- 1Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, Henan, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, Henan, China
- 3Anyang Institute of Technology, Anyang, Henan, China
Identification, evolution, and expression patterns of BSK (BR signaling kinase) family genes revealed that BSKs participated in the response of cotton to abiotic stress and maintained the growth of cotton in extreme environment. The steroidal hormone brassinosteroids (BR) play important roles in different plant biological processes. This study focused on BSK which were downstream regulatory element of BR, in order to help to decipher the functions of BSKs genes from cotton on growth development and responses to abiotic stresses and lean the evolutionary relationship of cotton BSKs. BSKs are a class of plant-specific receptor-like cytoplasmic kinases involved in BR signal transduction. In this study, bioinformatics methods were used to identify the cotton BSKs gene family at the cotton genome level, and the gene structure, promoter elements, protein structure and properties, gene expression patterns and candidate interacting proteins were analyzed. In the present study, a total of 152 BSKs were identified by a genome-wide search in four cotton species and other 11 plant species, and phylogenetic analysis revealed three evolutionary clades. It was identified that BSKs contain typical PKc and TPR domains, the N-terminus is composed of extended chains and helical structures. Cotton BSKs genes show different expression patterns in different tissues and organs. The gene promoter contains numerous cis-acting elements induced by hormones and abiotic stress, the hormone ABA and Cold-inducing related elements have the highest count, indicating that cotton BSK genes may be regulated by various hormones at different growth stages and involved in the response regulation of cotton to various stresses. The expression analysis of BSKs in cotton showed that the expression levels of GhBSK06, GhBSK10, GhBSK21 and GhBSK24 were significantly increased with salt-inducing. This study is helpful to analyze the function of cotton BSKs genes in growth and development and in response to stress.
1 Introduction
With a habit of sessile growth, plants need to evolve a variety of regulatory methods to adapt to changes in the environment and deal with stress. Brassinosteroids (BR) has been reported as an important plant hormone involved in plant cell proliferation, senescence, male sterility, inducing flowering and fruit ripening and resisting abiotic stress (Clouse and Sasse, 1998; Nakaya et al., 2002; Symons et al., 2006; Bajguz and Hayat, 2009; Li et al., 2010; Ye et al., 2010; Fàbregas and Caño-Delgado, 2014). In the BR signaling pathway model that has been explored and established, many regulatory elements play an important role. The upstream elements include BRI (BR insensitive), BKI (BRI kinase inhibitor 1), BAK (BRI associated receptor kinase) and BSK (BR signaling kinase), and the downstream elements are jointly regulated by BIN2 (BR insensitive 2), BSU (BRI1 suppressor), BZR (BR resistant transcription factor) and BES (BRI-EMS suppressor) (Choudhary et al., 2012).
BSK is a class of receptor-like cytoplasmic kinases involved in BR signaling transduction. BSK is a phosphorylated substrate of BRI1, and after activation, it transmits signals downstream and is involved in regulating the expression of BR responsive genes (He et al., 2000). BSKs belong to the RLCK-XII superfamily, and their encoded proteins contain two typical kinase domains, PKc (Putative kinase catalytic) and TPR (Tetra-tri-co peptide repeat domains) domains. PKc can play a role in cell division, proliferation, apoptosis and differentiation, and TPR can mediate the interaction between proteins.
As an important cash crop, cotton inevitably suffers from adversity during the growth process, which affects its yield and quality. Therefore, revealing the components of cotton BR signal and its transduction pathway is of great significance for the response to stress and the regulation of growth and development. There are 12 BSKs in the model plant Arabidopsis thaliala (Li et al., 2019). Except for the bsk3-1 mutant which significantly reduced sensitivity to BR, the other single mutants showed no obvious phenotypic changes (Tang et al., 2008). However, quadruple mutants bsk3, 4, 7, 8 and quintuple mutants bsk3, 4, 6, 7, 8 significantly reduced the sensitivity and growth phenotype of BR, indicating that plant BSKs have functional redundancy (Kim et al., 2009; Kim et al., 2011; Sreeramulu et al., 2013). BSKs regulate BR signaling via an allosteric mechanism constitutively inactive protein kinases (Grütter et al., 2013).
Recent studies have pointed out that Arabidopsis BSK1 is physiologically related to pathogen-related molecular patterns PAMP (Pathogen-associated molecular patterns) and FLS2 (Flagellin-sensitive 2), and participates in the positive regulation of PAMP-triggered immunity (PAMP-triggered immunity), preventing the spread of pathogen infection and improving plant disease resistance (Stout et al., 2006; Bent and Mackey, 2007; Schwessinger and Zipfel, 2008; Boller and Felix, 2009; Shi et al., 2013). So far, there have been no reports on the functional studies of cotton BSKs genes in hormone signal transduction, growth and development, and response to stress. The expression level of two BSKs in transgenic ScALDH21 cotton was higher than that of non-transgenic cotton under salt stress (Yang et al., 2023). Overexpression of BSKs in crops such as rice and maize has been found to enhance salt tolerance (Li et al., 2022; Liu et al., 2022; Zhang et al., 2022). However, the detailed characterization of BSK family proteins and their important function in plants remains unclear.
In this study, bioinformatics methods were used to analyze the gene structure, evolution, and expression patterns of cotton BSKs gene family. It lays the foundation for the transduction pathway, thereby providing new genetic resources for cotton breeding.
2 Materials and methods
2.1 Data download
The genome annotation files, protein sequence data, the genome sequence information (fasta format file) and genome gene structure annotation information (gff3 format file) of the four cotton species Gossypium arboreum (CRI), Gossypium raimondii (JGI), Gossypium hirsutum (ZJU) and Gossypium barbadense (ZJU) were obtained from Cotton Functional Genomics Database (CottonFGD) (Zhu et al., 2017) (http://www.cottonfgd.org/). The latest Genome annotation files and protein sequence data of other seven species Arabidopsis thaliana. TAIR10, Glycine max_v2.1, Oryza sativa. IRGSP-1.0, Populus trichocarpa. Pop_tri_v3, Theobroma cacao_20110822, Vitis vinifera.12X and Zea mays. B73_RefGen_v4 were provided by JGI (https://gold.jgi.doe.gov/).
These softwares were used: TBtools (https://github.com/CJ-Chen/TBtools/releases), BLAST+ (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), HMMER3.0 (http://www.hmmer.org/) and MEGA7 (https://www.megasoftware.net/). The Hidden Markov Models (HMMs) profile of PF07714 (PKc) and PF07719 (TPR) were downloaded from Pfam (https://pfam.xfam.org/).
2.2 Identification of BSK family genes
The BSK family members of Arabidopsis have been identified, with a total of 12 members (Li et al., 2019). The protein sequences of the 12 AtBSK family members were downloaded from the Tair website (https://www.arabidopsis.org/) as seed sequences to perform multiple sequence alignments in the plant protein database with BLAST + to identify the BSK family members of G. hirsutum, G. barbadense, G. arboreum, G. raimondii, G. max, O. sativa, P. trichocarpa, T. cacao, V. vinifera and Z. mays. The e value was less than 1e-5. The protein sequences of the members were queried with HMMER3.0 to obtain the correct family members (Zhang et al., 2021). TBtools helped to rename all the identified genes according to the gene ID to facilitate subsequent analysis.
2.3 Phylogenetic analysis and sequences alignments
In order to explore the evolutionary relationship between BSK family members, it is necessary to submit the protein sequences to MEGA7 (Kumar et al., 2016) to calculate the optimal evolutionary model and to construct a phylogenetic tree using the maximum likelihood method based on this model. The detailed parameters were as follows: maximum likelihood, Bootstrap replications 500, Jones-taylor-Thornton (JTT) model, Gamma distributed with Invariant sites (G + I), No of Discrete Gamma Categories 5, Partial deletion 80 (Huang et al., 2022). The Evolview website (https://www.evolgenius.info/evolview/) was used for visual analysis of the evolutionary tree (Wang et al., 2021).
2.4 Conserved protein motifs, gene structure analysis and prediction of subcellular localization
The identified BSK protein sequences of four cotton species were submitted to the Motif Elicitation (MEME) (https://meme-suite.org/) online website to obtain the conserved protein motifs and save the MAST xml file. The nwk file of cotton phylogenetic tree which was construct by MEGA7 was also needed. The MAST xml file, nwk file and gff3 genome file of cotton were placed to TBtools to gain a phylogenetic tree with conserved motifs and gene structures. BSK family protein sequences were submitted to the website WoLF PSORT: Protein Subcellular Localization Prediction (https://wolfpsort.hgc.jp/) for subcellular localization prediction.
2.5 Collinearity analysis of BSK genes in four cotton species
The collinearity between the BSK duplicate gene pairs of the four cotton species was analyzed by MCScanX software on the four cotton protein sequence data files, and the graphical results were visualized with TBtools software.
2.6 Calculation of selection pressure
The CDS sequences of the BSK genes in the four cotton species was downloaded from CottonFGD (http://www.cottonfgd.org/). The homologous gene pairs in the four cotton species from duplicated gene pairs were obtained by TBtools. In order to investigate the selection pressure, we calculated the ratio of the number of non-synonymous substitutions per non-synonymous site to the number of synonymous substitutions per synonymous site (Ka/Ks) for duplicated genes.
2.7 Cis-acting elements and expression pattern analysis
For the analysis of the promoter region of BSK, the upstream 2000 bp DNA sequence of BSK was downloaded from CottonFGD and uploaded to PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict the cis-acting elements (Wang et al., 2021). The nwk file of BSK phylogenetic tree, cis-acting element prediction results and CDS were submitted to TBtools for visual analysis.
For the analysis of the expression pattern (fragments per kilo base of exon per million mapped, FPKM) of BSK family genes, we downloaded RNA-Seq data from GRAND (Hu et al., 2019) (Gossypium Resource And Network Database) (http://grand.cricaas.com.cn/home) and draw heatmap based on the expression levels of BSK family genes via TBtools. The expression level change of GhBSKs under salt stress was shown in bar graph.
3 Results
3.1 Identification of BSK family members
After BLAST + sequence alignment, there are 28, 27, 13 and 15 members of BSK gene family in G. hirsutum, G. barbadense, G. arboretum and G. raimondii, a total of 83 were identified. The number of BSK members of tetraploid G. hirsutum and G. barbadense is twice that of diploid G. arboretum and G. raimondii. This became strong evidence that tetraploid cotton was an allotetraploid formed by hybridizing A-genome diploid and D-genome diploid. As for the other species, G. max, O. sativa, P. trichocarpa, T. cacao, V. vinifera and Z. mays were identified 13, 5, 14, 8, 7 and 10 BSK members. A total of 140 members were identified from the 10 species, and only one complete splice mutant was retained for each genomic locus. The total 140 BSK genes identified in these 10 species were sorted by gene ID and renamed with TBtools as GhBSK01 - GhBSK28, GbBSK01 - GbBSK27, GaBSK01 - GaBSK13, GrBSK01 - GrBSK15, GmBSK01 - GmBSK13, OsBSK01 - OsBSK05, PtBSK01 - PtBSK14, TcBSK01 - TcBSK08, VvBSK01 - VvBSK07 and ZmBSK01 - ZmBSK10 (Supplementary Table S1). The names of AtBSK family members were consistent with Tair website. The BSKs of tetraploid cotton has more BSK members than diploid plants such as rice and grapes, indicating that BSK has gene duplication in evolution.
3.2 Phylogenetic analysis and evolution analysis
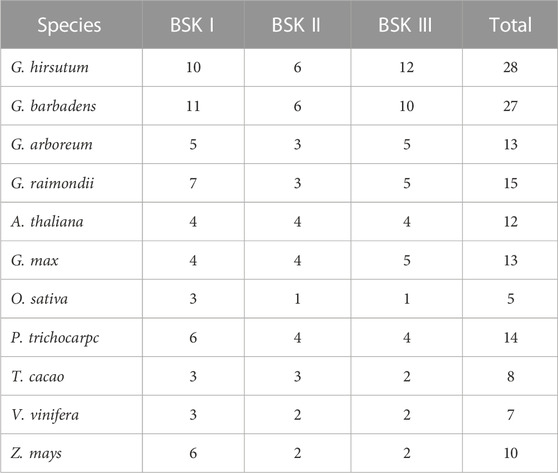
The phylogenetic relationship between BSK proteins is shown in the phylogenetic tree shown in Figure 1. The phylogenetic tree shows that BSKs have three distinct evolutionary branches, classified as BSK I - BSK III. ZmBSKs and OsBSKs proteins were more closely related. The number of different classes of BSKs from different species were shown in Table 1. There were more BSK I and BSK III present in cotton, twice as much as BSK II. Arabidopsis thaliana, G. max, V. vinifera and T. cacao have an even distribution. O. sativa, P. trichocarpa and Z. mays have more BSK I, but less BSK II and BSK III.
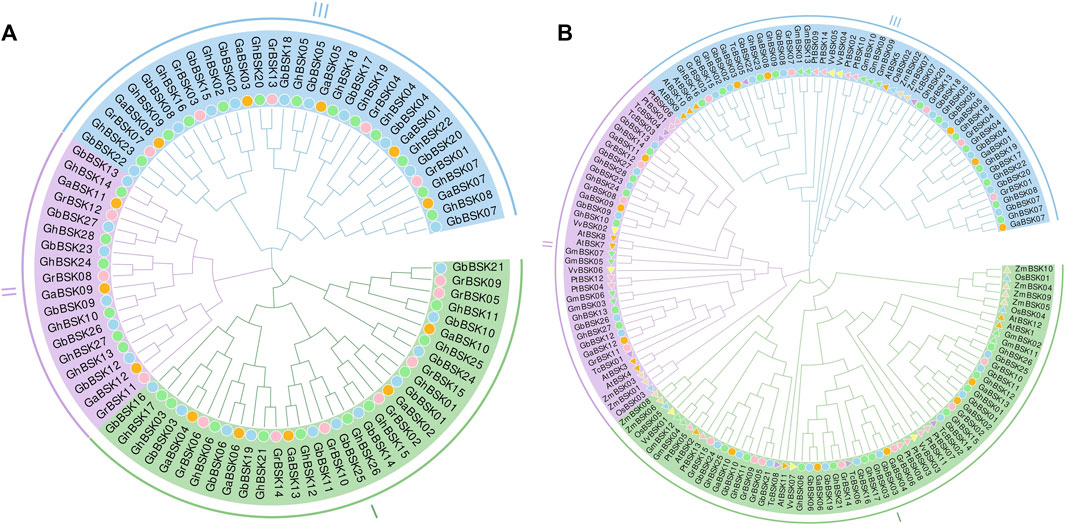
FIGURE 1. Phylogenetic analysis of plant BSKs. (A) The ML tree of identified BSKs from four cotton species. (B) The ML tree of BSKs from four cotton species, Arabidopsis thaliana and other six plant species.
3.3 Chromosomal localization of four cotton species
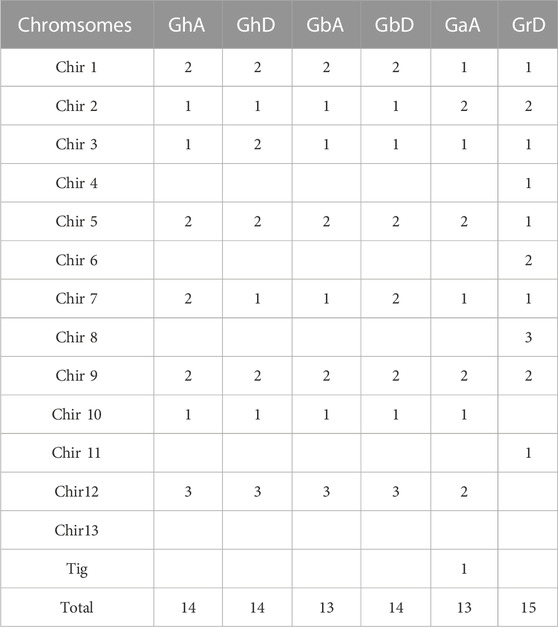
Chromosome distribution maps show the distribution of cotton BSK members on chromosomes in detail, and counted the number of genes on each chromosome (Table 2). The distribution in G. hirsutum, G. barbadense, and G. arboreum is highly conserved, and the fourth, sixth, eighth (excepted GbD), 11th and 13th chromosomes in each group of chromosomes have no BSK distribution (Figure 2). While G. raimondii is different from the other three species. There is no gene distribution on the 10th, 12th, and 13th chromosomes. Only one BSK gene of G. arboretum is located on Tig, which means the evolution of BSK gene family is mature.
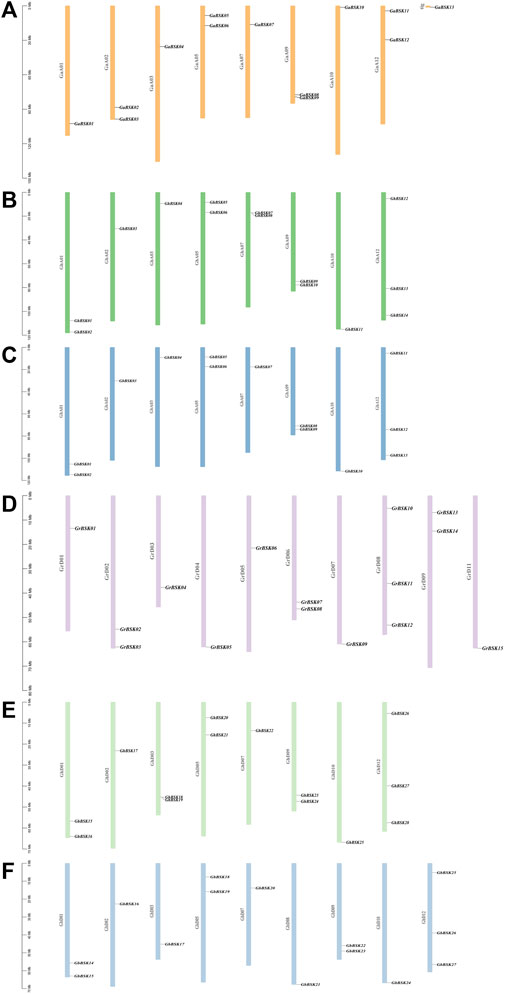
FIGURE 2. Chromosomal locations of the four cotton species BSK gene family members. The scale on the left is in mega-bases. The gene ID on the right side of each chromosome corresponds to the location of each BSK gene. (A) G. arboreum A-genome “Ga”, (B) G. hirsutum At-sub genome “GhA”, (C) G. barbadense At-sub genome “GbA”, (D) G. raimondii D-genome “Gr”, (E) G. hirsutum Dt-sub genome “GhD”, (F) G. barbadense Dt-sub genome “GbD”.
3.4 Conserved protein motifs, gene structure analysis and prediction of subcellular localization
Motif architecture, exon-intron structure analysis was performed on 83 BSKs of cotton, and their evolutionary relationships were visualized and drawn into a map (Figure 3). GhBSK08, GhBSK18 and GbBSK07 only lack PKc, and GhBSK19 and GhBSK22 only lack TPR. Both domains are missing from GhBSK07, GbBSK21, GrBSK05, and GrBSK09. The remaining 74 BSKs contain two domains. GbBSK22 has two PKcs. The remaining 74 BSKs contain two domains, and their motifs are well conserved. Some BSK IIIs only retain a small number of motifs, which may be very critical to the function of BSK. Further, BSKs in the same clade were always predicted to have the same subcellular localization.
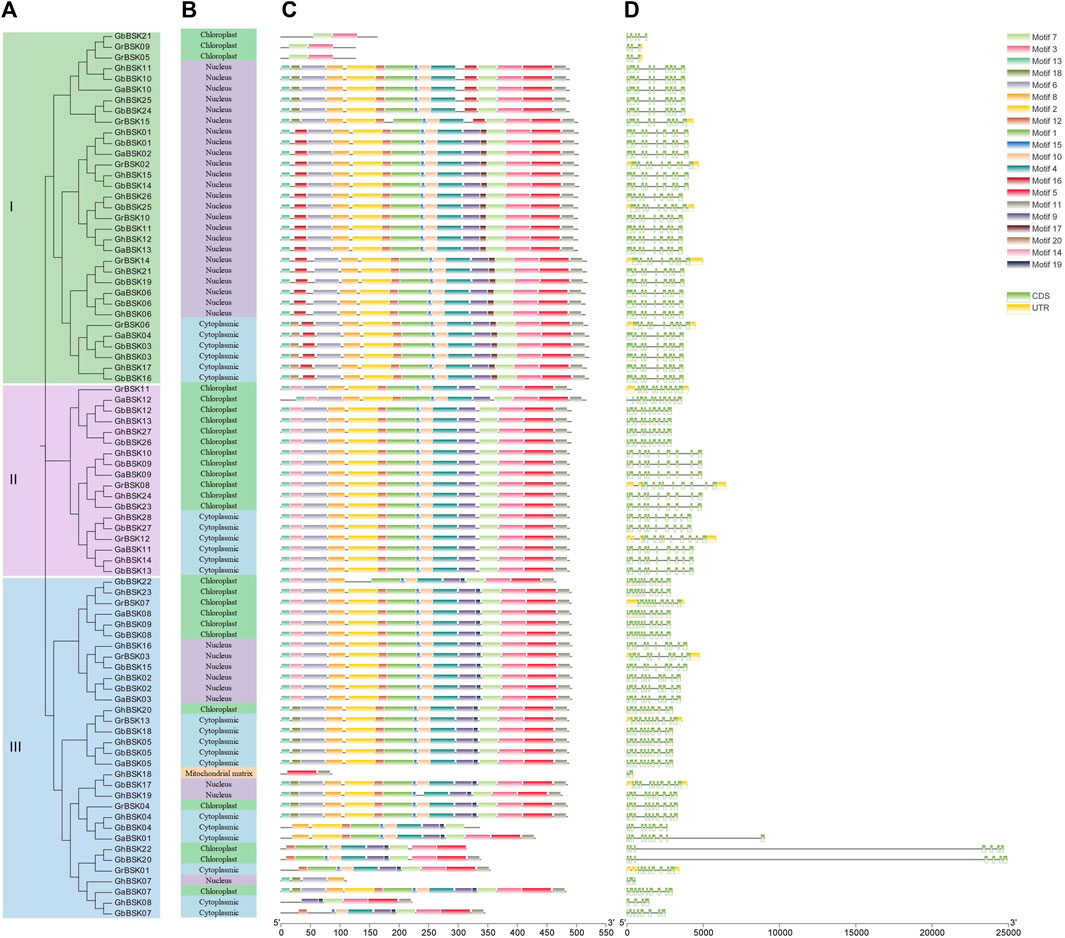
FIGURE 3. Conserved domain recognition and gene structure analysis of GhBSKs and GbBSKs. (A) Phylogenetic tree of GhBSKs and GbBSKs, (B) Prediction of subcellular localization of GhBSKs and GbBSKs, (C) Conserved motifs of GhBSKs and GbBSKs proteins, (D) Exon-intron structures of GhBSKs and GbBSKs.
3.5 Gene duplication and collinearity analysis
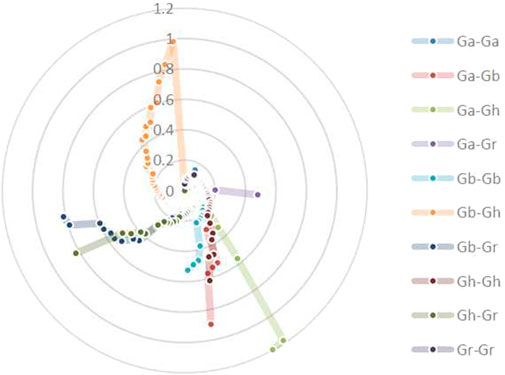
To explore the evolutionary process of BSK genes, MCscanX was used to analyze the gene duplication patterns of four cotton species and conduct genetic correlation analysis (Figure 4). A total of 385 gene pairs of genome-wide duplication were identified from 4 cotton species, of which 94 were segmental duplication and no tandem duplication was found. Based on this result, it is speculated that parallel homologous genes are generated through whole-genome duplication and segmental duplication and whole-genome duplication is an important driving force for species differentiation. Gh and Gb collinearity gene pairs are the most among the 10 groups, with 81 pairs, while Ga and Ga have the least, with only 7 pairs, which is in line with the comparison of the number of diploid and tetraploid genes. The other collinear pairs of Ga-Gb, Ga-Gh, Ga-Gr, Gb-Gb, Gb-Gr, Gh-Gh, Gh-Gr and Gr-Gr were 39, 35, 22, 41, 81, 59, 38, 55 and 8.
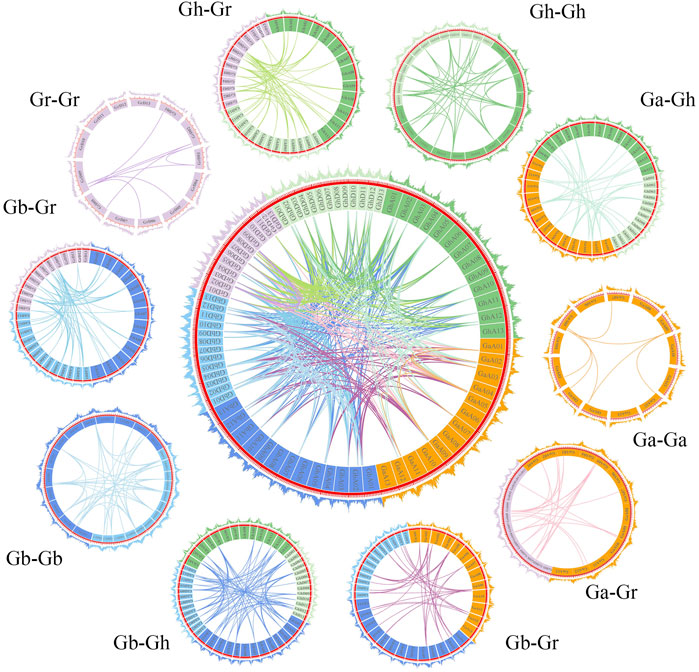
FIGURE 4. The collinearity relationship of repeated BSK gene pairs in the four cotton species of Gossypium hirsutum, Gossypium barbadense, Gossypium arboreum and Gossypium raimondii. The inner ring uses different colors to represent the chromosomes of different cotton species, the inner lines represent the collinearity of BSK genes, the outer ring represents the density of genes on the chromosome.
3.6 Selection pressure Ka/Ks is analyzed
To investigate the relationship between Darwinian positive selection and BSK gene duplication, the non-synonymous levels (Ka) and synonymous levels (Ks) were calculated for 385 duplicated gene pairs in 10 combinations from four cotton species. These combinations include Ga-Ga, Ga-Gb, Ga-Gh, Ga-Gr, Gb-Gb, Gb-Gh, Gb-Gr, Gh-Gh, Gh-Gr, Gr-Gr. Inference of selection pressure for duplicate gene pairs was based on Ka/Ks values. It is generally acknowledged that Ka/Ks = 1 indicates neutral selection (pseudogenes), Ka/Ks < 1 indicates purification or negative selection, which means that the gene evolution has a trend towards purification, and Ka/Ks > 1 indicates positive selection. Gene pairs with Ka/Ks > 1 appeared in all groups except Ga-Ga and Gr-Gr, indicating that these genes evolved faster. Most gene pairs have Ka/Ks values within 1, centered between 0 and 0.5 (Figure 5; Supplementary Table S2).
3.7 Cis-acting elements and expression pattern analysis
Cis-acting elements analysis of cotton showed that BSKs had a large number of hormone-responsive elements and stress-responsive elements induced by cold and drought, indicating that BSKs may be regulated by various hormones at different growth stages and participate in the response regulation of cotton to various stresses (Figure 6). Tissue-specific expression analysis showed that BSKs were highly expressed in roots and stems, and some BSK I and BSK II genes were highly expressed in pistil. BSK III had a high express level in leaf. Gene expression analysis under cold, heat, salt and PEG stresses showed that BSKs were differentially expressed under different stress treatments, which were associated with stress response elements.

FIGURE 6. Cis-acting elements and expression pattern analysis. (A) Evolutionary relationship of GhBSK and GbBSK. (B) Prediction of cis-acting elements of GhBSK and GbBSK. (C) Expression pattern of GhBSK and GbBSK in different tissues. (D) Expression pattern of GhBSK and GbBSK under cold, heat, salt, and PEG. The values used for drawing the heat map were normalized FPKM.
AtBSK5 and ZmBSK1 acted as a positive regulator in plant salt tolerance (Li et al., 2012; Liu et al., 2022). In order to explore the expression patterns of cotton BSKs under salt stress, the significance analysis of the expression levels of upland cotton BSK was carried out (Figure 7). The expression levels of GhBSK03 and GhBSK17 decreased significantly after salt stress and increased gradually with the increase of stress time. GhBSK06 and GhBSK21 increased at 1 h after salt stress and then decreased, while GhBSK06 increased again at 12 h after treatment. GhBSK24 decreased at 1 h after salt stress and increased at 12 h after treatment. GhBSK10 did not change significantly at the initial treatment but increased significantly 12 h after treatment. Taken together, it implies that 12 h is an intentional node, and some BSKs cannot respond to salt stress in time.
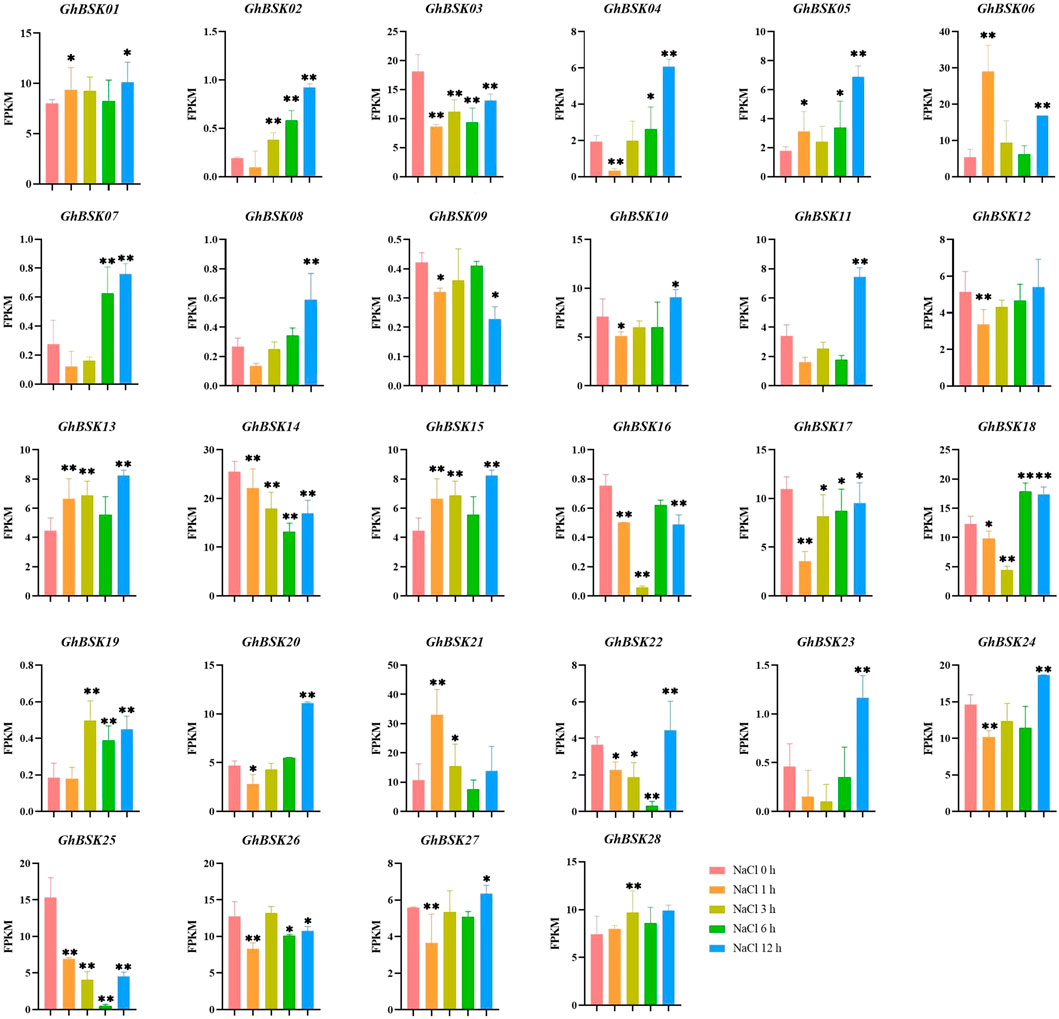
FIGURE 7. Expression analysis of GhBSKs under salt. The treatment groups were compared to NaCl 0 h group. Statistical significance was counted using one-way ANOVA. * mean p < 0.05, ** mean p < 0.01.
4 Discussion
Several studies have reported the roles of BSK proteins from Arabidopsis and rice in BR signaling and immunity, as well as in abiotic stress responses (Mora-García et al., 2004; Tang et al., 2008; Bayer et al., 2009; Li et al., 2012; Shi et al., 2013; Sreeramulu et al., 2013; Wang et al., 2017; Yan et al., 2018; Yu et al., 2018; Ren et al., 2019). However, the knowledge of BSK proteins in cotton is still quite limited. In this study, a total of 152 BSKs were identified in four cotton species and other 11 plant species by a genome-wide search, and bioinformatics methods were used to analysis their evolution, gene structure, promoter elements, protein structure and properties, gene expression patterns. Whole-genome duplication (WGD) has been reported in the two tetraploid cotton species studied (Paterson et al., 2012), and approximately two-fold amplification of the BSK gene was found. Genome duplication likely contributed the most to the expansion of the BSK gene family in many plant lineages. Approximately two-fold expansion of the BSK gene was found in plants with WGD, including A. thaliana, P. trichocarpa, and Z. mays. Furthermore, more than three-fold expansion of BSK was observed in B. rapa, possibly due to WTD (Li et al., 2019).
There are many similarities between cotton BSK genes. Except incomplete genes, BSKs possess an N-terminal PKc domain and a C-terminal TPR domain, consistent with previous studies (Tang et al., 2008). Although the expression patterns of BKSs were different, the homologous cotton BSKs proteins were basically identical in expression in tissues and organs, indicating that homologous proteins may have similar functions and redundancy.
The origin and development of the BR signaling system seems to be highly relevant with the evolution from aquatic to terrestrial plants, which have been observed in the ABA signaling system (Umezawa et al., 2010). A large number of ABA-responsive elements were also found in the promoter analysis of cotton BSKs (Figure 6). Besides ABA, a large number of hormone-responsive elements such as MeJA, GA, ethylene, SA and auxin were also found. It has been reported that BR interacts with ABA, Auxin, GA, JA and other hormones to regulate various growth and development processes of plants (Steber and McCourt, 2001; Assmann, 2004; Nemhauser et al., 2004; Perfus-Barbeoch et al., 2004; Hardtke et al., 2007; Finkelstein et al., 2008; Choudhary et al., 2012; Vandenbussche et al., 2013). BR is related to Auxin hypocotyl extension, root development, cell elongation, vascular bundle differentiation, lamellar tilt and shoot geotropism. Mutants of BR, which synergize with Auxin to promote cell elongation, often exhibit similar developmental defects, including an extremely short phenotype (Halliday, 2004; Teale et al., 2008). BR may affect plant stress response by promoting JA synthesis. BR also interacts with ethylene and auxin to regulate Arabidopsis shoot geotropism (Polko et al., 2013; Vandenbussche et al., 2013). Subcellular localization prediction of cotton BSKs revealed that no BSKs were anchored in the plasma membrane system.
Tissue-specific expression analysis of cotton BSKs suggests that different BSK proteins may function broadly in special tissues. The expression patterns of BSKs in the same subgroup were not always the same, and there were also differences in the expression patterns of G. hirsutum and G. barbadense. BR has been reported to play an important role in regulating root meristem maintenance, root elongation and promoting pollen germination and growth (González-García et al., 2011; Vogler et al., 2014). Significantly high expression of a large number of BSKs in the roots was observed in Figure 6. GhBSK14, GbBSK01, GbBSK05, GbBSK08, GbBSK11, GbBSK12, GbBSK22, and GhBSK28 were only highly expressed in pistils, while GbBSK02, GbBSK03, GbBSK13, and GbBSK21 were only highly expressed in anthers. Downstream genes of BSKs such as Arabidopsis BES1 and rice OsBZR1 have been reported to be involved in anther and pollen development, revealing that BSK members may be involved in male gametophyte development (Ye et al., 2010; Zhu et al., 2015). BRs also regulate photomorphogenesis through BES1 and associate with light signaling (Wang et al., 2012; Wang et al., 2018). A large number of light-responsive cis-acting elements were found in cotton BSK family members, suggesting that they may have light-dependent transcriptional regulation. Lots of BSKs were repressed by cold, heat, salinity and drought, only a few escaped, which may be related to their low expression in leaves. Some genes showed broad responses to multiple stresses, such as GbBSK05 was highly expressed under heat and salt stress; GbBSK11 and GbBSK23 were highly expressed under cold and salt stress, and GbBSK01 was highly expressed under salt and drought stress, and a few genes only responded to one abiotic stress. The upregulated expression of individual genes at different time points indicated that cotton BSKs played a regulatory role in cotton growth, development, and stress response. Further research is required to identify the interacting proteins of cotton BSK and investigate the molecular mechanisms for improving plant salt tolerance. Furthermore, the study will explore the role of cotton BSK in resisting other abiotic stresses to cultivate multi-resistance cotton.
5 Conclusion
In this study, the BSK family members of G. hirsutum, G. barbadense, G. arboretum and G. raimondii were identified for the first time, and their evolutionary, gene structure, conservative structure and expression pattern were analyzed. The evolution of the BSK family gened were very conservative and were divided into three clades, with different distribution in different species. In cotton, the extended family is mainly replicated by the whole genome, without tandem repeat events. The expression of BSK gene is regulated by various environments, such as light, plant hormones and stress. In conclusion, GhBSKs and GbBSKs may play an important regulatory role in cotton growth and development and stress response, but how to regulate hormone and stress response through BR signaling pathway needs further experiments to prove.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
WY and YL conceived these experiments. YL performed these experiments and completed the manuscript with the assistance of all authors. YC, RC, XC, JuW, XuL, DW, SW, LG, YZ, CR, YF, MH, LZ, HZ, XiL, NX, JiW, HH, XF, YX, KN, MZ, and TJ provided critical feedback and revised manuscript. WY read and confirmed the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, China Agriculture Research System of MOF and MARA, and supported by National Natural Science Foundation of China (Grant No. 31901509).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1169104/full#supplementary-material
References
Assmann, S. M. (2004). Plant G proteins, phytohormones, and plasticity: Three questions and a speculation. Science's STKE 2004 (264), re20. doi:10.1126/stke.2642004re20
Bajguz, A., and Hayat, S. (2009). Effects of brassinosteroids on the plant responses to environmental stresses. Plant physiology Biochem. 47 (1), 1–8. doi:10.1016/j.plaphy.2008.10.002
Bayer, M., Nawy, T., Giglione, C., Galli, M., Meinnel, T., and Lukowitz, W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323 (5920), 1485–1488. doi:10.1126/science.1167784
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. doi:10.1146/annurev.phyto.45.062806.094427
Boller, T., and Felix, G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann. Rev. Plant Biol. 60, 379–406. doi:10.1146/annurev.arplant.57.032905.105346
Choudhary, S. P., Yu, J.-Q., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.-S. P. (2012). Benefits of brassinosteroid crosstalk. Trends plant Sci. 17 (10), 594–605. doi:10.1016/j.tplants.2012.05.012
Clouse, S. D., and Sasse, J. M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. plant Biol. 49 (1), 427–451. doi:10.1146/annurev.arplant.49.1.427
Fàbregas, N., and Caño-Delgado, A. I. (2014). Turning on the microscope turret: A new view for the study of brassinosteroid signaling in plant development. Physiol. Plant. 151 (2), 172–183. doi:10.1111/ppl.12130
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi:10.1146/annurev.arplant.59.032607.092740
González-García, M.-P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-García, S., Russinova, E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138 (5), 849–859. doi:10.1242/dev.057331
Grütter, C., Sreeramulu, S., Sessa, G., and Rauh, D. (2013). Structural characterization of the RLCK family member BSK8: A pseudokinase with an unprecedented architecture. J. Mol. Biol. 425 (22), 4455–4467. doi:10.1016/j.jmb.2013.07.034
Halliday, K. J. (2004). Plant hormones: The interplay of brassinosteroids and auxin. Curr. Biol. 14 (23), R1008–R1010. doi:10.1016/j.cub.2004.11.025
Hardtke, C. S., Dorcey, E., Osmont, K. S., and Sibout, R. (2007). Phytohormone collaboration: Zooming in on auxin–brassinosteroid interactions. Trends Cell Biol. 17 (10), 485–492. doi:10.1016/j.tcb.2007.08.003
He, Z., Wang, Z.-Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., et al. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288 (5475), 2360–2363. doi:10.1126/science.288.5475.2360
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51 (4), 739–748. doi:10.1038/s41588-019-0371-5
Huang, H., He, Y., Cui, A., Sun, L., Han, M., Wang, J., et al. (2022). Genome-wide identification of GAD family genes suggests GhGAD6 functionally respond to Cd stress in cotton. Front. Genet. 13, 965058. doi:10.3389/fgene.2022.965058
Kim, T.-W., Guan, S., Burlingame, A. L., and Wang, Z.-Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43 (4), 561–571. doi:10.1016/j.molcel.2011.05.037
Kim, T.-W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J.-X., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11 (10), 1254–1260. doi:10.1038/ncb1970
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Li, J., Li, Y., Chen, S., and An, L. (2010). Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 61 (15), 4221–4230. doi:10.1093/jxb/erq241
Li, Y., Zhang, H., Zhang, Y., Liu, Y., Li, Y., Tian, H., et al. (2022). Genome-wide identification and expression analysis reveals spinach brassinosteroid-signaling kinase (BSK) gene family functions in temperature stress response. BMC genomics 23 (1), 453. doi:10.1186/s12864-022-08684-5
Li, Z.-Y., Xu, Z.-S., He, G.-Y., Yang, G.-X., Chen, M., Li, L.-C., et al. (2012). A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. biophysical Res. Commun. 426 (4), 522–527. doi:10.1016/j.bbrc.2012.08.118
Li, Z., Shen, J., and Liang, J. (2019). Genome-wide identification, expression profile, and alternative splicing analysis of the brassinosteroid-signaling kinase (BSK) family genes in Arabidopsis. Int. J. Mol. Sci. 20 (5), 1138. doi:10.3390/ijms20051138
Liu, L., Sun, Y., Di, P., Cui, Y., Meng, Q., Wu, X., et al. (2022). Overexpression of a Zea mays brassinosteroid-signaling kinase gene ZmBSK1 confers salt stress tolerance in maize. Front. plant Sci. 13, 894710. doi:10.3389/fpls.2022.894710
Mora-García, S., Vert, G., Yin, Y., Caño-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes and Dev. 18 (4), 448–460. doi:10.1101/gad.1174204
Nakaya, M., Tsukaya, H., Murakami, N., and Kato, M. (2002). Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiology 43 (2), 239–244. doi:10.1093/pcp/pcf024
Nemhauser, J. L., Mockler, T. C., Chory, J., and Dangl, J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2 (9), e258. doi:10.1371/journal.pbio.0020258
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492 (7429), 423–427. doi:10.1038/nature11798
Perfus-Barbeoch, L., Jones, A. M., and Assmann, S. M. (2004). Plant heterotrimeric G protein function: Insights from Arabidopsis and rice mutants. Curr. Opin. plant Biol. 7 (6), 719–731. doi:10.1016/j.pbi.2004.09.013
Polko, J. K., Pierik, R., van Zanten, M., Tarkowská, D., Strnad, M., Voesenek, L. A., et al. (2013). Ethylene promotes hyponastic growth through interaction with ROTUNDIFOLIA3/CYP90C1 in Arabidopsis. J. Exp. Bot. 64 (2), 613–624. doi:10.1093/jxb/ers356
Ren, H., Willige, B. C., Jaillais, Y., Geng, S., Park, M. Y., Gray, W. M., et al. (2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15 (1), e1007904. doi:10.1371/journal.pgen.1007904
Schwessinger, B., and Zipfel, C. (2008). News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. plant Biol. 11 (4), 389–395. doi:10.1016/j.pbi.2008.06.001
Shi, H., Shen, Q., Qi, Y., Yan, H., Nie, H., Chen, Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25 (3), 1143–1157. doi:10.1105/tpc.112.107904
Sreeramulu, S., Mostizky, Y., Sunitha, S., Shani, E., Nahum, H., Salomon, D., et al. (2013). BSK s are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74 (6), 905–919. doi:10.1111/tpj.12175
Steber, C. M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant physiol. 125 (2), 763–769. doi:10.1104/pp.125.2.763
Stout, M. J., Thaler, J. S., and Thomma, B. P. (2006). Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 51, 663–689. doi:10.1146/annurev.ento.51.110104.151117
Symons, G. M., Davies, C., Shavrukov, Y., Dry, I. B., Reid, J. B., and Thomas, M. R. (2006). Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant physiol. 140 (1), 150–158. doi:10.1104/pp.105.070706
Tang, W., Kim, T.-W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321 (5888), 557–560. doi:10.1126/science.1156973
Teale, W., Ditengou, F., Dovzhenko, A., Li, X., Molendijk, A., Ruperti, B., et al. (2008). Auxin as a model for the integration of hormonal signal processing and transduction. Mol. Plant 1 (2), 229–237. doi:10.1093/mp/ssn006
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell physiology 51 (11), 1821–1839. doi:10.1093/pcp/pcq156
Vandenbussche, F., Callebert, P., Zadnikova, P., Benkova, E., and Van Der Straeten, D. (2013). Brassinosteroid control of shoot gravitropism interacts with ethylene and depends on auxin signaling components. Am. J. Bot. 100 (1), 215–225. doi:10.3732/ajb.1200264
Vogler, F., Schmalzl, C., Englhart, M., Bircheneder, M., and Sprunck, S. (2014). Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod. 27 (3), 153–167. doi:10.1007/s00497-014-0247-x
Wang, J., Shi, H., Zhou, L., Peng, C., Liu, D., Zhou, X., et al. (2017). OsBSK1-2, an orthologous of AtBSK1, is involved in rice immunity. Front. plant Sci. 8, 908. doi:10.3389/fpls.2017.00908
Wang, J., Zhang, Y., Xu, N., Zhang, H., Fan, Y., Rui, C., et al. (2021). Genome-wide identification of CK gene family suggests functional expression pattern against Cd2+ stress in Gossypium hirsutum L. Int. J. Biol. Macromol. 188, 272–282. doi:10.1016/j.ijbiomac.2021.07.190
Wang, W., Lu, X., Li, L., Lian, H., Mao, Z., Xu, P., et al. (2018). Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30 (9), 1989–2005. doi:10.1105/tpc.17.00994
Wang, Z.-Y., Bai, M.-Y., Oh, E., and Zhu, J.-Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi:10.1146/annurev-genet-102209-163450
Yan, H., Zhao, Y., Shi, H., Li, J., Wang, Y., and Tang, D. (2018). BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176 (4), 2991–3002. doi:10.1104/pp.17.01757
Yang, H., Yang, Q., Zhang, D., Wang, J., Cao, T., Bozorov, T. A., et al. (2023). Transcriptome reveals the molecular mechanism of the ScALDH21 gene from the desert moss syntrichia caninervis conferring resistance to salt stress in cotton. Int. J. Mol. Sci. 24 (6), 5822. doi:10.3390/ijms24065822
Ye, Q., Zhu, W., Li, L., Zhang, S., Yin, Y., Ma, H., et al. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. U. S. A. 107 (13), 6100–6105. doi:10.1073/pnas.0912333107
Yu, M.-H., Zhao, Z.-Z., and He, J.-X. (2018). Brassinosteroid signaling in plant–microbe interactions. Int. J. Mol. Sci. 19 (12), 4091. doi:10.3390/ijms19124091
Zhang, H., Zhang, Y., Xu, N., Rui, C., Fan, Y., Wang, J., et al. (2021). Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton. Int. J. Biol. Macromol. 192, 1058–1074. doi:10.1016/j.ijbiomac.2021.10.038
Zhang, S., Hu, X., Dong, J., Du, M., Song, J., Xu, S., et al. (2022). Identification, evolution, and expression analysis of OsBSK gene family in Oryza sativa Japonica. BMC plant Biol. 22 (1), 565. doi:10.1186/s12870-022-03905-1
Zhu, T., Liang, C., Meng, Z., Sun, G., Meng, Z., Guo, S., et al. (2017). CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 17 (1), 101. doi:10.1186/s12870-017-1039-x
Keywords: BR, BSK, brassinosteroid-signaling kinase, abiotic stress, salt stress
Citation: Lei Y, Cui Y, Cui R, Chen X, Wang J, Lu X, Wang D, Wang S, Guo L, Zhang Y, Rui C, Fan Y, Han M, Zhao L, Zhang H, Liu X, Xu N, Wang J, Huang H, Feng X, Xi Y, Ni K, Zhang M, Jiang T and Ye W (2023) Characterization and gene expression patterns analysis implies BSK family genes respond to salinity stress in cotton. Front. Genet. 14:1169104. doi: 10.3389/fgene.2023.1169104
Received: 29 March 2023; Accepted: 15 May 2023;
Published: 07 June 2023.
Edited by:
Rupesh Deshmukh, Central University of Haryana, IndiaReviewed by:
Balpreet Kaur Dhatt, University of Nebraska-Lincoln, United StatesJun Tang, Jiangsu Academy of Agricultural Sciences, China
Copyright © 2023 Lei, Cui, Cui, Chen, Wang, Lu, Wang, Wang, Guo, Zhang, Rui, Fan, Han, Zhao, Zhang, Liu, Xu, Wang, Huang, Feng, Xi, Ni, Zhang, Jiang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wuwei Ye, eWV3MTU4QDE2My5jb20=
†These authors have contributed equally to this work
 Yuqian Lei1,2†
Yuqian Lei1,2† Yupeng Cui
Yupeng Cui Xiugui Chen
Xiugui Chen Xuke Lu
Xuke Lu Delong Wang
Delong Wang Yuexin Zhang
Yuexin Zhang Cun Rui
Cun Rui Wuwei Ye
Wuwei Ye