
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Genet., 05 June 2023
Sec. Genetics of Common and Rare Diseases
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1165019
 Lingping Li1,2,3
Lingping Li1,2,3 Xijing Liu1,2,3
Xijing Liu1,2,3 Qinqin Li1,2,3
Qinqin Li1,2,3 Lili Zhang1,2,3
Lili Zhang1,2,3 Yueyue Xiong1,2,3
Yueyue Xiong1,2,3 Shanling Liu1,2,3
Shanling Liu1,2,3 He Wang1,2,3
He Wang1,2,3 Hongmei Zhu1,2,3*
Hongmei Zhu1,2,3* Xuemei Zhang1,2,3*
Xuemei Zhang1,2,3*Objective: We described a unique case of near-negative chromosome mosaicism in chorionic villi but complete monosomy X in amniotic fluid.
Methods: Chorionic villus sampling and amniocentesis were performed separately in the first and second trimesters. Chromosomal microarray (CMA) and rapid aneuploidy detection (QF-PCR and FISH) were performed on placental villi and uncultured amniotic fluid. After pregnancy termination, the placenta, umbilical cord, and fetal muscle tissues were sampled for FISH detection.
Results: The CMA revealed a lower signal from chromosome X in chorionic villi, with a copy number of 1.85, implying the presence of mosaic monosomy X. However, the QF-PCR and FISH results were nearly normal. In uncultured amniotic fluid, CMA and rapid aneuploidy detection indicated complete monosomy X. Across different sampling points on the aborted fetus, the FISH results varied from normal, to mosaic, and then complete monosomy X.
Conclusion: This case presents a rare and complex situation where sampling from uncultured chorionic villi indicated low-level chromosome mosaicism, while sampling from amniotic fluid revealed complete monosomy X. Although some of these discordant outcomes may be due to methodological limitations, we conclude that prenatal consultation should be combined with fetal ultrasound phenotype and genetic testing for a comprehensive evaluation of fetal genetic abnormalities.
Chromosome mosaicism is defined as the presence of two or more genetically distinct cell types in one individual developed from a single zygote. This phenomenon is considered an abnormal chromosomal event stemming from postzygotic errors (Phillips et al., 1996; Grati, 2014; Taylor et al., 2014). The mechanisms of mosaicism include chromosome non-disjunction, anaphase lag, or endoreplication. Fetal chromosome mosaicism is broadly categorized into two types: general mosaicism (presence of two or more cell lines throughout the entire organism) and confined mosaicism. The latter includes confined placental mosaicism (CPM, in which a chromosomally abnormal cell line is restricted to the placenta while the fetal chromosomes are normal) and confined fetal mosaicism (the presence of an abnormal chromosome cell line in a particular area of the fetus) (Taylor et al., 2014; Toutain et al., 2018). Where and when an error occurs from zygote to fetus may cause genetic testing to yield discordant results, depending on if the sample was obtained from chorionic villi (CV), amniotic fluid (AF), or fetal blood (FB). These three sample origins correspond to the predominant testing procedures during prenatal diagnosis: chorionic villus sampling (CVS), amniocentesis (AC), and fetal blood sampling (FBS). Clinically, the most common form of mosaicism is CPM (Phillips et al., 1996; Taylor et al., 2014), whereas the opposite (abnormal fetal chromosome but normal or near-normal placental chromosome) is rare.
Herein, we present a case study of the rarer form, with samples from uncultured chorionic villi indicating a near-normal X chromosome, but the amniotic fluid showing complete monosomy X. We postulated that this case represented low-level mosaicism of X monosomy in the placenta with the fetus exhibiting complete X monosomy and explored the potential causes of these discordant outcomes.
A 28-year-old woman (gravidity 1, parturition 0) underwent CVS at 13 + 2 gestational weeks (GW) because the first-trimester ultrasound showed an increased nuchal translucency of 5.5 mm, accompanied by an a-wave reversal in the venous catheter. Genomic DNA was extracted from uncultured CV, then subjected to aneuploidy detection via quantitative fluorescent PCR (QF-PCR) and Chromosomal microarray analysis (CMA) (Affymetrix 750K). The latter method involves array comparative genomic hybridization and detection of single-nucleotide polymorphisms (SNPs). Additionally, genomic DNA extracted from the peripheral blood of this woman was used to test for maternal cell contamination (MCC) (Figure 1).
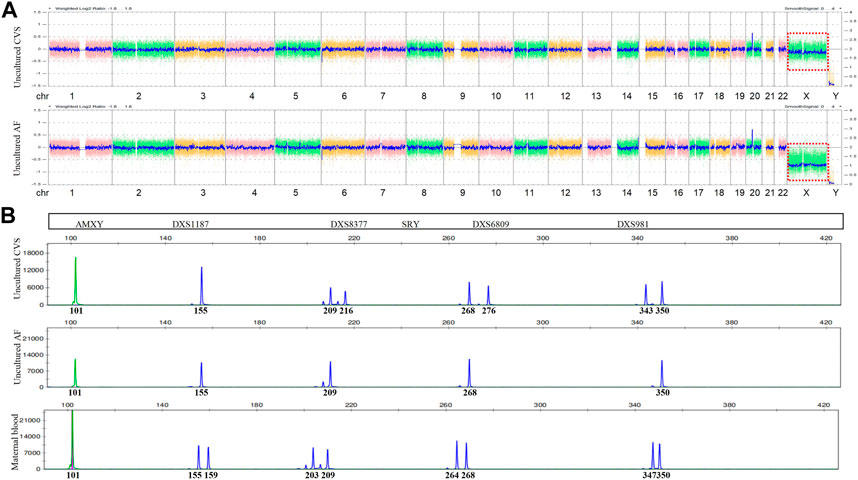
FIGURE 1. Chromosomal microarray (CMA) and quantitative fluorescent (QF)-PCR results. (A) Chromosome X of CMA (red box) in uncultured CVS (upper) and AF (lower). (B) Markers on chromosome X of QF-PCR in uncultured CVS (upper), AF (middle), and maternal blood (lower).
The results ruled out MCC, while QF-PCR showed that chromosomes 13, 18, 21, X, and Y were nearly normal. The only aberration was a slight deviation in the ratio of short tandem repeats (STR) on chromosome X from reference values (0.86–1.21) (Table 1). In contrast, CMA detected a decreased chromosome X signal with a copy number of 1.85, while the Y copy number was 0, a finding suggestive of monosomy X. We then verified these findings using fluorescence in situ hybridization (FISH) with centromere probes for the X and Y chromosomes. After counting 200 cell nuclei, we found an X/XX ratio of 4/196, practically ruling out a monosomy X cell line in uncultured CV (Figure 1).
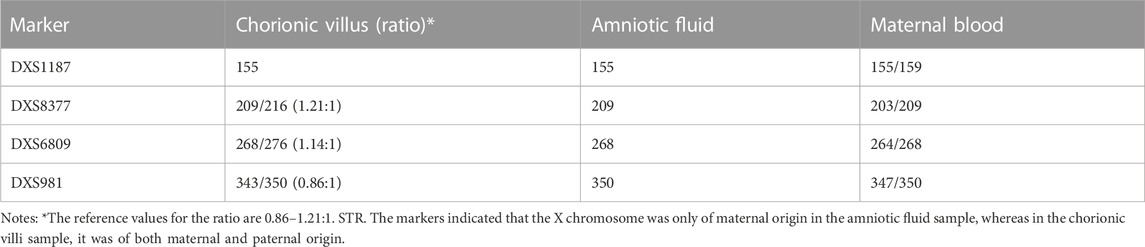
TABLE 1. Short tandem repeat (STR) markers of QF-PCR showing the suspected abnormal chromosome X in chorionic villi and amniotic fluid, suggestive of its maternal origin.
Given the conflicting results of FISH, QF-PCR, and CMA with CV, along with the observation of nuchal translucency in the fetus, we could not exclude the possibility of an abnormal chromosome X. We, therefore, performed more fetal ultrasounds at 18 GW and discovered a nuchal cystic hygroma (2.6 × 0.7 × 1.9 cm in size), along with coronary sinus dilatation and persistent left superior vena cava. After our advice that AC would provide additional information about the fetus, the couple decided to provide AF samples at 20 GW. We then performed QF-PCR and CMA again, followed by FISH verification. The results clearly indicated complete X monosomy in uncultured AF.
After further counseling, the woman requested pregnancy termination in our hospital. We then obtained informed consent to perform multiple biopsies on the aborted fetus, and FISH was performed on placental, amniotic sac, umbilical cord, skin, and muscle samples. Postnatal chorionic villi samples from the placenta yielded the same result as those for the prenatal chorionic villi, while the amniotic sac exhibited mosaic monosomy X (16%), and the fetal skin, muscle, and umbilical cord samples showed near-complete monosomy X. The details are presented in Table 2.
Chromosomal mosaicism is an important cause of cytogenetic variation across fetal tissues. This phenomenon is a major challenge for all cytogenetic laboratories concerned with prenatal diagnosis and clinical counseling (Capalbo and Laura, 2017; Cherry et al., 2017; Lund et al., 2020; Westenius et al., 2021). Samples for genetic testing in prenatal diagnosis are obtained from CVS, AC, and FBS. In particular, CVS is widely accepted as the diagnostic sample of choice in the first trimester (Emily et al., 2018), and CPM is usually the most common form of placental mosaicism. The present case exhibited a special mosaicism and was distinct from CPM, where the AF samples revealed completely abnormal chromosome X, but the CV samples suggested only low-level mosaicism.
Therefore, we analyzed the cell line of CV samples in the present case. The CV is composed of three cell types: syncytiotrophoblast, cytotrophoblast, and mesodermal core (Figure 2A). Syncytiotrophoblasts develop from cytotrophoblasts of the trophectoderm, and the mesodermal core develops from the extraembryonic mesoderm of the inner cell mass. None of these lines originate directly from the fetus proper. Therefore, the distribution of mosaicism between the fetus and placental cells depends on when and where the mutation occurred (Figure 2A) (Crane and Cheung, 1988; Bianch et al., 1993; Van Den Berg et al., 2006; Boss et al., 2018). Analysis of CV mosaics has shown that the mesenchyme core contributes nearly 50% to a DNA pool derived from uncultured dissociated CVS (Mann et al., 2007). In the present case, we performed QF-PCR and CMA on genomic DNA extracted from uncultured CVS, containing a mixture of cytotrophoblast and mesenchymal-core populations (Figure 2B). The chorionic villi samples used for FISH were uncultured and treated with a dissociation solution of methanol-glacial acetic acid without digestion; thus, the cell populations used for this test were probably mainly derived from the cytotrophoblast (Figure 2B). Thus, we hypothesized that the number of X chromosomes in the syncytiotrophoblast and cytotrophoblast was normal but that mosaic or complete monosomy of chromosome X was present in the mesenchymal core.
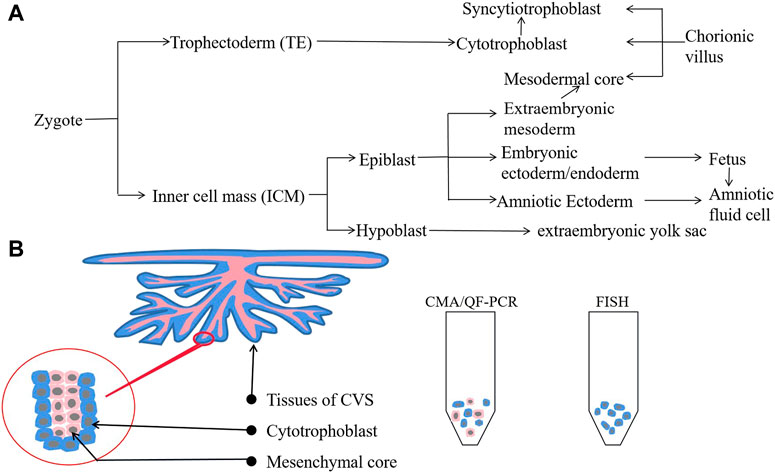
FIGURE 2. Cell lineage from zygote to fetus and populations of CVS. (A) Cell lineage from zygote to fetus. (B) Cell populations from uncultured CVS differ between CMA/QF-PCR (both cytotrophoblasts and mesenchymal core) and FISH (mainly cytotrophoblasts).
In addition, we performed FISH on samples taken from multiple biopsy sites (placenta, umbilical cord, amniotic sac, skin, and muscle) to better understand the cause of this special case. The results confirmed our speculation that the mosaicism increased gradually from the placenta to the fetal membrane, and finally to the umbilical cord. All eight placental samples revealed near-normal chromosome X, while results from fetal skin, muscle, and umbilical cord samples were consistent with the AF results showing complete monosomy X. The amniotic sac revealed mosaicism of monosomy X. The QF-PCR and SNP results showed that the two X chromosomes were respectively of maternal and paternal origin, meaning the phenomenon could not be the result of a monomic rescue event after an error during meiosis. Instead, mitotic errors may have been the cause of the mosaic cell lines in our case, with the genetic error occurring during the early stage of the inner cell mass, similar to the conclusions of previous studies on cell differentiation in human embryos (Guichet et al., 1995; Beverstock et al., 1998; Chen et al., 2013; Westenius et al., 2021; Taylor et al., 2014).
Regarding clinical significance, our findings indicate that if CVS reveals suspicious abnormalities, further genetic testing should be performed on AF as it contains more blastoderm-derived fetal cells and, thus, provides more information. Traditional karyotype analysis following cell culture targets cells from the mesenchymal core, which has been shown to be a very reliable predictor of fetal karyotype, whereas more recent molecular techniques are thought to test both mesenchymal core and cytotrophoblast (Mann et al., 2007). Moreover, the advent of newer molecular cytogenomic technologies such as CMA has brought about the prospect of greater diagnostic resolution than conventional cytogenetic methods. For over a decade, CMA has been broadly offered when multiple fetal malformations are detected including NT ≥ 3.5 mm (Wapner et al., 2012; Armour et al., 2018; Riggs et al., 2020). However, CMA and QF-PCR may not be sensitive enough to detect low-level mosaicism for aneuploidy, which can detect mosaicism as low as approximately 10%–20%, while FISH provides greater accuracy when estimating chromosome mosaicism (detection limit as low as ≤10%) as it can count more cells. Therefore, FISH is the more suitable method for unusual cases like the present case. Moreover, it is also important to consider the cell lines we test as this is also an important factor in avoiding false-negative or false-positive findings in clinical genetics (Figure 2B). Therefore, we recommend that multiple tests be performed simultaneously to reduce the risk of misdiagnosis in prenatal diagnosis.
Nevertheless, all our selected samples and methods have limitations, which should be considered to minimize false-negative or false-positive findings during diagnostic testing. Another important direction for future research is a comprehensive exploration of chromosome types across different fetal tissues, along with an investigation of the mechanisms underlying chromosomal abnormalities from the perspective of embryonic cell differentiation. The combination of improved detection technology and clarity on cell differentiation mechanisms should improve the accuracy of clinical genetic diagnoses and help with prenatal decision-making.
In conclusion, we recommend that prenatal diagnosis employs multiple, simultaneous techniques to reduce the risk of misdiagnosis. Prenatal consultation should combine genetic testing with ultrasounds to verify the phenotype to ensure a comprehensive evaluation of fetal health.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding authors.
This study was performed according to the principles of the Declaration of Helsinki. Approval was granted by the Medical Ethics Committee of West China Second University Hospital, Sichuan University (Approval Number: 2021-157). Informed consent was obtained from all subjects involved in this study. Written informed consent was obtained from the individual for the publication of any potentially identifiable images or data included in this article.
HZ, XZ, and LL contributed to the study conception and design. Material preparation, data collection, and analysis were performed by LL, XL, QL, LZ, YX, HZ, XZ, SL, and HW. The first draft of the manuscript was written by HZ, LL, and XZ. All authors contributed to the article and approved the submitted version.
This research was supported by the Sichuan Science and Technology Program (2022NSFSC0658).
The authors thank the patient for her involvement in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1165019/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | FISH results of the aborted fetus. The blue signal is chromosome 18, and the green signal is chromosome X.
Armour, C. M., Dougan, S. D., Brock, J. A., Chari, R., Chodirker, B. N., DeBie, I., et al. (2018). On-behalf-of the Canadian college of medical Geneticists.Practice guideline: Joint CCMG-SOGC recommendations for the use of chromosomal microarray analysis for prenatal diagnosis and assessment of fetal loss in Canada. J. Med. Genet. 55 (4), 215–221. doi:10.1136/jmedgenet-2017-105013
Beverstock, G. C., Hansson, K., Helderman-van den Enden, A. T., Bröcker-Vriends, A., Bartelings, M., Klumper, F., et al. (1998). A near false-negative finding of mosaic trisomy 21--a cautionary tale. Prenat. Diagn 18 (7), 742–746. PMID: 9706658.
Bianch, D. W., Wilkins-Haug, L. E., Enders, A. C., and Hay, E. D. (1993). Origin of extraembryonic mesoderm in experimental animals: Relevance to chorionic mosaicism in humans. Am. J. Med. Genet. 46 (5), 542–550. doi:10.1002/ajmg.1320460517
Boss, A. L., Chamley, L. W., and James, J. L. (2018). Placental formation in early pregnancy: How is the centre of the placenta made? Hum. Reprod. Update 24 (6), 750–760. doi:10.1093/humupd/dmy030
Capalbo, A., and Laura, R. (2017). Mosaicism between trophectoderm and inner cell mass. Fertil. Steril. 107 (5), 1098–1106. doi:10.1016/j.fertnstert.2017.03.023
Chen, C. P., Chen, Y. Y., Chern, S. R., Wu, P. S., Su, J. W., Chen, W. L., et al. (2013). Prenatal diagnosis of a distal 3p deletion associated with fetoplacental chromosomal discrepancy and confined placental mosaicism detected by array comparative genomic hybridization. Taiwan J. Obstet. Gynecol. 52 (2), 278–284. doi:10.1016/j.tjog.2013.04.0236
Cherry, A. M., Barr, K. M., Akkari, Y. M., Kearney, H. M., Rose, N. C., South, S. T., et al. (2017). Diagnostic cytogenetic testing following positive noninvasive prenatal screening results: A clinical laboratory practice resource of the American college of medical genetics and genomics (acmg). Genet. Med. 19 (8), 845–850.
Crane, J. P., and Cheung, S. W. (1988). An embryogenic model to explain cytogenetic inconsistencies observed in chorionic villus versus fetal tissue. Prenat. Diagn 8 (2), 119–129. doi:10.1002/pd.1970080206
Emily, L., Alice, P., Jane, H., and Lisa, H. (2018). Population-based trends in ultrasound-indicated prenatal diagnosis from 1994 to 2016: Two decades of change. Ultrasound Obstet. Gynecol. 53 (4), 503–511. doi:10.1002/uog.19107
Grati, F. R. (2014). Chromosomal mosaicism in human feto-placental development: Implications for prenatal diagnosis. J. Clin. Med. 24 (3), 809–837. doi:10.3390/jcm3030809
Guichet, A., Briault, S., Toutain, A., Paillet, C., Descamps, P., Pierre, F., et al. (1995). Prenatal diagnosis of trisomy 8 mosaicism in CVS after abnormal ultrasound findings at 12 weeks. Prenat. Diagn 15 (8), 769–772. doi:10.1002/pd.1970150815
Lund, I. C. B., Becher, N., Christensen, R., Petersen, O. B., Steffensen, E. H., Vestergaard, E. M., et al. (2020). Prevalence of mosaicism in UnculturedChorionic villus samples after chromosomal microarray and clinical outcome in pregnancies affected by confined placental mosaicism. Prenat. Diagn 40, 244–259. doi:10.1002/pd.5584
Mann, K., Kabba, M., Donaghue, C., Hills, A., and Ogilvie, C. M. (2007). Analysis of a chromosomally mosaic placenta to assess the cell populations in dissociated chorionic villi: Implications for QF-PCR aneuploidy testing. Prenat. Diagn 27 (3), 287–289. doi:10.1002/pd.1663
Phillips, O. P., Tharapel, A. T., Lerner, J. L., Park, V. M., Shulman, L. P., Wachtel, S. S., et al. (1996). Risk of fetal mosaicism when placental mosaicism is diagnosed by chorionic villus sampling. Am. J. Obstet. Gynecol. 174 (3), 850–855. doi:10.1016/S0002-9378(96)70312-5
Riggs, E. R., Andersen, E. F., Cherry, A. M., Kantarci, S., Kearney, H., Patel, A., et al. (2020). Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American college of medical genetics and genomics (ACMG) and the clinical genome resource (ClinGen). Genet. Med. 22 (2), 245–257. doi:10.1038/s41436-019-0686-8
Taylor, T. H., Gitlin, S. A., Patrick, J. L., JackCrain, L. J. L., J Michael Wilson, J. M., and Darren K Griffin, D. K. (2014). The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum. Reprod. Update 20 (4), 571–581. doi:10.1093/humupd/dmu016
Toutain, J., Goutte-Gattat, D., Horovitz, J., and Saura, R. (2018). Confined placental mosaicismrevisited: Impact on pregnancy characteristics and outcome. PLoS ONE 13 (4), e0195905. doi:10.1371/journal.pone.0195905
Van Den Berg, C., Van Opstal, D., Polak-Knook, J., and Galjaard, R. J. (2006). (Potential) false-negative diagnoses in chorionic villi and a review of the literature. Prenat. Diagn 26 (5), 401–408. doi:10.1002/pd.1421
Wapner, R. J., Martin, C. L., Levy, B., Ballif, B. C., Eng, C. M., Zachary, J. M., et al. (2012). Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 367 (23), 2175–2184. doi:10.1056/NEJMoa1203382
Keywords: chromosome mosaicism, prenatal diagnosis, nuchal cystic hygroma, monosomy X, genetic testing
Citation: Li L, Liu X, Li Q, Zhang L, Xiong Y, Liu S, Wang H, Zhu H and Zhang X (2023) Case report: Prenatal diagnosis of rare chromosome mosaicism: discordant results between chorionic villi and amniotic fluid samples. Front. Genet. 14:1165019. doi: 10.3389/fgene.2023.1165019
Received: 24 February 2023; Accepted: 22 May 2023;
Published: 05 June 2023.
Edited by:
Philip D. Cotter, University of California, San Francisco, United StatesReviewed by:
Ewelina Bukowska-Olech, Poznan University of Medical Sciences, PolandCopyright © 2023 Li, Liu, Li, Zhang, Xiong, Liu, Wang, Zhu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Zhu, emh1aG0zMzQ0QDEyNi5jb20=; Xuemei Zhang, eHVlbWVpemgwMDFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.