- School of Biological Sciences, Jining Medical University, Rizhao, China
The SlADH gene plays a key role in environmental stress response. However, limited studies exist regarding the tomato SlADH gene. In this study, we identified 35 SlADH genes in tomato by genome-wide identification. Among the 12 chromosomes of tomato, SlADH gene is distributed on 10 chromosomes, among which the 7th and 10th chromosomes have no family members, while the 11th chromosome has the most members with 8 family members. Members of this gene family are characterized by long coding sequences, few amino acids, and introns that make up a large proportion of the genetic structure of most members of this family. Moreover, the molecular weight of the proteins of the family members was similar, and the basic proteins were mostly, and the overall distribution was relatively close to neutral (pI = 7). This may indicate that proteins in this family have a more conserved function. In addition, a total of four classes of cis-acting elements were detected in all 35 SlADH promoter regions, most of which were associated with biotic and abiotic stresses. The results indicate that SlADH gene had a certain response to cold stress, salt stress, ABA treatment and PEG stress. This study provides a new candidate gene for improving tomato stress resistance.
1 Introduction
Alcohol dehydrogenase (ADH, EC 1.1.1.1), a part of the dehydrogenase superfamily, is a zinc-binding enzyme that catalyzes the mutual conversion between acetaldehyde and ethanol as a dimer, relying on the NAD(P) cofactor (Strommer, 2011). It is a well-studied member of the medium-length dehydrogenase/reductase (MDR) protein superfamily. And is found in a wide range of organisms (Speirs et al., 1998; Koch et al., 2000; Manriquez et al., 2006; Singh et al., 2010; Jin et al., 2016). The ADH gene family is a large family, mainly divided into three subfamilies: Short-chain dehydrogenase/Reductase (SDR) -ADH (containing about 250 amino acid residues), medium-chain dehydrogenase/reductase (MDR) -ADH (containing about 350 amino acid residues), and long-chain dehydrogenase/reductase (LDR) -ADH (600–750 amino acid residues or about 385–900 amino acid residues) (Alka et al., 2013). In plants, most ADHs contain zinc ligands and belong to the medium-chain protein subfamily (Yeganova et al., 2004). These enzymes are widely involved in metabolic processes and have a positive role in increasing resistance to biotic and abiotic stresses (Linskens and Schrauwen, 1966; Jacobs et al., 1988; Tadege et al., 1999; Garabagi et al., 2005; Bailey-Serres and Voesenek, 2008; Banach et al., 2009; Bolte et al., 2011).
ADH is ubiquitously present in both animals and plants, though it is more commonly known in the latter. Plants require respiration at all times to energize their physiological functions. However, when there’s insufficient oxygen for normal respiration, the aerobic process transitions to anaerobic respiration. Plants produce large amounts of alcohol during anaerobic respiration, and alcohol dehydrogenase (ADH) is a key enzyme in the ethanol fermentation pathway and subsequent adaptive anaerobic metabolism of plant tissues (Taneja and Mande, 1999). For example, during submergence stress, many plants alter their structure by elongating their stems to produce aerenchyma, which delivers oxygen from leaves exposed to air to submerged plant parts (Sasidharan and Voesenek, 2015). Concurrently, there’s an unavoidable transition from aerobic to anaerobic glycolysis, termed fermentation. Under these conditions, typical transcription and translation processes halt, and “anaerobic peptides” are preferentially synthesized. Although less energy is produced in this way, it is thought to play an important role in cell survival. Studies have shown that the submergence tolerance of plants is proportional to the change of ADH activity in response to submergence. ADH gene has been considered as an important candidate gene for genetic manipulation of submergence tolerance by improving the adaptability of plants to hypoxia (Jacobs et al., 1988; Takahashi et al., 2014). Research by Komatsu et al. revealed that soybean GmADH was particularly induced by waterlogging, yet these genes remained relatively stable under cold and drought stresses (Komatsu et al., 2010; Komatsu et al., 2012). In addition, ADH family genes have also been found to be involved in other abiotic stresses such as cold and drought. Studies have shown that in cereal crops and Arabidopsis, the ADH gene is a chilling-induced gene and plays a very important role in plant resistance to chilling stress (Kato-Noguchi and Yasuda, 2007; Zhang et al., 2023). The ADH gene is involved in the cold resistance of forest strawberry (Fragariavesca) and is a good molecular marker candidate for cold stress. When maize seedlings were exposed to low temperature stress, the activity of ADH was also found to increase. Conversely, in the ADH1-ADH2 double deletion mutant, membrane lipid peroxidation intensified, leading to pronounced cellular damage (Gao et al., 2022).
ADH also plays a vital role in regulating fruit flavor. The expression of the ADH gene is stringently regulated in mature fruits. Owing to ADH’s function in alcohol production, diminished ADH activity in fruits can result in a subdued and modified flavor profile (Jelski et al., 2014). Mature tomato fruit has a high content of ADH2, but does not contain ADH1. In transgenic tomato fruits with ADH2 overexpression, there was a notable rise in C6 alcohol levels compared to C6 aldehydes, leading to a richer tomato flavor. This enhancement was especially linked to elevated concentrations of Z-3-hexenol (Speirs et al., 1998). Similarly, in grapes with a small ADH gene family, fruits produce higher levels of ADH and exhibit increased ADH activity at late maturation (Yamauchi et al., 1990).
Tomato stands out as one of the paramount vegetable crops worldwide, serving as a crucial model crop, the tomato frequently features in plant science research. It has always been favored in the international and domestic markets, and is one of the horticultural crops with the highest economic benefits in the world. In recent years, people’s demand for tomato is increasing day by day, and the demand for planting area is also gradually increasing, but the resulting planting problems are also increasing significantly. These consequent problems caused a large reduction in tomato production, resulting in great economic losses. To counteract severe biotic and abiotic stresses, plants have naturally evolved a repertoire of defensive strategies, allowing them to swiftly adapt to their intricate surroundings, mitigate harm, and compete for vital resources (Atkinson and Urwin, 2012). Faced with complex environmental conditions, plants have a complex and comprehensive response, including the ADH gene. At the same time, as a characteristic of tomato fruit, the flavor of tomato is also regulated by ADH gene, so it is necessary to study the ADH gene family in depth. At present, there are few studies on tomato ADH gene family, and the specific contribution of each family to tomato stress resistance and flavor is relatively vague. Studying tomato SlADH gene can provide important information for tomato molecular breeding. In this study, we delved into the gene structure, chromosome location, phylogenetic tree and transcriptome-based expression data of tomato SlADH gene family, and constructed the expression profiles of this gene in different tissues under different biotic and abiotic stresses. In this study, the ADH gene family was analyzed in detail, which could provide a reference for the study of the role of ADH gene family in exploring the response of tomato to biotic and abiotic stress.
2 Materials and methods
2.1 Plant materials and treatments
Tomato cv. MoneyMaker was preserved and propagated in our laboratory. The setting of the plant growth chamber was as follows: with a photoperiod of 16 h/8 h (day/night), 22°C/18°C (day/night) with a humidity of 60%/50% (day/night). The leaves were collected when tomato seedlings grew to the five-leave stage after treatment at 0, 6, 12, 24, and 48 h, respectively.
For the cold stress treatment, the temperature of the chamber was set at 4°C. For the drought stress treatment, tomato plants were subjected to a 10% polyethylene glycol (PEG) solution. For the salinity stress treatment, tomato plants were subjected to a 200 mM NaCl solution. For the ABA treatment, a 0.1 mM ABA solution was evenly sprayed on the surface of tomato seedlings until droplets began to form at the edges of the leaves.
2.2 Identification and sequence analysis of SlADH genes
Tomato genome sequence version SL4.0 and the genome annotation file version ITAG4.0 were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). The seed alignment files of the ADH domain (PF00107: ADH_zinc_N and PF08240: ADH_N) were downloaded from the Pfam (http://www.ebi.ac.uk/interpro/entry/pfam/#table) database. HMMER3 software was used to search the protein containing the ADH_N and ADH_zinc_N (Altermann et al., 1999), with an e-value cutoff of 0.01 (Finn et al., 2011), then all the candidate protein sequences were submitted to Pfam, CDD and SMART databases to verify the domains. Only a protein containing two domains will be considered an ADH protein.
2.3 Chromosome location analysis of tomato SlADH genes
The chromosomal position data of SlADH genes were extracted from the genome annotation file. The figure showing the location of SlADH genes were generated with the Show Genes on Chromosome model of TBtools software (Chen et al., 2020).
2.4 Phylogenetic tree construction of SlADH proteins in tomato
The ADH proteins of the other species were downloaded from the NCBI database (Onodera et al., 2023), and the multiple sequence alignment was performed using MUSCLE with the default options (Kumar et al., 2016). The phylogenetic tree was constructed using the Neighbor-joining (NJ) method with 1,000 bootstraps.
2.5 Analysis of conserved motifs and gene structure of SlADH genes
The MEME software was used to identify the conserved motifs among tomato ADH genes with the common set value (Yum et al., 1991), the length of motifs were range 6–20, the maximum number of motifs was 10. The gene structure information of SlADH genes were obtained from the genome annotation file (version: ITAG4.0), containing the CDS (coding sequence) region, the UTR (untranslated region) region and the intron region.
2.6 Analysis of cis-acting elements of tomato SlADH genes
The 1.5 kb upstream sequence of SlADH genes were obtained from the genome sequence (version: SL4.0) by perl script. Meanwhile, PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database was used to predict the cis-elements of the promoter regions of SlADH genes, and a Python script was used to stat the cis-elements data, a heatmap was used to show the distribution of cis-elements.
2.7 Synteny analysis of SlADH genes
The gene duplication events were analyzed by the One step MCScanX model of Comparative Genomics function of TBtools, and the Advanced Circos model was used to show the duplication events of SlADH genes in tomato.
In addition, the genome sequence file and the genome annotation file of A. thaliana, Solanum tuberosum and Zea mays were downloaded from Phytozome database, then the OneStepMCScanX model of TBtools was used to analyze the synteny relationship of orthologous ADH genes between the tomato and A. thaliana, S. tuberosum, Z. mays.
2.8 Transcriptome expression pattern of SlADH genes
The transcriptome raw data with accession: PRJNA634438 was downloaded from the NCBI database, containing drought stress treatment and heat stress treatment were used in this study. The fastp (v0.23.3) software was used to filter the RNA-seq data (Chen et al., 2018), the hisat2 software was used to map the raw data to the tomato genome sequence (Kim et al., 2019), the featurecounts of Rsubread package in R was used to the genes quantification (Mo et al., 2021). The heatmap model of TBtools was used to display the heatmap of SlADH gene expression.
2.9 Quantitative real-time PCR analysis of SlADH genes
After stress treatment on tomato plants, we selected 16 genes for q-PCR to analyze the response of SlADH family members to stress. Primer5 (Supplementary Table S1) was used for primer design. Primers were ordered from Sogon Bioscience Co., Ltd., and the dye used for qRT-PCR was purchased from Biorun (BioRun Biosciences Co., Ltd., Wuhan, China). The system of qRT-PCR is referred to Mo’s study (Mo et al., 2021).
2.10 Statistical analyses
All the data are presented as the means ± standard deviations (SDs). Statistical analysis was carried out with one-way analysis of variance (ANOVA) in SPSS software (SPSS, Chicago, United States). For all comparisons, p < 0.05 was considered statistically significant, represented in the figures by asterisks. All experiments were repeated three times.
3 Results
3.1 SlADH gene identification and chromosomal distribution analysis
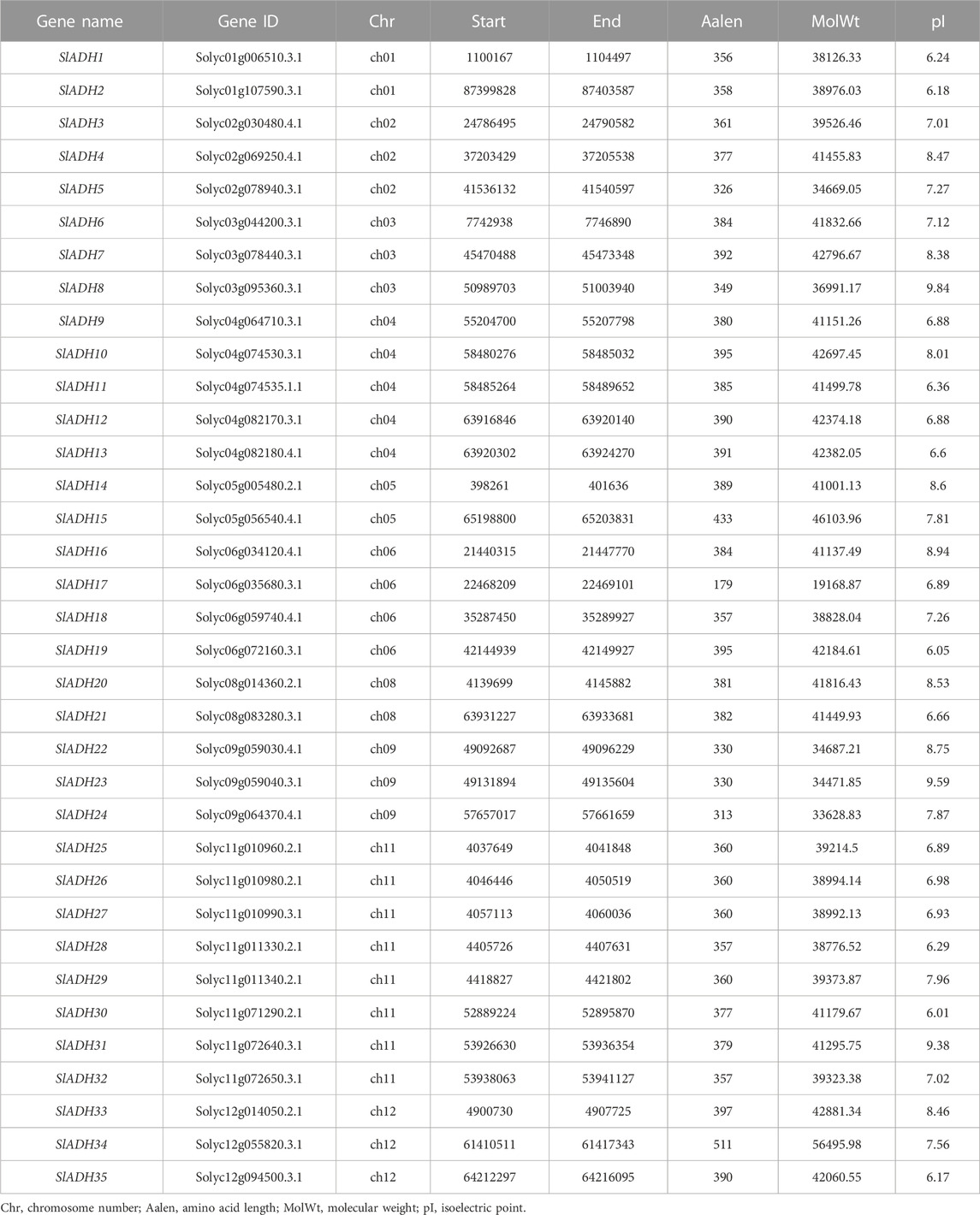
A total of 35 SlADH genes were identified in tomato genome (Table 1). The SlADH gene lengths varied, ranging from 892 bp (SlADH17) to 14,237 bp (SlADH8). Only two of the SlADH genes had genome lengths less than 2,000 bp (SlADH17, SlADH28). The number of amino acids varies from 179 (SlADH17) to 511 (SlADH34), but most members have between 300 and 400 amino acids, except for SlADH17, SlADH15, and SlADH34. The genome length of SlADH family members is generally large, but the corresponding number of encoded amino acids is not particularly large, indicating that introns account for a large proportion of the genetic structure of the gene family. The molecular weight of SlADH protein ranges from 19.16 kDa (SlADH17) to 56.49 kDa (SlADH34), and the theoretical pI ranges from 6.01 (SlADH30) to 9.84 (SlADH8). Furthermore, out of the total, 21 SlADH proteins are anticipated to be alkaline (with a pI > 7.0), while 15 are predicted to be acidic (pI < 7.0).
A total of 35 SlADH genes were identified on 12 tomato chromosomes, and their distribution on the chromosomes was uneven. All chromosomes, except for the 7th and 10th, contain the SlADH gene, as depicted in Figure 1. Among the 10 chromosomes, SlADH family members are mainly distributed on chromosomes 4 and 11. Chromosome 11 contains the most SlADH family members, 8 in total, and the positions of these members on chromosome 11 are very close to each end. In general, the distribution of members of this family on chromosomes is found that most members of this family are distributed at both ends of chromosomes, only three members exist in the middle of chromosomes 2 and 6.
3.2 Phylogenetic relationships among SlADH genes
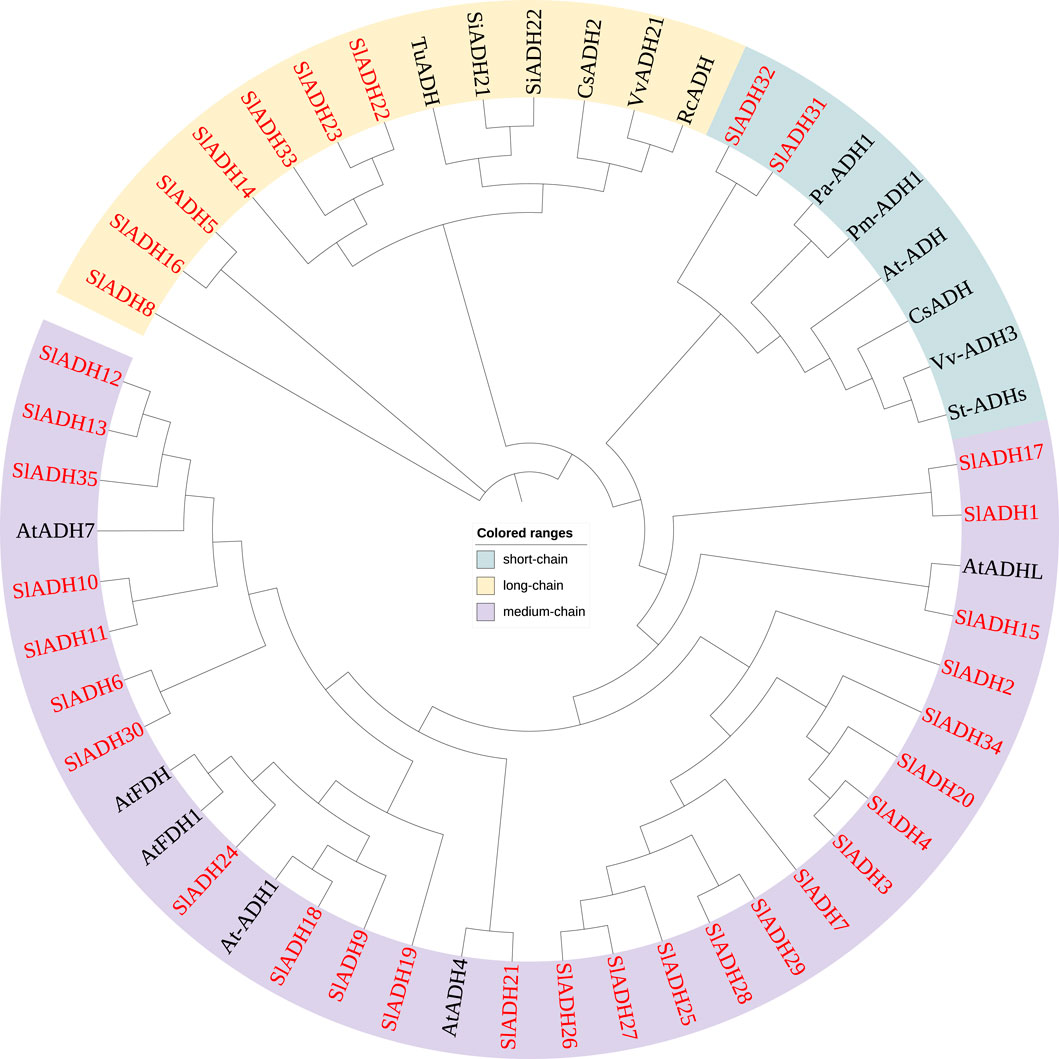
In order to understand the possible functional differences among individual members of tomato SlADH gene superfamily, the sequences of a set of ADH family members from other plant species, including Solanum lycopersicum (Sl), Triticum urartu (Tu), Setaria italica (Si), Cucumis sativus (Cs), Vitis vinifera (Vv), Rosa chinensis (Rc), Prunus armeniaca (Pa), Prunus mume (Pm), and A. thaliana (At), were analyzed using a Neighbor-joining (NJ) phylogenetic tree. As shown in Figure 2, the phylogenetic tree divided these ADHs into three clades: short-chain ADHs, medium-chain ADHs and long-chain ADHs. Among the 35 SlADH family members of tomato, most belong to the medium-chain ADH branch (26 members), while the short-chain ADH branch contains the least SlADH family members, only two. It is not difficult to see from the evolutionary relationship that members of the SlADH family are closer to A. thaliana than other species, but far from masson pine, rose and other woody plants, so they contain the least members in the short-chain ADH branch.
3.3 Conserved motifs and gene structure analyses of SlADH genes
We used MEME tool to analyze the conserved motif of SlADH protein. A total of 10 distinct motifs were detected in all 35 SlADH proteins. While some members showcased distinct motif variations, underscoring the diversity of SlADH proteins, a majority exhibited highly similar motifs. Meanwhile, through the analysis of conserved motifs, we speculated that the proteins expressed by SlADH family members might play a conservative and single function. Motif 4 is present in all proteins except SlADH18, suggesting that this motif may be a conserved structure of the SlADH family. Then, in order to study the exon-intron structure of SlADH gene, we used the gene structure display server program to visually analyze the SlADH gene genome and coding sequence. Of the 35 SlADH genes, all members contain introns. In addition, all except SlADH17 and SlADH27 contain UTRs (Figure 3). SlADH20, SlADH26, SlADH28, SlADH30, and SlADH32 contain only 3′ UTR, while SlADH4, SlADH7, and SlADH15 contain only 5′ UTR.
3.4 Duplication analysis of tomato SlADH genes
Analyzing gene duplication is pivotal, as it often unravels insights into evolutionary processes and the functional diversification or redundancy of genes, potentially driving the adaptability and evolution of the species. Duplication events among SlADH members were analyzed (Figure 4). There was only one duplicate gene pair in 35 SlADH genes (SlADH12/SlADH35), which reflected the functional specificity and diversity of the members of this family.
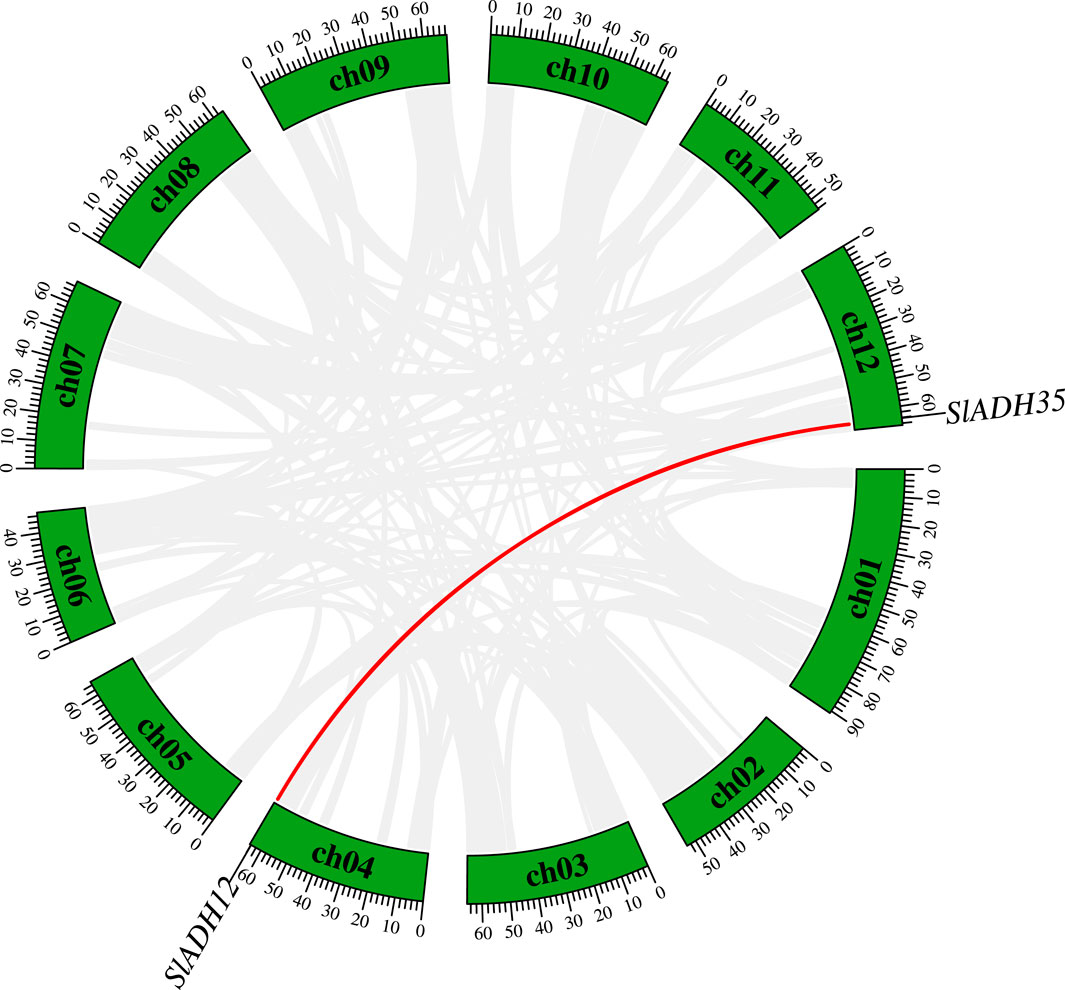
FIGURE 4. The duplication genes in SlADH gene family. Segmental duplication gene pairs inked by red line.
3.5 Promoter cis-element analysis of SlADH genes
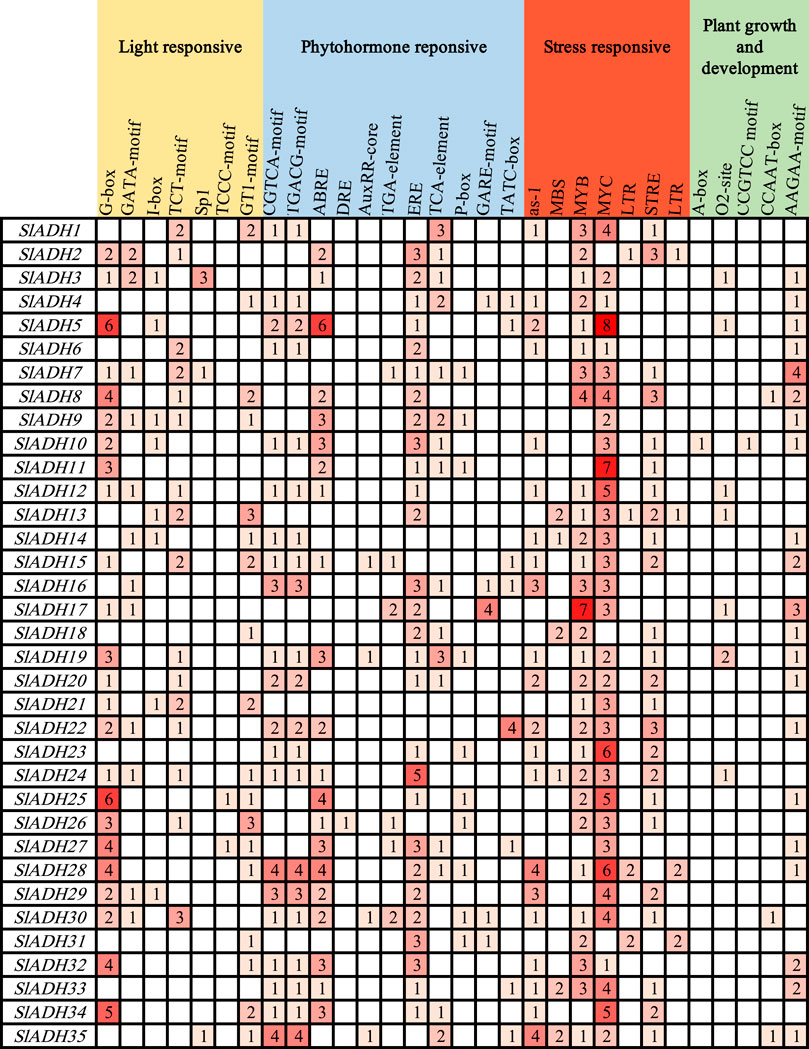
To reveal the diversity of cis-elements in the promoter region of the SlADH gene, we selected the 1,500 bp sequence upstream of the SlADH gene and submitted it to the PlantCARE database for promoter cis-element analysis (Figure 5). We then visualized the results, and found that there were seven elements related to light responsive and seven elements related to stress response. The largest number of elements related to phytohormone reponsive were 11. There were only five elements related to plant growth and development. In these homeopathic elements, MYC is widely distributed throughout the SlADH family species, with only three members lacking it. As a key component of plant response to various stresses, such a large amount of MYC also reflects the key role of SlADH family in plant.
3.6 Synteny analysis of ADH genes between tomato and arabidopsis, potato, maize
To support the presented information about the evolutionary process in tomato, we investigated the syntenic relationships of the ADH gene pairs in different species (Figure 6). We selected the model plant A. thaliana, the Solanaceae crop potato, and the monocotyledon crop maize to explore inter-species synteny relationship ADH genes. The results showed that there were 17 pairs of ADH genes between tomato and A. thaliana, 28 pairs of ADH genes between tomato and potato, and only two pairs of ADH genes between maize and tomato.
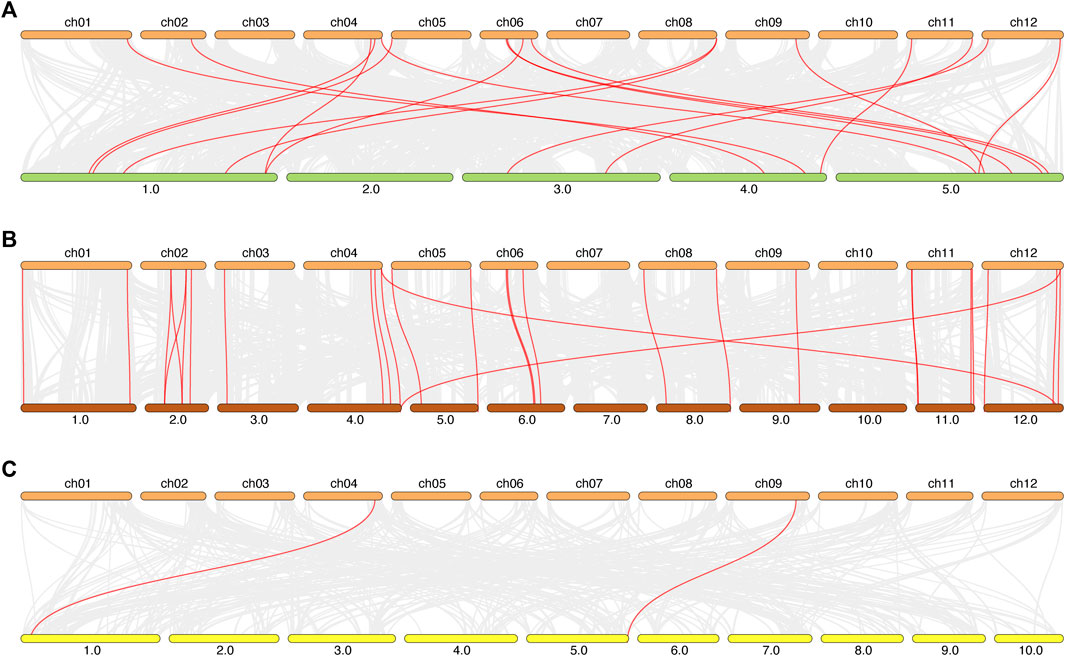
FIGURE 6. The synteny relationship between tomato and Ara, rice and potato. (A): tomato and Ara, (B): tomato and potato, (C): tomato and maize. All the synteny relationship ADH genes between different species were linked by red, the collinear blocks within the tomato and other plant genomes were linked by gray lines in the background.
3.7 Expression pattern analysis of tomato SlADH gene family members under stress
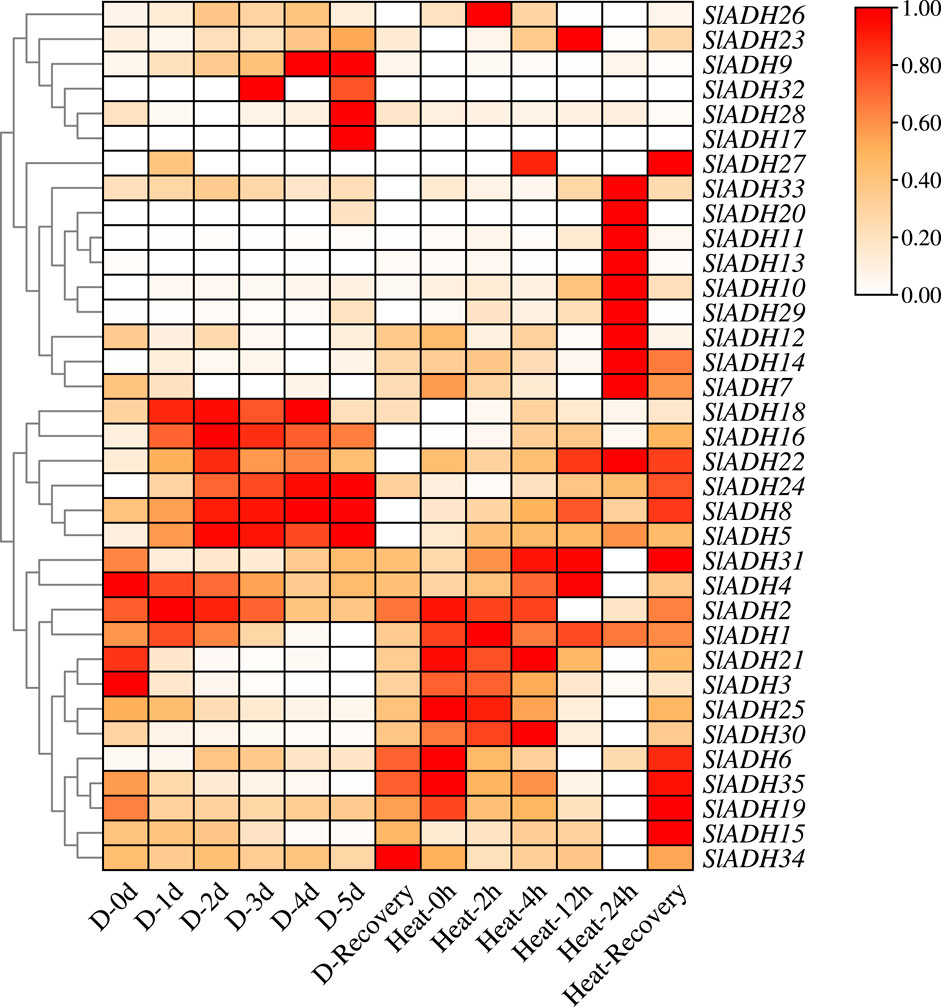
From the transcriptome data, following drought and heat stress, SlADH family members predominantly exhibited two patterns: first increase, then decrease and gradually increase (Figure 7), and only a few members did not respond strongly. In drought stress, most members reached the highest expression level of SlADH gene on the third day after drought stress, whereas, during heat stress, the highest expression levels of SlADH family members were observed at 12 and 24 h. For both stresses, SlADH family members showed an extremely significant response trend in the recovery treatment after stress. The level of expression increased dramatically. This suggests that the SlADH family members play a pivotal role not just in stress resistance but also in reestablishing plant homeostasis following the stress.
3.8 qRT-PCR analysis of SlADH gene under abiotic and ABA treatments
Subsequently, tomato plants were treated with salt stress (NaCl), cold stress, drought stress (PEG) and ABA treatment, and a total of 16 genes from different subbranches of the phylogenetic tree were selected for response analysis (Figure 8). All 16 selected family members exhibited responses to at least one of the stresses or treatment. Of the 16 selected gene family members, 13 SlADH genes responded to salt stress (except SlADH16, SlADH8, SlADH15), 11 SlADH genes responded to cold stress(except SlADH10, SlADH16, SlADH8, SlADH20, SlADH31), 11 SlADH genes responded to drought stress(except SlADH20), and 12 SlADH genes responded to ABA treatment(except SlADH34, SlADH33, SlADH24, SlADH20). Among them, SlADH6, SlADH26, SlADH24, SlADH32, SlADH12, SlADH9, and SlADH5 all responded to the four kinds of stress. Except for SlADH12, which showed no obvious trend, the expression levels of the other genes all showed a trend of gradual increase after stress. In addition, SlADH15 showed obvious response to cold stress, drought stress and ABA treatment except salt stress, and the expression level decreased gradually after a significant increase. However, SlADH20 was only responsive to salt stress, showing a trend of first increasing and then decreasing. SlADH16 and SlADH8 showed a good response to drought stress and ABA treatment, but no significant response to the other two stresses. These results suggest that the SlADH family members are responsive to a majority of stresses or treatments, underscoring their potential significance in plant reactions to diverse abiotic challenges. Concurrently, individual family members exhibit varied response patterns to distinct stresses, highlighting the functional diversity and specificity of these genes.
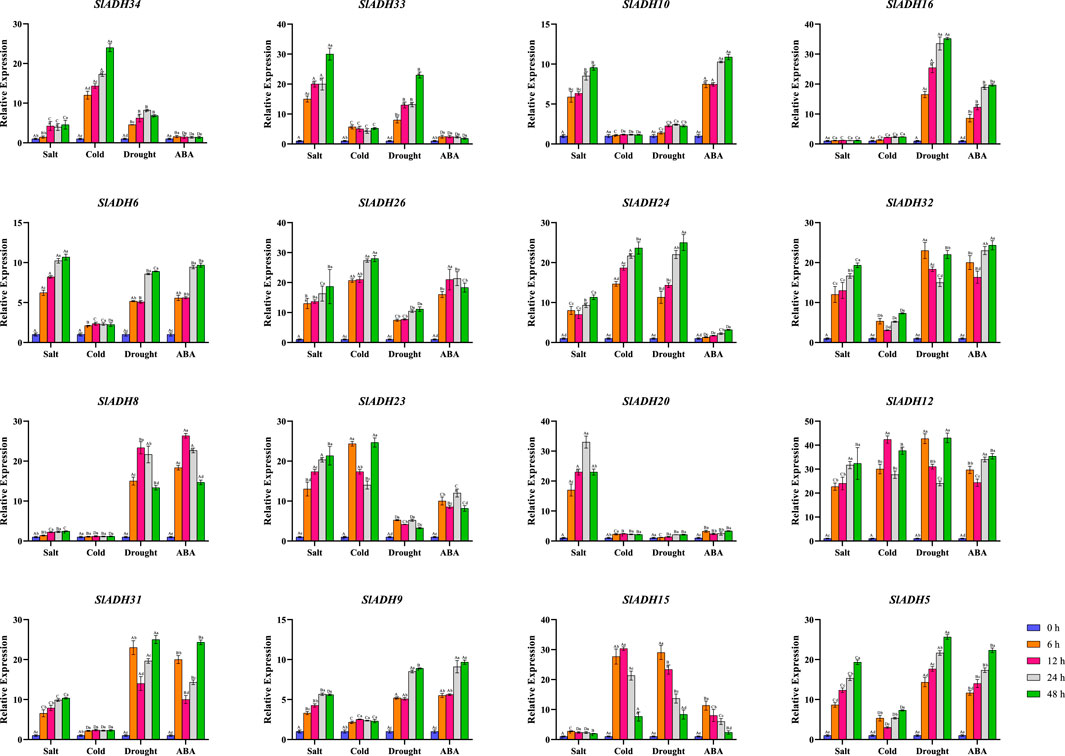
FIGURES 8. Expression levels of 16 selected SlADH genes in response to salt, cold, drought and ABA stress.
4 Discussion
In recent years, extreme weather has occurred frequently around the world. Drought, waterlogging, salinity and freezing damage are all environmental factors that threaten plant growth. Adverse environmental conditions have a negative impact on agricultural production and reduce both the qualitative and quantitative yields of crops. Climate change is expected to pose serious challenges to agricultural production worldwide by causing rising temperatures and water shortages. Exploring the molecular responses of plants to abiotic stresses and using this knowledge to develop crop plants that can adapt to these adverse environmental conditions and maintain high yields under these adverse environmental conditions has been a major goal of molecular breeders (Xiong et al., 2002; Umezawa et al., 2006; Seki et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Hirayama and Shinozaki, 2010; Zhu, 2016; Fontemaggi, 2023). Despite concerted efforts and significant findings, this challenge persists, highlighting its complexity and the need to develop new methods to mitigate the damage caused by environmental stress. As a result, identifying robust resistance genes in plants has emerged as a primary solution.
Over the past few decades, several studies have characterized ADH genes, and this work has greatly increased our understanding of plant responses to stress in many species. Many studies of ADH gene expression have linked it to both biotic and abiotic stresses. However, until now, the ADH gene family has not been reported in tomato, which has hindered the study on the function of tomato ADH gene to a certain extent. On this basis, we systematically analyzed the ADH gene family of tomato using bioinformatics. In this study, 35 ADH genes were identified from tomato, which had more ADH family members than maize (seven members) and grape (ten members) (Su et al., 2020). The members of this gene family are characterized by long coding sequence but few amino acids, and most members of this family have a gene structure with many introns. At the same time, the molecular weight of the total family members proteins were similar, and most of the proteins were alkaline proteins, and the overall distribution was relatively neutral (pI = 7). This may indicate that the proteins in this family have a more conservative function. Then, through duplication analysis of the tomato SlADH family members, we found that there was only one gene pair (SlADH12/SlADH35) among the 35 members, which indicated the functional diversity of the family members. In motif analysis, it is found that there are 10 motifs in this family, which are evenly distributed in different members of the family, forming a variety of different forms in the way of combination. We speculate that it is this relatively uniform combination distribution that causes the specificity and diversity among the members of this family, and they likely have more specific functions. The results of conserved motifs, gene characteristics and gene structure analysis of ADH family genes supported the classification results of phylogenetic analysis. We found that the signature of the ADH gene was similar within each subfamily and varied between different subfamilies.
In addition, four types of cis-acting elements were identified through the analysis of SlADH gene family, which were related to light response, phytohormone reponsive, stress response and plant growth and development. Among these elements, the types of elements related to phytohormone reponsive were the largest, but the total number of elements related to stress was the largest. Of these elements, MYC is the most numerous, with only three members without MYC element. MYC is one of the members of bHLH transcription factor superfamily, and MYC has been widely reported. MYC plays a very important role in plant growth and development, especially in stress resistance (Abe et al., 1997; Perez-Alonso et al., 2021). In addition, most family members also include MYB element and LTR element. Research has shown that, overexpression of MYB12 and MYB75 in transgenic plants significantly increased the accumulation of flavonoids with strong antioxidant activity, thus resulting in enhanced tolerance to abiotic stresses such as drought and oxidative stresses (Nakabayashi et al., 2014; Wang et al., 2016). And LTR elements may be potential molecular targets for temperature domestication (Wu et al., 2019). Meanwhile, stress-related elements such as MBS and STRE also make up a large portion. The presence of such a large number of stress-related response elements in the gene structure further demonstrated the key role of SlADH gene family in tomato resistance process.
Subsequently, we analyzed the expression level of tomato SlADH gene family after heat stress and drought stress by transcriptome data, and selected 16 genes for qRT-PCR. The results showed that the q-PCR trend of the selected genes was consistent with that of the transcriptome. This verified the authenticity and reliability of transcriptomic data. In the entire transcriptome, most members of the SlADH gene family showed a response to heat stress and showed an increasing tendency after the plants were treated with heat stress. Then, we subjected tomato to salt stress (NaCl), cold stress, drought stress (PEG) and ABA treatment, and selected 16 SlADH family members for expression level detection. Through the above experiments, we once again verified that SlADH gene family has a certain degree of response to a variety of abiotic stresses, indicating that SlADH gene family members play a key role in the process of tomato abiotic stress. Studies have shown that there are three ADH genes in Pygmy cattle, and two of them are induced by hypoxia (Garabagi et al., 2005). Similarly, ADHA of two ADH genes in cotton is induced by waterlogging (Estrada et al., 2022; Pan et al., 2023), indicating that ADH gene family plays a vital role in plant resistance to waterlogging. Low temperature can cause lipid peroxidation of plant cell membrane, and even death of some plant cells sensitive to low temperature (Kidokoro et al., 2022). Noguchi et al. found that low temperature stress (5°C, 7.5°C, 10°C) can significantly improve the activities of ADH and PDC in rice seedlings, and enhance the ethanol fermentation pathway. Owing to its unique gene function, ADH not only holds significance in diverse biotic and abiotic stress responses but also assumes a pivotal role across various domains. ADH, a zinc-binding enzyme, is a key enzyme in the metabolic pathway of fatty acids, which can reversibly convert aldehydes into corresponding alcohols and may play an important role in the synthesis of volatile esters. So the gene also plays a crucial role in regulating fruit flavor. In ADH, not only the medium-chain ADH gene is associated with the production of aromatic volatiles (Singh et al., 2010; Bajpai et al., 2018), the short chain ADH may contribute to the biosynthesis of plant aroma (Moummou et al., 2012). Other studies have shown that ADH genes are expressed in a developmentally regulated manner, especially during fruit ripening (Manriquez et al., 2006). LeADH2 is expressed in tomato fruits and its abundance increases during ripening, especially in late ripening (Longhurst et al., 1994). Similarly, VvADH expression is noted during the late ripening phase of grapes (Tesniere and Verries, 2000). A similar pattern was observed in Nanguo pear, especially in varieties with high expression of ADH6 and ADH7. The augmented transcriptional accumulation of ADH6 in ripe Nanguo Pear fruit correlates with heightened ADH activity, akin to the scenario with VvADH1 in grape (Tesniere and Verries, 2000). Qin et al. found that according to the transcription mode of ADH, ADH activity and hexyl ester synthesis in Dangshansu pear and Nanguo pear fruits are related to the transcription of ADH6, indicating that ADH plays a decisive role in the volatiles of pear fruits (Zeng et al., 2020). Both stress resistance and fruit quality will have a direct impact on the economic value of tomato and directly affect the income of farmers. Therefore, through bioinformatics analysis and omics data analysis of ADH gene, this paper reveals the important role of ADH gene on tomato stress resistance and fruit quality to some extent. It can provide a strong reference for further exploration of its function.
5 Conclusion
In conclusion, this study identified SlADH gene family members in tomato for the first time. Bioinformatics analysis and qRT-PCR analysis under abiotic stress revealed that tomato SlADH gene family members may play an important role in tomato growth and development and stress resistance. This investigation underscores the pivotal significance of the SlADH gene in tomato and establishes a foundational platform for subsequent inquiries into the broader role of the SlADH gene family in tomato breeding and augmenting resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
QZ and YH put forward conceptualization; YH and WY found the methodology; WY, HZ, and GL conducted the experiments; YH and WY analyzed the data; QZ wrote the draft preparation; YH prepared, edited and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Research project of undergraduate teaching reform in Shandong province (M2020311) and the National Natural Science Foundation of China (32000194).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1186192/full#supplementary-material
References
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 (10), 1859–1868. doi:10.1105/tpc.9.10.1859
Alka, K., Windle, H. J., Cornally, D., Ryan, B. J., and Henehan, G. T. (2013). A short chain NAD(H)-dependent alcohol dehydrogenase (HpSCADH) from Helicobacter pylori: a role in growth under neutral and acidic conditions. Int. J. Biochem. Cell Biol. 45 (7), 1347–1355. doi:10.1016/j.biocel.2013.04.006
Altermann, E., Klein, J. R., and Henrich, B. (1999). Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage (phi)adh. Gene 236 (2), 333–346. doi:10.1016/s0378-1119(99)00236-x
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63 (10), 3523–3543. doi:10.1093/jxb/ers100
Bailey-Serres, J., and Voesenek, L. A. (2008). Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339. doi:10.1146/annurev.arplant.59.032607.092752
Bajpai, A., Khan, K., Muthukumar, M., Rajan, S., and Singh, N. K. (2018). Molecular analysis of anthocyanin biosynthesis pathway genes and their differential expression in mango peel. Genome 61 (3), 157–166. doi:10.1139/gen-2017-0205
Banach, K., Banach, A. M., Lamers, L. P., De Kroon, H., Bennicelli, R. P., Smits, A. J., et al. (2009). Differences in flooding tolerance between species from two wetland habitats with contrasting hydrology: implications for vegetation development in future floodwater retention areas. Ann. Bot. 103 (2), 341–351. doi:10.1093/aob/mcn183
Bolte, S., Lanquar, V., Soler, M. N., Beebo, A., Satiat-Jeunemaitre, B., Bouhidel, K., et al. (2011). Distinct lytic vacuolar compartments are embedded inside the protein storage vacuole of dry and germinating Arabidopsis thaliana seeds. Plant Cell Physiol. 52 (7), 1142–1152. doi:10.1093/pcp/pcr065
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), 884–890. doi:10.1093/bioinformatics/bty560
Estrada, F., Flexas, J., Araus, J. L., Mora-Poblete, F., Gonzalez-Talice, J., Castillo, D., et al. (2022). Exploring plant responses to abiotic stress by contrasting spectral signature changes. Front. Plant Sci. 13, 1026323. doi:10.3389/fpls.2022.1026323
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Fontemaggi, G. (2023). Non-coding RNA regulatory networks in post-transcriptional regulation of VEGFA in cancer. IUBMB Life 75 (1), 30–39. doi:10.1002/iub.2620
Gao, N., Chen, J., Qi, B., Zhao, T., Guo, Y., Fang, Y., et al. (2022). Effects of gene polymorphisms, metabolic activity, and content of alcohol dehydrogenase and acetaldehyde dehydrogenases on prognosis of hepatocellular carcinoma patients. Turk J. Gastroenterol. 33 (7), 606–614. doi:10.5152/tjg.2022.21340
Garabagi, F., Duns, G., and Strommer, J. (2005). Selective recruitment of Adh genes for distinct enzymatic functions in Petunia hybrida. Plant Mol. Biol. 58 (2), 283–294. doi:10.1007/s11103-005-3545-8
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61 (6), 1041–1052. doi:10.1111/j.1365-313X.2010.04124.x
Jacobs, M., Dolferus, R., and Van den Bossche, D. (1988). Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of arabidopsis thaliana (L.) Heynh. Biochem. Genet. 26 (1-2), 105–122. doi:10.1007/BF00555492
Jelski, W., Laniewska-Dunaj, M., Orywal, K., Kochanowicz, J., Rutkowski, R., and Szmitkowski, M. (2014). The activity of alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase (ALDH) in the sera of patients with brain cancer. Neurochem. Res. 39 (12), 2313–2318. doi:10.1007/s11064-014-1402-3
Jin, Y., Zhang, C., Liu, W., Tang, Y., Qi, H., Chen, H., et al. (2016). The alcohol dehydrogenase gene family in melon (Cucumis melo L): bioinformatic analysis and expression patterns. Front. Plant Sci. 7, 670. doi:10.3389/fpls.2016.00670
Kato-Noguchi, H., and Yasuda, Y. (2007). Effect of low temperature on ethanolic fermentation in rice seedlings. J. Plant Physiol. 164 (8), 1013–1018. doi:10.1016/j.jplph.2006.06.007
Kidokoro, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 27 (9), 922–935. doi:10.1016/j.tplants.2022.01.008
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi:10.1038/s41587-019-0201-4
Koch, M. A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 (10), 1483–1498. doi:10.1093/oxfordjournals.molbev.a026248
Komatsu, S., Kuji, R., Nanjo, Y., Hiraga, S., and Furukawa, K. (2012). Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteomics 77, 531–560. doi:10.1016/j.jprot.2012.09.032
Komatsu, S., Sugimoto, T., Hoshino, T., Nanjo, Y., and Furukawa, K. (2010). Identification of flooding stress responsible cascades in root and hypocotyl of soybean using proteome analysis. Amino Acids 38 (3), 729–738. doi:10.1007/s00726-009-0277-0
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Linskens, H. F., and Schrauwen, J. (1966). Measurement of oxygen tension changes in the style during pollen tube growth. Planta 71 (1), 98–106. doi:10.1007/BF00384646
Longhurst, T., Lee, E., Hinde, R., Brady, C., and Speirs, J. (1994). Structure of the tomato Adh2 gene and Adh2 pseudogenes, and a study of Adh2 gene expression in fruit. Plant Mol. Biol. 26 (4), 1073–1084. doi:10.1007/BF00040690
Manriquez, D., El-Sharkawy, I., Flores, F. B., El-Yahyaoui, F., Regad, F., Bouzayen, M., et al. (2006). Two highly divergent alcohol dehydrogenases of melon exhibit fruit ripening-specific expression and distinct biochemical characteristics. Plant Mol. Biol. 61 (4-5), 675–685. doi:10.1007/s11103-006-0040-9
Mo, F. L., Zhang, N. A., Qiu, Y. W., Meng, L. J., Cheng, M. Z., Liu, J. Y., et al. (2021). Molecular characterization, gene evolution and expression analysis of the F-box gene family in tomato (Solanum lycopersicum). Genes-Basel 12 (3), 417. doi:10.3390/genes12030417
Moummou, H., Tonfack, L. B., Chervin, C., Benichou, M., Youmbi, E., Ginies, C., et al. (2012). Functional characterization of SlscADH1, a fruit-ripening-associated short-chain alcohol dehydrogenase of tomato. J. Plant Physiol. 169 (15), 1435–1444. doi:10.1016/j.jplph.2012.06.007
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77 (3), 367–379. doi:10.1111/tpj.12388
Onodera, W., Hara, N., Aoki, S., Asahi, T., and Sawamura, N. (2023). Phylogenetic tree reconstruction via graph cut presented using a quantum-inspired computer. Mol. Phylogenet Evol. 178, 107636. doi:10.1016/j.ympev.2022.107636
Pan, Y., Xu, X., Li, L., Sun, Q., Wang, Q., Huang, H., et al. (2023). Melatonin-mediated development and abiotic stress tolerance in plants. Front. Plant Sci. 14, 1100827. doi:10.3389/fpls.2023.1100827
Perez-Alonso, M. M., Sanchez-Parra, B., Ortiz-Garcia, P., Santamaria, M. E., Diaz, I., and Pollmann, S. (2021). Jasmonic acid-dependent MYC transcription factors bind to a tandem G-box motif in the YUCCA8 and YUCCA9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 22 (18), 9768. doi:10.3390/ijms22189768
Sasidharan, R., and Voesenek, L. A. (2015). Ethylene-Mediated acclimations to flooding stress. Plant Physiol. 169 (1), 3–12. doi:10.1104/pp.15.00387
Seki, M., Umezawa, T., Urano, K., and Shinozaki, K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10 (3), 296–302. doi:10.1016/j.pbi.2007.04.014
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 (2), 221–227. doi:10.1093/jxb/erl164
Singh, R. K., Sane, V. A., Misra, A., Ali, S. A., and Nath, P. (2010). Differential expression of the mango alcohol dehydrogenase gene family during ripening. Phytochemistry 71 (13), 1485–1494. doi:10.1016/j.phytochem.2010.05.024
Speirs, J., Lee, E., Holt, K., Yong-Duk, K., Steele Scott, N., Loveys, B., et al. (1998). Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiol. 117 (3), 1047–1058. doi:10.1104/pp.117.3.1047
Strommer, J. (2011). The plant ADH gene family. Plant J. 66 (1), 128–142. doi:10.1111/j.1365-313X.2010.04458.x
Su, W., Ren, Y., Wang, D., Su, Y., Feng, J., Zhang, C., et al. (2020). The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation. Bmc Genomics 21 (1), 521. doi:10.1186/s12864-020-06929-9
Tadege, M., Dupuis, I. I., and Kuhlemeier, C. (1999). Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci. 4 (8), 320–325. doi:10.1016/s1360-1385(99)01450-8
Takahashi, H., Greenway, H., Matsumura, H., Tsutsumi, N., and Nakazono, M. (2014). Rice alcohol dehydrogenase 1 promotes survival and has a major impact on carbohydrate metabolism in the embryo and endosperm when seeds are germinated in partially oxygenated water. Ann. Bot. 113 (5), 851–859. doi:10.1093/aob/mct305
Taneja, B., and Mande, S. C. (1999). Conserved structural features and sequence patterns in the GroES fold family. Protein Eng. 12 (10), 815–818. doi:10.1093/protein/12.10.815
Tesniere, C., and Verries, C. (2000). Molecular cloning and expression of cDNAs encoding alcohol dehydrogenases from Vitis vinifera L. during berry development. Plant Sci. 157 (1), 77–88. doi:10.1016/s0168-9452(00)00274-0
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17 (2), 113–122. doi:10.1016/j.copbio.2006.02.002
Wang, F., Kong, W., Wong, G., Fu, L., Peng, R., Li, Z., et al. (2016). AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics 291 (4), 1545–1559. doi:10.1007/s00438-016-1203-2
Wu, C., Zheng, C., Ji, G., and Jiang, P. (2019). Synergistic effects of HSE and LTR elements from hsp70 gene promoter of Ulva prolifera (Ulvophyceae, Chlorophyta) upon temperature induction(1). J. Phycol. 55 (3), 738–743. doi:10.1111/jpy.12854
Xiong, L., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi:10.1105/tpc.000596
Yamauchi, T., Hiraiwa, M., Kobayashi, H., Uda, Y., Miyatake, T., and Tsuji, S. (1990). Molecular cloning of two species of cDNAs for human alpha-N-acetylgalactosaminidase and expression in mammalian cells. Biochem. Biophys. Res. Commun. 170 (1), 231–237. doi:10.1016/0006-291x(90)91264-s
Yeganova, L., Smith, L., and Wilbur, W. J. (2004). Identification of related gene/protein names based on an HMM of name variations. Comput. Biol. Chem. 28 (2), 97–107. doi:10.1016/j.compbiolchem.2003.12.003
Yum, J. S., Starmer, W. T., and Sullivan, D. T. (1991). The structure of the adh locus of Drosophila mettleri: an intermediate in the evolution of the adh locus in the repleta group of Drosophila. Mol. Biol. Evol. 8 (6), 857–867. doi:10.1093/oxfordjournals.molbev.a040692
Zeng, W., Qiao, X., Li, Q., Liu, C., Wu, J., Yin, H., et al. (2020). Genome-wide identification and comparative analysis of the ADH gene family in Chinese white pear (Pyrus bretschneideri) and other Rosaceae species. Genomics 112 (5), 3484–3496. doi:10.1016/j.ygeno.2020.06.031
Zhang, R., Xuan, L., Ni, L., Yang, Y., Zhang, Y., Wang, Z., et al. (2023). ADH gene cloning and identification of flooding-responsive genes in taxodium distichum (L.) rich. Plants (Basel) 12 (3), 678. doi:10.3390/plants12030678
Keywords: ADH, gene family, bioinformatics, stress, tomato
Citation: Zhu Q, Han Y, Yang W, Zhu H, Li G, Xu K and Long M (2023) Genome-wide identification and characterization of ADH gene family and the expression under different abiotic stresses in tomato (Solanum lycopersicum L.). Front. Genet. 14:1186192. doi: 10.3389/fgene.2023.1186192
Received: 14 March 2023; Accepted: 22 August 2023;
Published: 01 September 2023.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Alsamman M. Alsamman, International Center for Agriculture Research in the Dry Areas (ICARDA), EgyptFirat Kurt, Mus Alparslan University, Türkiye
Copyright © 2023 Zhu, Han, Yang, Zhu, Li, Xu and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingdong Zhu, emh1cWluZ2RvbmcyMDIzQDE2My5jb20=
 Qingdong Zhu
Qingdong Zhu Yading Han
Yading Han