
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 09 June 2023
Sec. Genetics of Common and Rare Diseases
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1161696
This article is part of the Research TopicExploiting Genetics and Genomics to Improve the Understanding of Eye DiseasesView all 14 articles
 Giuseppe Giovanni Nardone1†
Giuseppe Giovanni Nardone1† Beatrice Spedicati1,2†
Beatrice Spedicati1,2† Maria Pina Concas2*
Maria Pina Concas2* Aurora Santin1
Aurora Santin1 Anna Morgan2
Anna Morgan2 Lorenzo Mazzetto1
Lorenzo Mazzetto1 Maurizio Battaglia-Parodi3
Maurizio Battaglia-Parodi3 Giorgia Girotto1,2
Giorgia Girotto1,2Introduction: Color vision defects (CVDs) are conditions characterized by the alteration of normal trichromatic vision. CVDs can arise as the result of alterations in three genes (OPN1LW, OPN1MW, OPN1SW) or as a combination of genetic predisposition and environmental factors. To date, apart from Mendelian CVDs forms, nothing is known about multifactorial CVDs forms.
Materials and Methods: Five hundred and twenty individuals from Silk Road isolated communities were genotyped and phenotypically characterized for CVDs using the Farnsworth D-15 color test. The CVDs traits Deutan-Protan (DP) and Tritan (TR) were analysed. Genome Wide Association Study for both traits was performed, and results were corrected with a False Discovery Rate linkage-based approach (FDR-p). Gene expression of final candidates was investigated using a published human eye dataset, and pathway analysis was performed.
Results: Concerning DP, three genes: PIWIL4 (FDR-p: 9.01*10–9), MBD2 (FDR-p: 4.97*10–8) and NTN1 (FDR-p: 4.98*10–8), stood out as promising candidates. PIWIL4 is involved in the preservation of Retinal Pigmented Epithelium (RPE) homeostasis while MBD2 and NTN1 are both involved in visual signal transmission. With regards to TR, four genes: VPS54 (FDR-p: 4.09*10–9), IQGAP (FDR-p: 6,52*10–10), NMB (FDR-p: 8.34*10–11), and MC5R (FDR-p: 2.10*10–8), were considered promising candidates. VPS54 is reported to be associated with Retinitis pigmentosa; IQGAP1 is reported to regulate choroidal vascularization in Age-Related Macular Degeneration; NMB is involved in RPE homeostasis regulation; MC5R is reported to regulate lacrimal gland function.
Discussion: Overall, these results provide novel insights regarding a complex phenotype (i.e., CVDs) in an underrepresented population such as Silk Road isolated communities.
Color vision defines an organism’s ability to differentiate objects by wavelengths of light that they reflect, emit, or transmit. Many differences exist between animal and human color vision. Human vision is trichromatic, while some animals, such as frogs, fish, marsupials and some types of rodents, can perceive ultraviolet. Birds are believed to possess the most sophisticated visual system among vertebrates, probably perceiving hues of colors inaccessible to humans. In humans, color vision involves several retina and brain mechanisms. In particular, light-absorbing molecules defined as visual pigments, consisting of integral membrane proteins in photoreceptor cells, mediate human vision. The energy carried by photons activates the pigments and eventually starts the photo transduction signal cascade, thus converting the light into an electric signal. Visual pigments reside in two types of photoreceptors, the cone-shaped and rod-shaped cells, and are classified according to the wavelength of maximum light absorption (λmax). Cone photoreceptors contain the three primary visual pigments, the short-wavelength sensitive (S; λmax ∼420 nm), the medium-wavelength sensitive (M; λmax ∼530 nm), and the long-wavelength sensitive (L; λmax ∼560 nm), while rod-shaped cells contain a fourth pigment, called rhodopsin, which mediates vision in dim light conditions and absorbs maximally at λmax ∼495 nm (Nathans, 1999). Humans, along with primates, possess three genes encoding for the three primary visual pigments: OPN1SW, located on chromosome 7, and OPN1MW and OPN1LW, both located on chromosome X. Trichromacy arises with the duplication of the OPN1LW gene resulting in the formation of the OPN1MW gene (Dulai et al., 1999), which confers the ability to discriminate red fruits on a green background. Interestingly, the acquisition of trichromatic vision concurred with the deterioration of the olfactory system in primates (Gilad et al., 2004).
Color Vision Defects (CVDs) are conditions caused by the lack of visual pigments or by their abnormal function. CVDs are divided into congenital and acquired forms and classified based on the affected cone: Deutan affects M-cones causing a defective perception of the red-yellow-green spectrum; Protan affects L-cones causing poor discrimination of the red-green spectrum; and Tritan affects S-cones leading to an alteration in the perception of blue, yellow and orange hues (Hasrod and Rubin, 2016). Congenital CVDs (Deuteranopia, Protanopia, and Tritanopia) are the most characterised forms. They are caused by alterations in: genes coding visual pigments; genes controlling their expression; genes coding components of the phototransduction cascade; genes coding for the α- or β-subunits of the cone cyclic guanosine monophosphate–gated cation channels. Conversely, acquired CVDs arise as a result of ocular, neurologic, or systemic diseases. They can be classified based on the primary disease and the type of color vision alteration encountered (Simunovic, 2016). Moreover, color discrimination ability starts gradually declining from the age of 30. In addition, CVDs can arise during the ageing process, may be influenced by pupil size, crystalline lens coloration, and macular pigment, suggesting the existence of multifactorial forms of CVDs (Ichikawa et al., 2021).
In recent years, Genome-Wide Association Studies (GWAS) proved to be a valid tool in the discovery of novel genes associated with complex diseases and multifactorial traits (Struck et al., 2018). Moreover, it has been proven that GWAS discovery power can be increased by investigating genetic isolates - small communities with reduced genetic variation and environmental homogeneity - offering the possibility to highlight variants underlying complex traits (Girotto et al., 2011). Here, we present the first GWAS analysis investigating multifactorial CVDs in genetic isolates from the Silk Road countries. For centuries, the Silk Road has been a crucial trading route directly linking Europe with Asia. Due to its geographical location and past socio-economic relevance, today, the Silk Road represents a unique collection of populations with admixed ancestries (Mezzavilla et al., 2014), languages and traditions, thus offering the possibility to study rare and unique patterns of genetic variation. Therefore, this study aims to identify novel variants and genes potentially involved in the determination of multifactorial CVDs. To reach this goal, we performed 1) GWAS analyses, 2) evaluation of the expression of candidate genes in a published database, and 3) up-to-date in silico pathway construction to evaluate interactions between candidate genes and known genes involved in eye physiology and pathology.
Eight hundred ninety-three subjects from 20 isolated communities spread across nine Central Asia and Caucasus countries (Afghanistan, Armenia, Azerbaijan, Crimea, Georgia, Kazakistan, Kirghizistan, Tajikistan, Uzbekistan) were recruited during the “Marco Polo” scientific expedition in 2010 (www.marcopolo2010.it). Biological samples, along with information about age, sex, lifestyle, eating habits, profession, and smoking and alcohol consumption, were collected. Moreover, phenotypic information about sensorial abilities and performance, such as hearing thresholds, olfactory performance, and food preferences were collected using several tests and questionnaires. Written informed consent was obtained from all enrolled subjects. The study was conducted in accordance with the Helsinki Declaration, and the research protocol was approved by the ethical committee of IRCCS “Burlo Garofolo” of Trieste, Italy.
Participants were tested for color discrimination ability using the Farnsworth D-15 saturated color test, administered to all subjects in natural daylight lighting conditions (Oli and Joshi, 2019). Briefly, 15 enumerated colored disks and a reference disk are used in the test. First, subjects select the disk that most closely matches the reference. Then, they select the next color disk matching the previous one and continue until the last disk has been positioned. Scoring is defined by reading the number on the rear of the disk and diagramming the subject’s results on a template sheet. Lines crossing the center of the diagram define the type and severity of the CVD (Supplementary Figure S1). To obtain a more precise classification, the tests’ results interpretation was carried out by a certified ophthalmologist. Individuals were finally classified in Deutan-Protan (DP, 31 cases) and Tritan (TR, 12 cases). All the individuals without color vision alteration were classified as controls.
Saliva samples were collected from all enrolled subjects using the Oragene DNA collection kit, and DNA extraction was performed (DNA Genotek, Ontario, Canada). Genotyping was carried out using the Omni Express 700k Illumina Chip, including only samples and sites with call rate ≥0.99 (43,655 variants excluded), Hardy-Weinberg Equilibrium p-value ≥1*10–6 (3,153 variants excluded), and minor allele frequency (MAF) ≥ 0.01 (18 variants excluded). Imputation was performed using MINIMAC v4 (Howie et al., 2012) to Haplotype Reference Consortium imputation panel version r1.1 (McCarthy et al., 2016). All data were aligned to the human reference genome build 37 (GRCh37). After imputation, SNPs with Info Score <0.4 (13626943 variants excluded) and MAF <0.01 (40,203 variants excluded) were discarded from statistical analyses. A total of 7783479 SNPs were analysed.
GWAS analyses for DP and TR traits were conducted using GEMMA v0.98 software (Zhou and Stephens, 2012). Linear mixed model regressions assuming an additive genetic model were performed. The genomic kinship matrix was used as random effects to take into account relatedness. Covariates included in the analyses were: age, sex, educational level and the first ten principal components (PCs). PCs were calculated from 203,099 genotyped variants using Plink v1.9 (Purcell et al., 2007). Results were then adjusted using a False Discovery Rate (FDR) linkage-based approach adapted from (Chen et al., 2021). Briefly, linkage disequilibrium-based clumping of regression results was performed using Plink v1.9, setting an r2 threshold of 0.3 and a variant distance of 0.5 Mb. Then, the most significant SNPs of each clump were collected in a new dataset, and FDR calculation was carried out using R (https://www.r-project.org/index.html). For each GWAS, SNPs with an FDR-adjusted p-value (FDR-p) ≤ 1 × 10−5 were annotated with the Variant Effect Predictor tool (VEP) (McLaren et al., 2016) to obtain information on the distance from the closest genes and their functional characteristics (i.e., whether they were contained in an intronic, exonic, or intergenic region). For each SNP, the nearest coding gene in a range of 250 kb was annotated. Long non-coding RNAs (LINC) genes, genes with unknown functions identified with LOC and FAM symbols, and pseudo genes were excluded. Finally, potential candidate genes were selected based on the following criteria:
• FDR-p ≤ 5*10–8;
• Number of SNPs in linkage with the top SNP ≥ the median of SNPs in linkage for each clump;
• Expression levels in the inquired database;
• Involvement in physiological and/or pathological processes in the eye.
Manhattan and QQplots were generated using R, while heatmap was generated using the ComplexHeatmap R package (Gu et al., 2016).
The final candidate genes’ expression was investigated using a published human eye transcriptome dataset - the “Eye in a Disk” (EiaD) (https://eyeintegration.nei.nih.gov). Briefly, the dataset integrates expression data from GTEx (GTEx Consortium, 2013) and public repositories through the use of a Snakemake-based pipeline to normalise expression values indicated as log2 [Transcript Per Million (TPM) +1)] and perform quality control. Expression data was extracted from healthy human adult and fetal eye tissues and eye stem cells. For this study, gene expression of candidate genes in the following tissues was investigated: Corneal Endothelium (CE); Corneal Fetal Endothelium (CFE); Corneal Endothelial Stem Cells (CSE); Cornea (Cn); Lens Stem Cell Line (LSC); Fetal Eye Retina (RFE); Retinal Fetal Tissue (RFT); Retinal Ganglion Stem Cells (RGS); Retinal Pigment Epithelium (RPE); Fetal RPE (RPFT); Retina (Rt).
Finally, the involvement of the candidate genes in eye physiology and pathology was investigated using Qiagen’s Ingenuity Pathway Analysis (IPA) from Ingenuity Systems (Redwood City, California, United States; http://www.ingenuity.com). Briefly, a list of promising candidate genes resulting from significant FDR-corrected associations for both traits was uploaded to verify their interaction with a set of genes involved in eye pathologies and physiology using Path Explorer. Finally, interaction maps were generated using IPA’s Path Designer. The gene list was obtained by combining manual bibliographic research and IPA databases, including genes involved in cone photoreceptor disorders and cone cell development.
Individuals with missing data regarding age, sex and color vision, or missing genetic data were excluded from the analyses. In the GWAS for DP, 514 individuals were included, specifically 31 with DP and 483 without color vision alteration. In the GWAS for TR, 520 subjects were considered, precisely 12 with TR and 508 without color vision alteration. The number, gender, age, and educational level of analyzed participants are reported in Table 1.
Manhattan and QQ plots of GWAS on DP and TR are shown in Supplementary Figure S2. A total of 1,263 associations between SNPs and DP phenotype with p < 1*10–5 were detected. Of those, 103 displayed genome-wide significant p-values (p < 5*10–8). Regarding TR, 2,968 associations with p < 1*10–5 were found; 273 with genome-wide significant p-values. After the FDR correction, 489 variants with FDR-corrected p-values (FDR-p) < 1*10–5 were retained for DP. Of those, nine showed genome-wide significant FDR-p (Supplementary Table S1). Eight hundred eighty-three variants with FDR-p < 1*10–5 were retained for the TR trait, 45 with genome-wide significant FDR-p (Supplementary Table S1). All variants showing genome-wide significant FDR-p for both DP and TR were not in linkage-disequilibrium. VEP annotation of the association results for both traits is available in Supplementary Tables S2, S3. Following the selection criteria described in the Materials and Methods section, our analyses allowed the identification of 18 promising candidates, as displayed in Table 2.
After FDR correction, three genes resulted significantly associated with the DP phenotype: PIWIL4 (top SNP rs118136100, FDR-p = 9.01*10–9), MBD2 (top SNP rs17292725, FDR-p = 4.98*10–8) and NTN1 (top SNP rs117797822, FDR-p = 4.97*10–8). Moreover, 15 genes resulted significantly associated with TR, with NMB (top SNP rs145079583, FDR-p = 8.34*10–11), IQGAP1 (top SNP rs117555778, FDR-p = 6.52*10–10), NPTN (top SNP rs192430987, FDR-p = 6.52*10–10) and ATF7IP2 (top SNP rs146729070, FDR-p = 6.52*10–10) being the most significantly associated genes.
The expression patterns in the human eye of the three genes for DP and the 15 for TR were investigated using expression data from EiaD and are shown in Figure 1. The corresponding values of log2 (TPM+1) are fully reported in Supplementary Table S4.
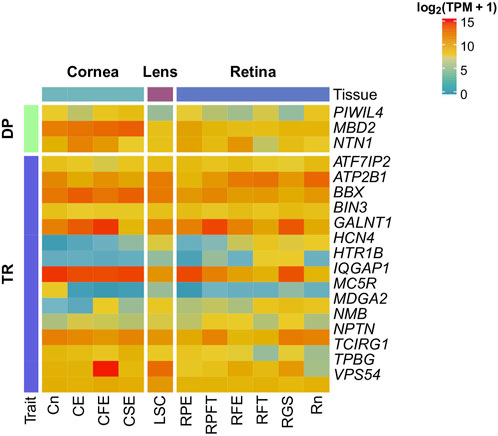
FIGURE 1. Gene expression of candidate genes in the eye. Gene expression of candidate genes was investigated in 11 healthy ocular tissues from Cornea, Lens, and Retina using the Eye on a Disk dataset. Candidate genes are at least weakly expressed in all the tissues. CE, Corneal Endothelium; CFE, Corneal Fetal Endothelium; CSE, Corneal Endothelial Stem Cells; Cn, Cornea; LSE, Lens Stem Cell Line; RFE, Fetal Eye Retina; RFT, Retinal Fetal Tissue; RGS, Retinal Ganglion Stem Cells; RPE, Retinal Pigment Epithelium; RPFT, Fetal RPE; Rt, Retina.
All genes displayed at least a weak expression in all inquired tissues. As regards DP, NTN1 and MBD2 showed high expression (log2 (TPM+1) > 10) throughout all the tissues, especially in corneal tissues. Conversely, PIWIL4 showed weak expression (log2 (TPM+1) < 5) in all tissues, excluding corneal tissues, RPE and Rn. Concerning TR, VPS54, BIN3, TCIRG1, ATF7IP2, ATP2B1, BBX, GALNT1 and IQGAP1 showed a high expression in all tissues. In contrast, MC5R, MDGA2, HTR1B and HCN4 showed weak expression in almost all investigated tissues.
Finally, the interaction between the resulting genes for both traits and a set of more than 500 genes involved in eye pathology and physiology was investigated using IPA’s Path Explorer. The interaction network is shown in Figure 2. The complete list of genes is reported in Supplementary Table S5.
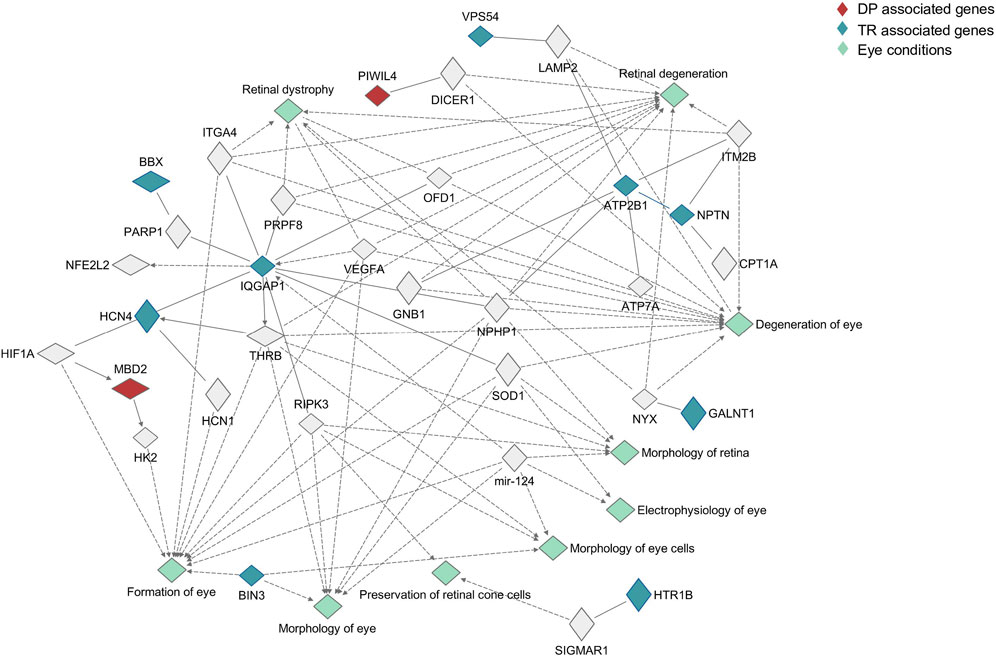
FIGURE 2. Pathway analysis of candidate genes. The role played in the eye by candidate genes was investigated using Ingenuity Pathway analysis, searching for interaction between candidate genes and a list of more than 500 genes involved in eye physiology and pathology. Genes associated with Deutan-Protan (DP) are highlighted in red, while genes associated with Tritan (TR) are highlighted in blue. Eye conditions associated with the interactions are highlighted in aquamarine.
With the exception of seven genes, TPBG, TCIRG1, MDGA2, ATF7IP2, NTN1, MC5R, and NMB, associations with genes involved in eye morphogenesis (formation of the eye, morphology of the eye and retina, morphology of eye cells), physiology (preservation of cone cells, electrophysiology of the eye) and degeneration (retinal dystrophy, retina degeneration, degeneration of the eye) have been found. Concerning DP, MBD2 was involved in the formation of the eye, interacting both with HK2 and HIF1A, while PIWIL4 displayed an interaction with DICER1, which was involved in retinal degeneration. With regards to TR, IQGAP1 and ATP2B1 showed the most complex networks of interaction, interacting both with other candidate genes and with genes involved in all the investigated processes.
In this study, we performed the first GWAS analyses investigating the molecular determinants of multifactorial forms of CVDs in the rare Silk Road cohort. Our analyses led to the identification of 18 candidate genes. Among them, seven genes proved to be particularly interesting, considering their expression data and pathway analysis results, as described below.
Concerning DP, three genes: PIWIL4, MBD2, and NTN1, were significantly associated after the FDR correction. The PIWIL4 gene encodes for a protein belonging to the Argonaute family of proteins. Piwi proteins are believed to play a crucial role in spermatogenesis, as their absence leads to male infertility. This gene showed strong expression values in corneal tissues (Cn, CFE, CSE), in the retinal pigmented epithelium, and in the retina. PIWIL4 is reported to regulate the expression of Alu RNA in response to oxidative stress, protecting RPE from degeneration (Hwang et al., 2019). Interestingly, the pathway analysis showed PIWIL4 interaction with DICER1, and deficiency of this gene in response to oxidative stress in RPE is reported to lead to the accumulation of Alu RNA and degeneration of the RPE (Kaarniranta et al., 2020), suggesting a role of both PIWIL4 and DICER1 in the preservation of RPE. Moreover, PIWIL4 modulates the expression of tight-junctions proteins in RPE, contributing to the structural integrity of the epithelium (Sivagurunathan et al., 2017).
MBD2 encodes for the methyl-CpG binding domain protein 2, a protein that binds specific methylated sequences, involved in transcription regulation, both repressing and activating the transcription of methylated sites. Moreover, MBD2 encoded protein is associated with many types of cancer, such as breast (Liu et al., 2021), renal (Li et al., 2020), colorectal (Yu et al., 2020) and prostate (Patra et al., 2003). Furthermore, MBD2 is reported to be involved in the T-cells development (Cheng et al., 2021). This gene showed a high expression in all investigated tissues, particularly in corneal tissues. MBD2 protein is reported to mediate apoptosis in retinal ganglion cells (Ge et al., 2020), and possibly play a role in the pathogenesis of age-related macular degeneration (Pan et al., 2014). In addition, our pathway analyses showed a link between MBD2 protein and hexokinase 2 (HK2) protein in the eye’s formation process. HK2 protein is reported to have several crucial roles in maintaining photoreceptors’ health and functionality (Weh et al., 2020).
NTN1 encodes for Netrin 1, a laminin-related secreted protein involved in axon guidance, cell migration, morphogenesis, angiogenesis, peripheral nerve regeneration and Schwann cell proliferation. NTN1 showed high expression values throughout all inquired tissues, especially in CE, CFE, RFE, and RPE. Although no association between NTN1 and genes involved in eye physiology and pathology was found by the pathway analysis, in the eye, netrin-1 is reported as a key molecule for the growth of axons directed to the optical nerve head (Mann et al., 2004) and for the migration of the glial-precursors from the brain to the retina (Tsai and Miller, 2002).
Notably, all the above genes could play a role in determining the DP phenotype. Alterations in PIWIL4 could lead to RPE degeneration, thus hindering its ability to support photoreceptors and correct color vision. Concerning MBD2 and NTN1, both of these genes play a role in the transmission of the visual signal, acting in retinal ganglion cells and in the optic nerve, respectively. We can therefore hypothesize that defects in these genes could alter the functionality of these tissues leading to an altered color vision.
With regards to TR, among those significantly associated after the FDR correction, four genes - VPS54, IQGAP1, NMB, and MC5R - stood out as promising candidates, considering the number of SNPs in linkage with the top SNP, their reported role in the eye, and their expression values. VPS54 encodes for a subunit of the GARP complex, a protein complex involved in retrograde transport from early and late endosomes to the trans-Golgi network (TGN). Moreover, VPS54 is reported to be part of RP28, a locus associated with recessive retinitis pigmentosa (Kumar et al., 2004). VPS54 showed a consistent expression in all investigated tissues, and the pathway analysis highlighted its interaction with LAMP2. Loss of function variants in LAMP2 cause Danon disease, a glycogen storage disease also known as X-linked vacuolar cardiomyopathy and myopathy. However, LAMP2 deficiencies are also reported to cause retinopathies and RPE degeneration independently (Kousal et al., 2021) or in association with Danon disease (Fukushima et al., 2020). Interestingly VPS54 knock-out and the resulting GARP deficiency are reported to alter the formation of lysosomal structures deputed to the accumulation of endolysosomal proteins, including Lamp2 (D’Souza et al., 2019).
IQGAP1 belongs to the IQGAP protein family, a family of scaffold proteins involved in the regulation of several cellular pathways and functions. In particular, the isoform encoded by IQGAP1 interacts with phosphatidylinositol phosphate kinase mediating cell motility and Akt phosphorylation. IQGAP1 showed high expression values throughout all tissues, especially in corneal tissues, in RPE and RGS, and showed a complex network of interactions in the pathway analysis. For instance, IQGAP1 interacts with THRB, a gene encoding for the thyroid hormone receptor (TRβ), and is reported to enhance transcriptional activation of TRβ isoform 2 (Hahm and Privalsky, 2013). The THRB gene is involved in cone photoreceptors’ development and function (Deveau et al., 2020). Indeed, THRB is reported to inhibit S opsin expression (Ma and Ding, 2016), and its mutation leads to an imbalance in the distribution of cone types in mice retina (Ng et al., 2001). Moreover, IQGAP1 is reported to regulate retinal neurite growth in chicken retina (Oblander and Brady-Kalnay, 2010) and to play a key role in the regulation of choroidal endothelial cells in the pathogenesis of Age-related Macular Degeneration (Ramshekar et al., 2021).
NMB encodes for neuromedin B, a member of the bombesin-related family of neuropeptides. Neuromedin B is expressed in the central nervous system and in the gastrointestinal tract and is involved in several physiological functions, including regulation of exocrine and endocrine secretions, smooth muscle contraction, feeding, blood pressure, blood glucose, body temperature, and cell growth. NMB showed a moderate expression in all tissues, with higher levels in RPFT, RFE, and RGS. Although no associations between the inquired genes and this gene were highlighted by the pathway analysis, NMB is reported to play a role in the induction of intracellular Ca2+ transient and phosphatidylinositol turnover in RPE cells (Kuriyama et al., 1992) and to constitute a marker of chicken retinal ganglion cells (Yamagata et al., 2006).
MC5R belongs to the family of melanocortin receptors, formed by five G-protein coupled receptors involved in a broad spectrum of physiological processes, including regulation of pigmentation, regulation of inflammation, and pain perception. MC5R has a wide peripheral distribution and is involved in many processes such as glucose uptake (Enriori et al., 2016), exocrine gland secretion (Chen et al., 1997) and fatty acids oxidation (An et al., 2007). This gene showed poor expression in all inquired tissues, excluding the corneal tissue, and no associations were found for this gene through the pathway analysis. However, MC5R is reported to protect from uveitis-derived retinal damage by regulating the activation of CD4+ regulatory T cells (Taylor et al., 2006) and to attenuate retinal degeneration in an experimental model of diabetic retinopathy (Rossi et al., 2016). Moreover, MC5R is reported to play a crucial role in the lacrimal gland, maintaining lacrimal function and secretion (Nguyen et al., 2004).
All the above genes proved to be potentially involved in TR determination: defects in VPS54 could lead to RPE degeneration, thus leading to color vision impairment; alterations in IQGAP1 can cause cone type imbalances, with a diminished number and functionality of S cones; defects in NMB may alter RPE’s ability to regulate fluid homeostasis, resulting in subretinal fluid accumulation and retinal function impairment; alterations in MC5R could affect lacrimal gland function leading to dry eye onset and progressive visual impairment.
In conclusion, this study highlighted, for the first time, the association of 15 new genes with the determination of multifactorial forms of CVDs. Notably, seven genes, PIWIL4, MBD2, NTN1, VPS54, IQGAP1, NMB, and MC5R, proved to be particularly promising considering their expression in the eye and their known implication in several physiologic and pathologic eye processes. It is worth noting how, in our study, the availability of phenotypic and genotypic data from individuals coming from isolated communities allowed us to enhance the discovery power of GWAS, offering the possibility to capture new and impactful polymorphisms associated with complex traits. Up to date, there are few to no studies investigating complex traits in Silk Road populations. Therefore, with this work, we provide new and insightful information on such underrepresented populations and a natural next step of future research could seek to replicate these findings in different cohorts.
Publicly available datasets were analyzed in this study. This data can be found here: European Variation Archive (EVA): https://www.ebi.ac.uk/eva/?eva-study=PRJEB60906.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Institute for Maternal and Child Health—I.R.C.C.S. “Burlo Garofolo” of Trieste (Italy). The patients/participants provided their written informed consent to participate in this study.
MB-P performed the phenotype classification. GGN analysed the data with the support of AS, MPC, BS and LM. GGN and BS wrote the manuscript with the contribution of MPC and AM. GG conceived the study and supervised the project. All authors contributed to the article and approved the submitted version.
This research was supported by D70-RESRICGIROTTO to GG.
The authors wish to thank all the volunteers for their keen participation in the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1161696/full#supplementary-material
An, J. J., Rhee, Y., Kim, S. H., Kim, D. M., Han, D.-H., Hwang, J. H., et al. (2007). Peripheral effect of α-Melanocyte-stimulating hormone on fatty acid oxidation in skeletal muscle. J. Biol. Chem. 282, 2862–2870. doi:10.1074/jbc.M603454200
Chen, W., Kelly, M. A., Opitz-Araya, X., Thomas, R. E., Low, M. J., and Cone, R. D. (1997). Exocrine gland dysfunction in MC5-R-deficient mice: Evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell. 91, 789–798. doi:10.1016/S0092-8674(00)80467-5
Chen, Z., Boehnke, M., Wen, X., and Mukherjee, B. (2021). Revisiting the genome-wide significance threshold for common variant GWAS. G3 Genes.|Genomes|Genetics 11, jkaa056. doi:10.1093/g3journal/jkaa056
Cheng, L., Zhou, K., Chen, X., Zhou, J., Cai, W., Zhang, Y., et al. (2021). Loss of MBD2 affects early T cell development by inhibiting the WNT signaling pathway. Exp. Cell. Res. 398, 112400. doi:10.1016/j.yexcr.2020.112400
D’Souza, Z., Blackburn, J. B., Kudlyk, T., Pokrovskaya, I. D., and Lupashin, V. V. (2019). Defects in COG-mediated golgi trafficking alter endo-lysosomal system in human cells. Front. Cell. Dev. Biol. 7, 118. doi:10.3389/fcell.2019.00118
Deveau, C., Jiao, X., Suzuki, S. C., Krishnakumar, A., Yoshimatsu, T., Hejtmancik, J. F., et al. (2020). Thyroid hormone receptor beta mutations alter photoreceptor development and function in Danio rerio (zebrafish). PLoS Genet. 16, e1008869. doi:10.1371/journal.pgen.1008869
Dulai, K. S., von Dornum, M., Mollon, J. D., and Hunt, D. M. (1999). The evolution of trichromatic color vision by opsin gene duplication in new world and old world primates. Genome Res. 9, 629–638. doi:10.1101/gr.9.7.629
Enriori, P. J., Chen, W., Garcia-Rudaz, M. C., Grayson, B. E., Evans, A. E., Comstock, S. M., et al. (2016). α-Melanocyte stimulating hormone promotes muscle glucose uptake via melanocortin 5 receptors. Mol. Metab. 5, 807–822. doi:10.1016/j.molmet.2016.07.009
Fukushima, M., Inoue, T., Miyai, T., and Obata, R. (2020). Retinal dystrophy associated with Danon disease and pathogenic mechanism through LAMP2-mutated retinal pigment epithelium. Eur. J. Ophthalmol. 30, 570–578. doi:10.1177/1120672119832183
Ge, Y., Zhang, R., Feng, Y., and Li, H. (2020). Mbd2 mediates retinal cell apoptosis by targeting the lncRNA mbd2-AL1/miR-188-3p/traf3 Axis in ischemia/reperfusion injury. Mol. Ther. Nucleic Acids 19, 1250–1265. doi:10.1016/j.omtn.2020.01.011
Gilad, Y., Wiebe, V., Przeworski, M., Lancet, D., and Pääbo, S. (2004). Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2, e5. doi:10.1371/journal.pbio.0020005
Girotto, G., Pirastu, N., Sorice, R., Biino, G., Campbell, H., d’Adamo, A. P., et al. (2011). Hearing function and thresholds: A genome-wide association study in European isolated populations identifies new loci and pathways. J. Med. Genet. 48, 369–374. doi:10.1136/jmg.2010.088310
GTEx Consortium (2013). The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585. doi:10.1038/ng.2653
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi:10.1093/bioinformatics/btw313
Hahm, J. B., and Privalsky, M. L. (2013). Research resource: Identification of novel coregulators specific for thyroid hormone receptor-β2. Mol. Endocrinol. 27, 840–859. doi:10.1210/me.2012-1117
Hasrod, N., and Rubin, A. (2016). Defects of colour vision: A review of congenital and acquired colour vision deficiencies. Afr. Vis. Eye Health 75, 6. doi:10.4102/aveh.v75i1.365
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J., and Abecasis, G. R. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959. doi:10.1038/ng.2354
Hwang, Y. E., Baek, Y. M., Baek, A., and Kim, D.-E. (2019). Oxidative stress causes Alu RNA accumulation via PIWIL4 sequestration into stress granules. BMB Rep. 52, 196–201. doi:10.5483/BMBRep.2019.52.3.146
Ichikawa, K., Yokoyama, S., Tanaka, Y., Nakamura, H., Smith, R. T., and Tanabe, S. (2021). The change in color vision with normal aging evaluated on standard pseudoisochromatic plates part-3. Curr. Eye Res. 46, 1038–1046. doi:10.1080/02713683.2020.1843683
Kaarniranta, K., Pawlowska, E., Szczepanska, J., and Blasiak, J. (2020). DICER1 in the pathogenesis of age-related macular degeneration (AMD) - Alu RNA accumulation versus miRNA dysregulation. Aging Dis. 11, 851–862. doi:10.14336/AD.2019.0809
Kousal, B., Majer, F., Vlaskova, H., Dvorakova, L., Piherova, L., Meliska, M., et al. (2021). Pigmentary retinopathy can indicate the presence of pathogenic LAMP2 variants even in somatic mosaic carriers with no additional signs of Danon disease. Acta Ophthalmol. 99, 61–68. doi:10.1111/aos.14478
Kumar, A., Shetty, J., Kumar, B., and Blanton, S. H. (2004). Confirmation of linkage and refinement of the RP28 locus for autosomal recessive retinitis pigmentosa on chromosome 2p14-p15 in an Indian family. Mol. Vis. 10, 399–402.
Kuriyama, S., Yoshimura, N., Ohuchi, T., Tanihara, H., Ito, S., and Honda, Y. (1992). Neuropeptide-induced cytosolic Ca2+ transients and phosphatidylinositol turnover in cultured human retinal pigment epithelial cells. Brain Res. 579, 227–233. doi:10.1016/0006-8993(92)90055-e
Li, L., Li, N., Liu, N., Huo, F., and Zheng, J. (2020). MBD2 correlates with a poor prognosis and tumor progression in renal cell carcinoma. Onco Targets Ther. 13, 10001–10012. doi:10.2147/OTT.S256226
Liu, Z., Sun, L., Cai, Y., Shen, S., Zhang, T., Wang, N., et al. (2021). Hypoxia-Induced suppression of alternative splicing of MBD2 promotes breast cancer metastasis via activation of FZD1. Cancer Res. 81, 1265–1278. doi:10.1158/0008-5472.CAN-20-2876
Ma, H., and Ding, X.-Q. (2016). Thyroid hormone signaling and cone photoreceptor viability. Adv. Exp. Med. Biol. 854, 613–618. doi:10.1007/978-3-319-17121-0_81
Mann, F., Harris, W. A., and Holt, C. E. (2004). New views on retinal axon development: A navigation guide. Int. J. Dev. Biol. 48, 957–964. doi:10.1387/ijdb.041899fm
McCarthy, S., Das, S., Kretzschmar, W., Delaneau, O., Wood, A. R., Teumer, A., et al. the Haplotype Reference Consortium (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283. doi:10.1038/ng.3643
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R. S., Thormann, A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17, 122. doi:10.1186/s13059-016-0974-4
Mezzavilla, M., Vozzi, D., Pirastu, N., Girotto, G., d’Adamo, P., Gasparini, P., et al. (2014). Genetic landscape of populations along the Silk Road: Admixture and migration patterns. BMC Genet. 15, 131. doi:10.1186/s12863-014-0131-6
Nathans, J. (1999). The evolution and physiology of human color vision: Insights from molecular genetic studies of visual pigments. Neuron 24, 299–312. doi:10.1016/S0896-6273(00)80845-4
Ng, L., Hurley, J. B., Dierks, B., Srinivas, M., Saltó, C., Vennström, B., et al. (2001). A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98. doi:10.1038/83829
Nguyen, D. H., Toshida, H., Schurr, J., and Beuerman, R. W. (2004). Microarray analysis of the rat lacrimal gland following the loss of parasympathetic control of secretion. Physiol. Genomics 18, 108–118. doi:10.1152/physiolgenomics.00011.2004
Oblander, S. A., and Brady-Kalnay, S. M. (2010). Distinct PTPmu-associated signaling molecules differentially regulate neurite outgrowth on E-, N-, and R-cadherin. Mol. Cell. Neurosci. 44, 78–93. doi:10.1016/j.mcn.2010.02.005
Oli, A., and Joshi, D. (2019). Efficacy of red contact lens in improving color vision test performance based on Ishihara, Farnsworth D15, and Martin Lantern Test. Med. J. Armed Forces India 75, 458–463. doi:10.1016/j.mjafi.2018.08.005
Pan, J.-R., Wang, C., Yu, Q.-L., Zhang, S., Li, B., and Hu, J. (2014). Effect of Methyl-CpG binding domain protein 2 (MBD2) on AMD-like lesions in ApoE-deficient mice. J. Huazhong Univ. Sci. Technol. Med. Sci. 34, 408–414. doi:10.1007/s11596-014-1292-2
Patra, S. K., Patra, A., Zhao, H., Carroll, P., and Dahiya, R. (2003). Methyl-CpG–DNA binding proteins in human prostate cancer: Expression of CXXC sequence containing MBD1 and repression of MBD2 and MeCP2. Biochem. Biophysical Res. Commun. 302, 759–766. doi:10.1016/S0006-291X(03)00253-5
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Ramshekar, A., Wang, H., and Hartnett, M. E. (2021). Regulation of Rac1 activation in choroidal endothelial cells: Insights into mechanisms in age-related macular degeneration. Cells 10, 2414. doi:10.3390/cells10092414
Rossi, S., Maisto, R., Gesualdo, C., Trotta, M. C., Ferraraccio, F., Kaneva, M. K., et al. (2016). Activation of melanocortin receptors MC1 and MC5 attenuates retinal damage in experimental diabetic retinopathy. Mediat. Inflamm. 2016, 7368389. doi:10.1155/2016/7368389
Simunovic, M. P. (2016). Acquired color vision deficiency. Surv. Ophthalmol. 61, 132–155. doi:10.1016/j.survophthal.2015.11.004
Sivagurunathan, S., Palanisamy, K., Arunachalam, J. P., and Chidambaram, S. (2017). Possible role of HIWI2 in modulating tight junction proteins in retinal pigment epithelial cells through Akt signaling pathway. Mol. Cell. Biochem. 427, 145–156. doi:10.1007/s11010-016-2906-8
Struck, T. J., Mannakee, B. K., and Gutenkunst, R. N. (2018). The impact of genome-wide association studies on biomedical research publications. Hum. Genomics 12, 38. doi:10.1186/s40246-018-0172-4
Taylor, A., Kitaichi, N., and Biros, D. (2006). Melanocortin 5 receptor and ocular immunity. Cell. Mol. Biol. (Noisy-le-Grand, France) 52, 53–59. doi:10.1170/T708
Tsai, H.-H., and Miller, R. H. (2002). Glial cell migration directed by axon guidance cues. Trends Neurosci. 25, 173–175. discussion 175-176. doi:10.1016/s0166-2236(00)02096-8
Weh, E., Lutrzykowska, Z., Smith, A., Hager, H., Pawar, M., Wubben, T. J., et al. (2020). Hexokinase 2 is dispensable for photoreceptor development but is required for survival during aging and outer retinal stress. Cell. Death Dis. 11, 422. doi:10.1038/s41419-020-2638-2
Yamagata, M., Weiner, J. A., Dulac, C., Roth, K. A., and Sanes, J. R. (2006). Labeled lines in the retinotectal system: Markers for retinorecipient sublaminae and the retinal ganglion cell subsets that innervate them. Mol. Cell. Neurosci. 33, 296–310. doi:10.1016/j.mcn.2006.08.001
Yu, J., Xie, Y., Liu, Y., Wang, F., Li, M., and Qi, J. (2020). MBD2 and EZH2 regulate the expression of SFRP1 without affecting its methylation status in a colorectal cancer cell line. Exp. Ther. Med. 20, 242. doi:10.3892/etm.2020.9372
Keywords: color vision defects, Deutan, Protan, Tritan, genome wide association study, genetic isolates, Silk Road, pathway analysis
Citation: Nardone GG, Spedicati B, Concas MP, Santin A, Morgan A, Mazzetto L, Battaglia-Parodi M and Girotto G (2023) Identifying missing pieces in color vision defects: a genome-wide association study in Silk Road populations. Front. Genet. 14:1161696. doi: 10.3389/fgene.2023.1161696
Received: 08 February 2023; Accepted: 31 May 2023;
Published: 09 June 2023.
Edited by:
Denis Plotnikov, Kazan State Medical University, RussiaReviewed by:
Michelle Grunin, Case Western Reserve University, United StatesCopyright © 2023 Nardone, Spedicati, Concas, Santin, Morgan, Mazzetto, Battaglia-Parodi and Girotto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Pina Concas, bWFyaWFwaW5hLmNvbmNhc0BidXJsby50cmllc3RlLml0
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.