) and scanning electron microscope (SEM). The following morphological characters were focused on: haustoria, stems, petals, and fruits. For stereoscopic microscope observation, the haustoria, stems, and flowers were placed upright on a flat, wet tissue paper and observed with Leica S8 APO, LAS v 4.8 collecting photographs. SEM observations of the materials proceeded in ZEISS EVO LS10 scanning electron microscope at the Public Technology Service Center, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences.
3 Results
3.1 Genome features
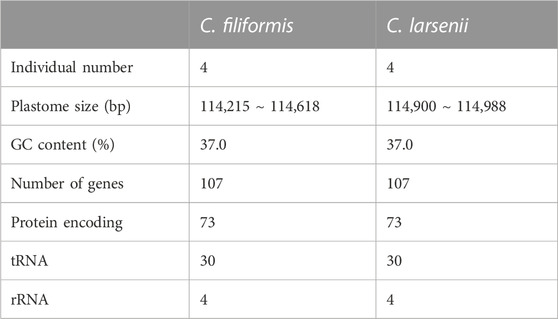
All eight newly sequenced Cassytha plastomes were assembled into single circular molecules lacking the typical inverted repeat (IR) region and quadripartite structure (Figure 2). The size of the plastomes varied from 114,215 bp in C. filiformis growing in Puerto Rico (P304) to 114,618 bp in C. filiformis growing in Guangdong (SY6130), while the sizes range was from 114,900 bp in C. larsenii growing in Yunnan (SY9917) to 114,988 bp in C. larsenii growing in Guangxi (SY6156). The GC content was similar in all eight plastomes, with a value of 37.0%. Each plastome contained a total of 107 functional genes, including 73 protein-coding genes, 30 tRNA genes, and four rRNA genes (Table 2). Notably, unlike the NADH dehydrogenase (ndh) genes found in other sequenced Lauraceae plastomes, five ndh genes including ѱndhB, ѱndhD, ѱndhE, ѱndhF, and ѱndhH are pseudogenes, and six ndh genes, including ndhA, ndhC, ndhG, ndhI, ndhJ, and ndhK are absent in all eight Cassytha plastomes. In addition, we identified three genes pafI, pafII, and pbf1 in all of these genomes.
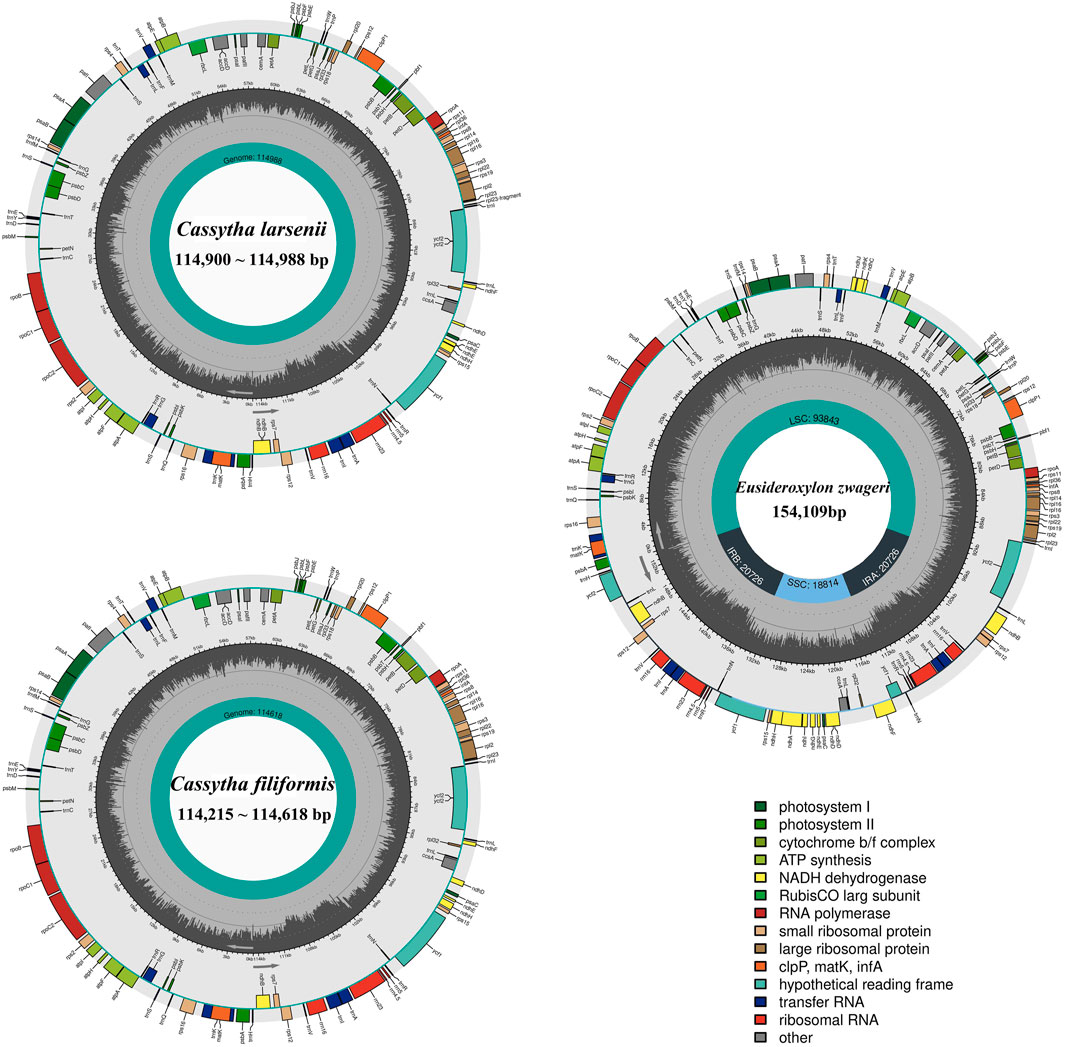
FIGURE 2. Plastome genome maps of two Cassytha species and Eusideroxylon zwageri. Genes displayed outside of the circle are transcribed counterclockwise, whereas those inside are transcribed clockwise. Diferent colors represent different functional gene groups. Eusideroxylon zwageri as a reference to IR/SSC/LSC.
3.2 Microstructural mutations
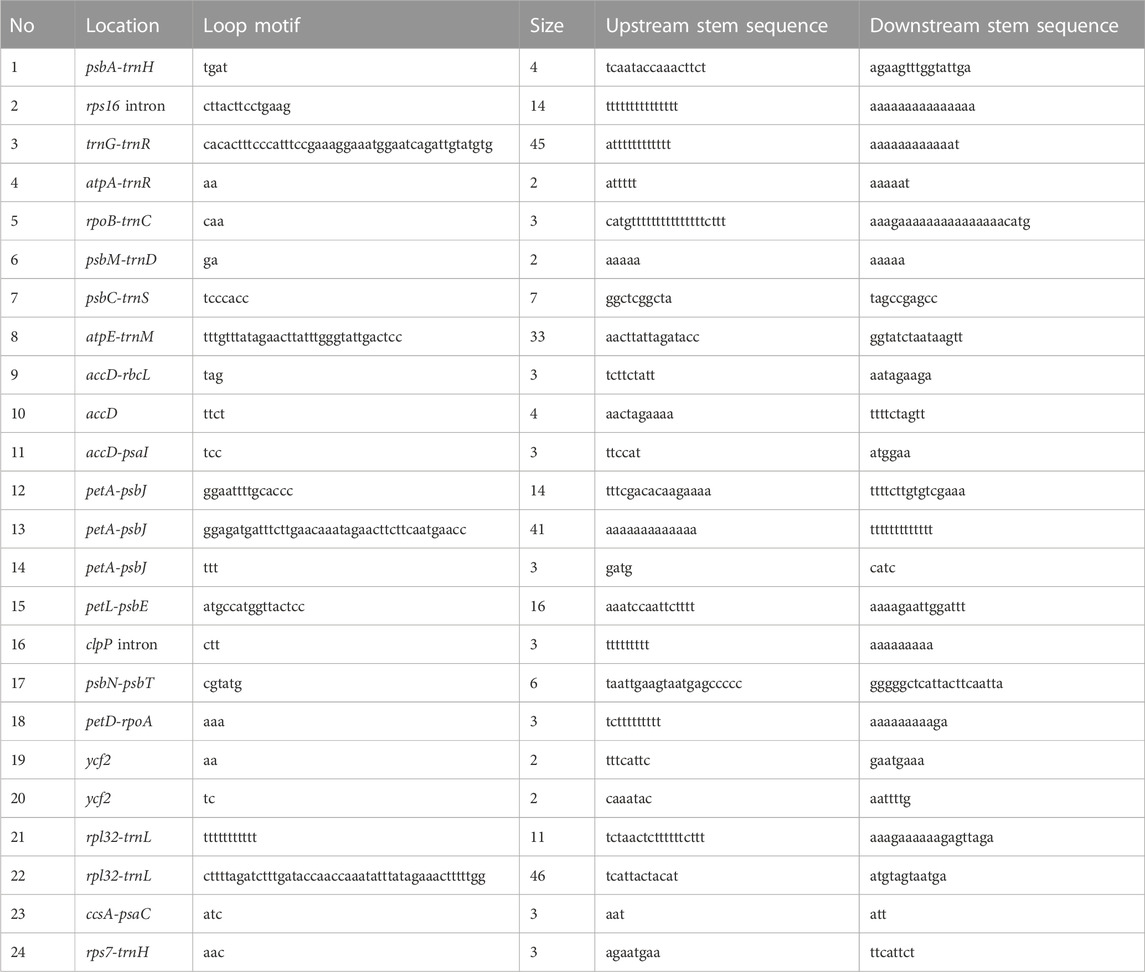
We assessed synteny and rearrangements in the eight Cassytha plastomes and found no large-scale recombination in the gene organization after verification. However, we manually detected 24 micro-inversions, ranging from 2 to 46 bp, in the regions of the accD, accD-psaI, accD-rbcL, atpA-trnR, atpE-trnM, ccsA-psaC, clpP intron, psbA-trnH, petA-psbJ (three regions), petD-rpoA, petL-psbE, psbC-trnS, psbM-trnD, psbN-psbT, rpl32-trnL (two regions), rpoB-trnC, rps7-trnH, rps16 intron, trnG-trnR, and ycf2 (two regions) (Table 3). Palindrome sequences in pairs with lengths of 3–23 bp were identified in the flanks of these inversions. Furthermore, we detected a total of 249 indels in the Cassytha species, which were classified into 195 simple sequences repeat (SSR) indels and 54 non-SSR indels (Supplementary Table S1).
3.3 Plastome comparisons
In comparison to the previous published plastome of Eusideroxylon zwageri Teijsm. & Binn. (LAU00006), which is an early divergent species in the Lauraceae family, the Cassytha plastomes have four missing segments (Figure 3). These missing segments include a 4 kb fragment containing three ndh genes (ndhC, ndhJ, and ndhK), a 16 kb fragment flanked by ndhB and ycf1, a 4 kb fragment containing three ndh genes (ndhA, ndhG, and ndhI), and a 10 kb fragment flanked by rpl2 and trnL-CAA. The missing segments with the length of 16 kb and 10 kb are located in the IRa and IRb regions of the E. zwageri plastome, respectively. The other two missing segments with a length of 4 kb are located in LSC and SSC regions of E. zwageri plastome, respectively. Additionally, when compared with the Cassytha plastome, a 1.5 kb segment containing the rpl2 gene is absent from the IRa region of the E. zwageri plastome.
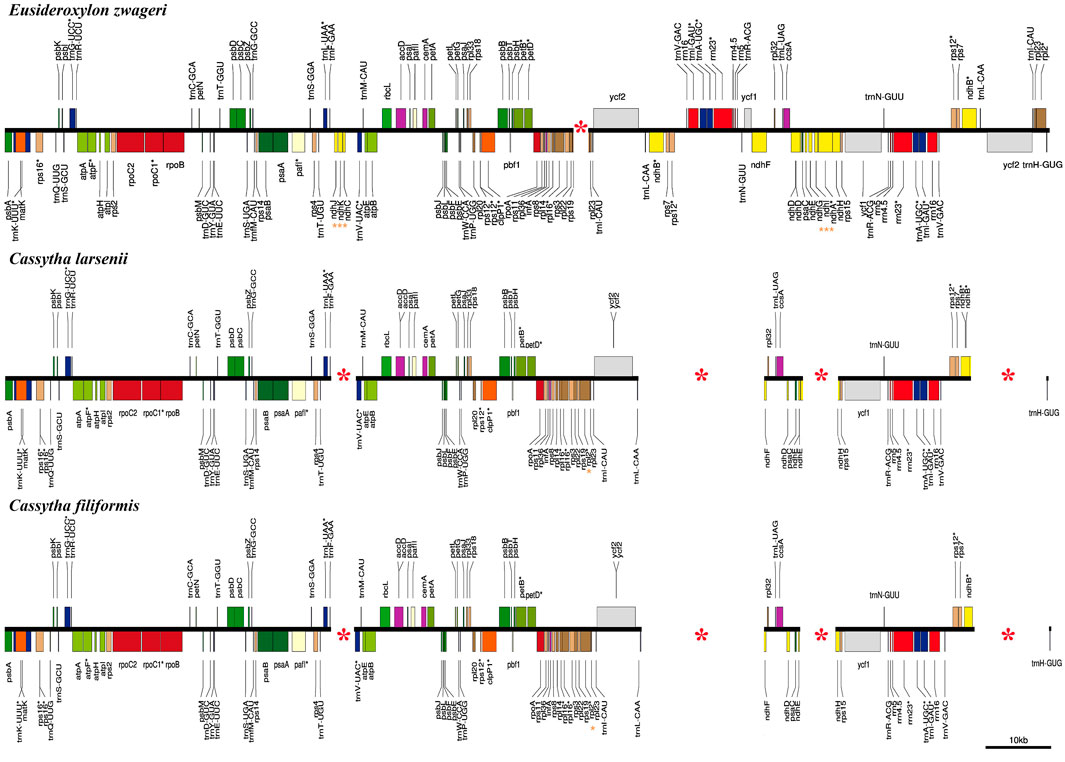
FIGURE 3. Comparison of the plastid genomes of Eusideroxylon zwageri, C. filiformis and C. larsenii. Missing segments and genes are marked with red asterisks.
3.4 Divergence hotspot regions
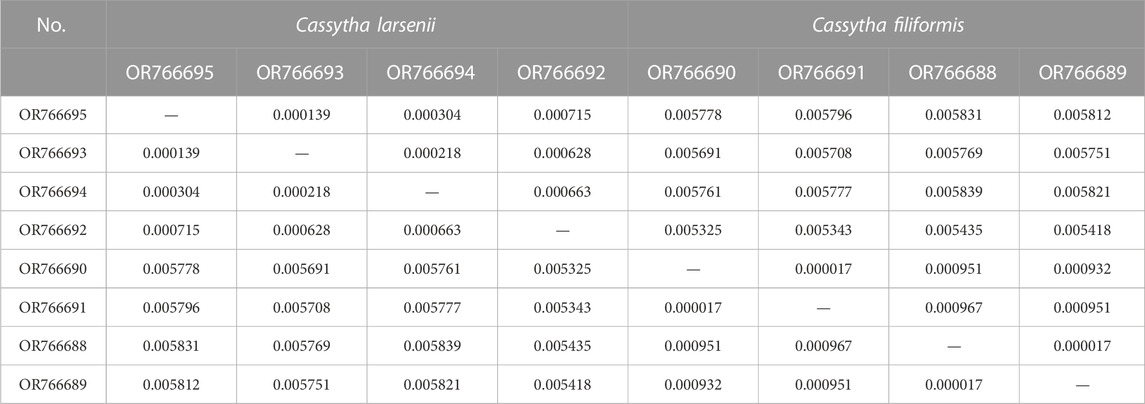
The mVISTA results show that the non-coding regions of the eight plastomes have higher levels of divergence than the coding regions. There are some gaps in the intergenic spacer regions of P304 and P306 of C. filiformis (Figure 4A). A total of 734, 200, and 193 SNP markers were detected in the plastomes of both Cassytha species, C. larsenii individuals, and C. filiformis individuals, respectively. The sequence divergence levels among the plastomes of C. filiformis and C. larsenii were determined (Figure 4B). Within the two Cassytha species, these values varied from 0 to 0.0168, with a mean of 0.0034. Within C. filiformis, these values varied from 0 to 0.0033, with a mean of 0.0004. Within C. larsenii, these values varied from 0 to 0.0053, with a mean of 0.0006. The pairwise nucleotide divergence values between two of the four plastomes varied from 0.000017 to 0.000967 in C. filiformis and from 0.000139 to 0.000715 in C. larsenii. The values between the two species varied from 0.005325 to 0.005839 (Table 4). These results indicate that the differences between the two species were more than six times higher than those among individuals. Five regions, namely, trnQ-psbK, trnP-psaJ, rpl23-ycf2, ndhE-ndhH, and trnN-rrn5 were particularly highly variable between C. filiformis and C. larsenii.
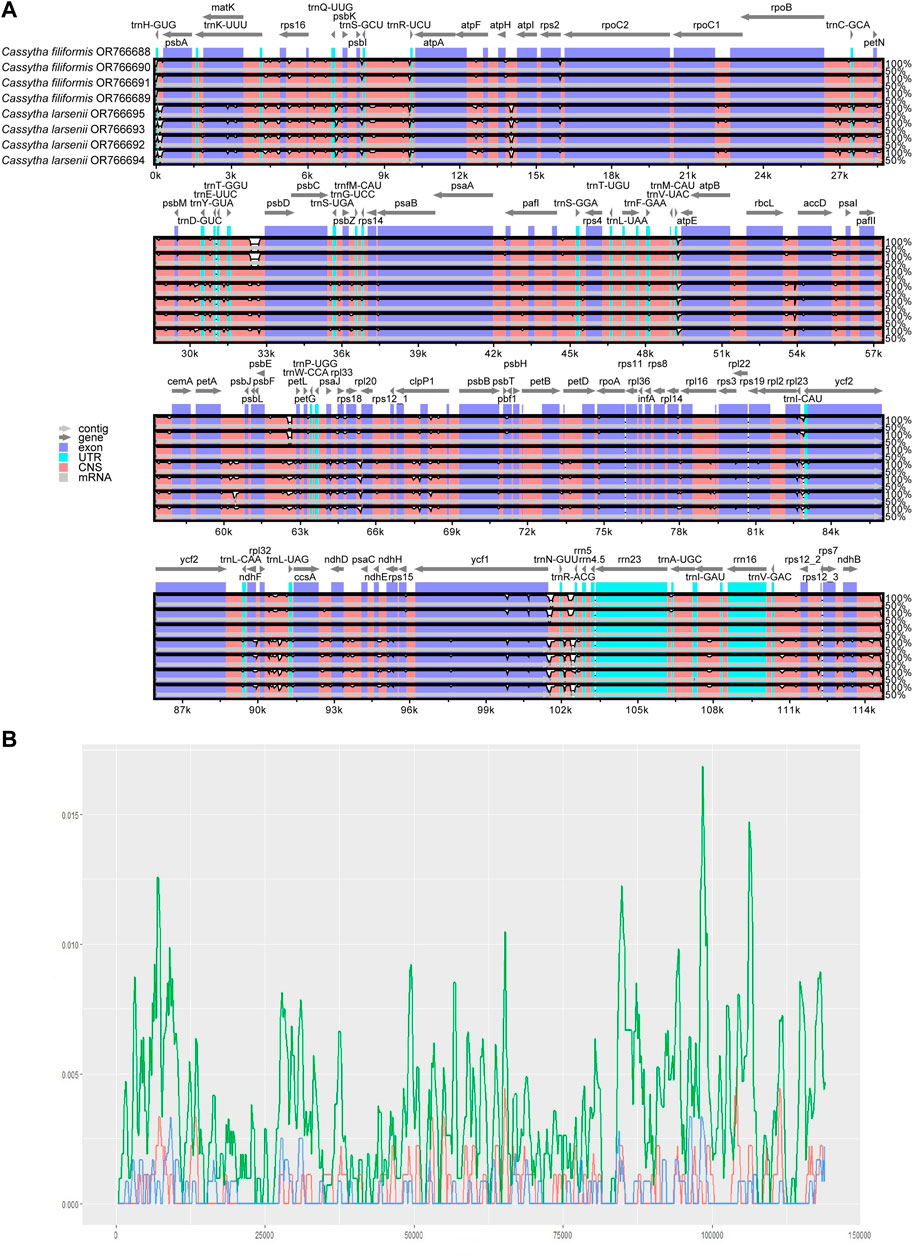
FIGURE 4. (A) Consistency alignment of the complete plastome of C. filiformis and C. larsenii using mVISTA.T-axis shows the percentage identity (50%–100%). Purple bars represent exon regions, blue bars represent untranslated regions (UTRs), pink bars represent noncoding sequences (CNS), gray bars represent mRNA. (B) Sliding-window analysis of the entire chloroplast genome of the two Cassytha species (green line), C. filiformis (blue line) and C. larsenii (red line). (window length: 600 bp, step size: 200 bp). X-axis: position of the window; Y-axis: nucleotide diversity of each window.
3.5 Phylogenetic reconstruction
To ascertain the phylogenetic placement of Cassytha species in relation to other members of the Lauraceae family with fully sequenced plastid genome sequences, we employed the complete plastomes of three Cassytha species to reconstruct phylogenetic relationships. We used two plastomes of Illigera species as out-groups. The phylogeny derived from the analysis of 61 complete plastid genome sequences is highly supported. Our phylogenetic analysis shows that the three Cassytha species form a sister clade to a group consisting of species belonging to the tribes Neocinnamomeae, Caryodaphnopsideae, and Laureae. The tribe Cryptocaryeae represents the next sister groups, followed by Illigera species (Figure 5A). The branch length in the maximum likelihood (ML) tree are 4.934 × 10−4 for four individuals of C. filiformis and 2.247 × 10−3 for four individuals of C. larsenii. To further investigate the phylogenetic relationships among the eight sequenced Cassytha individuals and other Cassytha taxa with reported barcoding data, we downloaded available trnK sequences from NCBI database. We included 91 Cassytha samples, with an outgroup accession of C. melantha, in the analysis of the data matrix with the length of 903 bp. The result of the Bayesian analysis shows that C. filiformis is sisters to C. larsenii, rather than C. ciliolata (Figure 5B). The branch lengths in the Bayesian inference (BI) tree are 1.997 × 10−3 for 57 individuals of C. filiformis and 5.902 × 10−4 for six individuals of C. larsenii. Finally, we downloaded available ITS sequences from GenBank and reconstructed the phylogeny consisted of 19 ITS sequences with a length of 579 bp. We used C. pubescens as an out-group (Figure 5C). The result of the Bayesian analysis shows that C. filiformis and C. larsenii individuals form two independent groups and the branch lengths are 1.204 × 10−2 for ten individuals of C. filiformis and 6.401 × 10−3 for eight individuals of C. larsenii.
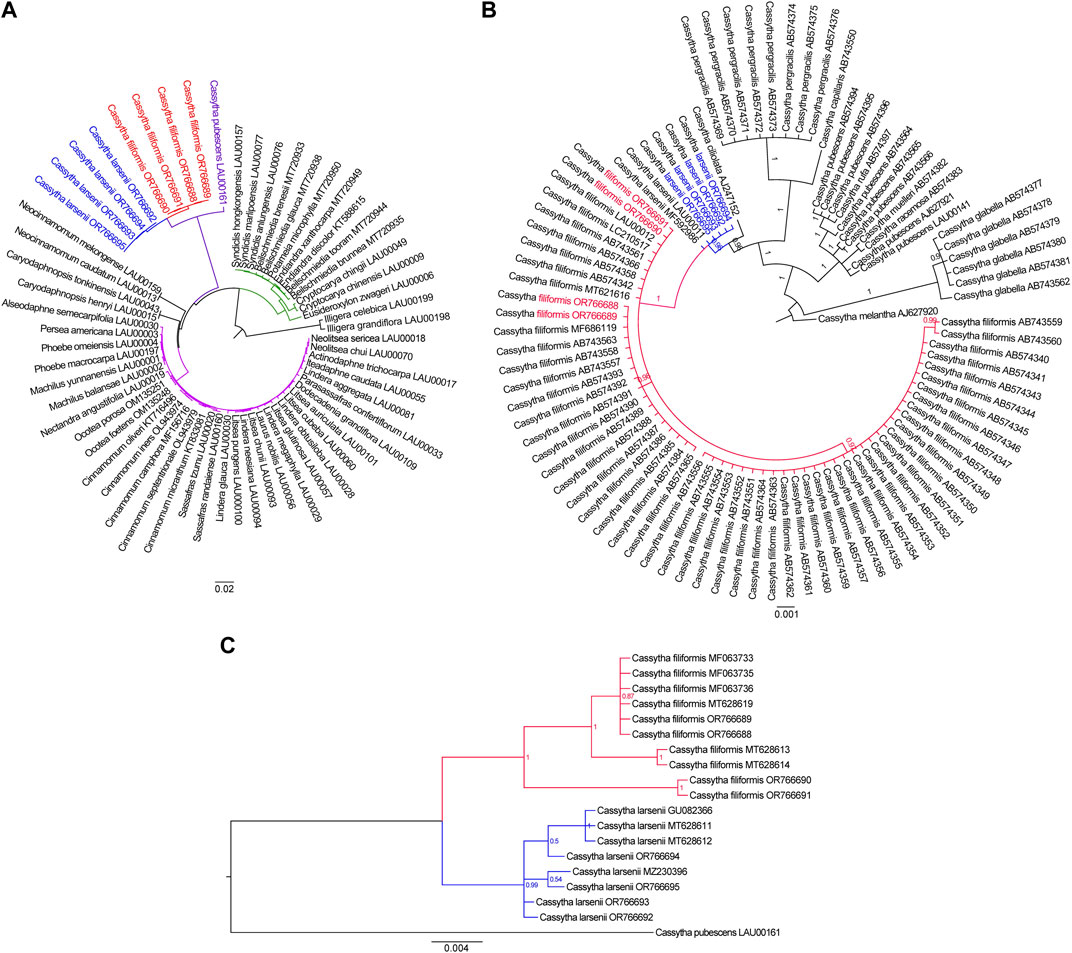
FIGURE 5. (A) The Maximum likelihood tree of 61 taxa of Laurales based on complete plastome sequences. Numbers at each node are bootstrap support values. (B) The Bayesian inference tree of 91 taxa of Cassytha species based on trnK sequences. The tree is rooted with trnK sequences of C. melantha. (C) The Bayesian inference tree of 19 taxa of Cassytha species based on ITS sequences. The tree is rooted with ITS sequences of C. pubescens.
3.6 Morphological characters
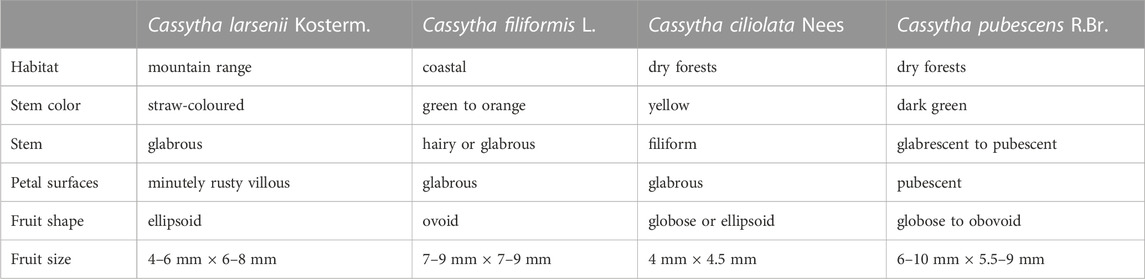
For C. larsenii, the scattered hairs on the haustoria were stable (Supplementary Figure S2A), and filiform hairs were observed on young stems (Supplementary Figure S2B) but not on annual and biennial stems (Supplementary Figure S2C), whereas stems with indumenta of simple, multicellular hairs were observed in two accessions of C. filiformis in China (Supplementary Figures S2G–I). For both C. larsenii and C. filiformis, the glabrous petal surfaces were observed with low magnification light microscopy (Supplementary Figures S2D, J), however, villous hairs were clearly observed on the edges of petals and petal surfaces in the electron microscopy photos (Supplementary Figure S2E, K). All members of genus Cassytha have fleshy fruits with a single seed, and the fruits of C. filiformis are oval with the mean size of 8 mm × 8 mm, while the fruits of C. larsenii are ellipsoid with the mean size of 5 mm × 7 mm. Compared with C. larsenii, C. filiformis has rounder and larger fruits (Supplementary Figure S2F, L).
4 Discussion
4.1 The extreme case of plastoma shrinkage in the family Lauraceae
This study produced eight complete plastid genomes for two species of the stem hemiparasitic genus Cassytha, which comprises nearly 23 species. The plastomes of Cassytha with the length of 114,215 ∼ 114,988 bp was significantly smaller than the other published plastomes in the family Lauraceae (Song et al., 2017b; Song et al., 2020). Three main reasons for these size differences were detected through comparative genomics analysis (Figure 3). First, one copy of the IR regions with the length of 24,717 bp was complete in E. zwageri but lost in the Cassytha plastomes with two segments, contributing almost 25 kb to the length difference. Second, the eight Cassytha plastomes have no six ndh genes including ndhA, ndhC, ndhG, ndhI, ndhJ, and ndhK. Two missing fragments with the length of 4 kb consist of three of the six genes, respectively, and their intergenic regions, which contributed around 8 kb to the length difference with Cassytha species. Third, five pseudogenes were detected in the eight Cassytha plastomes. Three ndh genes (ndhB, ndhD, and ndhE) were found to be pseudogenized, similar to C. filiformis (Wu et al., 2017). The length of ѱndhB, ѱndhD, ѱndhE, ѱndhF, and ѱndhH in C. filiformis are 1,191 bp, 428 bp, 159 bp, 302 bp, and 547 bp, respectively. In the plastome of E. zwageri, the length of the five ndh genes are 2,181 bp for ndhB, 1,508 bp for ndhD, 306 bp for ndhE, 2,229 bp for ndhF, and 1,182 bp for ndhH, which contributed around 7 kb to the length difference with Cassytha species.
4.2 The high sequence divergence among Lauraceae
Comparative genomic analysis indicated that there are 1,013 mutation sites including 24 micro-inversions, 249 indels, and 740 substitutions in the eight plastomes, which indicated that the nucleotide mutation sites in the plastomes of Cassytha species are more than that between species of Machillus (one micro-inversion, 65 indels, and 231 substitutions) and Phoebe (three micro-inversions, 73 indels, and 146 substitutions) (Song et al., 2015; Song et al., 2017a). The nucleotide variability values of the whole plastomes among the eight individuals from two Cassytha species were 0.34%, which approximates the nucleotide variability of five taxa (0.32%) (Song et al., 2016), 15 taxa (0.31%) (Liu et al., 2022), 18 taxa (0.37%) (Liu C. et al., 2021) in the tribe Laureae, and was much higher than the sequence divergence among three Alseodaphne species (0.12%) (Song et al., 2018), seven trinerved Lindera species (0.15%) (Tian et al., 2019), and seven Ocotea species (0.10%) (Trofimov et al., 2022).
4.3 Phylogeny of the sequenced Cassytha species and plastomes of Lauraceae
With species from 27 genera of Laurales, our phylogenomic analysis based on 61 plastid genomes supported a monophyletic Cassytha clade comprising species of C. filiformis, C. larsenii, and C. pubescens. Species of Beilschmiedia, Cryptocarya, Endiandra, Eusideroxylon, Potameia, Sinopora, and Syndiclis, formed the Beilschmiedia-Cryptocarya clade in the phylogeny, and the third clade including Neocinnamomeae, Caryodaphnopsideae, and Laureae species is separate from both the Beilschmiedia-Cryptocarya clade and Cassytha clade, as in previously published phylogenetic trees in the family Lauraceae (Song et al., 2017b; Song et al., 2020). The deep relationships of Cassytha taxa are separated into the following groups in our study. C. melantha, endemic to Australia, forms the first group in the phylogeny. C. glabella, endemic to Western Australia, forms the second group. The third group includes four Australia species C. muelleri, C. pubescens, C. racemose, and C. rufa. The fourth group includes two Asia species C. capillaris and C. pergracilis. And the last group includes an Africa species C. ciliolata, an Asia species C. larsenii, and the pantropical species C. filiformis. The phylogenetic placements of most groups are consistent with previously published phylogenetic relationships (Kokubugata et al., 2012), and the position of C. larsenii was firstly settled here in the way predicted from morphology.
4.4 Morphological difference among Cassytha specie
Although C. ciliolata, C. filiformis, and C. larsenii form the same group in the phylogeny, the persistence of hairs on stems and petal surfaces was used to distinguish the Cassytha species (Kokubugata et al., 2012). In C. ciliolata, filiform hairs on stems and glabrous or rufous-hispidulous twig tips were descripted on the basis of type specimens collected from mountains near Cape Town in Africa (Stapf, 1912). In C. larsenii, absence or sparsity of hairs on stems and glabrous or rufous-hispidulous twig tips were observed in all samples in China (Kostermans, 1994). In C. filiformis, it is remarkable that glabrous stems were observed not only in samples from Ryukyus of Japan and Taiwan of China but also in samples from Luzon Island and Rota Island in the Pacific (Kokubugata et al., 2012). However, stems with indumenta of simple, multicellular hairs were observed in C. filiformis samples from Australia, China, Japan, and Malaysia (Kokubugata et al., 2012). Therefore, glabrous stem is not a taxonomic key character for identifying C. ciliolata, C. filiformis and C. larsenii. Based on the type specimens, the glabrous petal surface was descripted as a common taxonomic character for C. ciliolata, C. filiformis and C. larsenii. However, villous petal surfaces and pubescent edge of petals were observed in the electron microscopy photos of C. filiformis and C. larsenii. Thus, presence or absence of hairs on petal surfaces is not appropriate as a taxonomic key character for identifying the three species. Finally, we suggest that, presence or absence of hairs on haustoria should be treated as key taxonomic evidence to distinguish C. filiformis and C. larsenii. Also, fruit shape or size can be a well distinction between the two species. Diversity of fruits may be related to growing in various geographical environments (Table 5).
4.5 New records for China
Cassytha larsenii Kosterm. was only known from Khun Yuam District, Thailand, this is the first record from China (Puer, Yunnan Province; Guangzhou, Guangdong Province; Linshui County, Hainan Province and Dongxing, Guangxi Province). It was found hosting as a hemiparasitic on the tree trunks of Fabaceae, Myrtaceae and Asteraceae at altitudes from 900 to 1900 m in the forest (Zhang et al., 2022). C. larsenii was observed flowering in June in Mojiang of Yunnan. Morphologically, it bears resemblance to C. filiformis due to hairs on petal surfaces, but differs from it in terms of the presence or absence of hairs on haustoria, shorter inflorescence, smaller flowers (0.5–0.75 mm) (Kostermans, 1994), and smaller fruits. Additionally, C. filiformis grows along coastal regions while C. larsenii is found in mountainous areas. Furthermore, strong evidence from our phylogenetic studies supports treating the new records of C. larsenii from China as a distinct species from C. filiformis. Using different analyses, in the present study we showed that the biodiversity of the genus Cassytha in China is underestimated, with more species than previously recognized (Li et al., 2008). And, evidences from plastid genome size, phylogenetics, and morphology characters suggest that at least two species partitions would require validation and formal description (Liu Z. F. et al., 2021). Our study provides important insights into the taxonomic, biodiversity, conservation biology, and phylogeographic of the genus Cassytha.
5 Conclusion
Cassytha is the only hemiparasitic vines in the Lauraceae family. Our study reports complete plastid genomes of two Cassytha species. 1,013 mutation sites, four large intragenomic deletions and five hotspots were found during comparative genomic research. Meanwhile, based on whole plastid, trnK, and ITS phylogenetic analyses respectively, confirmed a non-basal group comprising C. filiformis, C. larsenii, and C. pubescens. The position of C. larsenii was settled for the first time in accordance with presence or absence of hairs on the haustoria and the shape or size of fruits.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/, OR766688 to OR766695.
Author contributions
YS and Q-FY conceived and designed the study. Q-FY, S-TY, and CL analyzed the data. Y-HT, W-BY, J-PH, AZ, and MC-O discussed and interpreted the data. Q-FY, CL, and YS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 32260060 and 32060710) and the Special Program for Technology Bases and Talents of Guangxi (Grant No. 2022AC20002), and the Local Colleges Applied Basic Research Projects of Yunnan Province (Grant No. 202001BA070001-131).
Acknowledgments
The authors would like to thank Jun-Bo Yang, Jing Yang, Zheng-Shan He, and Ji-Xiong Yang at the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy Sciences (CAS), for sequencing technology. We sincerely thank Assistant Professor Athanasios Zervas for providing the raw data of Cassytha pubescens. We thank Hui Chen, Yun Juan Zou, and the Central Laboratory at Xishuangbanna Tropical Botanical Garden, CAS for their assistance in sampling and bioinformatics analysis. We sincerely thank three reviewers for critical and invaluable comments that greatly improved our manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1192170/full#supplementary-material
References
Adamu, A. A., Garba, F. N., Ahmed, T. M., and Abubakar, A. (2017). Pharmacognostic studies and elemental analysis of Cassytha filiformis Linn. J. Pharmacogn. Phytother. 9, 131–137. doi:10.5897/jpp2017.0448
Awang, K., Conran, J. G., and Waycott, M. (2018). Cuticular and ultrastructure characters on Cassytha L. (Lauraceae) stem. Available online at: https://www.researchgate.net/publication/325870821(accessed April 18, 2022).
Brophy, J. J., Goldsack, R. J., and Forster, P. I. (2009). The esential oils of some Australian Cassytha species (Lauraceae). J. Essent. Oil Res. 21, 543–546. doi:10.1080/10412905.2009.9700239
Burkill, H. M. (1995). The useful plants of west and tropical Africa. R. Bot. Gard. Kew (UK) Evol., 3, 39–41.
Carpenter, R. J., Macphail, M. K., Jordan, G. J., and Hill, R. S. (2015). Fossil evidence for open, proteaceae-dominated heathlands and fire in the Late Cretaceous of Australia. Am. J. Bot. 102, 2092–2107. doi:10.3732/ajb.1500343
Chanderbali, S. A., van der Werff, H., and Renner, S. S. (2001). Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Ann. Mo. Bot. Gard. 88, 104–134. doi:10.2307/2666133
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModeltest 2: more models, new heuristics, and parallel computing. Nat. Methods 9, 772. doi:10.1038/nmeth.2109
Dong, W., Li, E., Liu, Y, L., Xu, C., Wang, Y. S., Liu, K. J., et al. (2022b). Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 20, 92. doi:10.1186/s12915-022-01297-0
Dong, W., Liu, Y., Li, E., Xu, C., Sun, J., Li, W., et al. (2022a). Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol. Phylogenetics Evol. 166, 107330. doi:10.1016/j.ympev.2021.107330
Doyle, J. J., and Doyle, J. L. (1987). Deviations from michaelis-menten behaviour of plant glutamate dehydrogenase with ammonium as variable substrate. Phytochemistry 19, 11–13. doi:10.1016/0031-9422(80)85004-7
Hall, T., Biosciences, I., and Carlsbad, C. (2011). BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Hatusima, S. (1971). On some species of the Lauraceae from Japan and Taiwan(2). J. Geobotany 19, 25–28.
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi:10.1186/s13059-020-02154-5
Jo, S., Kim, Y. K., Cheon, S. H., Fan, Q., and Kim, K. J. (2019). Characterization of 20 complete plastomes from the tribe Laureae (Lauraceae) and distribution of small inversions. PLoS One 14, e0224622. doi:10.1371/journal.pone.0224622
Jordan, G. J., Carpenter, R. J., and Brodribb, T. J. (2014). Using fossil leaves as evidence for open vegetation. Palaeogeogr. Palaeoclimatol. Palaeoecol. 395, 168–175. doi:10.1016/j.palaeo.2013.12.035
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi:10.1038/nmeth.4285
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform 20, 1160–1166. doi:10.1093/bib/bbx108
Kearse, M., Moir, R., Wilson, A., Stones, H. S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kokubugata, G., Nakamura, K., Forster, P. I., Wilson, G. W., Holland, A. E., Hirayama, Y., et al. (2012). Cassytha pubescens and C. glabella (Lauraceae) are not disjunctly distributed between Australia and the Ryukyu Archipelago of Japan-evidence from morphological and molecular data. Aust. Syst. Bot. 25, 364–373. doi:10.1071/SB10040
Kostermans, A. J. G. H. (1994). Four new asiatic Lauraceae and a new species of Durio Adans. Bombacaceae) Rheedea Evol. 4, 14.
Li, H. T., Luo, Y., Gan, L., Ma, P. F., Gao, L. M., Yang, J. B., et al. (2021). Plastid phylogenomic insights into relationships of all flowering plant families. Bmc Biol. 19, 232. doi:10.1186/s12915-021-01166-2
Li, H. T., Yi, T. S., Gao, L. M., Ma, P. F., Zhang, T., Yang, J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. doi:10.1038/s41477-019-0421-0
Li, H. W., Li, J., Huang, P. H., Wei, F, N., Tsui, H, P., and van der Werff, H. (2008). Calycanthaceae–schisandraceae, flora of China. Beijing St. Louis Sci. press Mo. botanical Gard. press 7, 102–254.
Li, L., Madrinan, S., Li, J., Xu, Q., Song, S., Zuo, X., et al. (2016). Phylogeny and biogeography of Caryodaphnopsis (Lauraceae) inferred from low-copy nuclear gene and ITS sequences. Taxon 65, 433–445. doi:10.1016/j.bios.2015.09.003
Liu, C., Chen, H. H., Cai, J., Tian, X. Y., Han, L. H., and Song, Y. (2022). Characteristics of the complete plastid genome sequences of the monotypic genus Dodecadenia (Family: Lauraceae) and its phylogenomic implications. Forests 13, 1240. doi:10.3390/f13081240
Liu, C., Chen, H. H., Tang, L. Z., Khine, P. K., Han, L. H., Song, Y., et al. (2021a). Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae). Plant divers. 44, 377–388. doi:10.1016/j.pld.2021.11.009
Liu, Z. F., Ma, H., Ci, X. Q., Li, L., Song, Y., Liu, B., et al. (2021b). Can plastid genome sequencing be used for species identification in Lauraceae? Bot. J. Linn. Soc. 197, 1–14. doi:10.1093/botlinnean/boab018
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi:10.1093/molbev/msaa015
Nazar, A., Ayuning, F., and Ahmadin, A. (2019). The impact of Cassytha filiformis butanol fraction to the pregnancy and fetal development on mice. Int. J. Appl. Pharm. 11, 153–156. doi:10.22159/ijap.2019.v11s5.T1010
Rohwer, J. G., and Rudolph, B. (2005). Jumping genera. the phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Ann. Mo. Bot. Gard. 92, 153–178. doi:10.1093/aob/mci032
Rohwer, J. G. (2000). Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst. Bot. 25, 60–71. doi:10.2307/2666673
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi:10.1093/sysbio/sys029
Rozas, J., Ferrer, M. A., Sánchez-DelBarrio, J. C., Guirao, R. S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Song, Y., Dong, W. P., Liu, B., Xu, C., Yao, X., Gao, J., et al. (2015). Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 6, 662. doi:10.3389/fpls.2015.00662
Song, Y., Yao, X., Liu, B., Tan, Y. H., and Corlett, R. T. (2018). Complete plastid genome sequences of three tropical Alseodaphne trees in the family Lauraceae. Holzforschung 72, 337–345. doi:10.1515/hf-2017-0065
Song, Y., Yao, X., Tan, Y. H., Gan, Y., and Corlett, R. T. (2016). Complete chloroplast genome sequence of the avocado: Gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can. J. For. Res. 46, 1293–1301. doi:10.1139/cjfr-2016-0199
Song, Y., Yao, X., Tan, Y. H., Gan, Y., Yang, J. B., and Corlett, R. T. (2017a). Comparative analysis of complete chloroplast genome sequences of two subtropical trees, Phoebe sheareri and Phoebe omeiensis (Lauraceae). Tree Genet. Genomes 13, 120. doi:10.1007/s11295-017-1196-y
Song, Y., Yu, W. B., Tan, Y. H., Jin, J. J., Wang, B., Yang, J. B., et al. (2020). The predictive value of procalcitonin in ventilator-associated pneumonia after cardiac valve replacement. J. Syst. Evol. 58, 423–426. doi:10.1080/00365513.2020.1762242
Song, Y., Yu, W. B., Tan, Y. H., Liu, B., Yao, X., Jin, J. J., et al. (2017b). Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol. Evol. 9, 2354–2364. doi:10.1093/gbe/evx180
Tian, X. Y., Ye, J. W., and Song, Y. (2019). Plastome sequences help to improve the systematic position of trinerved Lindera species in the family Lauraceae. Peerj 7, e7662. doi:10.7717/peerj.7662
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi:10.1093/nar/gkx391
Trofimov, D., Cadar, D., Schmidt, C. J., de Moraes, P. L. R., and Rohwer, J. G. (2022). A comparative analysis of complete chloroplast genomes of seven Ocotea species (Lauraceae) confirms low sequence divergence within the Ocotea complex. Sci. Rep. 12, 1120. doi:10.1038/s41598-021-04635-4
Tsai, T. H., Wang, G. J., and Lin, L. C. (2008). Vasorelaxing alkaloids and flavonoids from Cassytha filiformis. J. Nat. Prod. 71, 289–291. doi:10.1021/np070564h
Wang, Z. H., Li, J., Conran, J. G., and Li, H. W. (2010). Phylogeny of the Southeast asian endemic genus Neocinnamomum H. Liu (Lauraceae). Plant Syst. Evol. 290, 173–184. doi:10.1007/s00606-010-0359-1
Weber, J. Z. (1981). A taxonomic revision of Cassytha (Lauraceae) in Australia. J. Adel. Bot. Gard. 3, 187–226.
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi:10.1093/bioinformatics/btv383
Wu, C. S., Wang, T. J., Wu, C. W., Wang, Y. N., and Chaw, S. M. (2017). Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol. Evol. 9, 2604–2614. doi:10.1093/gbe/evx177
Keywords: hemiparasitic genus, chloroplast, Cassytha, Eusideroxylon, Laurales
Citation: Yu Q-F, Tan Y-H, Yu W-B, Yang S-T, Huang J-P, Caraballo-Ortiz MA, Liu C and Song Y (2023) Comparative analyses of eight complete plastid genomes of two hemiparasitic Cassytha vines in the family Lauraceae. Front. Genet. 14:1192170. doi: 10.3389/fgene.2023.1192170
Received: 23 March 2023; Accepted: 06 June 2023;
Published: 13 December 2023.
Edited by:
Diego Hojsgaard, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Agostina B. Sassone, Instituto de Botánica Darwinion, ArgentinaWenpan Dong, Beijing Forestry University, China
Copyright © 2023 Yu, Tan, Yu, Yang, Huang, Caraballo-Ortiz, Liu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Liu, bGl1Y2hhb184MEAxNjMuY29t; Yu Song, c29uZ3l1QGd4bnUuZWR1LmNu
 Qun-Fei Yu
Qun-Fei Yu Yun-Hong Tan1,3
Yun-Hong Tan1,3 Wen-Bin Yu
Wen-Bin Yu Jie-Peng Huang
Jie-Peng Huang Marcos A. Caraballo-Ortiz
Marcos A. Caraballo-Ortiz Chao Liu
Chao Liu