- 1Department of Chemistry, College of Science, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
- 2Hera General Hospital, Makkah, Saudi Arabia
- 3Faculty—Clinical Laboratory Sciences Department, College of Applied Medical Sciences, King Saud University, Riyadh, Saudi Arabia
- 4Department of Medical Laboratory Technology, Faculty of Applied Medical Science at Taibah University, Al Madinah Al Munawarah, Saudi Arabia
- 5Faculty of Medicine, Bioscience and Nursing, MAHSA University, Selangor, Malaysia
Introduction: Hyperglycemia-induced endothelial dysfunction and the subsequent increase of oxidative stress could lead to aberrant regulation of various genes which are responsible for a range of functions. This study aims to find out how hyperglycemia affect oxidative stress and then the expression and methylation of endothelin 1 (ET-1) gene in in human umbilical vein endothelial cells (HUVEC).
Methods: Cells were cultured in growth medium and exposed to low and high glucose concentrations to mimic normal and diabetic condition respectively. Computational analysis were performed using UCSC genome browser and eukaryotic promoter database (EPD). The expression of ET-1 gene was investigated by real time PCR. Cytotoxicity and oxidative stress were determined by MTT and DCFH-DA assays respectively. Promoter methylation was assessed by the bisulfite sequencing method.
Results: DCFH-DA assay showed that hyperglycemia can significantly increase the regulation of reactive oxygen species synthesis. The relative expression of ET-1 gene was increased due to exposure to high glucose concentration. MTT assay revealed reduced viability of cells due to the glucose induced damage. Methylation analysis revealed hypomethylation of the promoter of ET-1 however the difference was not significant. Out of 175 CpGs at 25 CpG sites, only 36 CpGs were methylated (20.5% methylation) in cell treated with normal glucose. Upon exposure to high glucose only 30 CpGs were methylated in 175 CpGs at 25 CpG sites (17.1% methylation).
Discussion: Our study concludes a significantly high expression of ET-1 gene in response to high glucose exposure in HUVECs. It also reports that hyperglycemic condition leads to elevated oxidative stress. No significant change was found in methylation when cells were treated with high and low glucose concentrations.
Introduction
Diabetes mellitus (DM) is a multifactorial disease in which body fails to release or respond to insulin. The increasing incidence of diabetes is a growing concern, causing a huge death toll and disability worldwide (Safi et al., 2014a; Barysheva, 2015; Shestakova and Maioriv, 2019; Darenskaya et al., 2021). In diabetes, chronic hyperglycemia consequently contributes to the progression of several other diabetic complications including endothelial dysfunction, insulin resistance, nephropathy and retinopathy (Safi et al., 2014b; Cho et al., 2018). Vascular endothelium performs a range of functions including maintenance of vascular homeostasis and regulation of adhesion molecules. The condition in which endothelium fails to maintain vascular homeostasis is known as endothelial dysfunction (Sena et al., 2013; Cai et al., 2021).
Under hyperglycemic conditions, the normal vascular physiology changes to endothelial dysfunction, which leads to dysfunctional cellular redox and increased generation of oxidative stress (Meza et al., 2019). Increased ROS generation leads to aberrant regulation of a range of genes including inflammatory cytokines and adhesion molecules. In response to hyperglycemic environment, several pro-inflammatory pathways also participate during the process of oxidation and antioxidation. Consequently, the imbalance of enzymatic and non-enzymatic antioxidant and ROS generation induce endothelial dysfunction by augmenting endothelium permeability, necrosis and apoptosis in endothelial cells (Batumalaie et al., 2013; SZ Safi et al., 2015; Safi et al., 2016; Escobar-Morreale et al., 2017; Luna and Estévez, 2018; Safi et al., 2022; Wang et al., 2022).
Endothelin 1 (ET-1) gene is a very important vasoconstrictor gene which is primarily expressed in vascular endothelial cells. Due to its role as vasoconstrictor, this gene contributes to vascular remodeling in diabetes and other diabetic complications. ET-1 is reported to play a key role in endothelial dysfunction (du Plooy et al., 2017; Kruger et al., 2012; Akter et al., 2015). ET-1, TNF-α and angiotensin II are some of the inducing factors of superoxide radical (O2−) generation (Münzel et al., 2010; Montezano and Touyz, 2012). In endothelial dysfunction, various studies have demonstrated the role of ET-1 gene in augmenting ROS and increased production of O2- in human arteries and animal vessels (Elmarakby et al., 2005; Loomis et al., 2005; Böhm et al., 2007; Cerrato et al., 2012; Sánchez et al., 2014). Studies have also shown that blocking the ET receptor improves endothelial function in human coronary arteries (Verma et al., 2001; Romero et al., 2009).
Epigenetics is an emerging field in which epigenetic modification can regulate genes without changing their sequences. These are heritable and stable modifications which alter the gene function without changing the DNA sequence (Pradhan et al., 2016; Álvarez-Nava and Lanes, 2018; Ling and Rönn, 2019). In diabetic complications, a number of studies have found alterations in DNA methylation which could change the gene expression profiles, and consequently the fate of the disease (Rakyan et al., 2011; Volkmar et al., 2012). One study reported hypomethylation in DNA from liver tissue (Williams et al., 2008). Another study on diabetic rat model found increased DNA methylation in pancreatic tissue. This study concluded that methylation and demethylation of DNA in diabetes may attribute to various factors the local conditions and type of tissue exposed to the disease (Williams and Schalinske, 2012).
Keeping in view the above reports, and the role of epigenetics, the purpose of this study was to assess the level of methylation and its association with the expression of ET-1 gene. We also aimed to investigate the association of hyperglycemia and the level of reactive oxygen species in hyperglycemic human umbilical vein endothelial cells (HUVEC). To this end, we exposed the cells to high and low glucose concentrations and evaluated the expression of ET-1 gene. We also studies the methylation level in ET-1 gene to establish its role in hyperglycemic condition and probe the possibility of any association between gene expression and methylation in cells with high oxidative stress.
Methodology
Cell culture
Using 10% serum and 1% Penicillin/Streptomycin, human umbilical vein endothelial cells (HUVEC) were cultured in a complete growth medium (Cell Systems). Cells were grown in 25 and 75 ml flask. One set of cells was treated with normal glucose (5 mM, 0.9008 g/L) while the other was exposed to high glucose concentration (25 mM, 4.5 g/L). Upon 80% confluency, cells were passaged using the same formulation of growth medium. Cells were incubated for 24 and 48 hours in 5% CO2 at 37°C and media was changed every 2–3 days. Cells viability and growth was observed everyday using microscope. Cells were sub-cultured by detaching the cells by Trypsin-EDTA (Gibco® United States). RNA and DNA were extracted using commercially available kits (Gene JET and Qiagen). The cell lines present in this study were obtained from Thermo Fisher Scientific, United States (C0155C).
MTT assay
MTT assay was conducted following an already reported method (Kumar et al., 2018) with modifications. Briefly, 96 well plates were used to seed the cells containing experimental (24 and 48 h) and control (24 and 48 h) groups, treated with high and low glucose concentrations. In each well, a total of 1 × 104 human umbilical vein endothelial cells were cultured for 24 and 48 h followed by incubation at 37°C with 5% CO2. Experiments were initiated with addition of MTT reagents in triplicates, followed by incubation for 4 h. Cells were rendered to extensive wash, and solubilization solution was added to dissolve the formazan crystals. A BIO-RAD-PR 4100 microplate reader was used to take readings at 570 nm after 24 and 48 h incubation.
Real time PCR
RNA was isolated from high and low glucose treated cell lysates using commercially available Gene JET RNA Purification kit (Thermo Scientific, United States). RevertAid™ kit (Thermo Scientific, United States) was used to synthesize cDNA. ET-1 gene was amplified by real time PCR using Maxima SYBR Green PCR master mix from Thermo Scientific, United States. Ct and ΔΔCt method along with beta-actin as control, was employed to calculate the relative expression of ET-1 gene.
DCFH-DA assay
To investigate the level of reactive oxygen species (ROS), reagent 2′7′-dichloro-dihydro-fluorescin diacetate was added to the experimental and control cells with high and low glucose concentrations, respectively. Cells were seeded in 96-well plates and 5 μM DCFH-DA reagent was added, followed by incubation for 24 and 48 h. Fluorescence plate reader was used to take readings at 530 nm emission and 485 nm excitation. For the calculation of reactive oxygen species, the mean control was deducted from the mean experimental group.
Computational analysis–Primer design and selection of sequences
To evaluate the promoter methylation of ET-1 gene, the gene transcript which consisted of 2032 bp, was retrieved from NCBI and UCSC genome browser databases. The 2,032 bp transcript of ET-1 gene had a 636 bp coding region, in which 269 bp was 5′UTR and 1,127 bp 3′UTR. Relative to transcription start site (TSS), promoter sequences (from 180 downstream to −1000 upstream) were identified using Eukaryotic Promoter Database (EPD). Out of 1180 bp long promoter sequence (from 180 downstream to −1000 upstream) a 400 bp sequence, with appropriate number of CpGs was identified for primer designing. The 400 bp sequence contained a 180 pb downstream sequence, relative to TSS within the 5′ UTR. The remaining 220 bp sequence was from the upstream region (−220) relative to TSS. Subsequently, primers were designed within the 400 bp sequence using MethPrimer tool, as shown in Figures 1, 2.
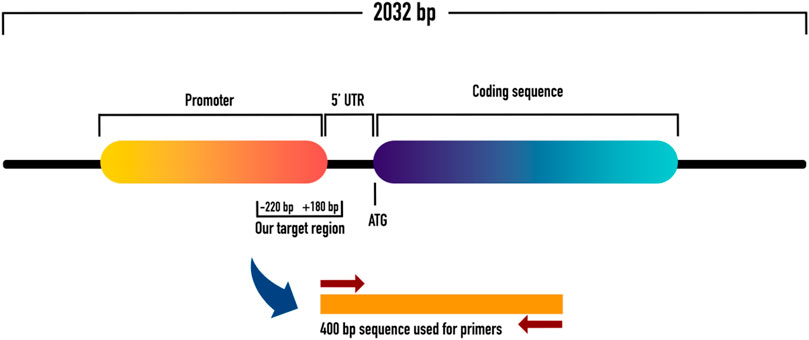
FIGURE 1. Shows the schematic representation of ET-1 with coding and promoter sequences. It also shows the 5′UTR with downstream and upstream regions from where we picked our sequence of interest for methylation analysis.

FIGURE 2. Shows the 400 bp sequence which contained a 180 pb downstream sequence (relative to TSS) within the 5′ UTR, and −220 bp upstream sequence (relative to TSS). This 400 bp sequence was used to design methylation primers for methylation study.
PCR amplification and bisulfite conversion
After quantification, a total of 500 ng of extracted DNA was bisulfite-converted using commercially available EZ DNA kit (Zymo United States). Bisulfite modified DNA was amplified using DNA polymerase (hot start taq) with specially designed primers for the promoter of ET-1 gene. PCR reactions conditions were set at 95°C for 5 minutes followed by 95°C for 45 sec and 60°C for 60°s. The final extension was given for 7 minutes at 72°C. Electrophoresis was carried out using agarose gel. A commercially available mini elute gel extraction kit (Qiagen) was used to purify the DNA from the agarose gel.
Cloning, transformation and sequencing
After purifying the amplified product from gel, ligation into pCR2.1 vector was initiated using a cloning kit from Invitrogen. A 4 μl of PCR product was thoroughly mixed with 1 μl of TOPO vector followed by a 5 minutes incubation at room temperature. The reaction was again incubated on ice for 10 minutes after adding chemically competent DH5α cells. It was followed by heat shock procedure for 30 s. To check the cloning, all samples were spread onto LB plates and incubated overnight at 37°C. After blue-white screening, 10 colonies of both types of samples (high and low glucose concentration for 48 h) were isolated from the LB plates. The isolated colonies were cultured using ImMedia Amp Liquid for 24 h. Plasmids were isolated using commercially available kit (Favorgen). The final samples were stored for sequencing. For methylation studies, we included only 48 h exposure (the maximum) and 24 h exposure was not included.
Methylation analysis
Out of 10 samples for each, we encountered cloning/sequencing problems in 3 samples. After deducting 3 samples, we were left with 7 samples for each control and high glucose treated sample. Different softwares and platforms including QUMA (Quantification tool for methylation analysis), NCBI and UCSC genome browser were used to analyze the data.
Statistical analysis
GraphPad prism and SPSS were used to for the statistical analysis. The results were calculated in mean SD. Statistical significance was determined using multiple t tests. Significance was set at a value of 0.05.
Results
Effect of high glucose on the proliferation and viability of cells
To see the effect of high glucose on HUVEC, MTT assay was performed in cells exposed to physiological glucose (5 mM), and High glucose (25 mM) concentrations. As shown in Figure 3, MTT assay revealed a significantly decreased cell proliferation index in cells treated with high glucose (25 mM) as compared to control (5 mM). Cells treated for 48 h showed far less viability (p-value 0.005) than cells treated for 24 h (p-value 0.010) It suggests that hyperglycemia creates a deleterious condition in the cells which doesn’t favor a healthy cell proliferation.
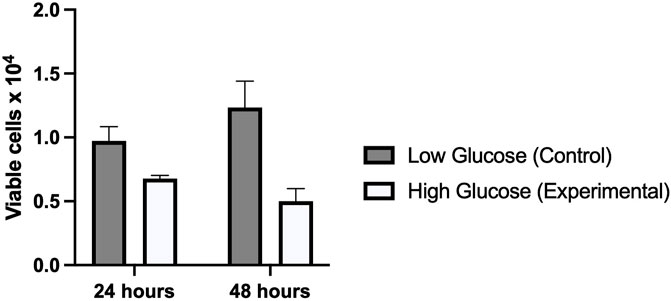
FIGURE 3. Graphical representation of MTT assay which shows a significantly reduced viability of cells treated with high glucose as compared to low glucose for 24 and 48 h. The viability further reduced in high glucose treated for 48 h.
Expression of ET-1 in hyperglycemic HUVECs
Results from real time PCR showed a significantly higher expression of ET-1 gene in HUVECs treated with high glucose concentration as compared to normal physiological concentration (Figure 4). Difference in expression was slightly higher in cells treated for 48 h (p-value 0.006) as compared to cells treated for 24 h (p-value 0.007). These results of cell viability from MTT assay, and ET-1 expression from real time PCR exhibited a similar time dependent pattern.
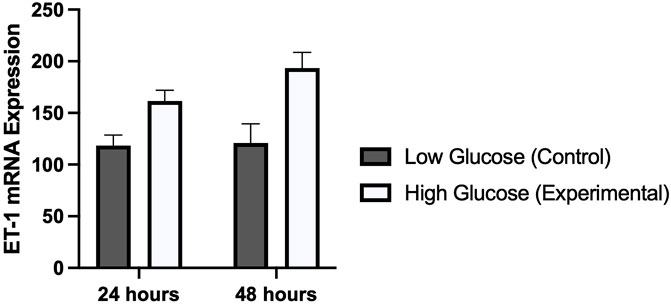
FIGURE 4. Graphical representation of the mRNA expression of ET-1 gene using real time PCR in HUVECs treated with normal/low glucose and high glucose concentrations for 24 and 48 h. There was a time dependent increased in the expression of ET-1.
Reactive oxygen species (ROS)
Several studies have reported a strong correlation between hyperglycemia and elevated levels of reactive oxygen species. We were interested to see this in human umbilical vein endothelial cells which were treated with high and low concentrations of glucose for different time periods (24 and 48 h). Our results demonstrated a significantly high oxidative stress in cells treated with high glucose as compared to those which were treated with normal glucose concentration. ROS levels were comparatively high in those cells which were exposed for 48 h (p-value 0.006) as compared to 24 h (p-value 0.014). This was consistent with the expression of ET-1 and cell viability in MTT assay (Figure 5).
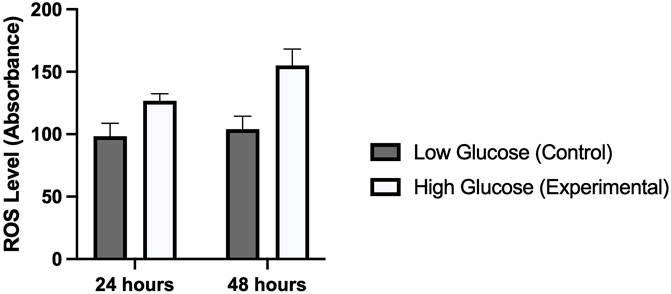
FIGURE 5. Shows the level of reactive oxygen species in HUVECs treated with normal/low glucose and high glucose concentrations for 24 and 48 h.
ET- 1 gene methylation and associated factors
After we detected a significantly high ET-1 expression in HUVECs, we presumed a hypomethylation in the promoter of ET-1. To check our hypothesis we carried out promoter methylation analysis. We evaluated 25 CpGs in each sample, where we had 7 samples for each high glucose and low glucose treated cells. So we investigated a total of 175 CpGs in high glucose treated cells and 175 CpGs in low glucose treated cells (control). As shown in Figures 6A, B, we found 36 methylated CpGs out of 175 CpGs in HUVECs treated with normal glucose concentration. In high glucose treated cells, the number of methylated CpGs was 30 (Figures 6C, D). Contrary to our expectations, the hypomethylation in glucose treated cells was very trivial and insignificant (Figure 6E). This demonstrates that hyperglycemia and high oxidative stress may regulate the expression of ET-1 gene without involving methylation at its promoter sequence. Cells with low glucose concentrations exhibited high methylation at CpG positions 22, 251, 236 and 380. At positions 92, 155, 247, 295 and 314, no methylation was found. Cells treated with high glucose concentrations, showed high methylation at positions 22, 74 and 260. No methylation was detected at positions 34, 100, 251, 289 and 305.

FIGURE 6. Shows the overall promoter methylation of ET-1 gene. (A) methylation of ET-1 gene at normal glucose concentration. (B) it summarizes the methylated vs. unmethylated CpGs in the promoter of ET-1. (C) methylation of ET-1 gene at high glucose concentration. (D) methylation vs. unmethylated CpGs in high cells treated with high glucose concentration. (E) the difference of methylation in HUVECs treated with normal and high glucose concentrations.
Discussion
Hyperglycemia and reactive oxygen species are the hallmarks of diabetes and diabetic complications. In this study we demonstrate that a 24 and 48 h exposure of HUVECs to high glucose significantly alter the ET-1 gene expression. It also negatively affect the viability of cells with increased level of reactive oxygen species. Hyperglycemia caused a significant reduction in cell viability which suggested that glucose has created a pro-oxidative stress environment. In such diabetic and high oxidative stress conditions, studies have shown that pro-diabetic, pro inflammatory and pro-oxidative stress genes are highly expressed. Several studies have reported altered gene expressions in such conditions (Shaik et al., 2010; Safi et al., 2018; Bima et al., 2022a; Aziz et al., 2022; Bima et al., 2022b). In a recent study, Hall et al. reported a significant impact of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets (Hall et al., 2018). Several genes including VAC14, RASD1, SYT16, CHRNA5 and TMED3 showed altered gene expression in response to high glucose exposure. Another study reported distinct patterns of gene expressions in primary human skin cells treated with normal and high glucose concentrations (Zhang et al., 2021).
A number of studies have reported altered methylation patterns in response to high glucose exposure. In 2020, Kandilya et al. reported varied levels of DNA methylation in human neural progenitor cells (Kandilya et al., 2020). Several other studies have also concluded that altered DNA methylation and other epigenetic mechanisms play an important role in the pathogenesis of diabetes and other diabetic complications (Ling et al., 2008; Yang et al., 2011; Wu et al., 2016; Ahmed et al., 2020; Chen et al., 2021). Similarly, elevated levels of both reactive oxygen species (ROS) and DNA methylation have been reported to be the characteristics of various diseases including cancer, diabetes, endothelial dysfunction and atherosclerosis (Wu et al., 2016). Several studies have reported a possible association of elevated oxidative stress and epigenetic modifications (Galligan et al., 2014; Wu and Ni, 2015; Khan et al., 2016; Kreuz and Fischle, 2016; Zhou et al., 2016; Zhou et al., 2018).
In diabetic patients, high blood glucose and elevated oxidative stress contribute to affect the epigenetic landscape, thus leading to persistent upregulation/downregulation of genes controlling vascular homeostasis. A significant shift in gene expression profiles also point to an altered methylation in the promoter sequences of the respective genes. After confirming varied cell viability and significant changes in oxidative stress toward normal and high glucose exposure, followed by a significantly high expression of ET-1 gene in high glucose samples, we hypothesized reduced methylation in the promoter of ET-1 gene. To check our hypothesis we analyzed 175 CpGs at 25 CpG sites in HUVECS treated with normal and high glucose concentrations. Out of 75 CpGs, we found only 36 methylated CpGs in cell treated with normal glucose (20.5% methylation). To our understanding it was quite low methylation however we were expecting further reduction after cells being exposed to high glucose concentration. Surprisingly, the outcome was not as we expected. In cells treated with high glucose, showed almost similar number of methylated CpGs (30 methylated CpGs out of 175) which makes 17.1% methylation in comparison to 20.5% methylation in normal glucose treated HUVECs (Figure 6). Cells with low glucose concentrations exhibited high methylation at CpG positions 22, 251, 236 and 380. At positions 92, 155, 247, 295 and 314, no methylation was found. Cells treated with high glucose concentrations, showed high methylation at positions 22, 74 and 260. No methylation was detected at positions 34, 100, 251, 289 and 305.
24 and 48 h exposure of cells to glucose demonstrated a time dependent effect on cell viability and oxidative stress. ET-1 gene expression was also affected in a time dependent manner. However we used only maximum (48 h) exposure for the methylation analysis, as it has shown the maximum effect on cell viability and reactive oxygen species.
Our data conclude that cells treated with high and low glucose concentrations demonstrate no significant difference in the promoter methylation of ET-1 gene. In hyperglycemic conditions, the significant changes in the expression of ET-1 gene might be due to other factors. Oxidative stress and hyperglycemia may regulate the gene expression by recruiting other proteins.
Conclusion
Our study demonstrate a significantly high expression of ET-1 gene in response to high glucose exposure in HUVECs. It also reports that hyperglycemic condition leads to elevated oxidative stress and reduce cell viability. Upon high glucose exposure, no significant changes were noticed in the promoter methylation of ET-1 gene.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R155), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, S. A. H., Ansari, S. A., Mensah-Brown, E. P. K., and Emerald, B. S. (2020). The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin. Epigenet 12, 104. doi:10.1186/s13148-020-00896-4
Akter, S., Jesmin, S., Iwashima, Y., Hideaki, S., Rahman, M. A., Islam, M. M., et al. (2015). Higher circulatory level of endothelin-1 in hypertensive subjects screened through a cross-sectional study of rural Bangladeshi women. Hypertens. Res. 38, 208–212. doi:10.1038/hr.2014.160
Álvarez-Nava, F., and Lanes, R. (2018). Epigenetics in turner syndrome. Clin. Epigenetics 10, 45. doi:10.1186/s13148-018-0477-0
Aziz, M. S., Aamir, A. H., Khan, A., Khan, Z., Shah, S. Q., Safi, S. Z., et al. (2022). Investigation of klotho G395A and C1818T polymorphisms and their association with serum glucose level and risk of type 2 diabetes mellitus. Genes 13 (09), 1532. doi:10.3390/genes13091532
Barysheva, E. V. (2015). Change parameters prooxidant-antioxidant system while reducing the concentration of deuterium in laboratory animals with alloxan diabetes. Fundam. Issled. (1-3), 457–461.
Batumalaie, K., Zaman Safi, S., Mohd Yusof, K., Devi Sekaran, S., and Qvist, R. (2013). Effect of gelam honey on the oxidative stress-induced signaling pathways in pancreatic hamster cells. Int. J. Endocrinol. 2013, 367312. doi:10.1155/2013/367312
Bima, A. I., Elsamanoudy, A. Z., Alamri, A. S., Felimban, R., Felemban, M., Alghamdi, K. S., et al. (2022). Integrative global co-expression analysis identifies key microRNA-target gene networks as key blood biomarkers for obesity. Minerva Med. 113 (3), 532–541. doi:10.23736/S0026-4806.21.07478-4
Bima, A. I. H., Elsamanoudy, A. Z., Albaqami, W. F., Khan, Z., Parambath, S. V., Al-Rayes, N., et al. (2022). Integrative system biology and mathematical modeling of genetic networks identifies shared biomarkers for obesity and diabetes. Math. Biosci. Eng. 19 (3), 2310–2329. doi:10.3934/mbe.2022107
Böhm, F., Settergren, M., and Pernow, J. (2007). Vitamin C blocks vascular dysfunction and release of interleukin-6 induced by endothelin-1 in humans in vivo. Atherosclerosis 190, 408–415. doi:10.1016/j.atherosclerosis.2006.02.018
Sena, C. M., Pereira, A. M., and Seiça, R. (2013), Endothelial dysfunction - a major mediator of diabetic vascular disease, Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 12, 2216–2231. doi:10.1016/j.bbadis.2013.08.006
Cai, Z., Yuan, S., Zhong, Y., Deng, L., Li, J., Tan, X., et al. (2021). Amelioration of endothelial dysfunction in diabetes: Role of takeda G protein–coupled receptor 5. Front. Pharmacol. 12, 637051. doi:10.3389/fphar.2021.637051
Cerrato, R., Cunnington, C., Crabtree, M. J., Antoniades, C., Pernow, J., Channon, K. M., et al. (2012). Endothelin-1 increases superoxide production in human coronary artery bypass grafts. Life Sci. 91, 723–728. doi:10.1016/j.lfs.2012.03.024
Chen, A. C. H., Huang, W., Fong, S. W., Chan, C., Lee, K. C., Yeung, W. S. B., et al. (2021). Hyperglycemia altered DNA methylation status and impaired pancreatic differentiation from embryonic stem cells. Int. J. Mol. Sci. 22 (19), 10729. doi:10.3390/ijms221910729
Cho, N. H., Shaw, J. E., Karuranga, S., Huang, Y., da Rocha Fernandes, J. D., Ohlrogge, A. W., et al. (2018). IDF diabetes atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138, 271–281. doi:10.1016/j.diabres.2018.02.023
Darenskaya, M. A., Kolesnikova, L. I., and Kolesnikov, S. I. (2021). Oxidative stress: Pathogenetic role in diabetes mellitus and its complications and therapeutic approaches to correction, Bull. Exp. Biol. Med. 171.
du Plooy, C., Martha Cornelia Mels, C., Huisman, H., and Kruger, R. (2017). The association of endothelin-1 with markers of oxidative stress in a biethnic South African cohort: The SABPA study. Hypertens. Res. 40, 189–195. doi:10.1038/hr.2016.128
Elmarakby, A. A., Loomis, E. D., Pollock, J. S., and Pollock, D. M. (2005). NADPH oxidase inhibition attenuates oxidative stress but not hypertension produced by chronic ET-1. Hypertension 45, 283–287. doi:10.1161/01.HYP.0000153051.56460.6a
Escobar-Morreale, H. F., Martínez-García, M., Montes-Nieto, R., Fernández-Durán, E., Temprano-Carazo, S., and Luque-Ramírez, M. (2017). Effects of glucose ingestion on circulating inflammatory mediators: Influence of sex and weight excess. Clin. Nutr. 36 (2), 522–529. doi:10.1016/j.clnu.2016.01.015
Galligan, J. J., Rose, K. L., Beavers, W. N., Hill, S., Tallman, K. A., Tansey, W. P., et al. (2014). Stable histone adduction by 4-oxo-2-nonenal: A potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 136, 11864–11866. doi:10.1021/ja503604t
Hall, E., Dekker Nitert, M., Volkov, P., Malmgren, S., Mulder, H., Bacos, K., et al. (2018). The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol. Cell. Endocrinol. 472, 57–67. doi:10.1016/j.mce.2017.11.019
Kandilya, D., Shyamasundar, S., Singh, D. K., Banik, A., Hande, M. P., Stunkel, W., et al. (2020). High glucose alters the DNA methylation pattern of neurodevelopment associated genes in human neural progenitor cells in vitro. Sci. Rep. 10, 15676. doi:10.1038/s41598-020-72485-7
Khan, M. A., Alam, K., Dixit, K., and Rizvi, M. M. (2016). Role of peroxynitrite induced structural changes on H2B histone by physicochemical method. Int. J. Biol. Macromol. 82, 31–38. doi:10.1016/j.ijbiomac.2015.10.085
Kreuz, S., and Fischle, W. (2016). Oxidative stress signaling to chromatin in health and disease. Epigenomics 8, 843–862. doi:10.2217/epi-2016-0002
Kruger, R., Schutte, R., Huisman, H. W., Van Rooyen, J. M., Malan, N. T., Fourie, C. M. T., et al. (2012). Associations between reactive oxygen species, blood pressure and arterial stiffness in black South Africans: The SABPA study. J. Hum. Hypertens. 26, 91–97. doi:10.1038/jhh.2010.134
Kumar, P., Nagarajan, A., and Uchil, P. D. (2018). Optical transfection. Cold Spring Harb. Protoc. 2018 (6), pdb.top096222. doi:10.1101/pdb.top096222
Ling, C., and Rönn, T. (2019). Epigenetics in human obesity and type 2 diabetes. Cell Metab. 29 (5), 1028–1044. doi:10.1016/j.cmet.2019.03.009
Ling, C., Del Guerra, S., Lupi, R., Ronn, T., Granhall, C., Luthman, H., et al. (2008). Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 51, 615–622. doi:10.1007/s00125-007-0916-5
Loomis, E. D., Sullivan, J. C., Osmond, D. A., Pollock, D. M., and Pollock, J. S. (2005). Endothelin mediates superoxide production and vasoconstriction through activation of NADPH oxidase and uncoupled nitric-oxide synthase in the rat aorta. J. Pharmacol. Exp. Ther. 315, 1058–1064. doi:10.1124/jpet.105.091728
Luna, C., and Estévez, M. (2018). Oxidative damage to food and human serum proteins: Radical-mediated oxidation vs. glyco-oxidation. Food Chem. 267, 111–118. doi:10.1016/j.foodchem.2017.06.154
Meza, C. A., La Favor, J. D., Kim, D. H., and Hickner, R. C. (2019). Endothelial dysfunction: Is there a hyperglycemia-induced imbalance of NOX and NOS? Int. J. Mol. Sci. 20 (15), 3775. doi:10.3390/ijms20153775
Montezano, A. C., and Touyz, R. M. (2012). Reactive oxygen species and endothelial function – role of nitric oxide synthase uncoupling and Nox family nicotinamide adenine dinucleotide phosphate oxidases. Basic Clin. Pharmacol. Toxicol. 110, 87–94. doi:10.1111/j.1742-7843.2011.00785.x
Münzel, T., Gori, T., Bruno, R. M., and Taddei, S. (2010). Is oxidative stress a therapeutic target in cardiovascular disease? Eur. Heart J. 31, 2741–2748. doi:10.1093/eurheartj/ehq396
Pradhan, P., Upadhyay, N., Tiwari, A., and Singh, L. P. (2016). Genetic and epigenetic modifications in the pathogenesis of diabetic retinopathy: A molecular link to regulate gene expression. New Front. Ophthalmol. 2 (5), 192–204. doi:10.15761/NFO.1000145
Rakyan, V. K., Beyan, H., Down, T. A., Hawa, M. I., Maslau, S., Aden, D., et al. (2011). Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLOS Genet. 7, e1002300. doi:10.1371/journal.pgen.1002300
Romero, M., Jiménez, R., Sánchez, M., López-Sepúlveda, R., Zarzuelo, M. J., O'Valle, F., et al. (2009). Quercetin inhibits vascular superoxide production induced by endothelin-1: Role of NADPH oxidase, uncoupled eNOS and PKC. Atherosclerosis 202, 58–67. doi:10.1016/j.atherosclerosis.2008.03.007
Safi, S. Z., Batumalaie, K., Qvist, R., Yusof, K. M., and Ismail, I. S. (2016). Gelam honey attenuates the oxidative stress-induced inflammatory pathways in pancreatic hamster cells Evidence-Based Complementary and Alternative Medicine.
Safi, S. Z., Batumalaie, K., Mansor, M., Chinna, K., Mohan, S., Qvist, R., et al. (2015). Glutamine treatment attenuates hyperglycemia-induced mitochondrial stress and apoptosis in umbilical vein endothelial cells. Clinics (Sao Paulo) 70 (8), 569–576. doi:10.6061/clinics/2015(08)07
Safi, S. Z., Qvist, R., Kumar, S., Batumalaie, K., and Ismail, I. S. B. (2014). Molecular mechanisms of diabetic retinopathy, general preventive strategies, and novel therapeutic targets. BioMed Res. Int. 2014, 801269. doi:10.1155/2014/801269
Safi, S. Z., Qvist, R., Yan, G. O. S., and Ismail, I. S. B. (2014). Differential expression and role of hyperglycemia induced oxidative stress in epigenetic regulation of β1, β2 and β3-adrenergic receptors in retinal endothelial cells. BMC Med. genomics 7 (1), 29–13. doi:10.1186/1755-8794-7-29
Safi, S. Z., Saeed, L., Shah, H., Latif, Z., Ali, A., Imran, M., et al. (2022). Mechanisms of β-adrenergic receptors agonists in mediating pro and anti-apoptotic pathways in hyperglycemic Müller cells. Mol. Biol. Rep. 49, 9473–9480. doi:10.1007/s11033-022-07816-0
Safi, S. Z., Shah, H., Qvist, R., Bindal, P., Mansor, M., Yan, G. O. S., et al. (2018). Beta adrenergic receptors stimulation attenuates phosphorylation of NF-κB and IκBα in hyperglycemic endothelial cells. Cell Physiol. Biochem. 51 (3), 1429–1436. doi:10.1159/000495591
Sánchez, A., Martínez, P., Muñoz, M., Benedito, S., García-Sacristán, A., Hernández, M., et al. (2014). Endothelin-1 contributes to endothelial dysfunction and enhanced vasoconstriction through augmented superoxide production in penile arteries from insulin-resistant obese rats: Role of ET(A) and ET(B) receptors. Br. J. Pharmacol. 171 (24), 5682–5695. doi:10.1111/bph.12870
Shaik, N. A., Shaik, J. P., Ali, S., Imran, A., and Rao, D. K. (2010). Increased frequency of micronuclei in diabetes mellitus patients using pioglitazone and glimepiride in combination. Food Chem. Toxicol. 48 (12), 3432–3435. doi:10.1016/j.fct.2010.09.016
Shestakova, M. V., and Maioriv, A. Y. (2019). Algorithms of specialized medical care for patients with diabetes mellitus. Dedov II. Moscow: Russian.
Verma, S., Lovren, F., Dumont, A. S., Mather, K. J., Maitland, A., Kieser, T. M., et al. (2001). Endothelin receptor blockade improves endothelial function in human internal mammary arteries. Cardiovas Res. 49, 146–151. doi:10.1016/s0008-6363(00)00244-3
Volkmar, M., Dedeurwaerder, S., Cunha, D. A., Ndlovu, M. N., Defrance, M., Deplus, R., et al. (2012). DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 31, 1405–1426. doi:10.1038/emboj.2011.503
Wang, M., Li, Y., Li, S., and Lv, J. (2022). Endothelial dysfunction and diabetic cardiomyopathy. Front. Endocrinol. (Lausanne) 13, 851941. doi:10.3389/fendo.2022.851941
Williams, K. T., Garrow, T. A., and Schalinske, K. L. (2008). Type I diabetes leads to tissue-specific DNA hypomethylation in male rats. J. Nutr., 138 2064–2069.
Williams, K. T., and Schalinske, K. L. (2012). Tissue-specific alterations of methyl group metabolism with DNA hypermethylation in the Zucker (type 2) diabetic fatty rat. Diabetes/Metabolism Res. Rev. 28, 123–131. doi:10.1002/dmrr.1281
Wu, H., Deng, X., Shi, Y., Su, Y., Wei, J., and Duan Pgc-1alpha, H. (2016). PGC-1α, glucose metabolism and type 2 diabetes mellitus. J. Endocrinol. 229, R99–R115. doi:10.1530/joe-16-0021
Wu, Q., and Ni, X. (2015). ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. drug targets 16 (1), 13–19. doi:10.2174/1389450116666150113121054
Yang, B. T., Dayeh, T. A., Kirkpatrick, C. L., Taneera, J., Kumar, R., Groop, L., et al. (2011). Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 54, 360–367. doi:10.1007/s00125-010-1967-6
Zhang, S., Ke, Z., Yang, C., Zhou, P., Jiang, H., Chen, L., et al. (2021). High glucose causes distinct expression patterns of primary human skin cells by RNA sequencing. Front. Endocrinol. 12, 603645. doi:10.3389/fendo.2021.603645
Zhou, X., Zhuang, Z., Wang, W., He, L., Wu, H., Cao, Y., et al. (2016). OGG1 is essential in oxidative stress induced DNA demethylation. Cell Signal 28, 1163–1171. doi:10.1016/j.cellsig.2016.05.021
Keywords: ET-1, gene expression, methylation, hyperglycemia, oxidative stress
Citation: Binjawhar DN, Alhazmi AT, Bin Jawhar WN, MohammedSaeed W and Safi SZ (2023) Hyperglycemia-induced oxidative stress and epigenetic regulation of ET-1 gene in endothelial cells. Front. Genet. 14:1167773. doi: 10.3389/fgene.2023.1167773
Received: 28 February 2023; Accepted: 31 March 2023;
Published: 17 April 2023.
Edited by:
Noor Ahmad Shaik, King Abdulaziz University, Saudi ArabiaReviewed by:
Nasreen Sultana, Acharya Nagarjuna University, IndiaMuhammad Farrukh Nisar, Cholistan University of Veterinary and Animal Sciences, Pakistan
Copyright © 2023 Binjawhar, Alhazmi, Bin Jawhar, MohammedSaeed and Safi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alaa T. Alhazmi, ZHIuYS5hbGhhem1pQGdtYWlsLmNvbQ==; Wejdan Nasser Bin Jawhar, d2JpbmpvaGFyQGtzdS5lZHUuc2E=; Walaa MohammedSaeed, d21vaGFtbWVkc2FlZWRAdGFpYmFodS5lZHUuc2E=; Sher Zaman Safi, ZHIuc3pzYWZpQGdtYWlsLmNvbQ==
 Dalal Nasser Binjawhar1
Dalal Nasser Binjawhar1 Walaa MohammedSaeed
Walaa MohammedSaeed Sher Zaman Safi
Sher Zaman Safi