- 1Key Laboratory for Quality Control of Characteristic Fruits and Vegetables of Hubei Province, College of Life Science and Technology, Hubei Engineering University, Xiaogan, China
- 2Department of Biology and Chemical Engineering, Weihai Vocational College, Weihai, China
- 3Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou University, Guiyang, China
WRKY transcription factors have been demonstrated to influence the anthocyanin biosynthesis in many plant species. However, there is limited knowledge about the structure and function of WRKY genes in the major ornamental plant azalea (Rhododendron simsii). In this study, we identified 57 RsWRKY genes in the R. simsii genome and classified them into three main groups and several subgroups based on their structural and phylogenetic characteristics. Comparative genomic analysis suggested WRKY gene family has significantly expanded during plant evolution from lower to higher species. Gene duplication analysis indicated that the expansion of the RsWRKY gene family was primarily due to whole-genome duplication (WGD). Additionally, selective pressure analysis (Ka/Ks) suggested that all RsWRKY duplication gene pairs underwent purifying selection. Synteny analysis indicated that 63 and 24 pairs of RsWRKY genes were orthologous to Arabidopsis thaliana and Oryza sativa, respectively. Furthermore, RNA-seq data was used to investigate the expression patterns of RsWRKYs, revealing that 17 and 9 candidate genes may be associated with anthocyanin synthesis at the bud and full bloom stages, respectively. These findings provide valuable insights into the molecular mechanisms underlying anthocyanin biosynthesis in Rhododendron species and lay the foundation for future functional studies of WRKY genes.
1 Introduction
The Rhododendron genus (Ericaceae), which has more than 1,000 species and 30,000 cultivars, is mostly found in the Northern Hemisphere and is renowned for the exceptional beauty and variety of its corollas (Yang F. S. et al., 2020; Olech et al., 2020). Rhododendron species such as Rhododendron simsii and Rhododendron delavayi Franch are globally famous as ornamental plants. Flower color is the primary consideration in the breeding and creation of new ornamental varieties. The majority of earlier studies on pigment analysis for azalea flowers concentrated on the pigment type and composition percentage (Yang F. S. et al., 2020). Their underlying genetic and regulatory mechanisms, however, are little understood.
Anthocyanins, a subclass of flavonoids, are crucial secondary metabolites in plants and are regarded as one of the primary pigments in floral color development, causing its red, pink, blue, purple and dark color (Yoshida et al., 2009; Xu et al., 2015). The biosynthesis of anthocyanins is mediated by two categories of genes. The first category consists of structural genes that code for the enzymes that catalyze each stage of the biosynthesis process, including phenylalanine-ammonia lyase (PAL), anthocyanidin synthase (ANS), UDP-glucose:flavonoid-3-O-glucosyltransferase (UFGT), dihydroflavonol-4-reductase (DFR), chalcone synthase (CHS), rhamnosyl transferase (RT), chalcone isomerase (CHI), 4-coumaryl-CoA ligase (4CL), flavonoid 3′-hydroxylase (F3′H) and flavanone 3-hydroxylase (F3H) (Forkmann and Martens, 2001; Debeaujon et al., 2003; Tanaka et al., 2009; Chen et al., 2012; Zhang B. et al., 2021). The majority of the structural genes associated with the biosynthesis of anthocyanins have been thoroughly studied and characterized in numerous plants. The second category consists of regulatory genes that encode different transcription factors (TFs), including WD40, bHLH, and MYB, which could control the transcription of structural genes (Zhao and Tao, 2015; Yan et al., 2021). These TFs could combine to form the ternary MYB-bHLH-WD40 (MBW) complex, which is widely recognized as the primary controller of anthocyanin deposition in many plants (Ramsay and Glover, 2005; Li, 2014; Cui et al., 2021). Recently, it was discovered that additional TFs, like WRKY, contribute to the synthesis of anthocyanins (Yang et al., 2015; Lloyd et al., 2017; An et al., 2019; Li C. et al., 2020; Cong et al., 2021).
The WRKY TF family is one of the largest transcription factor families in higher plants. According to earlier research, the WRKY family is essential for controlling plant development, growth, and responses to both biotic and abiotic stresses (Rushton et al., 2010; Rinerson et al., 2015). Recent investigations further demonstrated the role of WRKY TFs in the regulation of anthocyanin biosynthesis. For example, AtWRKY41 and its ortholog BnWRKY41-1 suppress the production of anthocyanins in Arabidopsis thaliana (Duan et al., 2018). In red pear (Pyrus L.), PpWRKY44 could directly activate the PpMYB10 promoter to positively control light-induced anthocyanin biosynthesis (Alabd et al., 2022). PbWRKY75 might enhance the expression of PbMYB10b and the genes responsible for the late biosynthesis of anthocyanins (PbDFR and PbUFGT), increasing the biosynthesis of anthocyanin in pears (Cong et al., 2021). Moreover, PyWRKY26 and PybHLH3 co-targeted the PyMYB114 promoter to control anthocyanin production and transport (Li C. et al., 2020). In apple (Malus domestica L.), the accumulation of anthocyanins could be greatly aided by MdWRKY11, and the expression of several MYB TFs and structural genes also rose dramatically (Liu W. et al., 2019). The TF cascade MdHY5-MdWRKY41-MdMYB controls the synthesis of proanthocyanins and anthocyanins (Mao et al., 2021). When exposed to ultraviolet light, MdWRKY72 may enhance the synthesis of anthocyanins in transgenic apple calli (Hu et al., 2020). Additionally, in collaboration with MdMYB1, MdWRKY40 was discovered to facilitate wounding-induced anthocyanin synthesis. (An et al., 2019). Overexpressing of McWRKY71 increased the resistance of Malus crabapple to ozone stress and promoted the accumulation of anthocyanins and proanthocyanidins (Zhang et al., 2022). In eggplant (Solanum melongena L.), SmWRKY44 and SmMYB1 can interact to enhance the production of anthocyanins (He et al., 2021). In potato (Solanum tuberosum L.) tubers, StWRKY13 promotes the production of anthocyanins (Zhang H. et al., 2021). These findings show that WRKY TFs are important for anthocyanin synthesis and accumulation.
WRKY TFs are characterized by the presence of one or two WRKY domains. The WRKY domain is around 60 amino acids long and contains the highly conserved sequence “WRKYGQK” and a zinc-finger motif (Eulgem et al., 2000). The heptapeptide motif ‘WRKYGQK’ and the zinc finger motif are both necessary for the WRKY protein to bind to the consensus cis-acting element W-box (C/TTGACC/T) (Zhang and Wang, 2005). WRKY TFs can be separated into 3 main groups and 7 subgroups according to the quantity of WRKY domains and zinc-finger motif characteristics. Typically, Group I contain two WRKY domains and one C2H2 motif (C–X4-5–C–X22-23–H–X–H, where “X” is an amino acid). Group II were found to possess a single WRKY domain and a C2H2 motif, this group has been further divided into five subgroups (IIa–IIe): IIa (CX5CPVKKK(L/V) Q), IIb (CX5CPVRKQVQ), IIc (CX4C), IId (CX5CPARKHVE) and IIe (CX5CPARK (Q/M) V (E/D)). Group III possesses a WRKY domain and a C2HC motif (C–X7–C–X23–H–X–C) (Eulgem et al., 2000; Rushton et al., 2010). Numerous plants, including Arabidopsis (Eulgem et al., 2000), wheat (Triticum aestivum L.) (Ning et al., 2017), tomato (Solanum lycopersicum L.) (Huang et al., 2012), cotton (Gossypium raimondii and Gossypium hirsutum) (Dou et al., 2014), poplar (Populus trichocarpa L.) (He et al., 2012), pineapple (Ananas comosus L. Merrill) (Xie et al., 2018), soybean (Glycine max L. Merrill) (Yu et al., 2016), peanut (Arachis hypogaea L.) (Song et al., 2016), and maize (Zea mays L.) (Hu et al., 2021) have had the full genome level identification of their WRKY TFs.
There is, however, limited knowledge, particularly on the WRKY genes of the azalea’s role in the synthesis of anthocyanins. Clarifying the evolution and potential roles of WRKY genes in azalea requires more research. Recently, the generation of azalea genome has presented an exceptional opportunity to identify the WRKY TF family across the entire genome (Yang F. S. et al., 2020). In this study a genome-wide analysis of azalea WRKY genes was accomplished, 57 WRKY genes in all were identified. Subsequently, we examined the categorization, phylogenetic relationships and gene duplications of the azalea WRKY (RsWRKY) genes. Moreover, the candidate RsWRKYs that linked to anthocyanin biosynthesis were identified according to RNA-seq data in three Rhododendron species with various flower colors. The findings of this research improve our knowledge of the RsWRKY gene family and will facilitate the genetic improvement of flower color in azalea.
2 Materials and methods
2.1 Investigation of WRKY gene family in azalea and other plant species
The whole protein sequence data of azalea were retrieved from RPGD (Rhododendron Plant Genome Database, http://bioinfor.kib.ac.cn/RPGD/) (Yang F. S. et al., 2020). The WRKY HMM profile (PF03106) was used as a query to search predicted RsWRKY proteins using the HMMER 3.0 software (Eddy, 2011). Subsequently, putative RsWRKY proteins were submitted to SMART (Letunic et al., 2021) and Pfam (Mistry et al., 2021) databases to confirm the presence of typical WRKY domains, only sequences with at least one WRKY characteristic amino acid sequence can be retained.
The WRKY proteins of following species were retrieved from prior studies: Arabidopsis thaliana (Eulgem et al., 2000), Brassica napus (He et al., 2016), Malus domestica (Qin et al., 2022), Camellia sinensis (Wang et al., 2019), Cucumis sativus (Chen et al., 2020b), Physcomitrella patens (Rensing et al., 2008), Theobroma cacao (Silva Monteiro de Almeida et al., 2017), Medicago truncatula (Kumar et al., 2016), Cyanidioschyzon merolae (Dou et al., 2014), rice (Oryza sativa) (Ramamoorthy et al., 2008), Ginkgo biloba (Cheng et al., 2019), Chlamydomonas reinhardtii (Rinerson et al., 2015), Vitis vinifera (Guo et al., 2014), Solanum melongena (Yang Y. et al., 2020), Azolla filiculoides (Zhao et al., 2021), Salvinia cucullate (Zhao et al., 2021), Selaginella moellendorffii (Rinerson et al., 2015), Moringa oleifera (Zhang et al., 2019), Solanum lycopersicum (Huang et al., 2012), Sorghum bicolor (Baillo et al., 2020) and Populus trichocarpa (Jiang et al., 2014). The evolutionary relationships of these species were determined according to the PGDD database (Lee et al., 2013) and previous study (Liu et al., 2021).
2.2 Classification of the RsWRKY TFs and phylogenetic analysis
According to the quantity of WRKY domains and zinc-finger motif features, WRKY TFs can be divided into 3 main groups and 7 subgroups (Eulgem et al., 2000; Rushton et al., 2010). Group I contain two WRKY domains and one C2H2 motif (C–X4-5–C–X22-23–H–X–H). Group II were found to possess a single WRKY domain and a C2H2 motif, this group has been further divided into five subgroups (IIa–IIe): IIa members contain CX5CPVKKK (L/V) Q structure, IIb members contain CX5CPVRKQVQ structure, IIc members contain CX4C structure, IId members contain CX5CPARKHVE structure, and IIe members contain CX5CPARK (Q/M) V (E/D) structure. Group III possesses a WRKY domain and a C2HC motif (C–X7–C–X23–H–X–C).
A phylogenetic tree was built using the following five species: Azalea, Rhododendron ovatum (a related asteroid eudicot), A. thaliana (a rosid eudicot), Aquigelia coerulea (a basal eudicot), and Amborella trichopoda (a basal angiosperm). The WRKY proteins from R. ovatum, A. coerulea and A. trichopoda were obtained using the same method as described in Section 2.1, whereas the AtWRKY proteins from A. thaliana were collected from prior investigations (Eulgem et al., 2000). The WRKY protein sequences from five species were aligned using T-Coffee (Notredame et al., 2000). The alignment sequences were then edited with the Jalview program (Waterhouse et al., 2009), with all non-WRKY domain sequences removed. A phylogenetic tree was constructed based on the WRKY domain sequences using IQ-TREE (Minh et al., 2020) with a maximum-likelihood approach and a bootstrap value of 5,000. Finally, iTOL (Letunic and Bork, 2021) was used to visualize and embellish the phylogenetic tree.
2.3 Gene duplication analysis
DupGen_finder (Qiao et al., 2019) was utilized to distinguish between whole-genome duplication (WGD), tandem duplication, transposed duplication, proximal duplication and dispersed duplication with the default settings. Duplicated RsWRKY gene pairs were visualized using TBtools software (Chen et al., 2020a). Using the KaKs_Calculator (version 2.0) (Wang et al., 2010), Ka, Ks and Ka/Ks values were calculated for all duplication gene pairs. The synteny of the WRKY genes between Azalea and other plants was examined using MCScanX (Wang et al., 2012), and the findings were presented visually using TBtools software (Chen et al., 2020a).
2.4 Expression analysis of RsWRKY genes in three different colors of azaleas by RNA-seq
The expression patterns of the RsWRKY genes were investigated using RNA-seq data obtained from azalea flower samples of three different colors and at two different stages of floral development. The RNA-seq data have been deposited in the SRA database (http://www.ncbi.nlm.nih.gov/sra) under the identifier PRJNA952839. The three azalea varieties are Rhododendron wardii var. puralbum (possessing white color flower, WF), Rhododendron hybridum Ker Gawl (possessing red color flower, RF), and Rhododendron simsii Planch (possessing pink color flower, PF). The flower samples for RNA-seq were collected at the bud and full bloom stages from plants grown in a controlled-environment growth chamber with a temperature of 25/18°C (day/night), 16/8 light/dark cycle, and 60%–70% relative humidity. Fragments per kilobase of exon model per million mapped reads (FPKM) was used to measure the transcript abundance of RsWRKY genes. The gene expression data for each RsWRKY were normalized by log2 (FPKM) and shown as heat maps using the TBtools software (Chen et al., 2020a). Genes that showed absolute |fold change| ≥ 2 (log2 |fold change| ≥ 1) for differential expression between two azalea varieties were defined as differentially expressed genes (DEGs).
The following comparative strategy was used to identify candidate genes associated with flower coloration. First, the comparison between colored and white flowers (WF versus RF, WF versus PF) was performed using gene expression data from the same stages of floral development to generate two DEG sets (both up- and downregulated DEGs were included). The two DEG sets were then intersected to produce the final DEGs. The candidate genes at the bud stage and full bloom stage were derived using this comparative strategy, respectively.
2.5 Confirmation of RNA-seq analysis in the flowers of three azalea varieties
The expression levels of 9 candidate RsWRKY genes at the full bloom stage were assessed by qRT-PCR to confirm the validity of RNA-seq results. The RNA used in the RT-PCR experiments was the same as the RNA used in the RNA-seq analysis mentioned above. The qRT-PCR Primers were designed by Beacon Designer 8 (Supplementary Table S1). According to our RNA-seq data and a previous study (De Keyser et al., 2013), the expression levels of RsGAPDH (Rhsim12G0106200) and RsEF1α (Rhsim02G0008200) were stable in different varieties with different flower colors and at different developmental stages, therefore these two genes were used as internal reference genes. The StepOnePlus™ System (Applied Biosystems, Foster City, CA, United States) was used to perform the qRT-PCR with the following cycling profile: 95°C for 30°s, 95°C for 5 s (40 cycles), 60°C for 30 s. Each qRT-PCR test employed three biological and three technical replicates. The 2−ΔΔCt method was used to calculate the outcomes (Livak and Schmittgen, 2001). The qRT-PCR reaction mixture was performed with a TB Green® Premix Ex Taq™ kit (TaKaRa, Dalian, China).
2.6 GO enrichment analysis of candidate RsWRKY genes
GOATOOLS (http://github.com/tanghaibao/GOatools) (Klopfenstein et al., 2018) was utilized to annotate candidate RsWRKY genes and Fisher’s exact test was employed for biological function enrichment analysis. To minimize false positives, the Bonferroni multiple testing correction was applied, with significance defined as a corrected p-value (Padjust) of less than 0.05. The results were presented using a bubble dot diagram, which was generated using the website (www.bioinformatics.com.cn).
2.7 Three-dimensional protein structure prediction
Three-dimensional (3D) structures of candidate RsWRKY proteins were modeled on the basis of homology modeling using SWISS-Model (https://swissmodel.expasy.org/) (Waterhouse et al., 2018). In addition, the model quality was evaluated by global model quality estimation (GMQE) (Waterhouse et al., 2018) and QMEANDisCo global (Studer et al., 2020). The GMQE and QMEANDisCo global score were between 0 and 1, and the higher score indicates the model is more reliable.
3 Result
3.1 Identifcation of the RsWRKYs
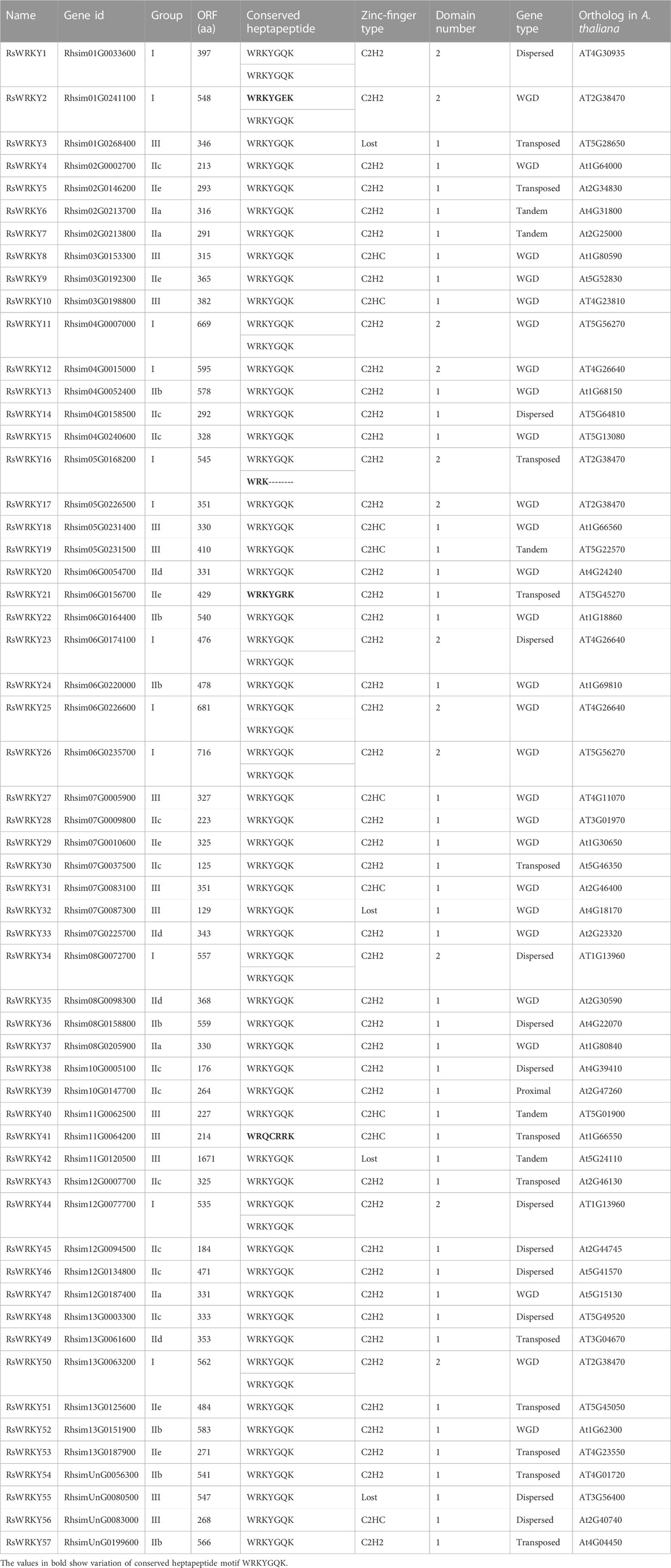
We totally identified 57 RsWRKY gene members, which were further renamed as RsWRKY1 to RsWRKY57 according to their chromosomal positions on Chr1-13 and the order of scaffolds (Table 1). The identification outcomes showed one gene that was different from the study by Wan et al. (2022). Our findings contain the RhsimUnG0199600 gene, while their findings include the Rhsim10G0148600 gene. Due to the presence of a complete WRKY domain, the RhsimUnG0199600 gene is undoubtedly a WRKY gene. However, the WRKY domain in the Rhsim10G0148600 gene had almost entirely been gone, therefore we removed it throughout the screening process.
3.2 Classification of the RsWRKY TFs and phylogenetic analysis
Except for four RsWRKY proteins (RsWRKY3, RsWRKY32, RsWRKY42, and RsWRKY55), all other RsWRKYs were categorized into 3 main groups and 7 subgroups based on structural characteristics (Supplementary Figure S1, Table 1). These four RsWRKY proteins cannot be assigned to either group since they lack a zinc-finger motif or only have a partial one.
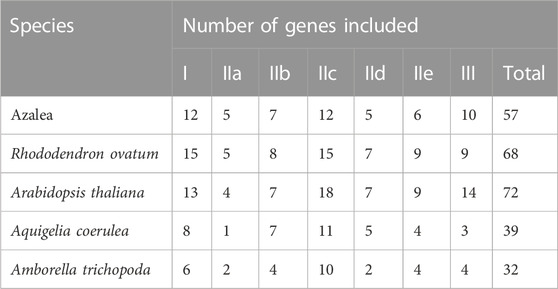
In the present study, we identified 68, 39, and 32 WRKY genes in R. ovatum, A. coerulea, and A. trichopoda, respectively (Supplementary Table S2). According to the topology of the phylogenetic tree, as well as the categorization and nomenclature of WRKY genes in A. thaliana, 268 members of the WRKY family from five diverse species were divided into 3 main groups and 7 subgroups (Figure 1; Table 2). As shown in Figure 1, all groups in our study were supported by previous studies in A. thaliana, and all grouping outcomes in azalea based on structure characteristic were supported by the phylogenetic analysis.
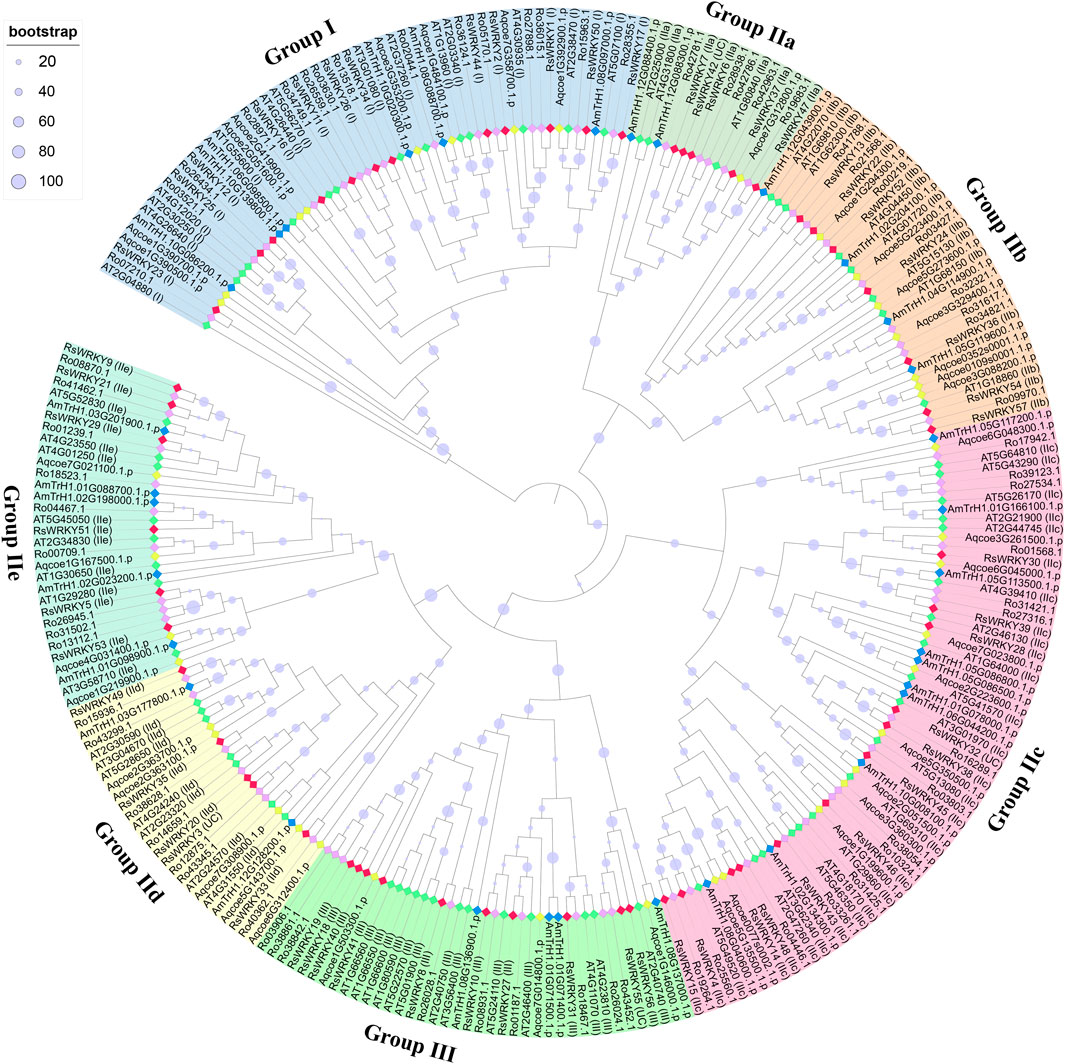
FIGURE 1. Phylogenetic analysis of WRKY proteins in azalea, R. ovatum, A. thaliana, A. coerulea, and A. trichopoda. The phylogenetic tree was constructed using maximum likelihood method based on the 268 WRKY domain sequences. The letters in parentheses that follow the names of the RsWRKY genes denote categorization outcomes based on structural characteristic, while the letters that follow AtWRKY genes expressed the previous grouping in Arabidopsis. The WRKY proteins from azalea, R. ovatum, A. thaliana, A. coerulea, and A. trichopoda are represented by the red, purple, green, yellow, and blue rhombuses, respectively. UC: unclassified.
According to the topology of the phylogenetic tree, the four unclassified members (RsWRKY3, RsWRKY 32, RsWRKY 42 and RsWRKY 55) were classified into different groups, with RsWRKY3 classified into group IId, RsWRKY 32 classified into group IIc, RsWRKY 42 classified into group IIa, and RsWRKY 55 classified into group III (Figure 1). In summary, Groups I, II, and III in azalea had 12, 35, and 10 RsWRKY members, respectively. Group II comprised of five subgroups, and each of subgroup IIa, IIb, IIc, IId, and IIe contained 5, 7, 12, 5, and 6 members, respectively.
Compared with a previous study (Wan et al., 2022), a better phylogenetic analysis and grouping outcomes was achieved in our study. For instance, the previous research mixed two different clades into group III in phylogenetic tree, which results in a categorization error. Rhsim04G0158500 (RsWRKY14) and Rhsim13G0003300 (RsWRKY48), which both have the typical “WRKYGQK” structure and CX4C structure, should both be in Group IIc but have been put in Group III in the previous research.
3.3 Comparative analysis of WRKY genes
To explore the evolution of the WRKY gene family, a comparative genomic study of WRKY genes was performed in 22 typical plants. Figure 2 depicts the evolutionary relationships of these species as well as the number of WRKY genes present in each genome. Our research revealed that WRKY gene numbers in lower plants are far less than that in higher plants, which indicated that the WRKY genes expanded considerably during the evolution from lower plant species to higher plant species. The genome size of C. sinensis (3020.0 MB) was larger than all other angiosperm species. However, compared to all other angiosperm plants, it possesses a genome with a reduced density of WRKY genes, which suggests that many CsWRKYs were lost during polyploid speciation. In addition, G. biloba exhibited an exceptionally low WRKY gene density compared to all other species, including lower plants. This phenomenon can be explained by the expansion of the G. biloba genome, which was accompanied by a significant extension of introns, mainly caused by the insertion of long terminal repeats rather than the recent occurrence of whole-genome duplication events (Liu et al., 2021).
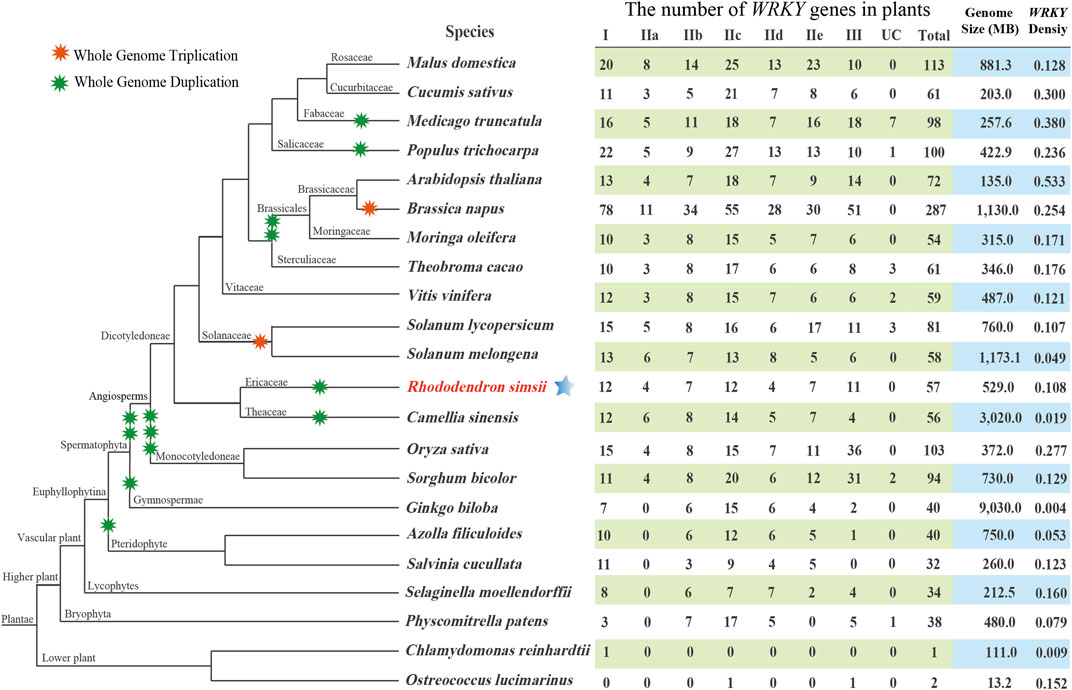
FIGURE 2. The evolutionary relationships of the 22 plant species and the specific information of the WRKY gene family in each genome. UC: unclassified.
3.4 Gene duplication and Ka/Ks analysis
Using DupGen finder, the 57 RsWRKY genes were divided into the following five groups in the current study: 27 WGD (47.37%), 5 tandem duplication (8.77%), 1 proximal tandem duplication (1.75%), 12 transposed tandem duplication (21.05%), and 12 dispersed tandem duplication (21.05%) (Table 1; Supplementary Figure S2). 52 duplication gene pairs in all were found in RsWRKYs (Supplementary Table S2; Figure 3A). Then, synonymous (Ks) and non-synonymous (Ka) mutations were examined in these gene pairs (Supplementary Table S3; Figure 3B). Genes were frequently subjected to purifying selection (Ka/Ks < 1), positive selection (Ka/Ks > 1), and neutral selection (Ka/Ks = 1) during the course of evolution. In the current study, all 52 RsWRKY duplication gene pairs showed Ka/Ks rates lower than 1, which suggests that purifying selection and less divergence occurred among them.
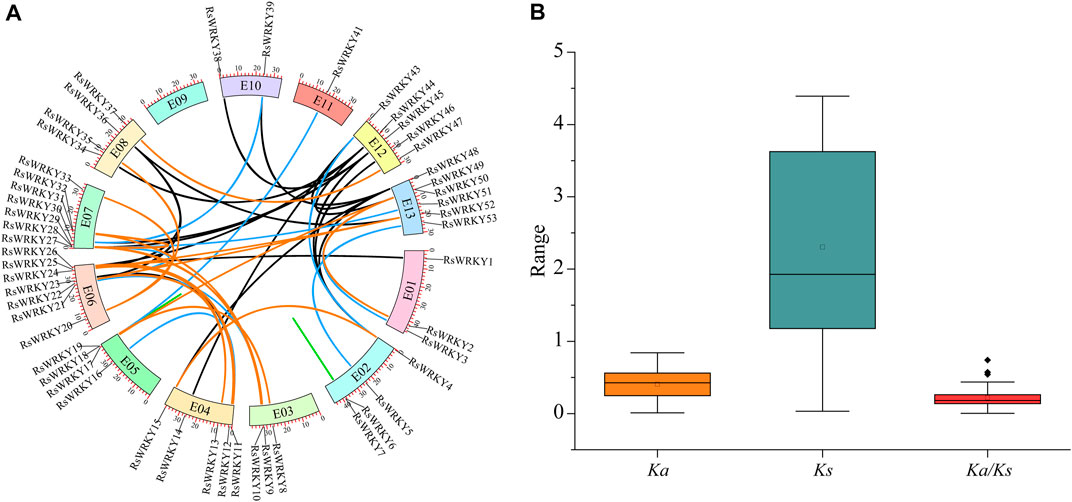
FIGURE 3. (A) Genomic locations of RsWRKYs and all duplicated gene pairs in the azalea genome. Duplicated RsWRKY gene pairs were indicated by the colored lines: WGD pairs (orange), tandem duplication pairs (green), transposed duplication pairs (blue), dispersed duplication pairs (black). Color boxes with a number inside it represents chromosomes. (B) Ka, Ks, and Ka/Ks ratio of all duplication gene pairs. The box plots are exhibiting the distributions of Ka, Ks, and Ka/Ks values. The small square and the line in the box represent average and median values of the Ka, Ks, and Ka/Ks values, respectively.
In order to explore the potential evolutionary processes of the WRKY gene family, collinearity analysis was employed to examine the orthologous relationships of WRKY family genes in azalea, Arabidopsis, and rice (Figure 4). A total of 63 pairs of orthologs were discovered between azalea and Arabidopsis (Supplementary Table S4), while 24 pairs of orthologs were found between azalea and rice (Supplementary Table S5), these RsWRKY genes and the corresponding AtWRKY or OsWRKY genes shared a common ancestor. Some ortholog relationships involved one RsWRKY corresponding to one ortholog gene (e.g., RsWRKY20-AT2G23320 and RsWRKY18-LOC_Os05g25770), while others had one RsWRKY corresponding to multiple ortholog genes (e.g., RsWRKY22-AT1G62300/AT4G22070/AT4G04450 and RsWRKY17-LOC_Os01g61080/LOC_Os05g39720/LOC_Os05g27730). These results suggest that duplication events have played a significant role in the evolution and functional diversification of the WRKY family.
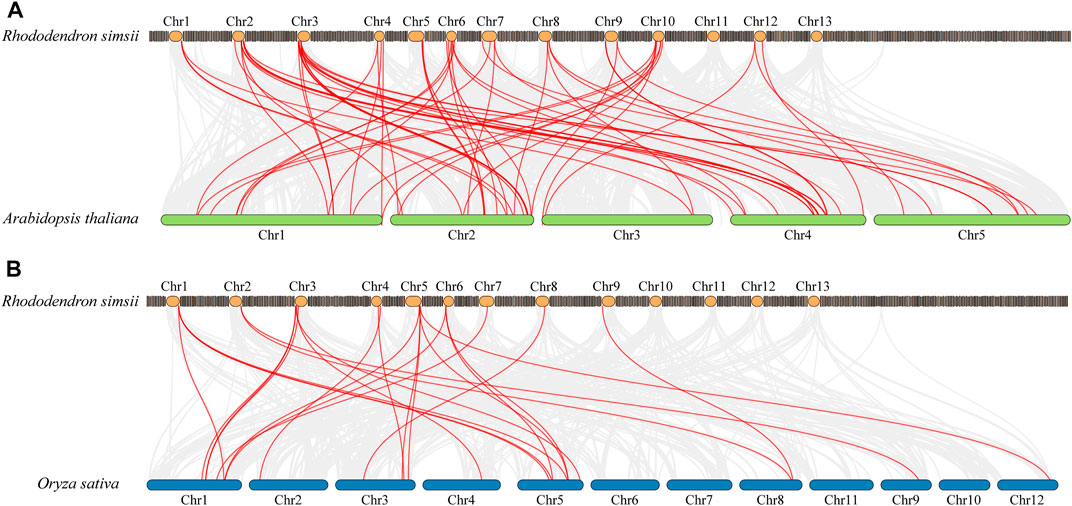
FIGURE 4. Comparative physical mapping showing the orthologous relationships of RsWRKY genes with Arabidopsis (A) and rice (B).
3.5 Expression profiling of RsWRKY genes in three azalea varieties
The expression profiles of RsWRKY genes in three azalea cultivars with different flower colors were analyzed using RNA-seq data to identify WRKY genes involved in anthocyanin biosynthesis in azaleas (Figures 5, 6; Supplementary Tables S6, S7). Genes that showed absolute |fold change| ≥ 2 (log2 |fold change| ≥ 1) for differential expression between two azalea varieties were defined as differentially expressed genes (DEGs). As a result, there are 6 upregulated DEGs and 21 downregulated DEGs between red flower (RF) and white flower (WF) azaleas at the bud stage. In addition, 8 upregulated DEGs and 14 downregulated DEGs were identified between pink flower (PF) and white flower (WF) azaleas at the bud stage. Combined with the two DEG sets ((WF versus RF) and (WF versus PF)), the candidate RsWRKYs associated with anthocyanin biosynthesis were screened. The expression level of 4 DEGs (RsWRKY21, 24, 33 and 39) was upregulated in colored-flower azaleas (PF and RF), indicating that these genes might play a role in the positive control of anthocyanin production at the bud stage. While the expression level of 13 DEGs (RsWRKY2, 7, 17, 18, 26, 35, 36, 37, 38, 45, 47, 50 and 52) was downregulated in colored-flower azaleas (PF and RF), indicating that these genes may have a role in suppressing the synthesis of anthocyanins at the bud stage (Figure 5; Supplementary Table S6). Using the same comparative strategy, we identified 4 candidate genes (RsWRKY10, 27, 41 and 51) that might play a role in the positive control of anthocyanin production at the full bloom stage, and 5 candidate genes (RsWRKY13, 25, 26, 29 and 49) that might be involved in negatively regulating the anthocyanin biosynthesis at the full bloom stage (Figure 6; Supplementary Table S7).
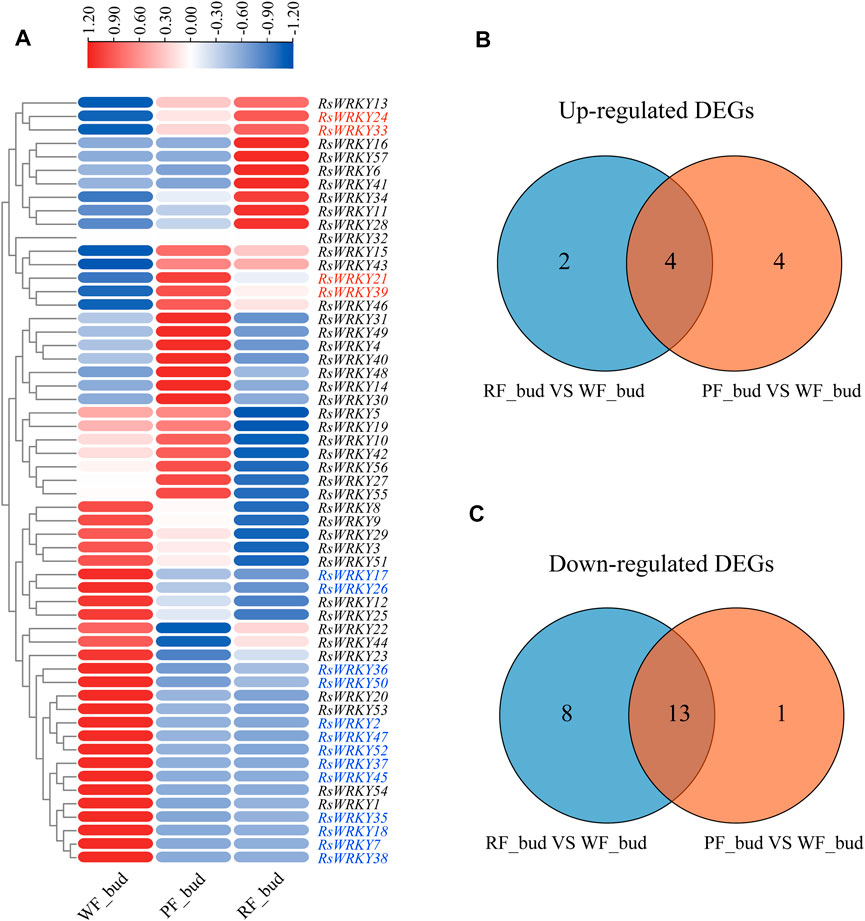
FIGURE 5. Identification of candidate RsWRKYs associated with anthocyanin biosynthesis among white flowering (WF), red flowering (RF) and pink flowering (PF) azaleas at the bud stage. (A) Heatmap of RsWRKYs at the full bloom stage, the scale bar represents relative expression value. (B) Venn diagram indicates the unique and common upregulated DEGs are found in set “RF_bud VS. WF_bud” and set “PF_bud VS. WF_bud.” (C) Venn diagram indicates the unique and common downregulated DEGs are found in set “RF_bud VS. WF_bud” and set “PF_bud VS. WF_bud.”
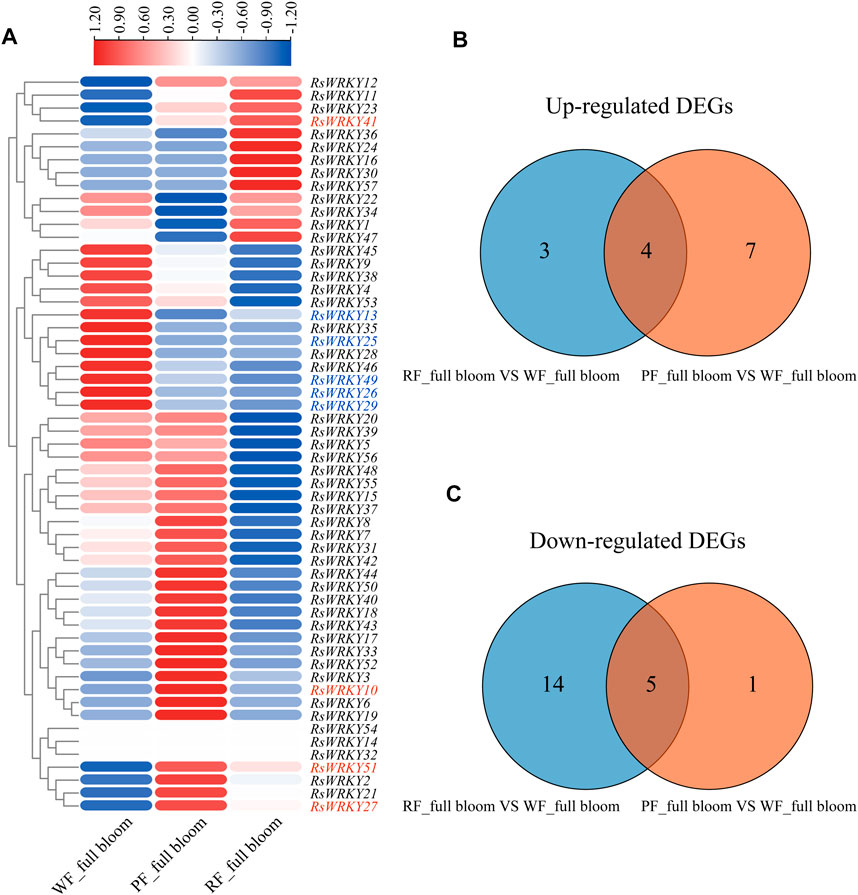
FIGURE 6. Identification of candidate RsWRKYs associated with anthocyanin biosynthesis among white flowering (WF), red flowering (RF) and pink flowering (PF) azaleas at the full bloom stage. (A) Heatmap of RsWRKYs at the full bloom stage, the scale bar represents relative expression value. (B) Venn diagram indicates the unique and common upregulated DEGs are found in set “RF_ full bloom VS. WF_ full bloom” and set “PF_ full bloom VS. WF_ full bloom.” (C) Venn diagram indicates the unique and common downregulated DEGs are found in set “RF_ full bloom VS. WF_ full bloom” and set “PF_ full bloom VS. WF_ full bloom.”
3.6 Validation of RNA-seq-based gene expression
To confirm the accuracy of the RNA-seq results, 9 candidate genes related to anthocyanin biosynthesis at the full bloom stage were chosen and examined using qRT-PCR. In RT-PCR tests, the genes that were up- or downregulated in RNA-seq experiments were likewise up- or downregulated. Even though the values of the gene fold change differ between the RNA-seq and RT-PCR data, the expression trend of gene is the same. For instance, RNA-seq data revealed that the expression of RsWRKY10 was 19.8 and 2 times higher in red and pink flowers than in white flowers, respectively, in RT-PCR tests, the corresponding fold change values were 8.4 and 3.3. According to RNA-seq data, the expression of RsWRKY29 in red and pink flowers was 0.38 and 0.26 less than that of white flowers, whereas the corresponding values in RT-PCR tests were 0.52 and 0.29. In conclusion, RT-PCR results agree well with the RNA-Seq data in terms of expression trends (Supplementary Table S7; Figure 7).
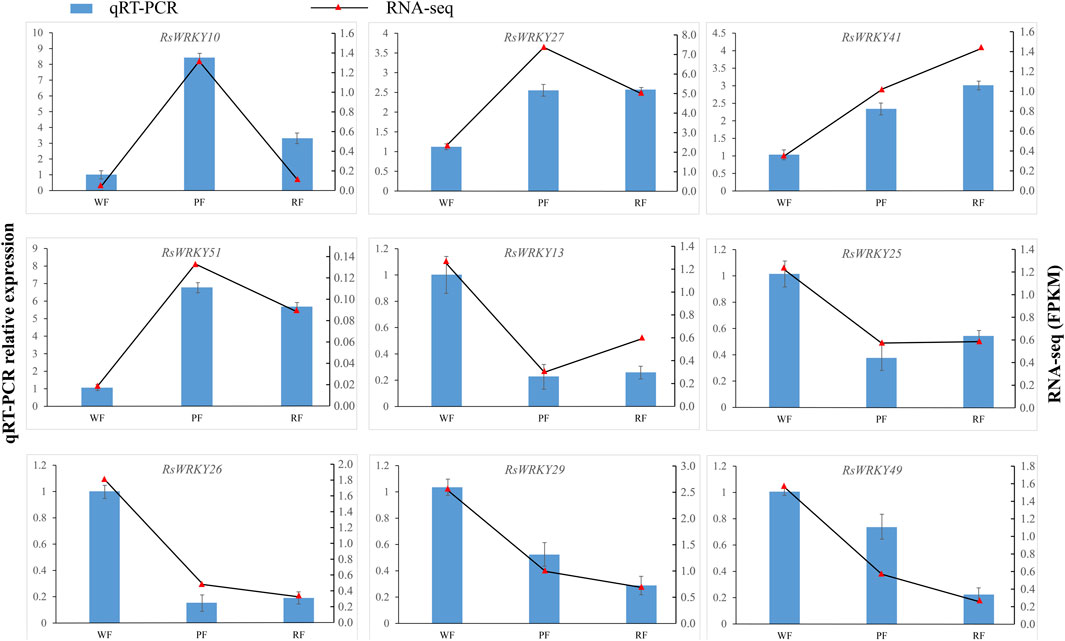
FIGURE 7. qRT-PCR validation of 9 candidate RsWRKY genes at the full bloom stage. WF, PF, and RF represent white-flowering variety, pink-flowering variety, the red-flowering variety, respectively.
3.7 GO enrichment analysis of candidate genes
In order to have a comprehensive understanding of the biological function of the candidate genes, gene ontology (GO) annotation and enrichment analysis of the 25 candidate RsWRKY genes were performed in present study (Figure 8; Supplementary Table S8). 123 GO terms showed significant enrichment, with 9 representing molecular functions, 5 representing cellular components, and 109 representing biological processes. In the molecular function category, “transcription regulator activity,” “sequence-specific DNA binding” and “DNA-binding transcription factor activity” were the most enriched terms. In the cellular component category, “nucleus,” “intracellular membrane-bounded organelle” and “membrane-bounded organelle” were the most enriched. In the biological process category, “response to organonitrogen compound,” “regulation of cellular metabolic process,” “response to nitrogen compound” and “gene expression” were the most enriched terms. GO enrichment results indicate that the candidate RsWRKY TF has primarily functions centered around common transcription factor functions such as regulation of gene expression and transcription regulatory.
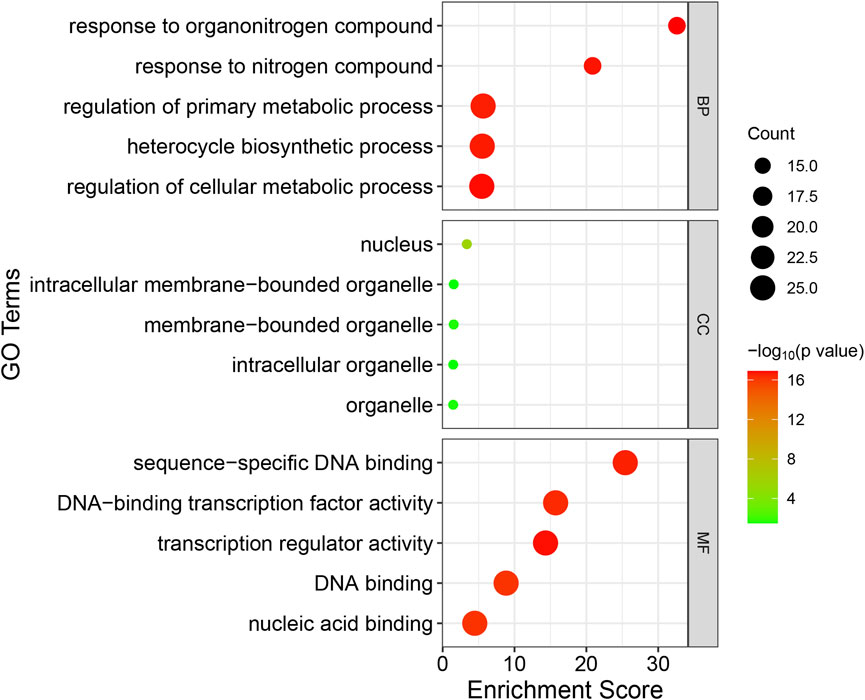
FIGURE 8. GO enrichment analysis of the candidate RsWRKY genes. GO was performed with three main categories: molecular function (MF), cellular component (CC) and biological process (BP). GO terms with p-value < 0.05 were identified as significant.
3.8 Three-dimensional structure prediction of candidate genes
We utilized SWISS-Model to predict the 3D protein structures of 25 candidate RsWRKYs (Figure 9). Out of the 25 models, 25 were successfully defined by at least 30% identity of the target to the template, which is widely accepted as a threshold for successful modeling (Xiang, 2006). The majority of QMEANDisCo global values were higher than 0.50, indicating good quality models, except for RsWRKY18 and RsWRKY41, which had QMEANDisCo global values of 0.49 and 0.44, respectively, indicating lower quality models (Supplementary Table S9). The GMQE values ranged from 0.05 to 0.27, which were very low because the models covered only WRKY domain sequences of the target proteins. All 25 models were monomer oligo-state, and different 3D structures were observed in RsWRKY41. Tertiary structure prediction results indicated that most RsWRKYs contained a zinc ion and the RsWRKY proteins domain were mainly composed of β folds.
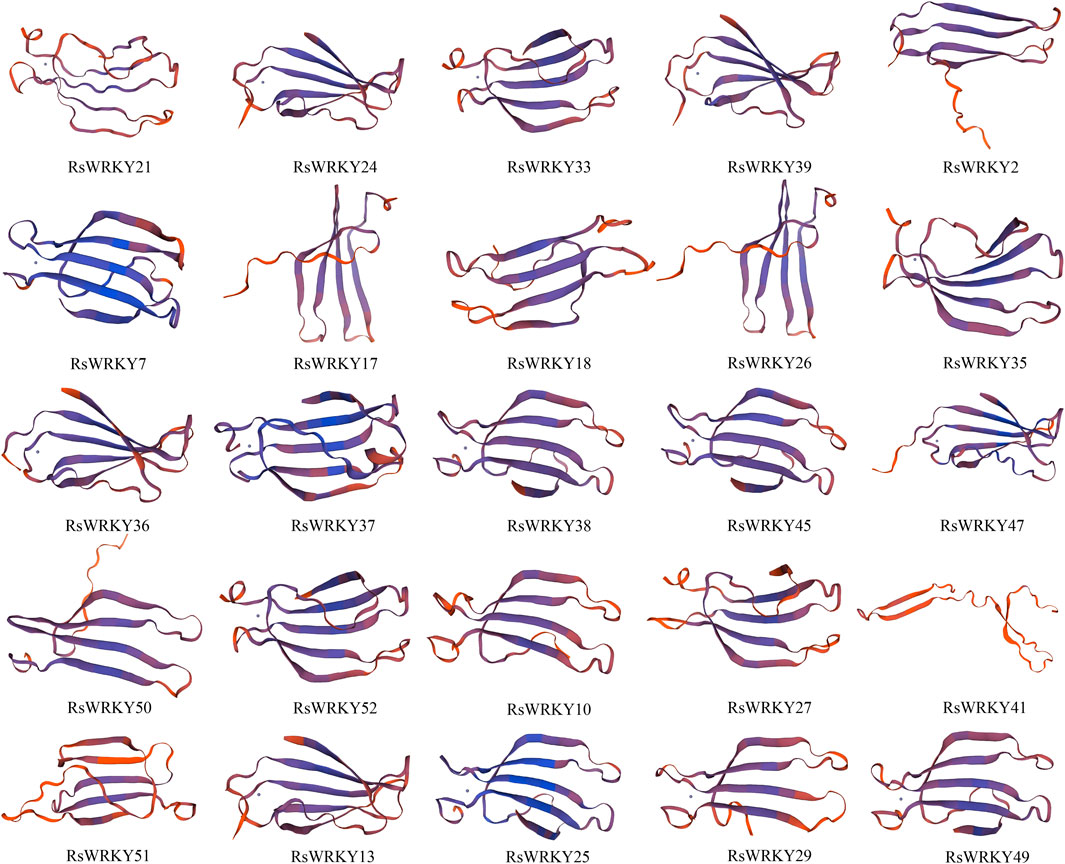
FIGURE 9. Three-dimensional (3D) protein structures of 25 candidate RSWRKYs. Broad strips are β-sheets and dots are zinc ions.
4 Discussion
WRKY genes are a vast family of TFs that are present in almost all plant species and serve key roles in a variety of physiological functions (Rinerson et al., 2015; Li W. et al., 2020; Wani et al., 2021). Numerous plant species have had their WRKY TFs identified at the entire genome level (Guo et al., 2014; He et al., 2016; Kumar et al., 2016; Wang et al., 2019; Chen et al., 2020b; Qin et al., 2022). In this research, we reported a genome-wide analysis of the WRKY family in azalea and 57 RsWRKY genes was identified. The WRKY domain is the crucial element that controls how the WRKY protein binds specifically to the cis-element W-box. According to sequence alignment analysis, the majority of RsWRKY proteins shared exceptionally heptapeptide motif WRKYGQK at their N-termini. But this analysis also uncovered other variants, including WRKYGEK (RsWRKY2), WRKYGRK (RsWRKY21), and WRQCRRK (RsWRKY41) (Supplementary Figure S1). The Arabidopsis (Eulgem et al., 2000), maize (Hu et al., 2021), apple (Qin et al., 2022), carrot (Daucus carota subsp. sativus L.) (Nan and Gao, 2019), and duckweed (Spirodela polyrhiza L.) (Zhao et al., 2021) WRKY members have also shown this occurrence. Additionally, it was revealed that one protein (RsWRKY16) possessing an incomplete WRKYGQK sequence and four RsWRKY proteins (RsWRKY3, 32, 42 and 55) that lacked the entire zinc finger motif were also identified as RsWRKY family members. Previous studies have demonstrated that the presence of both the heptapeptide motif WRKYGQK and the zinc finger motif is necessary for the WRKY TFs' significant binding affinity to their homologous cis-acting W-box element (TTGACC/T) (Wani et al., 2021). Due to this, modifications to the heptapeptide motif and the absence of the zinc finger motif may impact the regular interactions of RsWRKYs with their downstream target genes. Therefore, it may be essential to further explore the role and binding specificity of these eight RsWRKY proteins.
An ML phylogenetic tree based on conserved WRKY domains was constructed in this study using the WRKY proteins from azalea, R. ovatum (a related asteroid eudicot), Arabidopsis (a rosid eudicot), A. coerulea (a basal eudicot), and A. trichopoda (a basal angiosperm). As shown in Figure 1, the RsWRKY proteins from Groups IIa and IIb were more closely related, while Groups IId and IIe had a close relationship. Since Groups IIa and IIb and Groups IId and IIe split off from the progenitor of terrestrial plants considerably later than the other Groups (Zhang and Wang, 2005), it was assumed that these two Groups would merge to form two new subfamilies, IIa + b and IId + e, respectively (Rushton et al., 2010; Chen et al., 2017). We investigated the WRKY gene family in azalea and 21 other species, including 19 higher plants and 2 lower plants. Comparative genomic analysis revealed that single-celled C. reinhardtii and O. lucimarinus have 1 and 2 WRKY genes respectively. The moss P. patens' genome, however, encodes up to 38 WRKY members (Figure 2), showing a substantial increase following this species diverged from green algae. P. patens has a role in evolutionary analysis, which makes it possible to compare genomes with aquatic algae and vascular plants in order to reconstruct evolutionary changes brought on by land conquest (Rensing et al., 2008). The growth of the WRKY family in P. patens illustrates the significance of WRKY proteins in numerous physiological processes, particularly the complex functions held by higher plants. Furthermore, G. biloba had fewer WRKY genes than most species while having a larger genome than all other species, proving that genome size is not correlated with the amount of WRKY genes.
Gene duplication events are crucial in the rapid expansion and evolution of gene families (Maher et al., 2006). The most significant factors in the growth of gene families during evolution are WGD and tandem duplication events (Wang et al., 2012). Many transcription factor gene families, such as MADS-box, bHLH, and bZIP, most likely expanded through WGD duplication (Liu et al., 2014; Wang B. et al., 2022; Wang X. J. et al., 2022). While other transcription factor gene families, such as NBS-LRR and ERF, were expanded as a result of tandem duplication (Liu Z. et al., 2019; Li P. et al., 2020; Zhong et al., 2022). Since 47.37% of the RsWRKY genes were duplicated and retained through WGD duplication, our research suggested that WGD duplication was the major causes for the expansion of the RsWRKY family of genes in azalea. This outcome was consistent with previous research on apple (Qin et al., 2022), carrot (Nan and Gao, 2019), walnut (Juglans regia L.) (Hao et al., 2021), and potato (Zhang et al., 2017). Pseudogenization (loss of function), subfunctionalization, and neofunctionalization are possible outcomes of duplicated genes (Lynch and Conery, 2000). The WGD duplicated gene pair RsWRKY24/RsWRKY52 from Group IIb with varied levels of expression may be due to the fact that duplicated genes can be stably maintained if they differ in some aspects of their functions during selection and evolution. We also evaluated the rate of synonymous (Ks) to non-synonymous (Ka) mutation for all duplication gene pairs. It is well known that genes are generally subjected to three different types of selection—purifying selection (Ka/Ks < 1), positive selection (Ka/Ks > 1), and neutral selection (Ka/Ks = 1) (Hurst, 2002). All of the RsWRKY duplication gene pairs in our study had Ka/Ks ratios smaller than 1, indicating that they had undergone strong Darwinian purifying positive selection. Genomic comparison is an effective way to transfer knowledge about the genome of a well-studied taxonomic group to a less studied one (Lyons et al., 2008). In our research, we discovered that 63 pairs of R. simsii and Arabidopsis WRKY genes exhibit a collinear relationship, while 24 pairs of WRKY genes show a collinear relationship between R. simsii and rice (Supplementary Table S4, S5). This finding suggests that these gene pairs are orthologous, have common ancestors, and have been preserved throughout evolution, potentially performing similar functions. The identification of WRKY genes in Arabidopsis and rice serves as a reference for future studies on RsWRKY genes.
Azaleas are well-known decorative plants with major cultural and economic significance. Flower color is the primary trait in the breeding and creation of new ornamental cultivars. WRKY genes have been shown to be involved in the control of the anthocyanin biosynthesis (see “Introduction” section), and are thus key factors for the breeding of azaleas. In this study, we discovered 17 candidate RsWRKY genes that may be involved in the synthesis of anthocyanins at the bud stage and 9 candidate RsWRKY genes that may be involved in the synthesis of anthocyanins at the full bloom stage. Unexpectedly, there is little overlap between these two sets of genes (only RsWRKY26), which indicating that various RsWRKY genes are engaged in anthocyanin biosynthesis at various times of flowering. In this study, we utilized GO annotation and enrichment analysis to elucidate the functions of the potential RsWRKY genes, which are involved in sequence-specific DNA binding, biological process regulation, gene expression regulation, among others. However, our results solely revealed fundamental transcription factor functions, and did not provide any information related to anthocyanin synthesis. The main reason for this limitation is the insufficient research, particularly in terms of confirmed experimental investigations, on the role of WRKY in anthocyanin biosynthesis. As a result, further research is imperative to validate the candidate RsWRKY genes that were identified in this study to be associated with anthocyanin production.
5 Conclusion
In summary, we systematically identified and classified 57 WRKY genes from the R. simsii genome, revealing their structural and phylogenetic characteristics. Our comparative genomic analysis suggests that the WRKY gene family has significantly expanded during plant evolution, primarily through WGD duplication, and that selective pressure analysis indicates strong purifying selection during evolution. Synteny analysis further confirmed the conservation of RsWRKY genes with orthologs in A. thaliana and O. sativa. Additionally, we conducted expression profiling analysis of RsWRKY genes in three azalea varieties, providing a valuable resource for future investigation into the molecular control of anthocyanin synthesis in azalea. While further studies are needed to fully understand the functional mechanisms of candidate RsWRKY genes, our comprehensive analysis provides a basis for investigating the molecular mechanisms of anthocyanin biosynthesis in Rhododendron species and lay the foundation for future functional studies of WRKY genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
CW and XW designed the research. CW and DY performed experimental works and data analysis. YL and PH participated in data analysis. RX provided experimental materials. CW prepared the original draft. XW provided support for projects and funds, and revised the manuscript. All authors contributed to the article and approved the submitted version. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was funded by Scientific Research Project of Education Department of Hubei Province, grant number Q20222703.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1172321/full#supplementary-material
References
Alabd, A., Ahmad, M., Zhang, X., Gao, Y., Peng, L., Zhang, L., et al. (2022). Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 9. doi:10.1093/hr/uhac199
An, J., Zhang, X., You, C., Bi, S., Wang, X., and Hao, Y. (2019). Md WRKY 40 promotes wounding-induced anthocyanin biosynthesis in association with Md MYB 1 and undergoes Md BT 2-mediated degradation. New Phytol. 224 (1), 380–395. doi:10.1111/nph.16008
Baillo, E. H., Hanif, M. S., Guo, Y., Zhang, Z., Xu, P., and Algam, S. A. (2020). Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L) moench). PLoS One 15 (8), e0236651. doi:10.1371/journal.pone.0236651
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, C., Chen, X., Han, J., Lu, W., and Ren, Z. (2020b). Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC plant Biol. 20 (1), 443. doi:10.1186/s12870-020-02625-8
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36 (5-6), 311–335. doi:10.1080/07352689.2018.1441103
Chen, S., Li, C., Zhu, X., Deng, Y., Sun, W., Wang, L., et al. (2012). The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 56 (3), 458–464. doi:10.1007/s10535-012-0069-3
Cheng, S., Xiaomeng, L., Yongling, L., Zhang, W., Jiabao, Y., Shen, R., et al. (2019). Genome-wide identification of WRKY family genes and analysis of their expression in response to abiotic stress in Ginkgo biloba L. Not. Bot. Horti Agrobot. Cluj-Napoca 47 (4), 1100–1115. doi:10.15835/nbha47411651
Cong, L., Qu, Y., Sha, G., Zhang, S., Ma, Y., Chen, M., et al. (2021). PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plant 173 (4), 1841–1849. doi:10.1111/ppl.13525
Cui, D., Zhao, S., Xu, H., Allan, A. C., Zhang, X., Fan, L., et al. (2021). The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol. Biol. 106 (4-5), 407–417. doi:10.1007/s11103-021-01160-w
De Keyser, E., Desmet, L., Van Bockstaele, E., and De Riek, J. (2013). How to perform RT-qPCR accurately in plant species? A case study on flower colour gene expression in an azalea (Rhododendron simsii hybrids) mapping population. BMC Mol. Biol. 14, 13–15. doi:10.1186/1471-2199-14-13
Debeaujon, I., Nesi, N., Perez, P., Devic, M., Grandjean, O., Caboche, M., et al. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15 (11), 2514–2531. doi:10.1105/tpc.014043
Dou, L., Zhang, X., Pang, C., Song, M., Wei, H., Fan, S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289 (6), 1103–1121. doi:10.1007/s00438-014-0872-y
Duan, S., Wang, J., Gao, C., Jin, C., Li, D., Peng, D., et al. (2018). Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 268, 47–53. doi:10.1016/j.plantsci.2017.12.010
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7 (10), e1002195. doi:10.1371/journal.pcbi.1002195
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi:10.1016/s1360-1385(00)01600-9
Forkmann, G., and Martens, S. (2001). Metabolic engineering and applications of flavonoids. Curr. Opin. Biotechnol. 12 (2), 155–160. doi:10.1016/s0958-1669(00)00192-0
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L) WRKY gene family. J. Exp. Bot. 65 (6), 1513–1528. doi:10.1093/jxb/eru007
Hao, F., Yang, G., Zhou, H., Yao, J., Liu, D., Zhao, P., et al. (2021). Genome-wide identification and transcriptional expression profiles of transcription factor WRKY in common walnut (juglans regia L). Genes (Basel) 12 (9), 1444. doi:10.3390/genes12091444
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31 (7), 1199–1217. doi:10.1007/s00299-012-1241-0
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS One 11 (6), e0157558. doi:10.1371/journal.pone.0157558
He, Y., Wang, Z., Ge, H., Liu, Y., and Chen, H. (2021). Weighted gene co-expression network analysis identifies genes related to anthocyanin biosynthesis and functional verification of hub gene SmWRKY44. Plant Sci. 309, 110935. doi:10.1016/j.plantsci.2021.110935
Hu, J., Fang, H., Wang, J., Yue, X., Su, M., Mao, Z., et al. (2020). Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 292, 110377. doi:10.1016/j.plantsci.2019.110377
Hu, W., Ren, Q., Chen, Y., Xu, G., and Qian, Y. (2021). Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC plant Biol. 21 (1), 427. doi:10.1186/s12870-021-03206-z
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287 (6), 495–513. doi:10.1007/s00438-012-0696-6
Hurst, L. D. (2002). The ka/ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 18 (9), 486. doi:10.1016/s0168-9525(02)02722-1
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65 (22), 6629–6644. doi:10.1093/jxb/eru381
Klopfenstein, D. V., Zhang, L., Pedersen, B. S., Ramirez, F., Warwick Vesztrocy, A., Naldi, A., et al. (2018). Goatools: A Python library for gene ontology analyses. Sci. Rep. 8 (1), 10872. doi:10.1038/s41598-018-28948-z
Kumar, K., Srivastava, V., Purayannur, S., Kaladhar, V. C., Cheruvu, P. J., and Verma, P. K. (2016). WRKY domain-encoding genes of a crop legume chickpea (cicer arietinum): Comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s). DNA Res. 23 (3), 225–239. doi:10.1093/dnares/dsw010
Lee, T. H., Tang, H., Wang, X., and Paterson, A. H. (2013). Pgdd: A database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. Database issue). doi:10.1093/nar/gks1104
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi:10.1093/nar/gkab301
Letunic, I., Khedkar, S., and Bork, P. (2021). Smart: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi:10.1093/nar/gkaa937
Li, C., Wu, J., Hu, K. D., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020a). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7, 37. doi:10.1038/s41438-020-0254-z
Li, P., Chai, Z., Lin, P., Huang, C., Huang, G., Xu, L., et al. (2020b). Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L). BMC Genomics 21 (1), 685. doi:10.1186/s12864-020-07076-x
Li, S. (2014). Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 9 (1), e27522. doi:10.4161/psb.27522
Li, W., Pang, S., Lu, Z., and Jin, B. (2020c). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants (Basel) 9 (11), 1515. doi:10.3390/plants9111515
Liu, H., Wang, X., Wang, G., Cui, P., Wu, S., Ai, C., et al. (2021). The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 7 (6), 748–756. doi:10.1038/s41477-021-00933-x
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15 (1), 281. doi:10.1186/1471-2164-15-281
Liu, W., Wang, Y., Yu, L., Jiang, H., Guo, Z., Xu, H., et al. (2019a). MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5. J. Agric. Food Chem. 67 (32), 8783–8793. doi:10.1021/acs.jafc.9b02920
Liu, Z., Xie, J., Wang, H., Zhong, X., Li, H., Yu, J., et al. (2019b). Identification and expression profiling analysis of NBS–LRR genes involved in Fusarium oxysporum f. sp. conglutinans resistance in cabbage. 3 Biotech. 9 (5), 202–212. doi:10.1007/s13205-019-1714-8
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiology 58 (9), 1431–1441. doi:10.1093/pcp/pcx075
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 (5494), 1151–1155. doi:10.1126/science.290.5494.1151
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant physiol. 148 (4), 1772–1781. doi:10.1104/pp.108.124867
Maher, C., Stein, L., and Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16 (4), 510–519. doi:10.1101/gr.4680506
Mao, Z., Jiang, H., Wang, S., Wang, Y., Yu, L., Zou, Q., et al. (2021). The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 306, 110848. doi:10.1016/j.plantsci.2021.110848
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. 37 (5), 1530–1534. doi:10.1093/molbev/msaa015
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi:10.1093/nar/gkaa913
Nan, H., and Gao, L.-z. (2019). Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Genet. 10, 363. doi:10.3389/fgene.2019.00363
Ning, P., Liu, C., Kang, J., and Lv, J. (2017). Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L) and differential expression under water deficit condition. PeerJ 5, e3232. doi:10.7717/peerj.3232
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 (1), 205–217. doi:10.1006/jmbi.2000.4042
Olech, M., Lyko, L., and Nowak, R. (2020). Influence of accelerated solvent extraction conditions on the LC-ESI-MS/MS polyphenolic profile, triterpenoid content, and antioxidant and anti-lipoxygenase activity of Rhododendron luteum sweet leaves. Antioxidants (Basel) 9 (9), 822. doi:10.3390/antiox9090822
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20 (1), 38–23. doi:10.1186/s13059-019-1650-2
Qin, Y., Yu, H., Cheng, S., Liu, Z., Yu, C., Zhang, X., et al. (2022). Genome-wide analysis of the WRKY gene family in Malus domestica and the role of MdWRKY70L in response to drought and salt stresses. Genes (Basel) 13 (6), 1068. doi:10.3390/genes13061068
Ramamoorthy, R., Jiang, S.-Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell physiol. 49 (6), 865–879. doi:10.1093/pcp/pcn061
Ramsay, N. A., and Glover, B. J. (2005). MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends plant Sci. 10 (2), 63–70. doi:10.1016/j.tplants.2004.12.011
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 (5859), 64–69. doi:10.1126/science.1150646
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., and Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15 (1), 66. doi:10.1186/s12870-015-0456-y
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi:10.1016/j.tplants.2010.02.006
Silva Monteiro de Almeida, D., Oliveira Jordão do Amaral, D., Del-Bem, L.-E., Bronze dos Santos, E., Santana Silva, R. J., Peres Gramacho, K., et al. (2017). Genome-wide identification and characterization of cacao WRKY transcription factors and analysis of their expression in response to witches' broom disease. PLoS One 12 (10), e0187346. doi:10.1371/journal.pone.0187346
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7, 534. doi:10.3389/fpls.2016.00534
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, R., Haas, J., and Schwede, T. (2020). QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 36 (6), 1765–1771. doi:10.1093/bioinformatics/btz828
Tanaka, Y., Brugliera, F., and Chandler, S. (2009). Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 10 (12), 5350–5369. doi:10.3390/ijms10125350
Wan, Z., Li, X., Cheng, H., Zhang, J., Chen, Y., Xu, Y., et al. (2022). Comprehensive genomic survey, structural classification, and expression analysis of WRKY transcription factor family in Rhododendron simsii. Plants (Basel) 11 (21), 2967. doi:10.3390/plants11212967
Wang, B., Hu, W., Fang, Y., Feng, X., Fang, J., Zou, T., et al. (2022a). Comparative analysis of the MADS-box genes revealed their potential functions for flower and fruit development in longan (dimocarpus longan). Front. plant Sci. 12, 813798. doi:10.3389/fpls.2021.813798
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinforma. 8 (1), 77–80. doi:10.1016/S1672-0229(10)60008-3
Wang, P., Yue, C., Chen, D., Zheng, Y., Zhang, Q., Yang, J., et al. (2019). Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes genomics 41 (1), 17–33. doi:10.1007/s13258-018-0734-9
Wang, X. J., Peng, X. Q., Shu, X. C., Li, Y. H., Wang, Z., and Zhuang, W. B. (2022b). Genome-wide identification and characterization of PdbHLH transcription factors related to anthocyanin biosynthesis in colored-leaf poplar (Populus deltoids). BMC Genomics 23 (1), 244. doi:10.1186/s12864-022-08460-5
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 40 (7), 1071–1085. doi:10.1007/s00299-021-02691-8
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (1), W296–W303. doi:10.1093/nar/gky427
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25 (9), 1189–1191. doi:10.1093/bioinformatics/btp033
Xiang, Z. (2006). Advances in homology protein structure modeling. Curr. Protein Pept. Sci. 7 (3), 217–227. doi:10.2174/138920306777452312
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., and He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genomics 19 (1), 490. doi:10.1186/s12864-018-4880-x
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends plant Sci. 20 (3), 176–185. doi:10.1016/j.tplants.2014.12.001
Yan, H., Pei, X., Zhang, H., Li, X., Zhang, X., Zhao, M., et al. (2021). MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 22 (6), 3103. doi:10.3390/ijms22063103
Yang, F. S., Nie, S., Liu, H., Shi, T. L., Tian, X. C., Zhou, S. S., et al. (2020a). Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 11 (1), 5269. doi:10.1038/s41467-020-18771-4
Yang, Y., Liu, J., Zhou, X., Liu, S., and Zhuang, Y. (2020b). Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 8, e8777. doi:10.7717/peerj.8777
Yang, Y., Yao, G., Yue, W., Zhang, S., and Wu, J. (2015). Transcriptome profiling reveals differential gene expression in proanthocyanidin biosynthesis associated with red/green skin color mutant of pear (Pyrus communis L). Front. Plant Sci. 6, 795. doi:10.3389/fpls.2015.00795
Yoshida, K., Mori, M., and Kondo, T. (2009). Blue flower color development by anthocyanins: From chemical structure to cell physiology. Nat. Prod. Rep. 26 (7), 884–915. doi:10.1039/b800165k
Yu, Y., Wang, N., Hu, R., and Xiang, F. (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5 (1), 920. doi:10.1186/s40064-016-2647-x
Zhang, B., Zhu, Z. Z., Qu, D., Wang, B. C., Hao, N. N., Yang, Y. Z., et al. (2021a). MdBBX21, a B-box protein, positively regulates light-induced anthocyanin accumulation in apple peel. Front. Plant Sci. 12, 774446. doi:10.3389/fpls.2021.774446
Zhang, C., Wang, D., Yang, C., Kong, N., Shi, Z., Zhao, P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12 (7), e0181573. doi:10.1371/journal.pone.0181573
Zhang, H., Zhang, Z., Zhao, Y., Guo, D., Zhao, X., Gao, W., et al. (2021b). StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 49 (1), 102–114. doi:10.1071/FP21109
Zhang, J., Wang, Y., Mao, Z., Liu, W., Ding, L., Zhang, X., et al. (2022). Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple. Ecotoxicol. Environ. Saf. 232, 113274. doi:10.1016/j.ecoenv.2022.113274
Zhang, J., Yang, E., He, Q., Lin, M., Zhou, W., Pian, R., et al. (2019). Genome-wide analysis of the WRKY gene family in drumstick (Moringa oleifera Lam). PeerJ 7, e7063. doi:10.7717/peerj.7063
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5 (1), 1. doi:10.1186/1471-2148-5-1
Zhao, D., and Tao, J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6, 261. doi:10.3389/fpls.2015.00261
Zhao, X., Yang, J., Li, G., Sun, Z., Hu, S., Chen, Y., et al. (2021). Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 113 (4), 1761–1777. doi:10.1016/j.ygeno.2021.03.035
Keywords: azalea, WRKY, phylogenetic analysis, anthocyanin biosynthesis, expression pattern
Citation: Wang C, Ye D, Li Y, Hu P, Xu R and Wang X (2023) Genome-wide identification and bioinformatics analysis of the WRKY transcription factors and screening of candidate genes for anthocyanin biosynthesis in azalea (Rhododendron simsii). Front. Genet. 14:1172321. doi: 10.3389/fgene.2023.1172321
Received: 23 February 2023; Accepted: 02 May 2023;
Published: 10 May 2023.
Edited by:
Himanshu Sharma, National Agri-Food Biotechnology Institute, IndiaCopyright © 2023 Wang, Ye, Li, Hu, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojing Wang, xjwang8@gzu.edu.cn
 Cheng Wang
Cheng Wang Dan Ye1
Dan Ye1 Xiaojing Wang
Xiaojing Wang