
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 31 August 2023
Sec. Cancer Genetics and Oncogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1183528
Background: Lung cancer is one of the most common human malignant diseases. In this study, we aimed to explore the association between IL1RL1 genetic polymorphisms and lung cancer risk in the Chinese Han population.
Methods: We selected and genotyped six SNPs in the IL1RL1 gene using the Agena MassARRAY system in 507 lung cancer patients and 507 healthy controls. The association between IL1RL1 variants and lung cancer risk was assessed using logistic regression to calculate odds ratios (ORs) and 95% confidence intervals (CIs). Multi-factor dimensionality reduction (MDR) was used to analyze the impact of SNP-SNP interactions on the risk of lung cancer.
Results: The results of overall analysis indicated that rs12479210 (T vs. C: OR = 1.42, FDR-p = 0.002; TC vs. CC: OR = 1.70, FDR-p < 0.0001; TT vs. CC: OR = 1.77, FDR-p = 0.032; TT-TC vs. CC: OR = 1.71, FDR-p = 0.001; additive: OR = 1.44, FDR-p = 0.001) and rs1420101 (T vs. C: OR = 1.31, FDR-p = 0.036; TT-TC vs. CC: OR = 1.42, FDR-p = 0.031; additive: OR = 1.30, FDR-p = 0.030) were associated with an increased the risk of lung cancer among the Chinese Han population. Stratified analysis also found the association between these two SNPs and lung cancer risk. However, there were no significant association observed between the other four SNPs (rs3771180, rs3771175, rs10208293, and rs10197862) in IL1RL1 and lung cancer risk. Furthermore, MDR analysis showed that rs12479210 was the best single model with the highest testing accuracy (0.566) and perfect CVC (10/10) for predicting lung cancer risk. The expression level of the IL1RL1 gene is lower in lung cancer tissue than normal tissue, and there are significant differences in the expression levels of IL1RL1 between rs12479210 and rs1420101 genetypes in lung cancer tissue (p < 0.05).
Conclusion: Our findings suggest that IL1RL1 genetic variants (rs12479210 and rs1420101) are associated with an increased lung cancer risk in the Chinese Han population. These risk variants may serve as biomarkers for the prevention and treatment of lung cancer.
The incidence and mortality rates of lung cancer are increasing globally (Bade and Dela Cruz, 2020). In 2020, lung cancer was the second most commonly diagnosed cancer and the leading cause of cancer death, with an estimated 2.2 million new cancer cases (11.4%) and 1.8 million deaths (18.0%) (Sung et al., 2021). The incidence and mortality rates of lung cancer are approximately two times higher in men than in women. In China, lung cancer is the leading cause of death in both males and females, with an estimated 870,982 new cases and 766,898 deaths expected in 2022 (Xia et al., 2022). Lung cancer is a complex pathological process influenced by multiple factors, including cigarette smoking, which is reported in about one-third of adults worldwide and has a strong relationship between cigarette smoke exposure and lung cancer has been proven (Taucher et al., 2022). Other nontobacco risk factors include poor diet, occupational exposures, air pollution, chronic lung disease, and lung infections (Malhotra et al., 2016; Huang et al., 2021; Wei et al., 2021; Moayedi-Nia et al., 2022). Despite recent advances in treatments, lung cancer remains a disease with a poor prognosis, and it is often not diagnosed until the cancer is at an advanced stage. Therefore, early diagnosis of lung cancer is crucial. Genetic factors have been shown to play a crucial role in the development of lung cancer. In a Spanish study describing the characteristics of female lung cancer patients, 42.5% of patients had a family history of tumors (Garrido et al., 2019). Single nucleotide polymorphisms (SNPs) of candidate genes have also been found to be associated with lung cancer risk (Khadhraoui et al., 2020; Chongtham et al., 2022; Zhang et al., 2022).
The Interleukin 1 receptor like 1 (IL1RL1) gene, also known as ST2, ST2L, and IL33R, encodes a protein that belongs to the family of IL1R, and it is only known ligand is IL-33 (13). The IL-33/ST2L signal induces transcription of downstream inflammatory and anti-inflammatory genes by activating diverse intracellular kinases and factors to mount an adequate immune response (14). The IL-33/ST2 axis affects tumor growth by regulating mitophagy in macrophages and reprogramming their polarization (15). Xu et al. (16) demonstrated that the IL33-IL1RL1 pathway influenced tumor growth by regulating autophagy and reprogramming of macrophages. ST2 was significantly downregulated in human lung cancer tissues and cells compared to normal lung tissues and cells (17). Furthermore, Wang et al. (18) showed that the IL33-IL1RL1 signaling pathway is involved in the growth and metastasis of lung cancer. However, little is known about the correlation between IL1RL1 polymorphisms and lung cancer.
Therefore, in this study we aimed to assess the possible association between six IL1RL1 gene polymorphisms (rs12479210, rs3771180, rs1420101, rs3771175, rs10208293, and rs10197862) and susceptibility to lung cancer in the Chinese Han population. This research may provide a new biomarker for the prevention and development of lung cancer.
The sample size of this study was estimated using G*Power (3.1.9.7) software with the following parameters: tails = two, effect size = 0.2, a = 0.05, power = 0.889, and allocation ratio = 1. A total of 1014 participants were randomly enrolled from Xuanwei city, including 507 newly histologically diagnosed lung cancer patients in the case group. Patients who underwent chemotherapy, surgery, radiotherapy, had other tumors or inflammatory diseases were excluded. The control group consisted of 507 healthy individuals recruited from medical examination centers, with no history of cancers, lung dysfunction, or related diseases. Basic information, such as age, gender, body mass index (BMI), smoking and drinking status, and clinical test indicators (tumor type and stage), were collected from questionnaires and clinical data.
First, we searched the physical location (2:102928023-102968497) of the IL1RL1 gene (GRCh37. p13) through the NCBI database (https://www.ncbi.nlm.nih.gov/gene/). Second, we downloaded ped and info files of mutation sites in IL1RL1 in the Chinese Han Beijing (CHB) population from 1000 Genome Project through VCF to PED Converter window (http://grch37.ensembl.org/Homo Sapiens/Tools/VcftoPed), and we used Haploview software to screen SNPs with Hardy-Weinberg equilibrium (HWE) > 0.05, minor allele frequency (MAF) > 0.05, and min genotype frequency >75%, r2 > 0.8. Finally, based on previous studies associated with respiratory diseases (Ferreira et al., 2011; Shrine et al., 2019; Sun et al., 2019; Wu et al., 2021; Li et al., 2022; Rojo-Tolosa et al., 2023) and primer design, a total of six SNPs in the IL1RL1 gene were randomly selected (Supplementary Table S1). The potential role of SNPs was predicted using RegulomeDB (https://regulomedb.org/), HaploReg (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), and Genotype-Tissue Expression (GTEx) Portal databases (https://gtexportal.org). Peripheral blood samples were collected from each subject, and genomic DNA was extracted using the GoldMag DNA extraction Kit (GoldMag Co. Ltd., Xi’an, China) according to the manufacturer’s instructions. The concentration and purity of DNA were measured by a spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA), with an OD260/OD280 ratio between 1.7 and 2.0 and a concentration greater than 20 ng/μL. The primers for polymerase chain reaction (PCR) and unique base extension for the six SNPs in IL1RL1 were designed by Agena Bioscience Assay Design Suite version 2.0 software and synthesized by Bioengineering (Shanghai Co., Ltd.). The primer sequence is shown in Table 1. Genotyping of IL1RL1 variants was conducted by the Agena MassARRAY system (Agena Bioscience, San Diego, CA, USA), and data management and analysis were performed using AgenaTyper 4.0 software.
The differences between six SNPs in IL1RL1 and the expression level of IL1RL1 in lung cancer tissues were analyzed using GTEx Portal database (https://gtexportal.org/). The expression level of IL1RL1 in normal and lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) tissues was analyzed using GEPIA online analysis software based on TCGA database (http://gepia.cancer-pku.cn/). We predicted the signaling pathways involved in the IL1RL1 gene through the PathCards database (https://pathcards.genecards.org/).
Statistical analysis was performed using IBM SPSS software (version 25.0, SPSS Inc., Chicago, Illinois, USA) and PLINK software (version 1.07). The distribution of age and gender in the case and control group was evaluated using Student’s t-test and Pearson’s chi-square test, respectively. The p-value of HWE in controls was calculated by the chi-square test. The association between IL1RL1 variants and lung cancer risk was assessed using odds ratio (OR) and 95% confidence interval (95% CI) calculated by logistic regression under multiple genetic models (allele, codominant, dominant, recessive, and additive). To reduce the influence of confounding factors (age, gender, and smoking status) on the statistical results, stratified analyses were performed, and forest plots of stratified results were drawn using Sangerbox software (version 3.0). The Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) values were calculated using SNPStats software to select the optimal model. False discovery rate (FDR) correction (q = p *(n/k); n is the total number of p-values; k is the order in which p-values are sorted from smallest to largest) was performed for p-values to reduce false positives in the results. The multifactor dimensionality reduction (MDR) method was used to further evaluate the effect of SNP-SNP interactions on lung cancer risk by MDR 3.0.2 software. To determine the reliability of significant correlation results, statistical power and false positive report probability (FPRP) values of SNPs were calculated using the Excel spreadsheet offered on Wacholder’s website (Wacholder et al., 2004). All statistical analyses were two-sided, and p < 0.05 was considered statistically significant.
Table 2 describes the basic characteristics of the study participants. This study included 507 patients diagnosed with lung cancer (average age: 61.12 ± 10.03 years, 350 males and 157 females) and 507 healthy controls (average age: 60.97 ± 9.25 years, 349 males and 158 females). The results showed that the control and case groups were well-matched in terms of age (p = 0.810) and gender (p = 0.946) distribution. However, there were significant differences in the distribution of BMI, smoking, and alcohol consumption between the case group and the control group (p < 0.05). Therefore, in order to reduce the impact of these confounding factors on association analysis results, we conducted correction and stratified analysis.
This research selected six SNPs (rs12479210, rs3771180, rs1420101, rs3771175, rs10208293, and rs10197862) in the IL1RL1 gene as candidate SNPs. As shown in Table 3, rs3771175 is located in the 3′-UTR of IL1RL1 gene, and the rest loci are located in the intron region. RegulomeDB and HaploReg analysis predicted the potential functions of these SNPs. The call rates of all SNPs genotype were greater than 95%, and were consistent with HWE, indicating that our samples satisfied random distribution and the SNP genotyping technique was reliable (p > 0.05, Table 3).
Table 4 presents the correlation between IL1RL1 variants and lung cancer susceptibility in multiple genetic models. The results showed that rs12479210 was associated with an increased risk of lung cancer under the allele (T vs. C: OR = 1.42, 95% CI: 1.18-1.71, p = 0.0001, FDR-p = 0.002), codominant (TC vs. CC: OR = 1.70, 95% CI: 1.30-2.22, p < 0.0001, FDR-p < 0.0001; TT vs. CC: OR = 1.77, 95% CI: 1.17-2.69, p = 0.007, FDR-p = 0.032), dominant (TT-TC vs. CC: OR = 1.71, 95% CI: 1.33-2.21, p = 0.0001, FDR-p = 0.001), and additive models (OR = 1.44, 95% CI: 1.20-1.75, p = 0.0001, FDR-p = 0.001). The dominant model was the optimal model for rs12479210 with the minimum AIC (1393.2) and BIC (1412.9) values. Additionally, rs1420101 was significantly associated with an increased risk of lung cancer under the allele (T vs. C: OR = 1.31, 95% CI: 1.09-1.57, p = 0.005, FDR-p = 0.036), dominant (TT-TC vs. CC: OR = 1.42, 95% CI: 1.10-1.82, p = 0.006, FDR-p = 0.031), and additive models (OR = 1.30, 95% CI: 1.08-1.56, p = 0.005, FDR-p = 0.030). The additive model was the optimal model for rs1420101 with the minimum AIC (1404.5) and BIC (1424.1) values. Furthermore, the statistical power and FPRP of rs12479210 and rs1420101 were analyzed under multiple genetic models (Table 5), and the results showed that the correlation between these two SNPs and lung cancer risk was reliable. However, there were no significant differences between the other four SNPs (rs3771180, rs3771175, rs10208293, and rs10197862) in IL1RL1 and lung cancer risk.
To reduce the impact of confounding factors on the analysis results, we conducted stratified analysis based on mean age (>60 and ≤60), gender (male and female), and smoking status (yes and no). Gender stratified analysis (Figure 1 and Supplementary Table S2) showed that rs12479210 (T vs. C: OR = 1.49, 95% CI: 1.20-1.86, p < 0.0001, FDR-p = 0.007; CT vs. CC: OR = 1.62, 95% CI: 1.18-2.23, p = 0.003, FDR-p = 0.021; TT vs. CC: OR = 2.20, 95% CI: 1.31-3.70, p = 0.003, FDR-p = 0.026; CT-TT vs. CC: OR = 1.71, 95% CI: 1.26-2.32, p = 0.001, FDR-p = 0.006; additive: OR = 1.53, 95% CI: 1.22-1.93, p < 0.0001, FDR-p = 0.011) and rs1420101 (T vs. C: OR = 1.40, 95% CI: 1.12-1.75, p = 0.003, FDR-p = 0.018; TT vs. CC: OR = 1.98, 95% CI: 1.20-3.25, p = 0.007, FDR-p = 0.032; TC-TT vs. CC: OR = 1.49, 95% CI: 1.10-2.01, p = 0.010, FDR-p = 0.040; additive: OR = 1.40, 95% CI: 1.12-1.75, p = 0.003, FDR-p = 0.017) were associated with an increased risk of lung cancer in males. The dominant model and the additive model were the optimal models for rs12479210 (AIC = 960.1 and BIC = 973.7) and rs1420101 (AIC = 964.8 and BIC = 978.5) with the minimum AIC and BIC values, respectively. However, no significant association was found between IL1RL1 polymorphisms (rs12479210 and rs1420101) and the risk of lung cancer in females.

FIGURE 1. Association between rs12479210 and rs1420101 and lung cancer risk stratified by gender. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; FDR, false discovery rate; AIC, Akaike Information Criterion; BIC, Bayesian Information Criterion; p < 0.05 indicates statistical significance.
Age stratified analysis (Figure 2 and Supplementary Table S2) revealed that rs12479210 (T vs. C: OR = 1.60, 95% CI: 1.21-2.12, p = 0.001, FDR-p = 0.020; TT vs. CC: OR = 2.59, 95% CI: 1.33-5.08, p = 0.005, FDR-p = 0.048; CT-TT vs. CC: OR = 1.86, 95% CI: 1.26-2.73, p = 0.002, FDR-p = 0.019; additive: OR = 1.65, 95% CI: 1.23-2.22, p = 0.001, FDR-p = 0.031) was associated with an increased risk of lung cancer in participants aged >60. The additive model was the optimal model for rs12479210 with the minimum AIC (602.7) and BIC (619.1) values in participants aged >60. However, no significant association was found between IL1RL1 polymorphisms (rs12479210 and rs1420101) and the risk of lung cancer in participants aged ≤60.
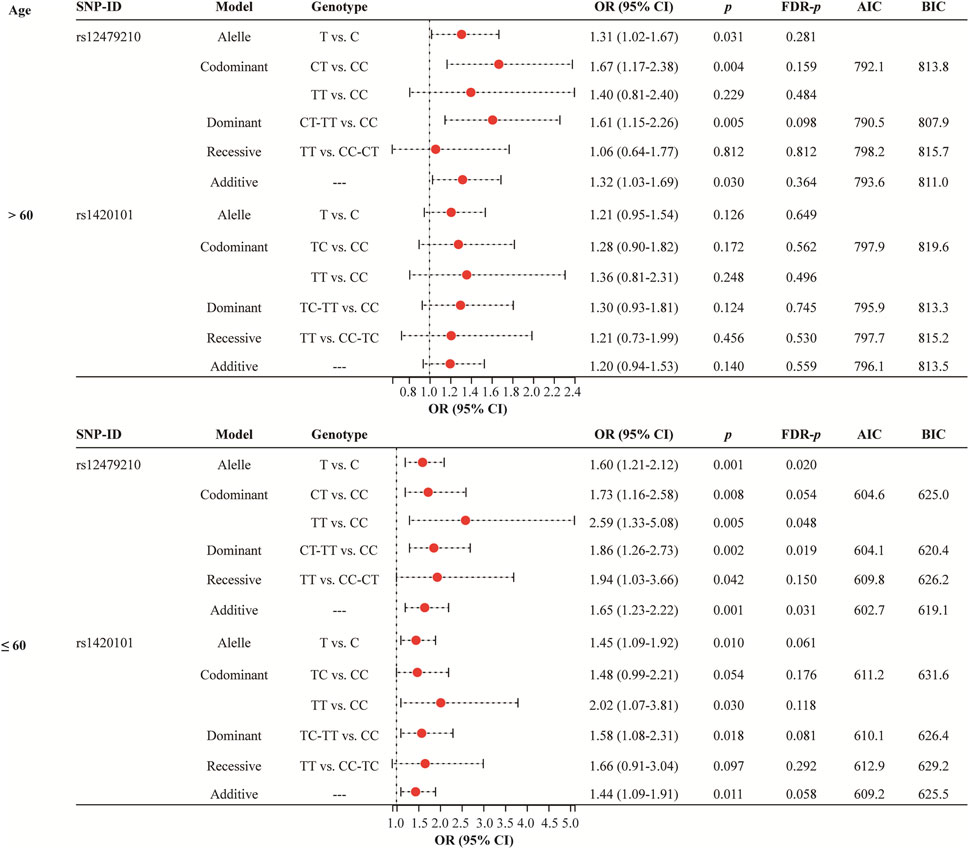
FIGURE 2. Association between rs12479210 and rs1420101 and lung cancer risk stratified by mean age. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; FDR, false discovery rate; AIC, Akaike Information Criterion; BIC, Bayesian Information Criterion; p < 0.05 indicates statistical significance.
Smoking status stratified analysis (Figure 3 and Supplementary Table S2) indicated that rs12479210 (CT vs. CC: OR = 2.25, 95% CI: 1.48-3.42, p < 0.0001, FDR-p = 0.005; CT-TT vs. CC: OR = 2.02, 95% CI: 1.36-2.99, p < 0.0001, FDR-p = 0.009) was associated with an increased risk of lung cancer in somking subjects. However, no significant association was found between IL1RL1 polymorphisms (rs12479210 and rs1420101) and risk of lung cancer in non-smoking. No significant association was found between the four SNPs (rs3771180, rs3771175, rs10208293, and rs10197862) in IL1RL1 and risk of lung cancer in stratified analysis (Supplementary Table S2).
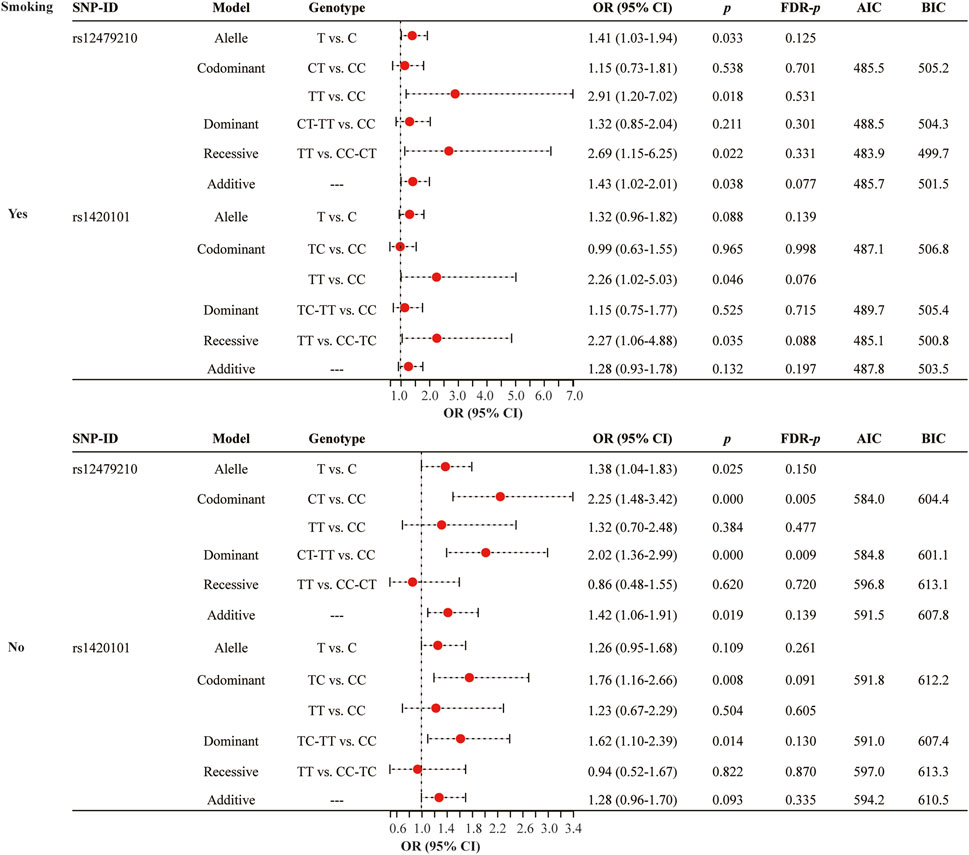
FIGURE 3. Association between rs12479210 and rs1420101 and lung cancer risk stratified by smoking status. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; FDR, false discovery rate; AIC, Akaike Information Criterion; BIC, Bayesian Information Criterion; p < 0.05 indicates statistical significance.
Moreover, we conducted the MDR analysis to assess the impact of SNP-SNP interactions on risk of lung cancer. The dendrogram of SNP-SNP interactions is displayed in Figure 4, and the results showed a strong negative correlation between rs12479210 and rs1420101 in terms of their impact on lung cancer risk. Moreover, the one loci model (rs12479210) was the best single model with the highest testing accuracy (0.566) and CVC (10/10) to predict lung cancer risk (Table 6).
We found statistically significant differences between six SNPs in IL1RL1 and the expression level of IL1RL1 in lung cancer tissues using GTEx Portal database (p < 0.001, Figure 5A). In addition, we found that the expression level of IL1RL1 was significantly different between normal and LUAD and LUSC tissues (p < 0.001, Figure 5B). We predicted the signaling pathways involved in the IL1RL1 gene through the PathCards database (Figure 5C), and found that IL1RL1 was mainly involved in 11 signaling pathways, such as IL-1 family signaling pathways, Th2 differentiation, cytokine signaling in immune system, and innate lymphoid cells differentiation.
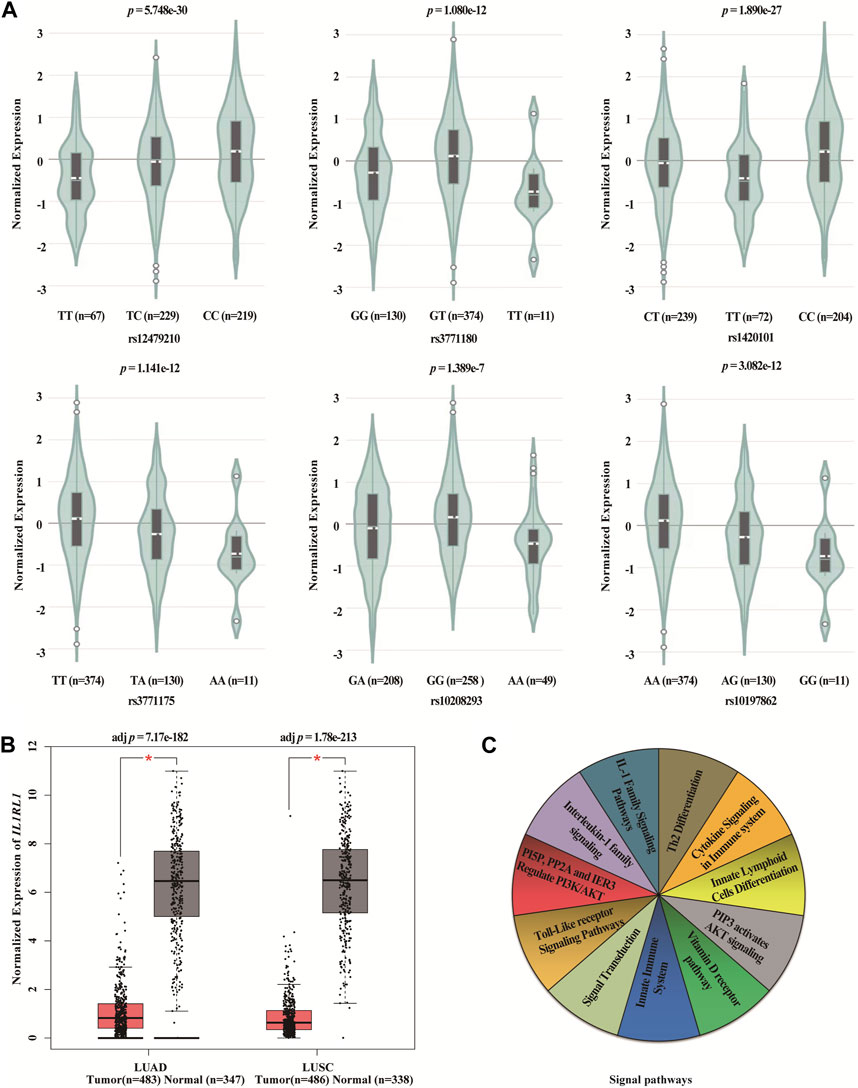
FIGURE 5. Bioinformatics analysis of IL1RL1 and polymorphisms (A) The differences between six SNPs in IL1RL1 and the expression level of IL1RL1 in lung cancer tissues. (B) The expression level of IL1RL1 in normal and LUAD and LUSC tissues. (C) The signaling pathways involved in the IL1RL1 gene. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; *p < 0.001, p < 0.05 indicates statistical significance.
This study is the first to explore IL1RL1 polymorphisms (rs12479210, rs3771180, rs1420101, rs3771175, rs10208293, and rs10197862) and lung cancer susceptibility in the Chinese Han population. The results showed that rs12479210 and rs1420101 in IL1RL1 were associated with an increased risk of lung cancer in the Chinese Han population. Moreover, bioinformatics analysis results revealed that the expression level of IL1RL1 was significantly different between normal and LUAD and LUSC tissues, and there were significant differences between six SNPs in IL1RL1 and the expression level of IL1RL1 in lung cancer tissues. These results suggest that IL1RL1 may be involved in the pathogenesis of lung cancer.
The IL1RL1 gene is located on chromosome 2q12.1 and contains 13 exons, also known as IL33R, ST2, and ST2L. An animal study has illustrated that IL1RL1 deficiency could reduce lung myeloid cell infiltration and repeatedly activate macrophage function (Dagher et al., 2020). Liu et al. have revealed that hypoxia significantly increased IL1RL1 expression by human pulmonary arterial endothelial cells in vitro (Liu et al., 2018). The expression levels of IL-33 and ST2 were found to be significantly downregulated in both adenocarcinoma and squamous cell carcinoma of the lung compared to adjacent normal lung tissues (Yang et al., 2018). The secretion of ST2 inhibited tumor growth of lung cancer patients, and low ST2 was associated with worse overall survival (Tzeng et al., 2018; Gezelius et al., 2022). sST2, a soluble form of ST2, has been shown to involve in the inflammatory tumor microenvironment and the progression of non-small cell lung cancer (Hong et al., 2019; Chang et al., 2020). A prvious study found that the IL-33/ST2 axis enhances lung-resident CD14+ monocyte function in patients with non-small cell lung cancer (Wang et al., 2023). In this study, the expression level of IL1RL1 was found to be significantly different between normal and LUAD and LUSC tissues based on TCGA database, and IL1RL1 was mainly involved in IL-1 family signaling pathways, cytokine signaling in immune system, and innate lymphoid cells differentiation. However, this study did not explore the specific mechanism of IL1RL1 in the occurrence and development of lung cancer.
Furthermore, IL1RL1 genetic variants have been reported to be associated with susceptibility to respiratorydiseases. A genome-wide association study indicated that rs13431828 and rs1041973 in IL1RL1 were associated with childhood asthma susceptibility in the Mexican population (Wu et al., 2010). Faber et al. found a genetic association between rs1921622 in IL1RL1 and disease severity in respiratory syncytial virus bronchiolitis (Faber et al., 2012). IL1RL1 polymorphisms rs72823628, rs950881 and rs3771175 were found to be associated with a reduced risk of allergic rhinitis risk in the Chinese Han population (Li et al., 2022). However, there are few studies on the correlation between IL1RL1 polymorphisms and lung cancer susceptibility. In this study, we explore the association betweewn IL1RL1 polymorphisms and lung cancer susceptibility in the Chinese Han population. This study reports for the first time a significant correlation between IL1RL1 polymorphisms (rs12479210 and rs1420101) and risk of lung cancer in the Chinese Han population. A previous study indicated that IL1RL1 rs1420101 was associated with high exhaled nitric oxide and blood eosinophil differentials among Japanese patients with asthma (Inoue et al., 2017). In addition, rs1420101 was strongly associated with IL1RL1 methylation and serum IL1RL1-a levels in asthma (Dijk et al., 2018). Bioinformatics analysis results revealed that there were significant differences between rs12479210 and rs1420101 and the expression level of IL1RL1 in lung cancer tissue. Therefore, we speculateed that these two SNPs (rs12479210 and rs1420101) may affect the risk of lung cancer by regulating the expression of IL1RL1.
In addition, smoking is indisputably linked to lung cancer, yet only a small fraction of smokers develops this disease (Schuller, 2019). A recent report showed that women have a higher risk of developing lung cancer upon smoking than men, and the odds ratio to develop lung cancer was almost three times greater for women than for men (Stapelfeld et al., 2020). The aging of the population is also one of the main reasons for the rise of the incidence rate of lung cancer (Jakobsen et al., 2021). The results of stratification analysis have shown that rs12479210 was related to the susceptibility of lung cancer in different subgroups, while rs1420101 was not significantly associated with lung cancer risk in males, age >60 years old, and smokers. Therefore, we speculated that the impacts of age, gender, and smoking on the susceptibility to lung cancer were stronger than rs1420101. These genetic variants of IL1RL1 may provide new biomarkers for early diagnosis of lung cancer, and it helps us to have a deeper understanding of the pathogenesis of lung cancer.
Of course, this study has some limitations. First, the significance of IL1RL1 polymorphisms and lung cancer susceptibility was only performed in the Chinese Han population. There are differences in genetic polymorphisms among different ethnic groups, a larger sample size case-control study can be conducted to further confirm the results of this study in different populations. Second, the occurrence of lung cancer is closely related to environmental factors, but this study lacks some information on BMI and alcohol consumption. Third, lung cancer is a heterogeneous disease, and different subtypes of lung cancer may be associated with different genetic variations. Therefore, in future research, we will collect more comprehensive data, including BMI and alcohol consumption, lung cancer subtypes, etc., to better evaluate the association between IL1RL1 polymorphisms and lung cancer risk. Finally, the specific molecular mechanism of IL1RL1 genetic variants in the occurrence and development of lung cancer risk is unclear. Therefore, in vitro experiments or animal models can be used to investigate the impact of these genetic variants on IL1RL1 expression and cell proliferation, apoptosis, and other biological processes. Moreover, the distribution of these genetic variants in lung cancer patients can be analyzed and correlated with clinical data such as tumor type, staging, treatment response, etc., to assess their potential clinical utility.
In conclusion, this study suggests that IL1RL1 genetic variants (rs12479210 and rs1420101) contribute to the susceptibility to lung cancer in the Chinese Han population. These risk variants may be used as biomarkers for the prevention and treatment of lung cancer. However, further research is needed to confirm these findings and to better understand the molecular mechanisms underlying the association between IL1RL1 polymorphisms and lung cancer risk.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by the Ethics committee of Xuanwei City. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
QL drafted the manuscript; CZ performed the DNA extraction and genotyping; YC performed the data analysis; XY and WC performed the sample collection and information recording; KH revised the manuscript; MC conceived and supervised the study. All authors contributed to the article and approved the submitted version.
This work was supported by the Applied Basic Research Foundation of Yunnan Province (CN) (grant number: 2017FE468 (−125) and 202001AY070001-111 and KUST-KH2022028Y), the Open Project of the Clinical Medicine Center of the First People’s Hospital of Yunnan Province (grant number: 2021LCZXXF-XY12 and 2022LCZXKF-XY04), CAMS Innovation Fund for Medical Sciences (CIFMS) (grant number: 2016-I2M), and Yunnan Province Clinical Center for Hematologic Disease and Yunnan Province Clinical Research Center for Hematologic Disease.
We would like to express our sincere gratitude to all participants who provided blood samples for this study, as well as the professionals responsible for blood collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1183528/full#supplementary-material
AIC, Akaike Information Criterion;; BIC, Bayesian Information Criterion; BMI, body mass index; CHB, Chinese Han Beijing; CI, confidence interval; FDR, false discovery rate; FPRP, false positive report probability; GTEx, Genotype-Tissue Expression; HWE, Hardy-Weinberg equilibrium; IL1RL1, interleukin 1 receptor like 1; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MAF, minor allele frequency; MDR, multifactor dimensionality reduction; OR, odds ratio; SNPs, single nucleotide polymorphisms.
Bade, B. C., and Dela Cruz, C. S. (2020). Lung cancer 2020: epidemiology, etiology, and prevention. Clin. chest Med. 41 (1), 1–24. doi:10.1016/j.ccm.2019.10.001
Chang, C. P., Hu, M. H., Hsiao, Y. P., and Wang, Y. C. (2020). ST2 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 1240, 83–93. doi:10.1007/978-3-030-38315-2_7
Chongtham, J., Pandey, N., Sharma, L. K., Mohan, A., and Srivastava, T. (2022). SNP rs9387478 at ROS1-DCBLD1 locus is significantly associated with lung cancer risk and poor survival in Indian population. Asian Pac. J. cancer Prev. APJCP 23 (10), 3553–3561. doi:10.31557/APJCP.2022.23.10.3553
Dagher, R., Copenhaver, A. M., Besnard, V., Berlin, A., Hamidi, F., Maret, M., et al. (2020). IL-33-ST2 axis regulates myeloid cell differentiation and activation enabling effective club cell regeneration. Nat. Commun. 11 (1), 4786. doi:10.1038/s41467-020-18466-w
Dijk, F. N., Xu, C., Melén, E., Carsin, A. E., Kumar, A., Nolte, I. M., et al. (2018). Genetic regulation of IL1RL1 methylation and IL1RL1-a protein levels in asthma. Eur. Respir. J. 51 (3), 1701377. doi:10.1183/13993003.01377-2017
Faber, T. E., Schuurhof, A., Vonk, A., Koppelman, G. H., Hennus, M. P., Kimpen, J. L., et al. (2012). IL1RL1 gene variants and nasopharyngeal IL1RL-a levels are associated with severe RSV bronchiolitis: A multicenter cohort study. PloS one 7 (5), e34364. doi:10.1371/journal.pone.0034364
Ferreira, M. A., McRae, A. F., Medland, S. E., Nyholt, D. R., Gordon, S. D., Wright, M. J., et al. (2011). Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur. J. Hum. Genet. EJHG 19 (4), 458–464. doi:10.1038/ejhg.2010.191
Garrido, P., Viñolas, N., Isla, D., Provencio, M., Majem, M., Artal, A., et al. (2019). Lung cancer in Spanish women: the WORLD07 project. Eur. J. cancer care 28 (1), e12941. doi:10.1111/ecc.12941
Gezelius, E., Bendahl, P. O., Gallo, W., de Oliveira, K. G., Ek, L., Bergman, B., et al. (2022). Circulating levels of the cardiovascular biomarkers ST2 and adrenomedullin predict outcome within a randomized phase III lung cancer trial (RASTEN). Cancers 14 (5), 1307. doi:10.3390/cancers14051307
Hong, J., Kim, S., and Lin, P. C. (2019). Interleukin-33 and ST2 signaling in tumor microenvironment. J. interferon and cytokine Res. 39 (1), 61–71. doi:10.1089/jir.2018.0044
Huang, Y., Zhu, M., Ji, M., Fan, J., Xie, J., Wei, X., et al. (2021). Air pollution, genetic factors, and the risk of lung cancer: A prospective study in the UK biobank. Am. J. Respir. Crit. care Med. 204 (7), 817–825. doi:10.1164/rccm.202011-4063OC
Inoue, H., Ito, I., Niimi, A., Matsumoto, H., Oguma, T., Tajiri, T., et al. (2017). Association of interleukin 1 receptor-like 1 gene polymorphisms with eosinophilic phenotype in Japanese adults with asthma. Respir. Investig. 55 (6), 338–347. doi:10.1016/j.resinv.2017.08.006
Jakobsen, E., Olsen, K. E., Bliddal, M., Hornbak, M., Persson, G. F., and Green, A. (2021). Forecasting lung cancer incidence, mortality, and prevalence to year 2030. BMC cancer 21 (1), 985. doi:10.1186/s12885-021-08696-6
Khadhraoui, C., Kaabachi, W., Tritar, F., Daghfous, H., Hamzaoui, K., and Hamzaoui, A. (2020). Association of BTLA rs1982809 polymorphism with lung cancer risk in Tunisian population. Int. J. immunogenetics 47 (6), 554–562. doi:10.1111/iji.12491
Li, Z., Ren, J., Zhang, J., Wang, X., Liu, Y., and Wang, Q. (2022). Association between IL1RL1 gene polymorphisms and allergic rhinitis risk in the Chinese Han population. J. Clin. laboratory analysis 36 (11), e24747. doi:10.1002/jcla.24747
Liu, J., Wang, W., Wang, L., Chen, S., Tian, B., Huang, K., et al. (2018). IL-33 initiates vascular remodelling in hypoxic pulmonary hypertension by up-regulating HIF-1α and VEGF expression in vascular endothelial cells. EBioMedicine 33, 196–210. doi:10.1016/j.ebiom.2018.06.003
Malhotra, J., Malvezzi, M., Negri, E., La Vecchia, C., and Boffetta, P. (2016). Risk factors for lung cancer worldwide. Eur. Respir. J. 48 (3), 889–902. doi:10.1183/13993003.00359-2016
Moayedi-Nia, S., Pasquet, R., Siemiatycki, J., Koushik, A., and Ho, V. (2022). Occupational exposures and lung cancer risk-an analysis of the CARTaGENE study. J. Occup. Environ. Med. 64 (4), 295–304. doi:10.1097/JOM.0000000000002481
Rojo-Tolosa, S., Sánchez-Martínez, J. A., Pineda-Lancheros, L. E., Gálvez-Navas, J. M., González-Gutiérrez, M. V., Jiménez-Gálvez, G., et al. (2023). Influence of genetics on the response to omalizumab in patients with severe uncontrolled asthma with an allergic phenotype. Int. J. Mol. Sci. 24 (8), 7029. doi:10.3390/ijms24087029
Schuller, H. M. (2019). The impact of smoking and the influence of other factors on lung cancer. Expert Rev. Respir. Med. 13 (8), 761–769. doi:10.1080/17476348.2019.1645010
Shrine, N., Portelli, M. A., John, C., Soler Artigas, M., Bennett, N., Hall, R., et al. (2019). Moderate-to-severe asthma in individuals of European ancestry: A genome-wide association study. Lancet Respir. Med. 7 (1), 20–34. doi:10.1016/S2213-2600(18)30389-8
Stapelfeld, C., Dammann, C., and Maser, E. (2020). Sex-specificity in lung cancer risk. Int. J. cancer 146 (9), 2376–2382. doi:10.1002/ijc.32716
Sun, Y., Wei, X., Deng, J., Zhang, J., He, Z., Yang, M., et al. (2019). Association of IL1RL1 rs3771180 and TSLP rs1837253 variants with asthma in the Guangxi Zhuang population in China. J. Clin. laboratory analysis 33 (6), e22905. doi:10.1002/jcla.22905
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Taucher, E., Mykoliuk, I., Lindenmann, J., and Smolle-Juettner, F. M. (2022). Implications of the immune landscape in COPD and lung cancer: smoking versus other causes. Front. Immunol. 13, 846605. doi:10.3389/fimmu.2022.846605
Tzeng, H. T., Su, C. C., Chang, C. P., Lai, W. W., Su, W. C., and Wang, Y. C. (2018). Rab37 in lung cancer mediates exocytosis of soluble ST2 and thus skews macrophages toward tumor-suppressing phenotype. Int. J. Cancer 143 (7), 1753–1763. doi:10.1002/ijc.31569
Wacholder, S., Chanock, S., Garcia-Closas, M., El Ghormli, L., and Rothman, N. (2004). Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 96 (6), 434–442. doi:10.1093/jnci/djh075
Wang, L., Mei, X., Liu, X., Guo, L., Yang, B., and Chen, R. (2023). The interleukin-33/ST2 Axis enhances lung-resident CD14+ monocyte function in patients with non-small cell lung cancer. Immunol. Investig. 52 (1), 67–82. doi:10.1080/08820139.2022.2130075
Wei, X., Zhu, C., Ji, M., Fan, J., Xie, J., Huang, Y., et al. (2021). Diet and risk of incident lung cancer: A large prospective cohort study in UK biobank. Am. J. Clin. Nutr. 114 (6), 2043–2051. doi:10.1093/ajcn/nqab298
Wu, H., Romieu, I., Shi, M., Hancock, D. B., Li, H., Sienra-Monge, J. J., et al. (2010). Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J. allergy Clin. Immunol. 125 (2), 321–327. doi:10.1016/j.jaci.2009.09.007
Wu, M., Zheng, X., Huang, J., and Hu, X. (2021). Association of IL33, IL1RL1, IL1RAP polymorphisms and asthma in Chinese han children. Front. Cell Dev. Biol. 9, 759542. doi:10.3389/fcell.2021.759542
Xia, C., Dong, X., Li, H., Cao, M., Sun, D., He, S., et al. (2022). Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med. J. 135 (5), 584–590. doi:10.1097/CM9.0000000000002108
Yang, M., Feng, Y., Yue, C., Xu, B., Chen, L., Jiang, J., et al. (2018). Lower expression level of IL-33 is associated with poor prognosis of pulmonary adenocarcinoma. PloS one 13 (3), e0193428. doi:10.1371/journal.pone.0193428
Keywords: IL1RL1, genetic, polymorphism, lung cancer, case-control study
Citation: Li Q, Zhang C, Cheng Y, Yang X, Chen W, He K and Chen M (2023) IL1RL1 polymorphisms rs12479210 and rs1420101 are associated with increased lung cancer risk in the Chinese Han population. Front. Genet. 14:1183528. doi: 10.3389/fgene.2023.1183528
Received: 10 March 2023; Accepted: 17 August 2023;
Published: 31 August 2023.
Edited by:
Anton A. Buzdin, European Organisation for Research and Treatment of Cancer, BelgiumReviewed by:
Itty Sethi, University of Jammu, IndiaCopyright © 2023 Li, Zhang, Cheng, Yang, Chen, He and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingwei Chen, bWluZ3dlaV9fY2hlbkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.