
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 23 March 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1151645
This article is part of the Research Topic Plant Secondary Metabolites and Their Effects on Environmental Adaptation Based on Functional Genomics View all 5 articles
Coptis chinensis Franch. (Huanglian in Chinese) is an important economic crop with medicinal value. Its rhizome has been used as a traditional herbal medicine for thousands of years in Asia. Protoberberine alkaloids, as the main bioactive component of Coptis chinensis, have a series of pharmacological activities. However, the protoberberine alkaloids content of C. chinensis is relatively low. Understanding the molecular mechanisms affecting the transcriptional regulation of protoberberine alkaloids would be crucial to increase their production via metabolic engineering. WRKY, one of the largest plant-specific gene families, regulates plant defense responses via the biosynthesis of specialized metabolites such as alkaloids. Totally, 41 WRKY transcription factors (TFs) related to protoberberine alkaloid biosynthesis were identified in the C. chinensis genome and classified into three groups based on phylogenetic and conserved motif analyses. Three WRKY genes (CcWRKY7, CcWRKY29, and CcWRKY32) may regulate protoberberine alkaloid biosynthesis, as suggested by gene-specific expression patterns, metabolic pathways, phylogenetic, and dual-luciferase analysis. Furthermore, the CcWRKY7, CcWRKY29, and CcWRKY32 proteins were specifically detected in the nucleus via subcellular localization. This study provides a basis for understanding the regulatory mechanisms of protoberberine alkaloid biosynthesis and valuable information for breeding C. chinensis varieties.
Coptis chinensis belongs to the Ranunculaceae family and is primarily distributed throughout Southern China, specifically in the Sichuan, Hubei, Hunan, and Guizhou Provinces. The rhizomes of Coptis chinensis (“Huanglian” in Chinese) are a traditional herbal medicine used to treat infectious and inflammatory diseases (Tseng et al., 2020; Yang et al., 2021; Tseng et al., 2022; Yan et al., 2023). The primary active ingredients in C. chinensis include protoberberine-type alkaloids such as berberine, palmatine, tetrahydropalmatine, coptisine, jatrorrhizine, and columbamine (Liu et al., 2016; Tan et al., 2016; Wu et al., 2019; Li et al., 2021). With the gradual development of society, people pay more attention to their health status, increasing the demand for C. chinensis in different degrees. In recent years, the amount of C. chinensis has continually increasingly reduced due to over-excavation or exploitation of this Huanglian. Synthetic biology provides a new approach to the acquisition of active ingredients in natural products, which is green and efficient characteristics. The biosynthesis of terpenoids, flavonoids and alkaloids has recently been achieved via synthetic biology (Paddon et al., 2013; Nakagawa et al., 2016; Wu et al., 2022). Therefore, further investigation into the underlying mechanisms of protoberberine alkaloid biosynthesis using a synthetic biology approach to increase production is the key to achieving industrialized production.
The WRKY transcription factors (TFs) family play a significant role in plant stress response, including regulation of specialized metabolite biosynthesis (Schluttenhofer and Yuan, 2015; Vashisht et al., 2016; Barco and Clay, 2019; Zhang et al., 2020). The WRKY family contains about 60 amino acid binding domains and covers Classes I-III according to the number of WRKY domains and the variety of zinc finger motifs (Rushton et al., 2010; Hussain et al., 2019). Several WRKY TFs have been shown to regulate terpenoid and alkaloid biosynthesis functionally (Li et al., 2019; Hao et al., 2021; Yamada et al., 2021). For example, the TF, AaWRKY1, positively regulates the biosynthesis of artemisinin via promoting the expression of the genes DBR2, CYP71AV1, and ADS in Artemisia annua (Han et al., 2014). CjWRKY1 regulates the biosynthesis of berberine in Coptis japonica (Yamada and Sato, 2016), and PsWRKY1 regulates the biosynthesis of morphine in Papaver somniferum (Mishra et al., 2013). These data suggest that potential WRKY genes were functionally diverse among protoberberine-producing medicinal plant species and secondarily obtained additional functions associated with protoberberine biosynthesis in C. chinensis.
This research investigated the WRKY family genes in the C. chinensis genome via bioinformatics. The conserved domains, expression profiles of the CcWRKY genes in various tissues, and the main components of C. chinensis were analyzed. CcWRKY7, CcWRKY29, and CcWRKY32 proteins were detected using subcellular localization. Finally, three CcWRKY genes were identified that could be involved in berberine biosynthesis, providing the foundation for a more in-depth regulatory study.
Gene annotation information using a typical WRKY domain was used to search the C. chinensis genome to identify the WRKY genes. 41 WRKY genes were identified from C. chinensis following the removal of incomplete sequences. The WRKY domain was further confirmed using TBtools software (Chen et al., 2020). The 41 putative CcWRKY genes were designated as CcWRKY1 to CcWRKY41. Table 1 contains the complete data for CcWRKY genes, including gene names, gene IDs, protein lengths, molecular weights (MW), and isoelectric points (pI). The protein lengths ranged from 118 to 931 amino acids (aa), the MW ranged from 13,358.29 to 103,620.60 Da, and the pI varied from 4.96 to 10.17. In order to further comprehend the conservation and diversity of the proteins, 10 motifs were identified by analyzing the CcWRKYs conserved motifs using the MEME program. Motif 1 and motif 2 were present in most genes (Figure 1) and most genes contained two or more motifs except CcWRKY22 and CcWRKY41. The same subgroups of CcWRKY members usually contained similar motifs (Supplementary Figure S1).
To classify the 41 CcWRKY proteins, an unrooted Maximum Likelihood (ML) phylogenetic tree was created using the CcWRKYs and Arabidopsis thaliana AtWRKYs proteins (Figure 2). According to the AtWRKYs classification, the CcWRKY proteins were separated into three groups (Group I-III). Group I had 6 members with two WRKY conserved domains (the N-terminal and C-terminal WRKY domains). Group II comprised twenty-five members, each harboring one WRKY domain (Supplementary Figure S1). According to the amino acid sequences, Group II was further categorized into subgroups IIa through IIe. Four CcWRKY proteins were classified in Group III, containing one WRKY domain. Group I and II had a C2H2 zinc finger motif, while Group III harbored the C2HC motif. Six CcWRKY proteins, including CcWRKY8, CcWRKY9, CcWRKY13, CcWRKY14, CcWRKY24, and CcWRKY41, did not cluster with any of the above Groups despite containing a WRKY domain.
Thirty-nine CcWRKY genes were distributed on nine chromosomes, and two genes (CcWRKY9 and CcWRKY29) did not belong to any chromosome (Figure 3). Most of the CcWRKY genes were abundant on chromosome 9 (nine genes, 21.95%) and chromosome 1 (six genes, 14.63%), whereas they were only two WRKY genes on chromosome 6. In addition, four CcWRKY genes were on chromosome 2, five on chromosome 3, and three on chromosome 4. Three and four CcWRKYs were on chromosomes 5 and 7, respectively, and four genes were identified on chromosome 8. The uneven distribution of CcWRKY genes may be due to the variation in the sizes and structures of C. chinensis chromosomes.
To investigate the tissue-specific accumulation patterns of alkaloids in C. chinensis, HPLC-QTOF-MS/MS was performed on samples, including the root, rhizome, petiole, and leaf tissues. Numerous protoberberine alkaloids found primarily in C. chinensis have been identified. These included significantly high levels of berberine, followed by jatrorrhizine, palmatine, and coptisine. Interestingly, the predominantly alkaloids and berberine content in tissues was as follows: rhizome > root > petiole > leaf (Figure 4A). However, palmatine and jatrorrhizine content was as follows: root > rhizome > petiole > leaf. These results suggest that the protoberberine biosynthetic enzymes may be tissue-specific.
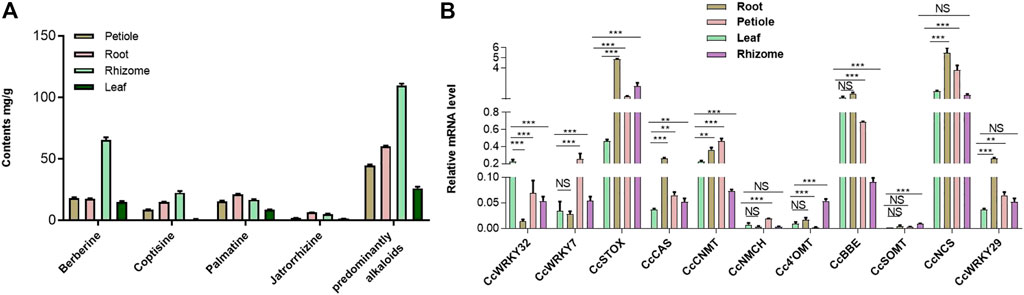
FIGURE 4. The main protoberberine alkaloids content of C. chinensis in different tissues and the expression profile of CcWRKY genes. (A) The main protoberberine alkaloid content of Coptis chinensis in different tissues. (B) Expression profile of CcWRKY genes and their candidate target genes involved in protoberberine alkaloids biosynthesis in different tissues including, root, petiole, leaf, and rhizome.
The expression profiles of genes in different tissues can provide essential information for for screening functional genes. Previous research demonstrated that CjWRKY1 overexpression in Eschscholzia californica cells strongly enhances benzylisoquinoline alkaloid (BIA) biosynthesis (Yamada et al., 2017; Yamada et al., 2021). Homology search using CjWRKY1 amino acid sequences as queries against the genome database of C. chinensis revealed a similarity of CjWRKY1 with the CcWRKY7, CcWRKY29, and CcWRKY32. The expression levels of these three genes in C. chinensis were analyzed by qRT–PCR. Furthermore, identifying the expression pattern of protoberberine biosynthetic genes can aid in the understanding of berberine biosynthesis regulation. Subsequently, CcWRKY7, CcWRKY29, CcWRKY32, and eight protoberberines biosynthetic genes (CcNCS, Cc4′OMT, CcCNMT, CcNMCH, CcBBE, CcSOMT, CcSTOX, and CcCAS) were detected in the root, rhizome, petiole, and leaf tissues. The expressions of CcWRKY7, CcWRKY29, and CcWRKY32 were very similar to CcCNMT (the upstream genes in protoberberine biosynthesis). The transcription factor CcWRKY29 and the biosynthetic genes CcBBE, CcCNMT, CcSTOX, and CcCAS, that act on the downstream enzymes for the biosynthesis of predominant protoberberines (berberine and palmatine), were significantly overexpressed in root compared with other tissues (Figure 4B, Supplementary Figure S2).
To analyze the prediction results of CcWRKY7, CcWRKY29, and CcWRKY32 subcellular localization, full-length CDSs of these three genes (lacking the stop codon) were cloned into the pCAMBIA2300 vector (containing the 35S promoter and GFP). Transient transformation assays were done using N. benthamiana Domin leaves. The results demonstrated that the GFP fused CcWRKY7, CcWRKY29, and CcWRKY32 proteins were specifically expressed in the nucleus of the Nicotiana benthamiana leaf. In contrast, the GFP control protein was detected in various organelles (Figure 5). The nuclear localization results were consistent with the expected role of CcWRKYs as transcription factors.
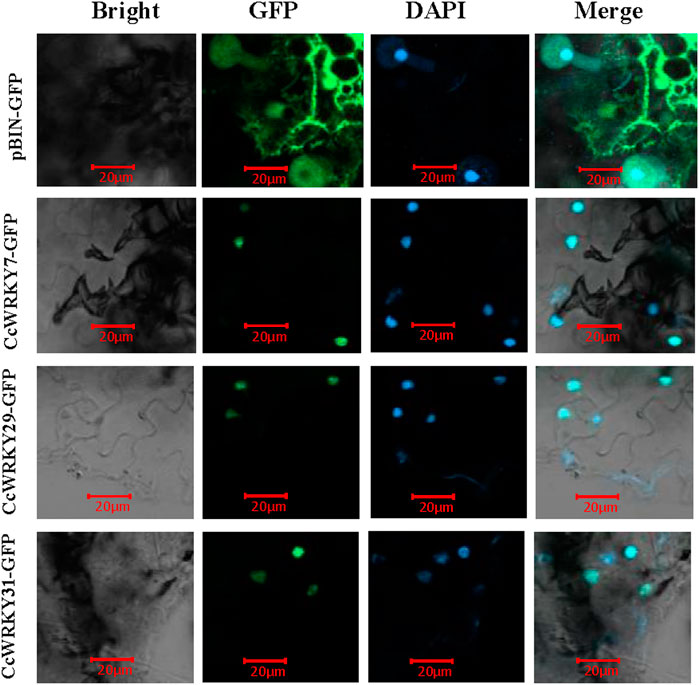
FIGURE 5. Subcellular localization of CcWRKY7, CcWRKY29 and CcWRKY32 proteins in Nicotiana benthamiana leaves. Laser scanning confocal microscope with wavelengths of 488 and 406 nm for GFP and DAPI, respectively.
The candidate target genes promoter elements of CcWRKYs transcription factors analyzed. It was showed that protoberberines biosynthetic genes (CcNCS, Cc4′OMT, CcCNMT, and CcSOMT contained W-box element, necessary binding site of WRKY. This suggests that CcWRKY may play a role in regulating protoberberines biosynthesis by regulating the expression of above target genes.To further verify CcWRKY7, CcWRKY29, and CcWRKY32 regulate the protoberberine biosynthetic gene, dual-luciferase assays were performed in N. benthamiana leaves. The results showed that CcWRKY7-pCcCNMT, CcWRKY29-pCcCNMT, and CcWRKY7-pCcCNMT significantly higher reporter activation than controls, respectively (Figure 6). Taken together, our results suggest that CcWRKY7, CcWRKY29, and CcWRKY32 can bind and activate the promoter of CcCNMT, respectively.
Previous studies reported that the medicinal plant C. chinensis is a diploid (2n = 18), and the genomic heterozygosity proportion is 0.77. The chromosome-level genome of C. chinensis was −958.20 Mb with a scaffold N50 of 4.53 Mb, and a contig N50 of 1.58 Mb (Chen DX. et al., 2021). A total of 34,109 protein-coding genes were identified in C. chinensis with an average CDS coding sequence length of 1031.26 bp. The genomic data of C. chinensis provides abundant gene resources for studying the biosynthetic pathway of berberine, especially the key enzyme genes and regulated transcription factors. Protoberberine alkaloids, the main ingredient of C. chinensis, have a series of biological activities (Qin et al., 2018; Chen Q. et al., 2021). As the demand for protoberberine alkaloids has grown in recent years, obtaining higher content of protoberberine alkaloids by controlling their biosynthetic pathway has attracted much attention. TFs can promote compound production by activating biosynthetic enzymes at the transcriptional level (Suttipanta et al., 2011). Therefore, this study investigated the WRKY TFs involved in regulating protoberberine alkaloid biosynthesis, and 41 WRKY genes were identified in the C. chinensis genome. Meanwhile, the conserved domains, the major components in C. chinensis, and expression profiles of the CcWRKY genes in different tissues were further analyzed. The phylogenetic, gene tissue-specific expression, dual-luciferase analysis suggested that three CcWRKY genes may regulate protoberberine alkaloid biosynthesis. The CcWRKY7, CcWRKY29, and CcWRKY32 proteins were specifically detected in the nucleus. The WRKY TFs are widely distributed throughout the plant kingdom, playing an essential role in plant growth and stress response (Liu et al., 2020). Moreover, developing the CcWRKY gene profile is useful for future morphogenesis and resistance formation studies in C. chinensis. Although we preliminarily found that three CcWRKYs may play a role in regulating berberine biosynthesis via the target gene CcCNMT, the molecular mechanism remains to be clarified, which is also our future focus.
The diploid cultivar of Coptis chinensis (two-year-old), which was originally collected from Mianyang, Sichuan. The plants were transferred to our laboratory greenhouse to acclimate for 2 weeks before the experiment began. The root, rhizome, petiole, and leaf of the C. chinensis were collected in three biological replicates. All samples were frozen in liquid nitrogen after incubating in RNAlater and then stored at −80°C for RNA extraction. Samples were freeze-dried under a vacuum and powdered ground for metabolic analysis.
The whole genome sequence of C. chinensis (Accession Number PRJNA649082) was downloaded from NCBI, and AtWRKY protein sequences from TAIR (https://www.arabidopsis.org) (Supplementary Text S1). Firstly, the CcWRKY protein sequences were queried using Auto BLAST and HMM to search for potential CcWRKY proteins. Secondly, the potential CcWRKY annotation information was compared with the Swiss-Port database, and then, candidate CcWRKY proteins were submitted to NCBI-CDD. Pfam database was used to verify whether candidate CcWRKY proteins contained the conserved domains. Next, EXPASy-ProtParam was used to predict the coding sequence (CDS) length, the isoelectric point and molecular weights of all CcWRKY proteins. The Plant-mPLo website was used to predict the subcellular localization.
Phylogenetic analysis was conducted using the Maximum Likelihood method (ML) with 1,000 bootstrap replicates (Jones-Taylor-Thornton model, site coverage cut off 95%). The conserved motifs were performed using Batch-Search and the TBtools software. To better understand the functions of CcWRKY proteins, the protein sequences were analyzed for conserved motifs using the MEME program (https://meme-suite.org/meme/). The CcWRKY gene distribution map was generated using the TBtools software.
Dry powder ground samples (0.1 g) with 5 mL methanol were sonicated for 30 min. Primary components of C. chinensis were determined using HPLC-QTOF-MS/MS (Agilent Technologies) with a C18 column (Agilent, 2.7 μm, 4.6 mm × 100 mm). The column temperature was 30°C, and the flow rate was set to 0.25 mL/min with a 5 μL injection volume. The mobile phase consisted of phase A (water containing 0.1% formic acid) and phase B (acetonitrile) with the following gradient program: 10% B at 0–2 min, 10%–20% B at 2–10 min, 20% B at 10–13 min, 20%–30% B at 13–16 min, 30% B at 16–17 min, 90%–90% B at 17–20 min, 90% B at 20–22 min, 90%–10% B at 22–23 min, 10% B at 23–25 min. Mass spectra were acquired using an electrospray ionization source, and the acquisition parameters in the positive mode were set as follows: drying gas flow rate, 10.0 L/min; temperature, 350°C; fragmentor voltage, 130 V and nebulizer, 35 psig; OCT RFV, 750 V; capillary voltage, 3500 V; collision energy 20 eV; Mass range from m/z 100 to 1700.
Total RNA was extracted with TRIzol reagent (Sigma-Aldrich), and cDNA was synthesized by reverse transcription with HiScript qRT SuperMix (Vazyme, China). QRT-PCR assays were performed with a LC480 Light Cycler (Roche, Germany). The Ccβ-actin gene from C. chinensis was used as an internal standard to calculate relative fold differences based on the comparative cycle threshold values. The RT-qPCR procedure was performed on the following parameters: 95°C for 10 min; 40 cycles of 95°C for 15 s; 60°C for 60 s. All primers of the qRT–PCR are listed in Supplementary Table S1.
The CcWRKYs CDS were constructed into a transient expression pCAMBIA2300 vector with a GFP fluorescent label and a CaMV 35S promoter. All amplification primers for plasmid construction were shown in Supplementary Table S1. The recombinant plasmid was transferred to Agrobacterium MSU440 via the conventional freezing-thawing method. Next, empty pCAMBIA2300-GFP or pCAMBIA2300-CcWRKY-GFP MSU440 were instantly infiltrated into 4-week-old N. benthamiana leaves with expression buffer (10 mM MgCl2, 10 mM MES pH 5.6, 100 µM acetosyringone). After the infiltrated N. benthamiana was in the dark for 24 h and then in low light for another 24 h, the leaf samples were detected using a Laser scanning confocal microscope (Zeiss LSM900).
The reporter constructs wt-pCcCNMT:fLUC and mutant-pCcCNMT:fLUC were obtained by inserting the promoter of CcCNMT and CcCNMT promoter with mutant W-box into the pGreenII 0800-LUC vector, to drive expression of the firefly luciferase gene. The Renilla luciferase (CaMV 35S promoter-driven) was used as an internal standard. The assembled vectors were co-transformed with the helper plasmid pSoup19 into A. tumefaciens strain EHA105. The A. tumefaciens strain EHA105 harboring pHBCcWRKY-YFP and pHB-YFP were used as the effector and the negative control, respectively. The reporter strains containing wt-pCcCNMT:fLUC or mutant-pCcCNMT:fLUC were mixed with effector strains harboring either pHBCcWRKY-YFP or pHB-YFP (a ratio of 1:1). The A. tumefaciens suspension was infiltrated into N. benthamiana leaves, while negative controls were infiltrated into the opposite parts of the same leaves (Hao et al., 2019). Leaves were collected after 48 h in the dark, and luciferase activities were detected using the Dual-Luciferase Reporter Assay System. All experiments were performed with three biological transfections.
This study identified 41 CcWRKY genes in the genome of C. chinensis and classified them into Groups I, II, and III based on phylogenetic relationships, motif patterns, and gene structure analysis. Phylogenetic, gene expression pattern, qRT-PCR, and dual-luciferase analysis suggest that CcWRKY7, CcWRKY29, and CcWRKY32 candidate genes may be involved in regulating the protoberberine biosynthesis. CcWRKY7, CcWRKY29, and CcWRKY32 proteins were specifically detected in the nucleus. The information collected in this study provides a basis for further exploration regulation of berberine biosynthesis and may increase the production of valuable protoberberine in metabolic engineering.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Investigation, XH and AJ; Methodology, LW and TH; Conceptualization, Writing—review and editing, GY and WZ. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China [Grant Number: 32200326]; Natural Science Foundation of Jiangsu Province [Grant Number: BK20220752] and Nanjing Postdoctoral Science Foundation [Grant Number: 032105302].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1151645/full#supplementary-material
Barco, B., and Clay, N. K. (2019). Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front. Plant Sci. 10, 1775. doi:10.3389/fpls.2019.01775
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, D. X., Pan, Y., Wang, Y., Cui, Y. Z., Zhang, Y. J., Mo, R. Y., et al. (2021a). The chromosome-level reference genome of Coptis chinensis provides insights into genomic evolution and berberine biosynthesis. Hortic. Res. 8 (1), 121. doi:10.1038/s41438-021-00559-2
Chen, Q., Ren, R., Zhang, Q., Wu, J., Zhang, Y., Xue, M., et al. (2021b). Coptis chinensis Franch polysaccharides provide a dynamically regulation on intestinal microenvironment, based on the intestinal flora and mucosal immunity. J. Ethnopharmacol. 267, 113542. doi:10.1016/j.jep.2020.113542
Han, J., Wang, H., Lundgren, A., and Brodelius, P. E. (2014). Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102, 89–96. doi:10.1016/j.phytochem.2014.02.011
Hao, X., Xie, C., Ruan, Q., Zhang, X., Wu, C., Han, B., et al. (2021). The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Hortic. Res. 8 (1), 7. doi:10.1038/s41438-020-00437-3
Hao, X., Zhong, Y., Nï Tzmann, H. W., Fu, X., Yan, T., Shen, Q., et al. (2019). Light-Induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 60 (8), 1747–1760. doi:10.1093/pcp/pcz084
Hussain, A., Noman, A., Khan, M. I., Zaynab, M., Aqeel, M., Anwar, M., et al. (2019). Molecular regulation of pepper innate immunity and stress tolerance: An overview of WRKY TFs. Microb. Pathog. 135, 103610. doi:10.1016/j.micpath.2019.103610
Li, J., Xiong, Y., Li, Y., Ye, S., Yin, Q., Gao, S., et al. (2019). Comprehensive analysis and functional studies of WRKY transcription factors in Nelumbo nucifera. Int. J. Mol. Sci. 20 (20), 5006. doi:10.3390/ijms20205006
Li, L., Zhang, D., Wang, Y., Liu, F., Xu, Y., and Bao, H. (2021). Effective extraction of palmatine and berberine from Coptis chinensis by deep eutectic solvents-based ultrasound-assisted extraction. J. Anal. Methods Chem. 2021, 9970338. doi:10.1155/2021/9970338
Liu, A., Liu, C., Lei, H., Wang, Z., Zhang, M., Yan, X., et al. (2020). Phylogenetic analysis and transcriptional profiling of WRKY genes in sunflower (Helianthus annuus L.): Genetic diversity and their responses to different biotic and abiotic stresses. Industrial Crops Prod. 148, 112268. doi:10.1016/j.indcrop.2020.112268
Liu, L., Wang, Z. B., Song, Y., Yang, J., Wu, L. J., Yang, B. Y., et al. (2016). Simultaneous Determination of Eight Alkaloids in Rat Plasma by UHPLC-MS/MS after Oral Administration of Coptis deltoidea C. Y. Cheng et Hsiao and Coptis chinensis Franch. Molecules 21 (7), 913. doi:10.3390/molecules21070913
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M. M., Shanker, K., et al. (2013). Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS One 8 (1), e52784. doi:10.1371/journal.pone.0052784
Nakagawa, A., Matsumura, E., Koyanagi, T., Katayama, T., Kawano, N., Yoshimatsu, K., et al. (2016). Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 7, 10390. doi:10.1038/ncomms10390
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496 (7446), 528–532. doi:10.1038/nature12051
Qin, Z., Wang, W., Liao, D., Wu, X., and Li, X. (2018). UPLC-Q/TOF-MS-Based serum metabolomics reveals hypoglycemic effects of rehmannia glutinosa, Coptis chinensis and their combination on high-fat-diet-induced diabetes in KK-ay mice. Int. J. Mol. Sci. 19 (12), 3984. doi:10.3390/ijms19123984
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi:10.1016/j.tplants.2010.02.006
Schluttenhofer, C., and Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167 (2), 295–306. doi:10.1104/pp.114.251769
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157 (4), 2081–2093. doi:10.1104/pp.111.181834
Tan, H. L., Chan, K. G., Pusparajah, P., Duangjai, A., Saokaew, S., Mehmood Khan, T., et al. (2016). Rhizoma coptidis: A potential cardiovascular protective agent. Front. Pharmacol. 7, 362. doi:10.3389/fphar.2016.00362
Tseng, C. Y., Sun, M. F., Kao, T. C., Li, T. C., and Lin, C. T. (2022). Role of Coptis chinensis in antibiotic susceptibility of carbapenem-resistant Klebsiella pneumoniae. J. Microbiol. Immunol. Infect. 55 (5), 946–955. doi:10.1016/j.jmii.2021.07.003
Tseng, C. Y., Sun, M. F., Li, T. C., and Lin, C. T. (2020). Effect of Coptis chinensis on biofilm formation and antibiotic susceptibility in Mycobacterium abscessus. Evid. Based Complement. Altern. Med. 2020, 9754357. doi:10.1155/2020/9754357
Vashisht, I., Pal, T., Sood, H., and Chauhan, R. S. (2016). Comparative transcriptome analysis in different tissues of a medicinal herb, Picrorhiza kurroa pinpoints transcription factors regulating picrosides biosynthesis. Mol. Biol. Rep. 43 (12), 1395–1409. doi:10.1007/s11033-016-4073-0
Wu, J., Luo, Y., Deng, D., Su, S., Li, S., Xiang, L., et al. (2019). Coptisine from Coptis chinensis exerts diverse beneficial properties: A concise review. J. Cell Mol. Med. 23 (12), 7946–7960. doi:10.1111/jcmm.14725
Wu, X., Liu, J., Liu, D., Yuwen, M., Koffas, M. a. G., and Zha, J. (2022). Biosynthesis of eriodictyol from tyrosine by Corynebacterium glutamicum. Microb. Cell Fact. 21 (1), 86. doi:10.1186/s12934-022-01815-3
Yamada, Y., Nishida, S., Shitan, N., and Sato, F. (2021). Genome-wide profiling of WRKY genes involved in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica). Front. Plant Sci. 12, 699326. doi:10.3389/fpls.2021.699326
Yamada, Y., and Sato, F. (2016). Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis. Sci. Rep. 6, 31988. doi:10.1038/srep31988
Yamada, Y., Shimada, T., Motomura, Y., and Sato, F. (2017). Modulation of benzylisoquinoline alkaloid biosynthesis by heterologous expression of CjWRKY1 in Eschscholzia californica cells. PLoS One 12 (10), e0186953. doi:10.1371/journal.pone.0186953
Yan, W., Feng, Q., Li, Y., Lin, Y., Yao, J., Jia, Z., et al. (2023). Integrated network pharmacology and DSS-induced colitis model to determine the anti-colitis effect of Rheum palmatum L. and Coptis chinensis Franch in granules. J. Ethnopharmacol. 300, 115675. doi:10.1016/j.jep.2022.115675
Yang, Y., Vong, C. T., Zeng, S., Gao, C., Chen, Z., Fu, C., et al. (2021). Tracking evidences of Coptis chinensis for the treatment of inflammatory bowel disease from pharmacological, pharmacokinetic to clinical studies. J. Ethnopharmacol. 268, 113573. doi:10.1016/j.jep.2020.113573
Keywords: protoberberine alkaloids, berberine biosynthesis, Coptis chinensis, WRKY family, transcription factors
Citation: Huang X, Jia A, Huang T, Wang L, Yang G and Zhao W (2023) Genomic profiling of WRKY transcription factors and functional analysis of CcWRKY7, CcWRKY29, and CcWRKY32 related to protoberberine alkaloids biosynthesis in Coptis chinensis Franch. Front. Genet. 14:1151645. doi: 10.3389/fgene.2023.1151645
Received: 26 January 2023; Accepted: 13 March 2023;
Published: 23 March 2023.
Edited by:
Ake Liu, Changzhi University, ChinaReviewed by:
Yongqing Li, South China Botanical Garden (CAS), ChinaCopyright © 2023 Huang, Jia, Huang, Wang, Yang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guohua Yang, TWlzdGFrZXlhbmcyMDIxQDE2My5jb20=; Wanli Zhao, endsMjAxNTA3MjhAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.