
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Genet. , 20 March 2023
Sec. Evolutionary and Population Genetics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1179564
This article is a correction to:
Population genetics analysis of Tolai hares (Lepus tolai) in Xinjiang, China using genome-wide SNPs from SLAF-seq and mitochondrial markers
 Miregul Mamat†
Miregul Mamat† Wenjuan Shan*†
Wenjuan Shan*† Pengcheng Dong†
Pengcheng Dong† Shiyu Zhou
Shiyu Zhou Peng Liu
Peng Liu Yang Meng
Yang Meng Wenyue Nie
Wenyue Nie Peichen Teng
Peichen Teng Yucong Zhang
Yucong ZhangA Corrigendum on
Population genetics analysis of Tolai hares (Lepus tolai) in Xinjiang, China using genome-wide SNPs from SLAF-seq and mitochondrial markers
by Mamat M, Shan W, Dong P, Zhou S, Liu P, Meng Y, Nie W, Teng P and Zhang Y (2022). Front. Genet. 13:1018632. doi: 10.3389/fgene.2022.1018632
In the published article, there was an error in Figure 3C as published. A “B” is missing from the note representing the BRJ population.
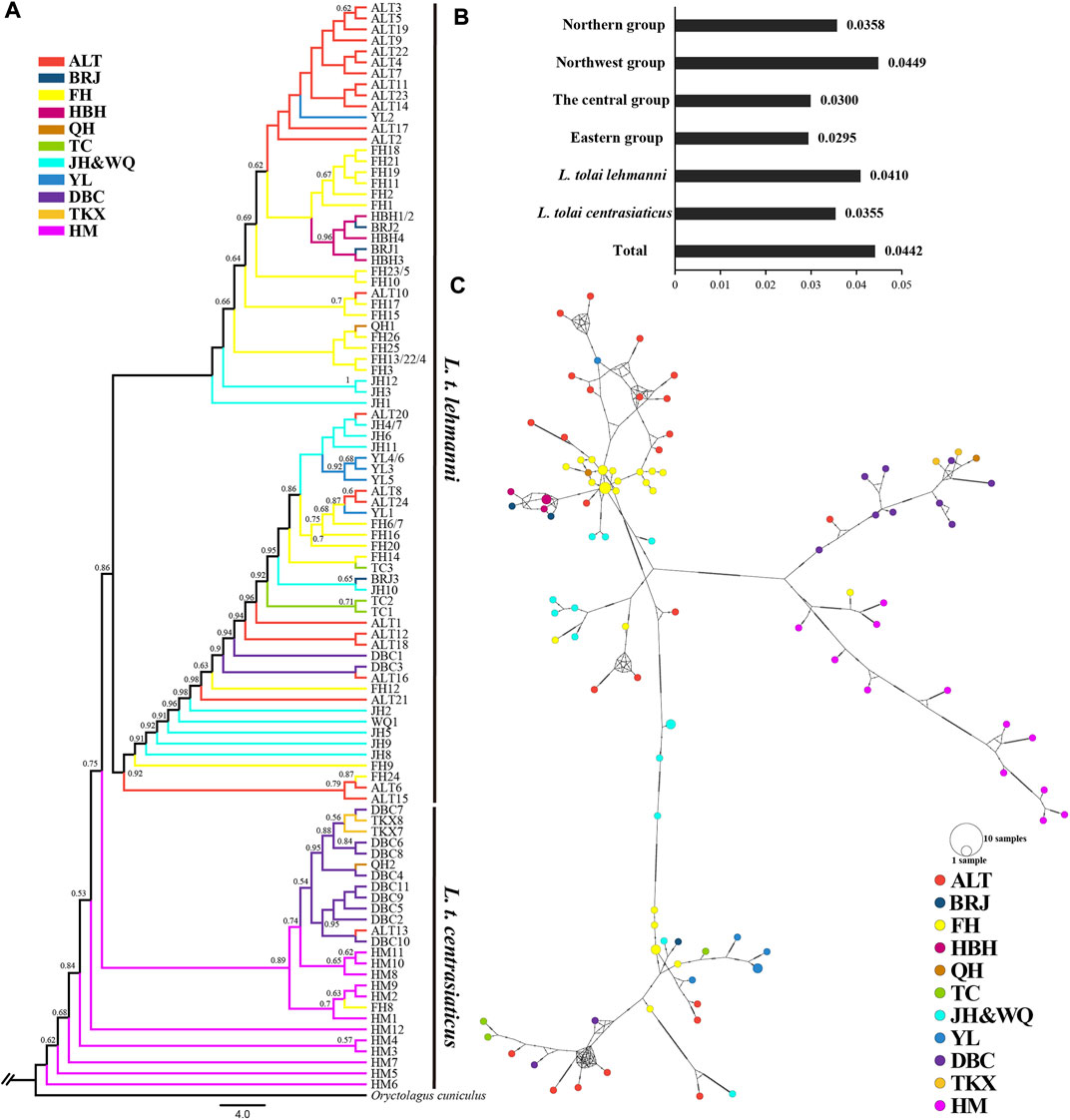
FIGURE 3. Genetic diversity and population structure of Tolai hares in Xinjiang based on mtDNA. (A). Phylogenctic tree constructed using ML method. (B). Nucleotide diversity (π). (C). Median-joining network of Tolai hare haplotypes. The hatch marks on the line indicates the mutation numbers.
The corrected Figure 3C and its caption “Genetic diversity and population structure of Tolai hares in Xinjiang based on mtDNA. (A). Phylogenctic tree constructed using ML method. (B). Nucleotide diversity (π). (C). Median-joining network of Tolai hare haplotypes. The hatch marks on the line indicates the mutation numbers.” appear below.
In the published article, there was an error. The sentence was misstated, one word was miswritten, ND4 was written as CYTB.
A correction has been made to Materials and methods, Mitochondrial DNA sequencing, First Paragraph. This sentence previously stated:
“PCR primers for CYTB were 5′-GCAAAGAATCATTACTACGCAAA-3′ (F) and 5′-TTGCGACGATTACTAAGGCTA-3′ (R) (Zhang Y. et al., 2020b)”
The corrected sentence appears below:
“PCR primers for ND4 were 5′-GCAAAGAATCATTACTACGCAAA-3′ (F) and 5′-TTGCGACGATTACTAAGGCTA-3′ (R) (Zhang Y. et al., 2020b)”
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Lepus tolai, SLAF-seq, mtDNA, genetic diversity, genetic structure
Citation: Mamat M, Shan W, Dong P, Zhou S, Liu P, Meng Y, Nie W, Teng P and Zhang Y (2023) Corrigendum: Population genetics analysis of Tolai hares (Lepus tolai) in Xinjiang, China using genome-wide SNPs from SLAF-seq and mitochondrial markers. Front. Genet. 14:1179564. doi: 10.3389/fgene.2023.1179564
Received: 04 March 2023; Accepted: 06 March 2023;
Published: 20 March 2023.
Edited and reviewed by:
Yongjie Wu, Sichuan University, ChinaCopyright © 2023 Mamat, Shan, Dong, Zhou, Liu, Meng, Nie, Teng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Shan, c3dqQHhqdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.