- 1Department 2 of Gynecology, Remin Hospital of Wuhan University, Wuhan, China
- 2Institute of Computing Science and Technology, Guangzhou University, Guangzhou, China
Nucleolin protein expression is higher on the ovarian cancer cell surface. AS1411, a DNA aptamer, can bind with nucleolin protein specifically. In this study, we developed HA and ST DNA tiles to assemble six AS1411 aptamers to deliver doxorubicin. In addition, to superior serum stability and drug loading, HA-6AS and ST-6AS outperformed TDN-AS in cellular uptake. HA-6AS and ST-6AS exhibited satisfactory targeted cytotoxicity and achieved resounding lysosomal escape. Moreover, when injected into nude mice subcutaneous xenograft models, HA-6AS reached the peak in tumor more quickly than ST-6AS, and better expressed the active targeting ability of AS1411. Our study suggests that designing appropriate DNA tiles to assemble different aptamers to deliver different chemotherapeutic drugs is a promising treatment for ovarian cancer.
1 Introduction
Epithelial ovarian cancer has the highest mortality rate among gynecological malignancies (Lheureux et al., 2019b; Colombo et al., 2019; Kuroki and Guntupalli, 2020). On one hand, due to the lack of early diagnosis methods and specific symptoms, most patients are at an advanced stage when they are diagnosed (Broekman et al., 2018; Lheureux et al., 2019a; Walker et al., 2019; Govindarajan et al., 2020; Wang et al., 2021); on the other hand, traditional platinum-taxane is still the main chemotherapeutic agent for ovarian cancer (Pujade-Lauraine et al., 2010; Pignata et al., 2011; Gourley and Bookman, 2019; García-Martínez et al., 2020). However, because of its indiscriminate toxicity, patients often cannot tolerate the adverse effects of chemotherapy.
Studies are increasingly exploring targeted therapies for ovarian cancer. Aptamers, which are screened from a random single-stranded DNA or RNA library (Ellington and Szostak, 1990; Tuerk and Gold, 1990), have recently become popular for guiding delivery materials, such as Au nanoparticles (Amouzadeh Tabrizi et al., 2018; Kim et al., 2021; Yang et al., 2021), quantum dots (Kim et al., 2019; Zheng et al., 2019; Adegoke et al., 2020; Jamei et al., 2021), and organic polymers (Hashemi et al., 2020; Chen et al., 2021; Dai et al., 2021), to their intended location. In addition, aptamers efficiently bind to targets with high affinity and are low immunogenic (Wu et al., 2015; Chen et al., 2017; Hoosen et al., 2018; Zhu and Chen, 2018; Yan et al., 2021; Zhao et al., 2022). AS1411 is a 26-nt DNA aptamer with a G-quadruplex structure that binds to nucleolin protein, whose expression in several cancer cells is much higher than that in normal cells, and is located on the surface of ovarian cancer cells, whereas it is normally located in the nucleus of other cancer cells (Bates et al., 2017; Li et al., 2017; Yazdian-Robati et al., 2020; Tong et al., 2022). Therefore, AS1411 can be used as a target signal in ovarian cancer cells.
Since metallic nanoparticles, graphene oxide, and carbon nanotubes are toxic, the encapsulation efficiency and stability of liposomes are unsatisfactory, and extracellular vesicles are heterogeneous and difficult to preserve (Cheng et al., 2021); therefore, biomaterials with biocompatibility and biodegradation have been developed, such as DNA origami or tiles. The DNA tile is a basic assembly unit that uses single strand DNA with sticky ends based on the complementary base pairing principle (Evans and Winfree, 2017). Over the last 2 decades, it has become increasingly complex and diverse and can be used to construct more complicated 2D patterns and 3D structures (Shi et al., 2014)). Based on the DNA tile motif, a computable program to assemble several aptamers could be an efficient targeting drug delivery unit. In addition, doxorubicin (DOX, also known as doxorubicin) is a common chemotherapeutic drug in cancer. In 2018, the National Comprehensive Cancer Network (NCCN) guidelines included pegylated liposomal doxorubicin in the treatment of patients with initial treatment of ovarian cancer and patients with platinum-sensitive and platinum-resistant recurrent ovarian cancer. At the same time, DOX can be loaded into the DNA nanomaterials simply by intercalation; therefore, we explored the feasibility of using a structured aptamer loaded with DOX to cure ovarian cancer.
In this study, we assembled six AS1411 aptamers to two hexangular DNA tiles for the first time and constructed two efficient but different structured aptamers; HA-6AS, which is 3D tubular and ST-6AS, which has a 2D six-star structure. We discovered that HA-6AS and ST-6AS could load more DOX than DNA tetrahedron and deliver DOX into ovarian cancer cells, allowing it to escape from lysosomes and reach the nucleus. HA-6AS-DOX and ST-6AS-DOX exhibited rapid cytotoxicity in ovarian cancer cells but low cytotoxicity in normal ovarian epithelial cells. Furthermore, we demonstrated that the internalization patterns of HA-6AS and ST-6AS might be different; HA-6AS was more actively targeted, whereas ST-6AS was more passively targeted. We speculated that tubular HA DNA tiles may be more appropriate for the assembly of AS1411 aptamers. In summary, DNA tiles can be a useful biocompatible candidate for targeted drug delivery and cancer treatment.
2 Materials and methods
2.1 Cell culture
The SKOV3 and IOSE80 cell lines were purchased from the China Center for Type Culture Collection (CCTCC, China). SKOV3 cells were maintained in RPMI-1640 medium (Hyclone, United States) containing 10% fetal bovine serum (FBS, Gibco, United States) and 1% penicillin-streptomycin (Biosharp, China), and IOSE80 cells were cultured in DMEM (Hyclone, United States) with the same content of FBS and penicillin-streptomycin. All cells were incubated at 37°C with 5% CO2 and 95% humidified atmosphere.
2.2 One-step preparation of HA-6AS, ST-6AS and TDN-AS
All single DNA strands and AS1411 aptamers (with or without fluorophores) were synthesized and purchased from Sangon Biotech (China). They were normalized to 10 μM with 1× TAE buffer containing 12.5 mM Mg2+ and stored at 4°C. Equal volumes of HAs1, HAs2, HAs3, HAs4, HAs5, and HAs6 and 6-fold AS1411 were mixed together and self-assembled during a PCR annel program, which was set to a slow rate with a 1°C drop every 8 min after for 15 min at 95°C. The synthesis of ST-6AS was the same as that of HA-6AS. The anchored strands are referred to as the “handle.” For TDN-AS, equal volumes of handle-S1, S2, S3, S4, and AS1411 were mixed and then self-assembled during the same PCR annealing program. All structured aptamers were validated using 4% acrylamide gel electrophoresis. The branch strand sequences from 5′to 3′of the two DNA tiles and the DNA tetrahedron are described in the supplementary materials. The sequence of AS1411 with the anchored strands and 4 T spacer was as follows: Handle-4T-AS1411: TCT TCT TTC TTA CTT TTT GGT GGT GGT GGT TGT GGT GGT GGT GG.
2.3 AFM characterization of HA, ST, HA-6AS and ST-6AS
AFM imaging was conducted in Scan Analyst-fluid mode (Multimode 8, Bruker) or AC water topography (Cipher ES, Oxford). A 5 μL sample was dropped onto newly prepared mica, left for adsorption for 10 min, and then observed under a microscope.
2.4 TEM characterization of HA, ST, HA-6AS and ST-6AS
Ten μL sample was spotted onto a continuous carbon support film (Zhongjingkeyi Film Technology Co., Ltd., Beijing). After deposition for 10 min, the excess sample was removed, and 2 μL 3% uranyl acetate was dropped for negative staining for 1 min. The samples were analyzed using TEM (Thermo Fisher Scientific Talos F200C).
2.5 Serum stability assay
HA, ST, TDN, HA-6AS, ST-6AS, and TDN-AS were incubated in equal volumes of 10% FBS medium for different durations at 37°C. The samples were collected and frozen at −20°C. A 4% PAGE was used to visualize degradation.
2.6 DOX loading
HA-6AS, ST-6AS, and TDN-AS were mixed with an equal volume of DOX, and the mixture was incubated for 3 h at room temperature with gentle shaking before it was centrifuged for 20 min at 14,000 g. A MicroSpectrophotometer (KAIAO K5500Plus, China) was used to measure the concentration of dissociative DOX in the supernatant at 495 nm absorption with a 195 extinction coefficient.
2.7 Flow cytometry
HA-6(FAM-AS), ST-6(FAM-AS), and TDN-(FAM-AS) were diluted to different concentrations (62.5, 125, 250, and 500 nM and 1 μM). After 3 h of incubation, SKOV3 or IOSE80 cells were trypsinized and centrifuged (1,500 rpm × 5 min), washed twice with PBS, and resuspended gently in 150 μL PBS. The fluorescence intensity of FAM was measured with a Beckman CytoFLEX by counting 15,000 events.
2.8 Co-localization of DOX and DNA origami
DOX (1.5 × 104) was seeded into 12-well plates and cultured overnight. Cells were incubated with HA-6(FAM-AS)-DOX, ST-6(FAM-AS)-DOX, or TDN-(FAM-AS)-DOX, which were added to a final concentration of 500 nM AS1411 in PBS buffer for 3 h. After washing three times with PBS, the cells were photographed using an inverted fluorescence microscope (Olympus IX71, Japan).
2.9 Cell viability assay
SKOV3 and IOSE80 cells (1×104) were seeded into 96-well plates and cultured overnight. The cells were treated with various concentrations of AS1411, HA, ST, HA-6AS, and ST-6AS for 24 h to examine the toxicity of free AS1411 and structured AS1411. After that, SKOV3 and IOSE80 cells (1.5×104) were seeded into 96-well plates and cultured overnight. For the cytotoxicity assay, DOX, HA-6AS-DOX, ST-6AS-DOX, and free DOX were diluted with complete medium to achieve a series of equidifferent concentrations from 6 to 30 μM, which were calculated using DOX. After 3 h of treatment, the cells were incubated for 2 h with CCK8. The absorbance was measured using a microplate reader at 450 nm. When the cytotoxicity assay with DOX was extended to 24 h, DOX was diluted in an equal ratio of 2.4 nM–24 μM.
2.10 Confocal imaging for lysosomal escape
SKOV3 cells (5×103) were seeded in 35 mm confocal dishes with a glass bottom (Biosharp, China) and cultured overnight. The cells were then incubated with 83.3 μM HA-6(FAM-AS) and ST-6(FAM-AS) for 3 h, incubated with Lyso Tracker Red (Beyotime, China), fixed with 4% paraformaldehyde, and stained with DAPI (Biosharp, China). After incubation with each reagent, the cells were washed 3 times with PBS. Fluorescence imaging was performed using a Nikon ECLIPSE TI (Tokyo, Japan).
2.11 In vitro imaging in nude mice
Six eight-week-old female nude mice were injected subcutaneously with SKOV3 cells (1×106), and maintained under SPF conditions until the subcutaneous tumor reached 10 mm. The mice were randomly divided into two groups, with three mice per group. HA-6(Cy5-AS) and HA-6(Cy5-AS-like) were injected into mice at a single dose (0.83 μM via the tail vein). The mice were anaesthetized at 1, 2, and 4 h after injection and imaged using a PerkinElmer IVIS Lumina S3 Imaging System (Shanghai, China). In vitro imaging of ST-6(Cy5-AS) and ST-6(Cy5-AS-like) was performed after complete metabolism of the HA group.
2.12 Statistical analysis
Statistical analyses were performed using the GraphPad Prism (8.0.2, for Windows, GraphPad Software, San Diego, California United States, www.graphpad.com). The data are presented as the mean ± SDs. Welch’s t-test was used to compare two groups when the data were normally distributed with uniform variance, and the following p values were used to determine statistical significance: n. s., p > 0.05; *, p < 0.05; **, p < 0.01; and ***, p < 0.001; ****, p < 0.0001.
3 Results
3.1 Design and characterization of structured aptamers
HA and ST, designed by NUPACK, are structures that are based on DNA tile self-assembly. Both consist of six DNA single strands with sticky ends that can combine with the handle of the AS1411 aptamers, and these DNA single strands contain repeatable sequences. Inside the anchored sequences of the AS1411 aptamer handle, there is a 4 T-deoxynucleotide spacer (Figure 1A). HA-6AS has a tubular structure with good rigidity, and ST-6AS has a six-star structure with good flexibility. At the turn point of each branch strand, two T-deoxynucleotide transitions provide flexibility to each strand. NUPACK confirmed that both the HA-6AS and ST-6AS structures were thermally stable. Polyacrylamide gel electrophoresis (PAGE) results (Figure 1B; Supplementary Figure S1) showed that HA, ST, and structured aptamers HA-6AS and ST-6AS were successfully self-assembled. The corresponding quantity of AS1411 aptamers that increased gradually from one to six was integrated to demonstrate that HA and ST were successfully connected with six AS1411 aptamers.
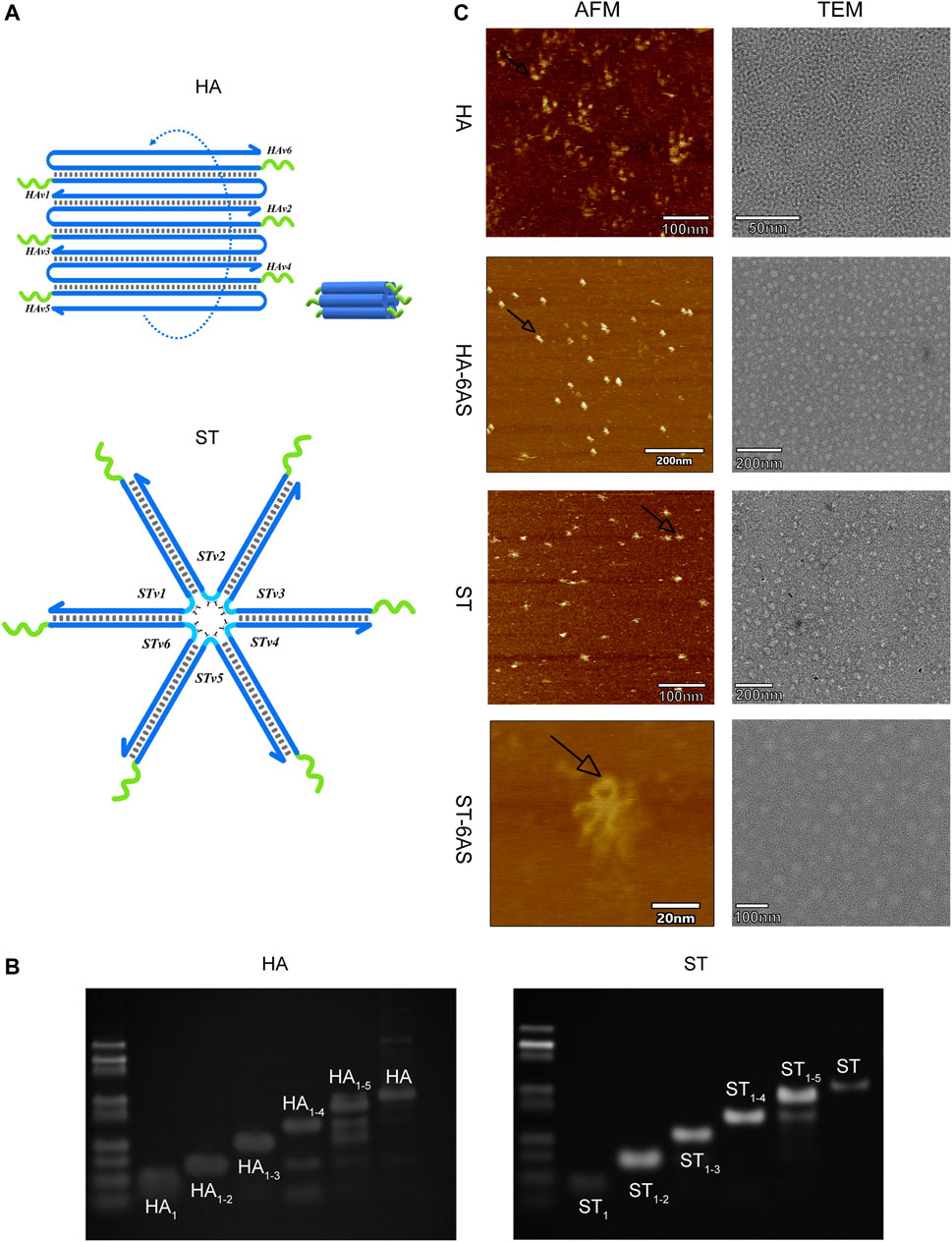
FIGURE 1. (A) Schematic diagram of the structure of HA and ST DNA tiles. The green curve represents the 4 T-deoxcnucleotide spacer and sequences anchored to the AS1411 aptamer handle. The blue curve represents each branch strand of HA or ST. (B) PAGE results show the integration of one to six strands of HA and ST branch strands. The most dominant band in the lane is the product. (C) AFM and TEM characterization of HA, ST, HA-6AS and ST-6AS. All samples had a concentration of 2 nM. The observed size of ST was about 15 nm, the observed size of HA was about 9 nm. The black arrows indicate the photographed dimer AS1411 aptamer.
The characterization of the two DNA tiles and structured aptamers was performed using AFM and TEM (Figure 1C). The theoretical particle size of ST should be 18 nm, and the theoretical cross-sectional diameter and length of HA should be 7–8 nm. The observed size of ST was approximately 15 nm, and the observed size of HA was approximately 9 nm. The AS1411 aptamers were closer to the HA DNA tiles than the ST tiles. The TEM images showed that HA-6AS and ST-6AS were more homogeneously dispersed than HA and ST. Surprisingly, dimeric AS1411 was photographed for the first time, which corroborated the existence of different forms of AS1411 aptamers.
3.2 Serum stability of structured aptamers
The ability of structured aptamers to remain stable in the serum is critical for effective drug delivery. The stability in medium containing 10% FBS at 37°C was evaluated. First, the structured aptamers were incubated in serum for 8 h, and no significant degradation was observed (Supplementary Figure S2). When the incubation time was extended to 24 h (Supplementary Figure S3), HA-6AS and ST-6AS were stable within 12 h, but began to degrade after 24 h.
Recently, DNA tetrahedra (TDN) have been widely used as delivery systems because of their rigid configuration, high flexibility, and ease of synthesis (Fan et al., 2020). Hence, our DNA tiles were compared with TDN (Supplementary Figures S2, S3). The TDN and TDN-AS were also treated in the same way, and the band of TDN nearly disappeared at 24 h, and the band of TDN-AS completely disappeared at 12 h. Consequently, the structured aptamers exhibited improved serum stability.
3.3 Determination and quantification of DOX loading
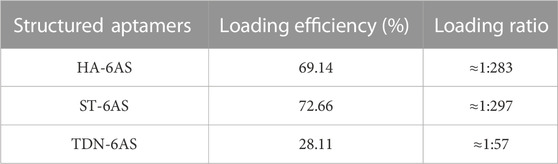
DOX can be loaded into DNA origami structures through intercalation (Zhao et al., 2012; Zhang et al., 2014; Song et al., 2017; Liu et al., 2018; Ge et al., 2020; Pan et al., 2020). After incubation at room temperature, DOX non-covalently conjugated with TDN-AS, HA-6AS, and ST-6AS. A concentration-absorbance standard curve of DOX was established first (Supplementary Figure S4). The loading efficiency and ratio were listed in Table 1. After centrifugation, the DOX concentration in the supernatant was calculated based on absorbance. The drug-loading rate was calculated using Formula 1 (Supplementary Formula S1). The relationship between incubation time and drug loading was then determined (Supplementary Figure S4). No bands were observed in lanes 5 or 8, which were incubated overnight. This could be because more DOX was bound to the structured aptamers, causing the overall positive charge to be excessive, even though the DNA was negatively charged. Structured aptamers loaded with DOX migrated at a slower rate than structured aptamers alone.
3.4 Co-localization of DNA carriers and DOX
To investigate whether HA-6AS and ST-6AS could successfully transport DOX into SKOV3 cells, fluorescence microscopy was performed. After incubating HA-6(FAM-AS) and ST-6(FAM-AS) with DOX overnight, SKOV3 cells were incubated with HA-6(FAM-AS)-DOX or ST-6(FAM-AS)-DOX for 3 h and washed three times before filming with the corresponding field. The fluorescence positions of DOX and FAM were exactly the same as the cellular position (Figure 2), verifying that HA-6AS and ST-6AS could deliver DOX into SKOV3 cells successfully as TDN-AS did.
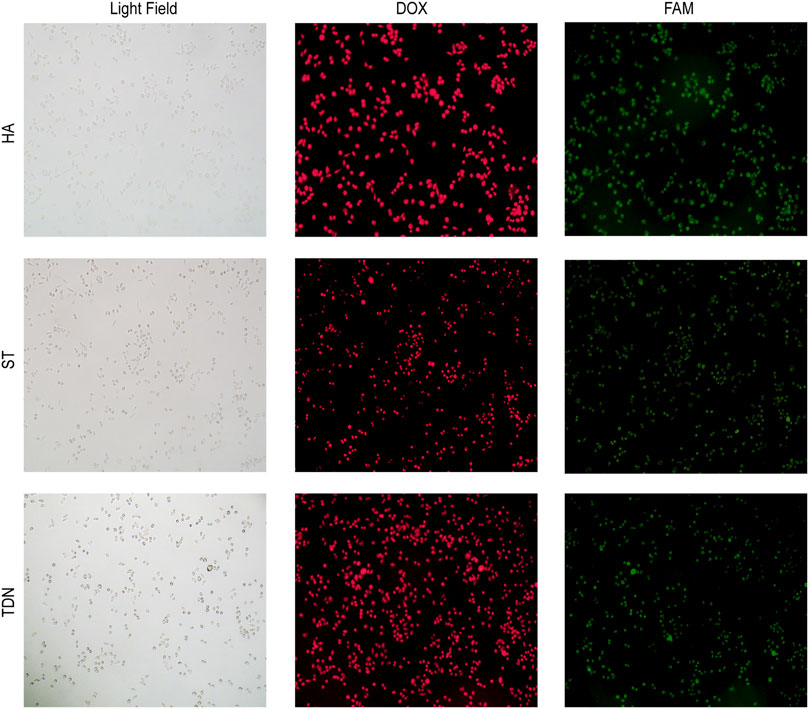
FIGURE 2. Co-localization results of HA-6AS-DOX, ST-6AS-DOX and TDN-AS-DOX in SKOV3 cells after incubation for 3 h at a concentration of 0.83 μM. The AS1411 aptamers were modified with FAM fluorophore. All images were taken under the same field of view with an inverted fluorescence microscope, the magnification was 100 times. DOX, doxorubicin; TDN, DNA tetrahedra.
3.5 Uptake of structured aptamers by cells
To compare the effects of different DNA molecular shapes on the cellular uptake of HA-6AS and ST-6AS, flow cytometry was performed. SKOV3 cells were incubated with HA-6(FAM-AS) and ST-6(FAM-AS) at the same concentration and their uptake was examined by detecting the intensity of FAM fluorescence in the FITC channel. Each sample was measured three times, and 15,000 events were counted. There was no significant difference in the uptake of HA-6(FAM-AS) and ST-6(FAM-AS) by the SKOV3 cells (Figure 3A). In addition, there was no significant uptake of HA-6FAM-AS or ST-6(FAM-AS) by IOSE80 cells. As shown in Figure 3B, the uptake of the two structured aptamers was concentration dependent in SKOV3 cells. To verify the specificity of AS1411 aptamers for cancer cells, a control AS1411 aptamer sequence called AS-like was designed. The length of the AS-like structure was the same as that of AS1411, but it could not form a G-quadruplet. Interestingly, the mean fluorescence intensity of HA-6(FAM-AS-like) was significantly lower than that of HA-6(FAM-AS); however, there were no differences between ST-6(FAM-AS) and ST-6(FAM-AS-like) when the concentration of AS1411 was below 500 nM. Furthermore, the mean fluorescence intensity of ST-6(FAM-AS) was lower than that of HA-6(FAM-AS) at all the concentrations tested.
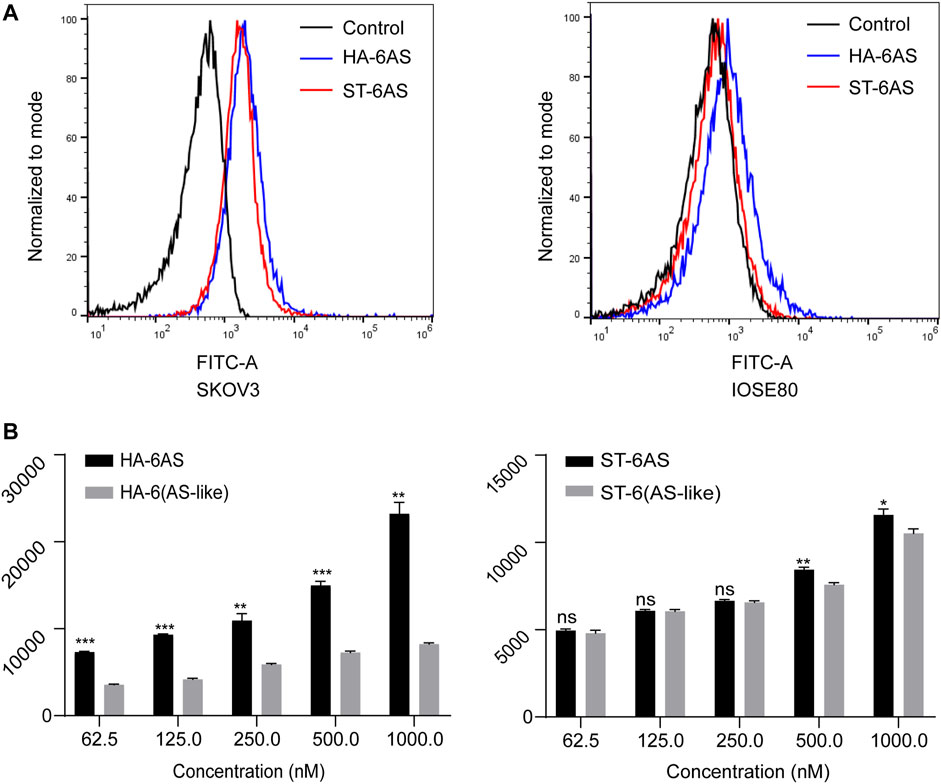
FIGURE 3. Flow cytometry analysis of the uptake in SKOV3 cells. (A) The uptake of HA-6AS and ST-6AS in SKOV3 cells, and the AS1411 aptamers were modified with FAM fluorophore. The SKOV3 cells were incubated with either HA-6AS and ST-6AS at a concentration of 0.83 μM for 3 h. (B) The concentration-dependent cellular uptake of the HA-6AS, HA-6(AS-like), ST-6AS, and ST-6(AS-like) in SKOV3 cells at a range of AS1411 concentrations fo r 3 h. The AS1411 aptamers were modified with FAM fluorophore. The data are presented as means ± SDs, n = 3, the following p values were used to determine statistical significance: ns, p > 0.05; *, p < 0.05; **, p < 0.01 and ***, p < 0.001.
3.6 Cell viability
To examine the cytotoxicity of free AS1411, HA, and ST, HA-6AS, ST-6AS, SKOV3, and IOSE80 cells were incubated with a series of concentrations of these DNA nanostructures for 24 h, and cell viability was detected using the CCK8 reagent (Figure 4A; Supplementary Figure S5). The concentration of free AS1411 ranged from 0 to 1 μM, and the concentration of HA and ST individuals was the same as that of HA-6AS and ST-6AS, of which AS1411 concentration was referred to as free AS1411. There were no obvious losses in the viability of IOSE80 cells when incubated with any of these compounds. To examine the targeting of HA-6AS-DOX, ST-6AS-DOX, SKOV3, and IOSE80 cells were incubated for 3 h with a range of concentrations of HA-6AS-DOX, ST-6AS-DOX, and AS1411 cells with DOX and free DOX. All DOX concentrations were maintained the same in each group (Figure 4B). The viability of SKOV3 cells was visibly lower when incubated with HA-6AS-DOX and ST-6AS-DOX than when incubated with AS1411 cells treated with DOX or free DOX, and there was no statistical difference between HA-6AS-DOX and ST-6AS-DOX. When IOSE80 cells were treated (Figure 4C), the results were completely different. When the incubation time was extended to 24 h, there were no significant differences between DOX-loaded HA-6AS-DOX, ST-6AS-DOX, and free DOX (Supplementary Figure S6). This might be due to the non-covalent binding of DOX and DNA structures; when the incubation time was prolonged, all DOX was fully released into the medium.
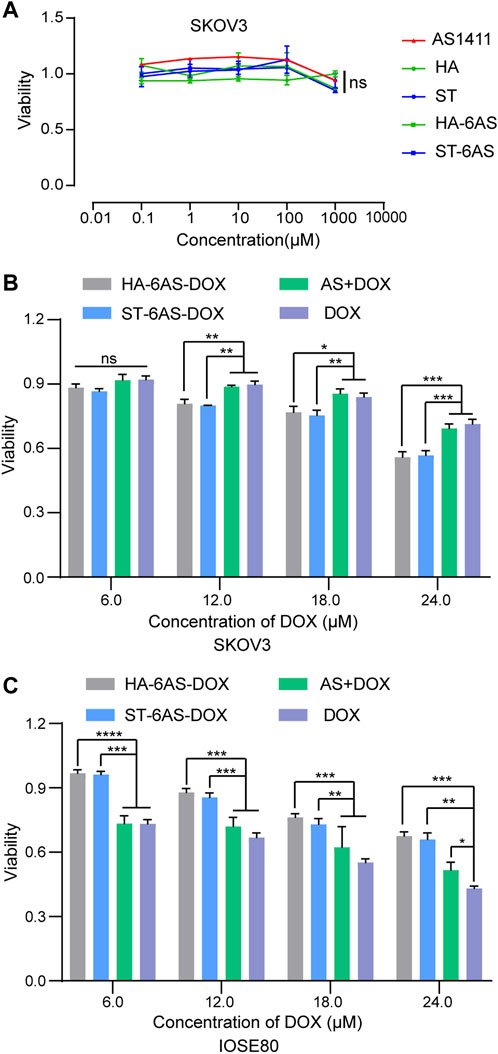
FIGURE 4. (A) Cell viability of SKOV3 cells after 24 h treatment with AS1411, HA, ST, HA-6AS, and ST-6AS evaluated using CCK8 assay. The concentration of each compound ranged from 0.01 to 10,000 μM. The data are presented as the means ± SDs. n = 3. (B)&(C) Cell viability of SKOV3 (B), IOSE80 (C) cells after 3 h treatment with HA-6AS-DOX, ST-6AS-DOX, DOX, AS + DOX evaluated using CCK8 assays. The DOX concentration of each compound ranged from 6 to 24 μM. The data are presented as the means ± SDs. n = 3, the following p values were used to determine statistical significance: ns, p > 0.05; *, p < 0.05; **, p < 0.01 and ***, p < 0.001, ****, p < 0.0001.
3.7 Lysosomal escape
Considering the therapeutic mechanism of DOX, it is not desirable for it to be engulfed by lysosomes. At the same time, nucleolin, the target of AS1411, is also expressed in cell nucleus. Therefore, to investigate whether HA-6AS and ST-6AS could deliver DOX into the nucleus and escape lysosomal phagocytosis, confocal laser imaging was performed to analyze the colocalization of HA-6AS or ST-6AS, nuclei, and lysosomes. After incubation with HA-6(FAM-AS) or ST-6(FAM-AS) for 3 h, SKOV3 cells were dyed with Lyso Tracker Red and DAPI, fixed with 4% paraformaldehyde, and imaged. Figure 5 shows the representative confocal images. There was no significant correlation between the position of HA-6(FAM-AS) or ST-6(FAM-AS) and the position of lysosomes. The Pearson’s R values of the fluorescence images were analyzed using Fiji, and the results demonstrated that they weren’t related to colocalization (Supplementary Figure S7).
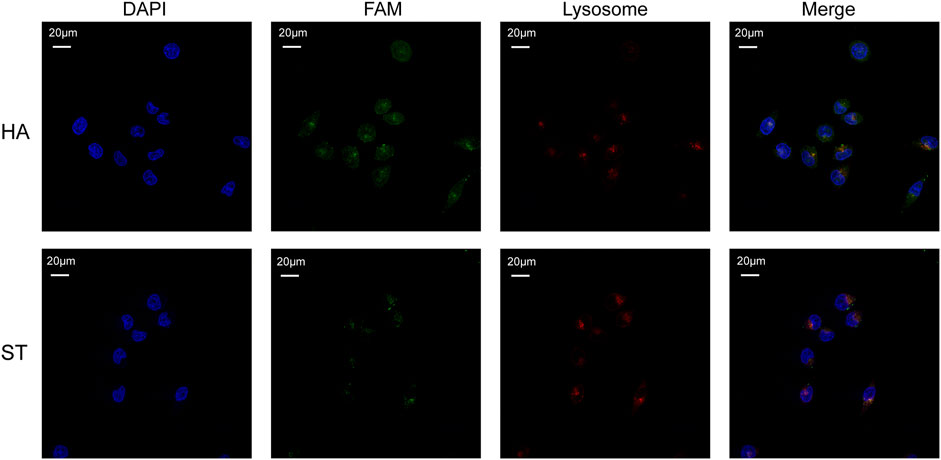
FIGURE 5. Representative confocal microscopy images showing the co-localization of nuclei (blue), HA-6AS or ST-6AS (green), lysosomes (red) in SKOV3 cells. The nuclei were dyed with DAPI, the AS1411 aptamers were modified with FAM fluorophore, the lysosomes were marked with Lyso Tracker Red. The SKOV3 cells were incubated with HA-6AS or ST-6AS for 3 h, marked with lysosomal tracker, and dyed with DAPI.
3.8 In vivo distribution of the structured aptamers
To examine the tumor targeting property of HA-6AS and ST-6AS, in vitro fluorescence imaging experiments were performed in nude mice. One, two, and 4 h after tail vein injection, the mice were anaesthetized for whole-body imaging. One hour after injection, the fluorescence intensity of HA-6(Cy5-AS) and HA-6(Cy5-AS-like) in the tumor reached its peak and then gradually declined, and 4 h after injection, the fluorescence completely disappeared in the tumor. The mean fluorescence intensity in the tumor region of HA-6(Cy5-AS) was obviously higher than that of HA-6(Cy5-AS-like) in the first 2 h, then became indistinct. After injection, the fluorescence intensity of ST-6(Cy5-AS) and ST-6(Cy5-AS-like) both reached a peak after 2 h, and ST-6(Cy5-AS) remained until 4 h, but ST-6(Cy5-AS-like) significantly decreased after 4 h. Additionally, the mean fluorescence intensity in the tumor region of ST-6(Cy5-AS) and ST-6(Cy5-AS-like) showed no difference at the beginning, but only became significantly different after 4 h (Figure 6; Supplementary Figure S7).
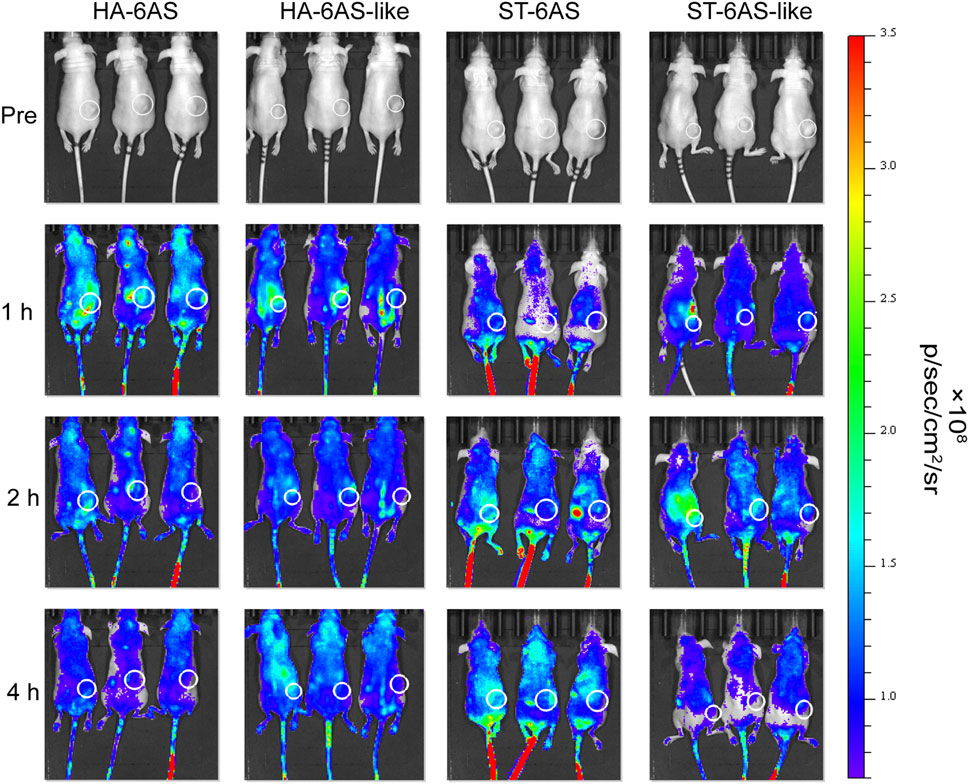
FIGURE 6. In vivo distribution of Cy5 fluorescence in nude mice 1, 2, and 4 h after tail vein injection of HA-6Cy5-AS, HA-6C-AS-like, ST-6Cy5-AS, and ST-6Cy5-AS-like at a concentration of 0.83 μM. The white circles indicate the subcutaneous tumors in nude mice. The scale of fluorescence intensity was consistent for all pictures.
4 Discussion
First, we designed two DNA tiles that could assemble six AS1411 aptamers into specific structures. The experimental results showed that the targeting and drug-loading capabilities of an HA or ST individual significantly outperformed those of TDN. The synthesis of HA-6AS and ST-6AS was simple; only one-pot annealing after mixing was required, and AFM and PAGE results showed that the yield was high. Serum stability experiments also showed that HA-6AS and ST-6AS were more stable for a longer time than TDN-AS. DOX is an anthracycline drug that is beneficial for improving the survival rate in patients with advanced ovarian cancer (45) and can be easily loaded into DNA. Therefore, we compared the DOX-loading ability of our DNA tiles and TDN. The experimental results revealed the outstanding loading efficiency and ratio of HA-6AS and ST-6AS. Furthermore, colocalization experiments verified that our structured aptamers could deliver DOX into SKOV3 cells successfully. To explore whether structured AS1411 affects the efficiency of cellular uptake, flow cytometry was performed. SKOV3 cells showed uptake of two types of structured aptamers, but IOSE80 cells didn’t, and cellular uptake of HA and ST was six times more efficient than that of TDN. The uptake of HA-6AS and ST-6AS showed concentration dependence. In addition, the mean fluorescence intensity of ST-6(FAM-AS) was lower than that of HA-6(FAM-AS) at all concentrations tested. The cell viability assays showed that free AS1411 aptamers, HA and ST DNA tiles individuals, HA-6AS, and ST-6AS were non-toxic. Simultaneously, HA-6AS-DOX and ST-6AS-DOX killed SKOV3 cells more rapidly than AS1411 with DOX or free DOX and showed low toxicity to IOSE80 cells. Confocal laser imaging experiments verified that HA-6AS and ST-6AS escaped lysosomal phagocytosis. Finally, in vitro fluorescence imaging of nude mice revealed high specificity for HA-6AS, which reached the tumor tissue more quickly than ST-6AS.
The reason for the flow cytometry and in vivo fluorescence imaging results may be that the ST structure is prone to passive uptake by ovarian cancer cells. This shows that, even at the same molecular weight, the two structures can differ. The smaller particle size of HA-6AS makes it easier to reach the tumor tissue, whereas ST-6AS takes an unfolded form; therefore, it did not play a good role in the targeting of AS1411 aptamers as the HA structure. Moreover, the mean fluorescence intensity of HA-6AS was higher than that of ST-6AS in these two experiments. From our experimental results, we can easily conclude that the HA structure is more suitable for targeted therapy than the ST structure, which may be because the distance of AS1411 aptamers on the HA DNA tiles is more suitable for the formation of G-quadruplexes. In conclusion, we hope that structured aptamers can further improve the therapeutic effect of chemotherapeutic drugs, such as DOX, and that different aptamers need to be designed with different structures to improve their efficiency.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the Institutional Review Board (or Ethics Committee) of Remin Hospital of Wuhan University.
Author contributions
Conceptualization: LR, XC, and XS. Methodology: LR, LH, and XC. Investigation: LR, LH, XC, GS, XW, YL, and CG. Visualization: LR, CX, and LH. Supervision: XL. Writing—original draft: LR and XC. Writing—review and editing: XL, XS, LR, and XC.
Funding
This study was supported by the National Natural Science Fund of China (Grant No. 62172302).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1170260/full#supplementary-material
References
A'Hern, R. P., and Gore, M. E. (1995). Impact of doxorubicin on survival in advanced ovarian cancer. J. Clin. Oncol. 13, 726–732. doi:10.1200/JCO.1995.13.3.726
Adegoke, O., Pereira-Barros, M. A., Zolotovskaya, S., Abdolvand, A., and Daeid, N. N. (2020). Aptamer-based cocaine assay using a nanohybrid composed of ZnS/Ag(2)Se quantum dots, graphene oxide and gold nanoparticles as a fluorescent probe. Mikrochim. Acta 187, 104. doi:10.1007/s00604-019-4101-6
Amouzadeh Tabrizi, M., Shamsipur, M., Saber, R., and Sarkar, S. (2018). Isolation of HL-60 cancer cells from the human serum sample using MnO(2)-PEI/Ni/Au/aptamer as a novel nanomotor and electrochemical determination of thereof by aptamer/gold nanoparticles-poly(3,4-ethylene dioxythiophene) modified GC electrode. Biosens. Bioelectron. 110, 141–146. doi:10.1016/j.bios.2018.03.034
Bates, P. J., Reyes-Reyes, E. M., Malik, M. T., Murphy, E. M., O'Toole, M. G., and Trent, J. O. (2017). G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 1861, 1414–1428. doi:10.1016/j.bbagen.2016.12.015
Broekman, K. E., Jalving, M., Van Tinteren, H., Sessa, C., and Reyners, A. K. L. (2018). Clinical benefit of controversial first line systemic therapies for advanced stage ovarian cancer - ESMO-MCBS scores. Cancer Treat. Rev. 69, 233–242. doi:10.1016/j.ctrv.2018.06.008
Chen, K., Liu, B., Yu, B., Zhong, W., Lu, Y., Zhang, J., et al. (2017). Advances in the development of aptamer drug conjugates for targeted drug delivery. Wiley Interdiscip. Rev. Nanomed Nanobiotechnol 9. doi:10.1002/wnan.1438
Chen, Q., Zhang, Y., Chen, H., Liu, J., and Liu, J. (2021). Enhancing the sensitivity of DNA and aptamer probes in the dextran/PEG aqueous two-phase system. Anal. Chem. 93, 8577–8584. doi:10.1021/acs.analchem.1c01419
Cheng, Z., Li, M., Dey, R., and Chen, Y. (2021). Nanomaterials for cancer therapy: Current progress and perspectives. J. Hematol. Oncol. 14, 85. doi:10.1186/s13045-021-01096-0
Colombo, N., Sessa, C., Du Bois, A., Ledermann, J., Mccluggage, W. G., Mcneish, I., et al. (2019). ESMO-ESGO consensus conference recommendations on ovarian cancer: Pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Ann. Oncol. 30, 672–705. doi:10.1093/annonc/mdz062
Dai, L., Wei, D., Zhang, J., Shen, T., Zhao, Y., Liang, J., et al. (2021). Aptamer-conjugated mesoporous polydopamine for docetaxel targeted delivery and synergistic photothermal therapy of prostate cancer. Cell Prolif. 54, e13130. doi:10.1111/cpr.13130
Ellington, A. D., and Szostak, J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. doi:10.1038/346818a0
Evans, C. G., and Winfree, E. (2017). Physical principles for DNA tile self-assembly. Chem. Soc. Rev. 46, 3808–3829. doi:10.1039/c6cs00745g
Fan, D., Wang, J., Wang, E., and Dong, S. (2020). Propelling DNA computing with materials' power: Recent advancements in innovative DNA logic computing systems and smart bio-applications. Adv. Sci. (Weinh) 7, 2001766. doi:10.1002/advs.202001766
GarcíA-MartíNEZ, E., Redondo, A., Piulats, J. M., RodríGUEZ, A., and Casado, A. (2020). Are antiangiogenics a good 'partner' for immunotherapy in ovarian cancer? Angiogenesis 23, 543–557. doi:10.1007/s10456-020-09734-w
Ge, Z., Guo, L., Wu, G., Li, J., Sun, Y., Hou, Y., et al. (2020). DNA origami-enabled engineering of ligand-drug conjugates for targeted drug delivery. Small 16, e1904857. doi:10.1002/smll.201904857
Gourley, C., and Bookman, M. A. (2019). Evolving concepts in the management of newly diagnosed epithelial ovarian cancer. J. Clin. Oncol. 37, 2386–2397. doi:10.1200/JCO.19.00337
Govindarajan, M., Wohlmuth, C., Waas, M., Bernardini, M. Q., and Kislinger, T. (2020). High-throughput approaches for precision medicine in high-grade serous ovarian cancer. J. Hematol. Oncol. 13, 134. doi:10.1186/s13045-020-00971-6
Hashemi, M., Shamshiri, A., Saeedi, M., Tayebi, L., and Yazdian-Robati, R. (2020). Aptamer-conjugated PLGA nanoparticles for delivery and imaging of cancer therapeutic drugs. Arch. Biochem. Biophys. 691, 108485. doi:10.1016/j.abb.2020.108485
Hoosen, Y., Pradeep, P., Kumar, P., Du Toit, L. C., Choonara, Y. E., and Pillay, V. (2018). Nanotechnology and glycosaminoglycans: Paving the way forward for ovarian cancer intervention. Int. J. Mol. Sci. 19, 731. doi:10.3390/ijms19030731
Jamei, H. R., Rezaei, B., and Ensafi, A. A. (2021). Ultra-sensitive and selective electrochemical biosensor with aptamer recognition surface based on polymer quantum dots and C(60)/MWCNTs-polyethylenimine nanocomposites for analysis of thrombin protein. Bioelectrochemistry 138, 107701. doi:10.1016/j.bioelechem.2020.107701
Kim, J., Park, H., Saravanakumar, G., and Kim, W. J. (2021). Polymer/aptamer-integrated gold nanoconstruct suppresses the inflammatory process by scavenging ROS and capturing pro-inflammatory cytokine TNF-α. ACS Appl. Mater Interfaces 13, 9390–9401. doi:10.1021/acsami.0c15727
Kim, M. W., Jeong, H. Y., Kang, S. J., Jeong, I. H., Choi, M. J., You, Y. M., et al. (2019). Anti-EGF receptor aptamer-guided Co-delivery of anti-cancer siRNAs and quantum dots for theranostics of triple-negative breast cancer. Theranostics 9, 837–852. doi:10.7150/thno.30228
Kuroki, L., and Guntupalli, S. R. (2020). Treatment of epithelial ovarian cancer. Bmj 371, m3773. doi:10.1136/bmj.m3773
Lheureux, S., Braunstein, M., and Oza, A. M. (2019a). Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 69, 280–304. doi:10.3322/caac.21559
Lheureux, S., Gourley, C., Vergote, I., and Oza, A. M. (2019b). Epithelial ovarian cancer. Lancet 393, 1240–1253. doi:10.1016/S0140-6736(18)32552-2
Li, F., Lu, J., Liu, J., Liang, C., Wang, M., Wang, L., et al. (2017). A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat. Commun. 8, 1390. doi:10.1038/s41467-017-01565-6
Liu, J., Song, L., Liu, S., Zhao, S., Jiang, Q., and Ding, B. (2018). A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew. Chem. Int. Ed. Engl. 57, 15486–15490. doi:10.1002/anie.201809452
Pan, Q., Nie, C., Hu, Y., Yi, J., Liu, C., Zhang, J., et al. (2020). Aptamer-functionalized DNA origami for targeted codelivery of antisense oligonucleotides and doxorubicin to enhance therapy in drug-resistant cancer cells. ACS Appl. Mater Interfaces 12, 400–409. doi:10.1021/acsami.9b20707
Pignata, S., Scambia, G., Ferrandina, G., Savarese, A., Sorio, R., Breda, E., et al. (2011). Carboplatin plus paclitaxel versus carboplatin plus pegylated liposomal doxorubicin as first-line treatment for patients with ovarian cancer: The MITO-2 randomized phase III trial. J. Clin. Oncol. 29, 3628–3635. doi:10.1200/JCO.2010.33.8566
Pujade-Lauraine, E., Wagner, U., Aavall-Lundqvist, E., Gebski, V., Heywood, M., Vasey, P. A., et al. (2010). Pegylated liposomal Doxorubicin and Carboplatin compared with Paclitaxel and Carboplatin for patients with platinum-sensitive ovarian cancer in late relapse. J. Clin. Oncol. 28, 3323–3329. doi:10.1200/JCO.2009.25.7519
Shi, X., Lu, W., Wang, Z., Pan, L., Cui, G., Xu, J., et al. (2014). Programmable DNA tile self-assembly using a hierarchical sub-tile strategy. Nanotechnology 25, 075602. doi:10.1088/0957-4484/25/7/075602
Song, L., Jiang, Q., Liu, J., Li, N., Liu, Q., Dai, L., et al. (2017). DNA origami/gold nanorod hybrid nanostructures for the circumvention of drug resistance. Nanoscale 9, 7750–7754. doi:10.1039/c7nr02222k
Tong, X., Ga, L., Ai, J., and Wang, Y. (2022). Progress in cancer drug delivery based on AS1411 oriented nanomaterials. J. Nanobiotechnology 20, 57. doi:10.1186/s12951-022-01240-z
Tuerk, C., and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. doi:10.1126/science.2200121
Walker, M., Jacobson, M., and Sobel, M. (2019). Management of ovarian cancer risk in women with BRCA1/2 pathogenic variants. Cmaj 191, E886–e893. doi:10.1503/cmaj.190281
Wang, Z., Meng, F., and Zhong, Z. (2021). Emerging targeted drug delivery strategies toward ovarian cancer. Adv. Drug Deliv. Rev. 178, 113969. doi:10.1016/j.addr.2021.113969
Wu, X., Chen, J., Wu, M., and Zhao, J. X. (2015). Aptamers: Active targeting ligands for cancer diagnosis and therapy. Theranostics 5, 322–344. doi:10.7150/thno.10257
Yan, J., Gao, T., Lu, Z., Yin, J., Zhang, Y., and Pei, R. (2021). Aptamer-targeted photodynamic platforms for tumor therapy. ACS Appl. Mater Interfaces 13, 27749–27773. doi:10.1021/acsami.1c06818
Yang, Y., Han, Y., Sun, Q., Cheng, J., Yue, C., Liu, Y., et al. (2021). Au-siRNA@ aptamer nanocages as a high-efficiency drug and gene delivery system for targeted lung cancer therapy. J. Nanobiotechnology 19, 54. doi:10.1186/s12951-020-00759-3
Yazdian-Robati, R., Bayat, P., Oroojalian, F., Zargari, M., Ramezani, M., Taghdisi, S. M., et al. (2020). Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 155, 1420–1431. doi:10.1016/j.ijbiomac.2019.11.118
Zhang, Q., Jiang, Q., Li, N., Dai, L., Liu, Q., Song, L., et al. (2014). DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 8, 6633. doi:10.1021/nn502058j
Zhao, J., Tan, W., Zheng, J., Su, Y., and Cui, M. (2022). Aptamer nanomaterials for ovarian cancer target theranostics. Front. Bioeng. Biotechnol. 10, 884405. doi:10.3389/fbioe.2022.884405
Zhao, Y. X., Shaw, A., Zeng, X., Benson, E., NyströM, A. M., and HöGBERG, B. (2012). DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 6, 8684–8691. doi:10.1021/nn3022662
Zheng, Y., Wang, X., He, S., Gao, Z., Di, Y., Lu, K., et al. (2019). Aptamer-DNA concatamer-quantum dots based electrochemical biosensing strategy for green and ultrasensitive detection of tumor cells via mercury-free anodic stripping voltammetry. Biosens. Bioelectron. 126, 261–268. doi:10.1016/j.bios.2018.09.076
Keywords: nucleolin, AS1411 aptamer, DNA tile, doxorubicin, ovarian cancer
Citation: Ruan L, Han L, Li X, Chen X, Sun G, Wang X, Luo Y, Gu C and Shi X (2023) Computable structured aptamer for targeted treatment of ovarian cancer. Front. Genet. 14:1170260. doi: 10.3389/fgene.2023.1170260
Received: 20 February 2023; Accepted: 13 April 2023;
Published: 03 May 2023.
Edited by:
Guohua Wang, Northeast Forestry University, ChinaCopyright © 2023 Ruan, Han, Li, Chen, Sun, Wang, Luo, Gu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Li, eGluLmxpQHdodS5lZHUuY24=
†These authors have contributed equally to this work
 Luoshan Ruan
Luoshan Ruan Liting Han
Liting Han Xin Li
Xin Li Xin Chen
Xin Chen Gege Sun1
Gege Sun1 Xiaolong Shi
Xiaolong Shi