
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 18 April 2023
Sec. Cancer Genetics and Oncogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1150976
Background: Triple-negative breast cancer (TNBC) is the most malignant subtype of breast cancer, and studies have found an association between the Myb proto-oncogene like 2 (MYBL2) gene and TNBC development; however, the specific mechanisms underlying development remain unknown. Recent studies have reported the association of alternative splicing (AS) with cancer, providing new approaches to elucidate the carcinogenesis mechanism. This study aimed to identify MYBL2 AS-related genetic variants that influence the risk of developing TNBC, providing new ideas for probing the mechanism of TNBC and novel biomarkers for TNBC prevention.
Methods: We conducted a case-control study of 217 patients with TNBC and 401 cancer-free controls. The CancerSplicingQTL database and HSF software were used to screen for MYBL2 AS-related genetic variants. The association of sample genotypes with the risk of TNBC development and with clinicopathological features was analysed via unconditional logistic regression. Combining multiple platforms, the candidate sites were subjected to biological function analysis.
Results: Two AS-associated SNPs, rs285170 and rs405660, were identified using bioinformatics analysis. Logistic regression analysis showed that both rs285170 (OR = 0.541; 95% CI = 0.343–0.852; p = 0.008) and rs405660 (OR = 0.642; 95% CI = 0.469–0.879; p = 0.006) exhibited protective effects against TNBC under the additive model. Stratification analysis showed that these two SNPs had more significant protective effects in the Chinese population aged ≧50 years. Additionally, we found that rs405660 was associated with the risk of lymph node metastasis (OR = 0.396, 95% CI = 0.209–0.750, p = 0.005) in TNBC. Functional analysis revealed that both rs285170 and rs405660 are associated with splicing of exon 3 and that the exon 3-deleted spliceosome does not increase breast cancer risk.
Conclusion: We found for the first time that MYBL2 AS-related genetic variants are associated with reduced TNBC susceptibility in the Chinese population, especially in women aged ≧50 years.
In recent years, the incidence of breast cancer has been increasing annually; according to the American Cancer Society, the number of new cases of breast cancer worldwide was 2.26 million in 2020, which is the highest number of new cancer cases recorded to date (Sung et al., 2021). Breast cancer can be classified into four subtypes based on the expression of oestrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor-2 (HER2), including luminal A, luminal B, and HER-2-overexpressing, and triple-negative breast cancer (TNBC; Burguin et al., 2021). Among them, triple negative breast cancer accounts for 15%–20%, which also results in poor prognosis due to its special molecular phenotype and insensitivity to endocrine or molecular targeted therapy. In general, the five-year survival rate for breast cancer can reach 90%; however, it is only 12% for metastatic TNBC (Burguin et al., 2021). Although treatment options for TNBC, such as specific receptor targeted therapies, immunotherapies, and antibody drug conjugate therapies, have been intensely investigated by researchers worldwide in recent years, some of which have achieved staged results, they are still far from being applied in clinical use (Yin et al., 2020; Bianchini et al., 2022). Therefore, the most effective treatment method involves prevention of TNBC development. Breast cancer prevention is now possible owing to the administration of preventive drugs and genetic screening; however, studies have shown that breast cancer preventive drugs recommended by international guidelines do not favourably reduce breast cancer mortality and only offer a certain preventive benefit in ER-positive breast cancer (Hong and Xu, 2022). Thus, in addition to continuing vigorous efforts to develop effective preventive drugs, studies on genetic predisposition to breast cancer are necessary. Breast cancer is typically a heterogeneous disease, and genetic variants play a large role in individual susceptibility. Thus, identification of genetic variants that influence breast cancer susceptibility may help reduce the risk of breast cancer.
Genetic susceptibility to breast cancer is increasingly investigated by post-genome-wide association studies, so that current research focuses on identifying functional genetic variants that affect breast tissue carcinogenesis and elucidating the underlying mechanisms via molecular biology experiments. Alternative splicing (AS) is a key molecular event during gene expression, and it involves generation of RNA exons from pre-mRNA transcription binding via selection of different splice sites, resulting in different mRNA splicing isoforms. It is an intricate mechanism of coordinated transcriptional regulation that enables a single gene to generate different mRNAs, thereby greatly increasing the diversity of the transcriptome and proteome. This phenomenon is present in almost all genes encoding proteins. AS occurs widely in human tissues; however, abnormal splicing events may lead to the development of various neurological diseases and cancers (Ren et al., 2021). At least 15,000 splice variants affect cancer development across 27 cancer types (Kahles et al., 2018). For example, the BRCA1--Δ11q variant affects breast carcinogenesis and drug resistance. Compared to the full-length BRCA1 spliceosome, BRCA1--Δ11q clearly promotes cancer cell growth as well as resistance to PARP inhibitors and cisplatin (Nielsen et al., 2016; Wang et al., 2016). In addition, there is evidence for the existence of specific spliceosomes in BC tissues that are not found in normal tissues (Dutertre et al., 2010). Aversa et al. (2016) also found 12 novel AS transcripts in BC tissues based on RNA-seq data. These abnormal ASs were closely related to the occurrence of breast cancer (Yang et al., 2019). Further studies on AS have high potential to disentangle the biological mechanisms of breast cancer.
The MYB proto-oncogene like 2 (MYBL2) gene, a member of the MYB family of transcription factors and one of the key genes regulating cell cycle progression, is expressed in almost all proliferating cells and regulates cell survival and differentiation, while also mediating aberrant cell progression in cancer (Ness, 2003). Reddel, (2000) discovered that cell immortalisation can be achieved by dysregulated proliferation and that MYBL2 amplification is the earliest observed event during this process. In addition, bioinformatics data mining revealed that dysregulation of genes, as key drivers in breast cancer, controls the cell cycle, in which MYBL2 plays an important role (Lauss et al., 2008). Distinct amplification of MYBL2 is found in approximately 9%–13% of patients with breast cancer, a feature that is particularly evident in TNBC (Thorner et al., 2009). Multiple studies have found that single nucleotide polymorphisms (SNPs) create functional alterations in MYBL2, which in turn affect disease initiation and progression. For example, the rs285207 polymorphism disrupts binding of MYBL2 to transcription factor IKZF1, affects transcription of MYBL2, and in turn reduces susceptibility to childhood acute lymphoblastic leukaemia (Guo et al., 2021). MYBL2 is phosphorylated by cyclin A/Cdk2 in the S phase, which activates the protein. When MYBL2 is expressed and activated in the late G1 and S phases of the cell cycle, it directly binds to the promoter and expresses trans-activated genes in the G2/M phases (Musa et al., 2017). Several recognised transcription factor binding sites are located in the MYBL2 promoter region, and the rs619289, rs826943, and rs826944 polymorphisms located in the promoter region affect MYBL2 expression and confer susceptibility to breast cancer (Shi et al., 2011). Analysis of microarray data also found that the MYBL2 rs2070235 polymorphism increases the risk of basal like breast cancer, but the specific mechanism remains unknown (Thorner et al., 2009). Thus, the aberrant expression of MYBL2 caused by SNPs likely affects the occurrence and development of breast cancer.
Although multiple SNPs associated with breast carcinogenesis have been found, the association of MYBL2 AS-related SNPs with breast cancer susceptibility has not been explored to date. SNPs have been reported to represent a major cause of AS, and AS of certain genes has been found to influence breast cancer susceptibility (Montgomery et al., 2010). Schmid et al. (2002) found that SNPs cause four different splice variants in the mucin 1 (MUC1) gene, and in-vitro assays showed that variants B and C are associated with invasiveness in breast cancer cell lines. A causal relationship between breast cancer risk-associated splicing events and SNPs has been confirmed in minigene experiments (Caswell et al., 2015). Based on these previous results, SNPs likely affect breast cancer susceptibility by affecting gene AS, leading to differential expression of different spliceosomes among individuals. The search for functional SNPs that mediate AS of MYBL2 may provide a basis to explain individual differences in breast cancer susceptibility.
In this study, we systematically summarised all potential AS-causing SNPs of MYBL2 via the CancerSplicingQTL database (Tian et al., 2019) and Human Splicing Finder (HSF) online software (Desmet et al., 2009). Moreover, we used a case-control study design to discover functional SNPs of this gene associated with TNBC risk. Our study provides new research ideas and methods for systematically discovering functional genetic variations. Our results provide insights into the mechanisms involved in the development and progression of TNBC and shed new light on screening of and individualised prevention for individuals at high risk for TNBC.
All potential splicing-causing SNPs of MYBL2 associated with breast cancer risk were searched for and downloaded from CancerSplicingQTL. And the coding DNA positions of these variants were obtained in the VannoPortal database. Moreover, linkage disequilibrium (LD) analysis between SNPs was performed using the Haploview 4.2 software. The potential AS function of these SNPs was further predicted in the HSF online software, and the minor allele frequency (MAF) of these SNPs in the southern Chinese population was detected using the 1000 Genomes Project. SNPs with an MAF >0.1 were screened, and redundant SNPs were removed according to HSF score and LD analysis.
This study recruited 217 women with TNBC and 401 cancer-free women (controls) at the Affiliated Hospital of Zunyi Medical University from 2018 to 2021. All cases were histopathologically and immunohistochemically confirmed for TNBC; the immunohistochemical results met ER (-), PR (-), and HER-2 (-), and the patients had no previous history of other malignancies. The control group comprised cancer-free women attending the physical examination department of this hospital during the same period according to the table of random digit, and they were matched with the case group based on age (±5 years). Briefly, 2 ml of peripheral blood was drawn for subsequent testing. General information and pathological data of the study participants were obtained by querying medical records and conducting interviews. Data entry was completed using the EpiData 3.1 software. All data were entered in duplicate, and then, inter-input comparisons were performed to ensure the accuracy of data entry. Participants who had never smoked or smoked less than one cigarette per day were defined as “never smoked” and all other cases were defined as “smoked.” Alcohol consumption twice or more per week for at least one year was defined as “drinking” and other cases were considered as “no drinking.” The ethics committee of the Affiliated Hospital of Zunyi Medical University approved the study (No. KLLY-2021-200). All participants provided written informed consent.
The collected peripheral blood samples were used to extract genomic DNA using the BloodZol Kit (TransGen Biotech, Beijing, China). The concentration of the DNA solution was diluted to 50 ng/µL and stored at −20°C. Sample DNA was genotyped using the CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States) and TaqMan probes (SANGON, Shanghai, China). Duplicate experiments were performed using 5% of samples randomly selected from cases and controls; the results of the two experiments were compared for consistency.
We used the CancerSplicingQTL database to analyse candidate sites, the exon in which they were predicted to act, the splicing pattern, and the splice site, to determine what spliceosome the candidate site might affect. The expression differences in MYBL2 splice isoforms among different tissues were analysed using the TSVdb platform (http://tsvdb.com/index.html) to determine whether the splice isoforms are associated with breast cancer risk.
Calculation of power was performed using the following formula:
p0: Exposure rates of risk factor in controls; p1: Exposure rates of risk factors in the cases; r: Ratio of control sample size to case sample size
It was calculated that 200 cases and 400 controls would provide 91% test power when OR = 2, α = 0.05, and MAF = 0.1. Chi-square tests and two independent-sample t-tests were used to compare whether demographic statistical characteristics were significantly different between cases and controls. The chi-square goodness-of-fit test was used to test whether the genotype distribution in controls was consistent with the Hardy–Weinberg equilibrium, and a logistic regression model was used to calculate the odds ratio (OR) and 95% confidence interval (CI). Values of p < 0.05 were considered statistically different. All data were analysed using the SPSS 26.0 software (IBM, Armonk, NY, United States).
Through bioinformatics analysis (Figure 1), we obtained a total of two SNPs (rs285170 and rs405660) potentially associated with MYBL2 AS in BC, and the specific screening pipeline was as described below.
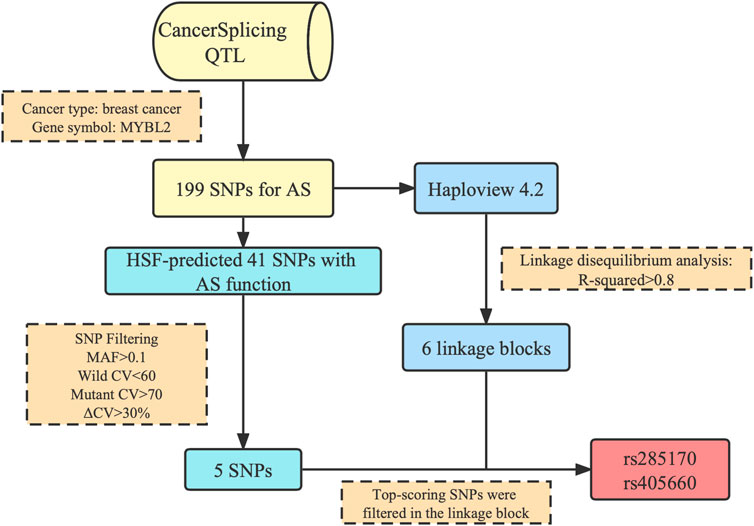
FIGURE 1. Flowchart of MYBL2 alternative splicing-related SNP screening. 199 MYBL2 AS-related genetic variants were acquired at CancerSplicingQTL. Five SNPs with potential functions in alternative splicing were further screened out in HSF. Six linkage blocks were simultaneously obtained by Haploview. Two SNPs, rs285170 and rs405660, were finally included in the follow-up study.
Analysing the association of splicing events with SNPs occurring in tumour tissues of TCGA breast cancer patients, we first selected SNPs within the breast cancer category among the 253767 SNPs potentially affecting AS in breast cancer. We then delimited the MYBL2 gene, which finally resulted in 199 breast cancer associated SNPs potentially affecting AS in MYBL2 in CancerSplicingQTL (Supplementary Table S1).
LD analysis was performed using Haploview to guarantee that the last SNP included in the study was genetically independent. After downloading the genotyping data of all SNPs on MYBL2 in Southern China from the Ensembl database, the data of the 199 SNPs from the first step were filtered out, collated into ped and info files (file format required for Haploview LD analysis), and input into Haploview. The r2 values among loci (Supplementary Table S2) and six linkage blocks (Figure 2) were obtained.
To further verify whether the above 199 SNPs have AS function, we employed HSF for AS function prediction. We used the Vannoportal database to query transcript IDs and HGV numbers corresponding to the 199 SNPs obtained in the first step and then predicted the data using HSF, which indicated that a total of 41 SNPs were likely to affect MYBL2 AS (Supplementary Table S3). To compare the strength of splicing activity at each locus, only SNP loci with HSF scores were included in this study. Of the 41 potentially AS functional sites, ten SNPs were predicted by HSF to affect splice donor or splice acceptor generation. Based on the scores given by the HSF predictions, we further filtered SNPs with wild-type CV values <60, mutant CV values >70, ΔCV >30%, and AS MAF >0.1, finally yielding a total of five SNPs. Combined with the results of the LD analysis, only two of the six linkage blocks contained SNPs that satisfied the filtering criteria, and two of the best scoring SNPs, rs285170 and rs405660, were finally screened out in each of these two linkage blocks, respectively (Table 1).
A total of 217 TNBC cases and 401 cancer-free controls were included in our case-control study, with a mean age of 50.41 years (±10.93 years) in the case group and 50.28 years (±11.57 years) in the control group. There was no significant difference in age (p = 0.889) between cases and controls. After stratification based on age, there was still no significant difference in age between cases and controls. There were also no significant differences in smoking and alcohol consumption between cases and controls. The TNM staging characteristics are described in Table 2.
Typing experiments were performed in which 5% of the samples were randomly selected for repeated experiments with an anastomosis rate of 100%. The distribution of rs285170 typing results of cases and controls is shown in Table 3. The genotype distributions of both SNPs in the control group were in accordance with HWE.
After logistic regression model analysis, the rs285170\C allele was found to be significantly associated with a decreased risk of TNBC. In addition, the dominant model (OR = 0.520; 95% CI = 0.323–0.839; p = 0.007) and additive model (OR = 0.541; 95% CI = 0.343–0.852; p = 0.008) both revealed that the C allele was a protective factor for TNBC; rs285170 (T > C) was a protective mutation for TNBC. Furthermore, we found that the rs405660GG genotype was significantly associated with a decreased risk of TNBC (OR = 0.142; 95% CI = 0.033–0.609; p = 0.009). The G allele exhibited protective effects against TNBC in the population under the dominant model (OR = 0.682; 95% CI = 0.476–0.976; p = 0.036) and additive model (OR = 0.642; 95% CI = 0.469–0.879; p = 0.006) (Table 3).
An age-stratified analysis was performed based on whether participants were aged ≧50 years. Notably, rs285170 was not significantly associated with TNBC susceptibility in participants aged <50 years; however, it showed a greater and significant difference in participants aged ≧50 years as compared to the whole population. In participants aged ≧50 years, the TC genotype was significantly associated with a decreased risk of TNBC (OR = 0.216; 95% CI = 0.088–0.529; p = 0.001), while the dominant (OR = 0.205; 95% CI = 0.084–0.503; p = 0.001) and additive (OR = 0.211; 95% CI = 0.087–0.510; p = 0.001) models similarly showed significant differences.
Similar to rs285170, we performed an age-based stratification analysis for rs405660. The G allele was not clearly associated with risk of TNBC in participants aged <50 years. However, in participants aged ≧50 years, the G allele showed a significant protective effect. The OR was significant under both dominant (OR = 0.485; 95% CI = 0.284–0.827; p = 0.008) and additive (OR = 0.461; 95% CI = 0.285–0.746; p = 0.002) models (Table 4).
We analysed the relationship of rs285170 and rs405660 with clinicopathological features of TNBC. No significant association between rs285170 and rs405660 was found with both tumour size (T) in TNBC (Table 5); however, rs405660 was associated with lymph node metastasis (N) under both dominant (OR = 0.396, 95% CI = 0.209–0.750, p = 0.005) and additive (OR = 0.390, 95% CI = 0.210-0.725, p = 0.003) models (Table 6). However, rs285170 was not associated with lymph node metastasis.
Functional prediction of our candidate SNPs in CancerSplicingQTL indicated that both rs285170 and rs405660 were associated with MYBL2 AS. The splice site was chr20:42310424, and both affected MYBL2 exon 3 skipping; that is, generation of a spliceosome with exon 3 skipping (Table 7). The expression landscape of MYBL2 splice isoforms in breast cancer was next analysed on the TSVdb platform. In this database, we performed an analysis of the exonic expression landscape of MYBL2 in breast cancer, with the final output shown in Figure 3. The figure illustrates the two splice isoforms of MYBL2 and the 14 exons on MYBL2 in normal, orthotopic, and metastatic breast cancer tissue, and the difference between the two splice isoforms is shown in Figure 3 as to whether exon 3 skipping occurs during the splicing process. The expression differences of the two splice isoforms between tissues are shown in Figures 4, 5. The expression of whole exon spliceosomes without AS was significantly higher in breast cancer than in normal tissues (Figure 4), while the expression of exon 3-skipped spliceosomes was not significantly different between breast cancer and normal tissues (Figure 5). This result suggests that the exon 3-skipped spliceosome does not increase breast cancer susceptibility and that this spliceosome reduces breast cancer susceptibility compared to whole exon spliceosome carriers.
TNBC is a highly proliferative subtype of breast cancer that is often associated with poor prognosis. The treatment strategy of breast cancer varies based on molecular typing; however, as TNBC lacks expression of ER, PR, and HER-2, it has low hormone sensitivity, and the prognostic markers and therapeutic targets remain unknown. Compared with other invasive breast cancers, TNBC has an earlier onset and a higher recurrence rate, and it is also more likely to spread and metastasise (Berger et al., 2021). TNBC is often difficult to treat owing to the lack of effective therapeutic agents, and to date, no targeted therapy has been approved by the US Food and Drug Administration (Zheng et al., 2020). Therefore, studies on TNBC susceptibility are particularly important. Only a few individuals with similar exposure to environmental risk factors for breast cancer eventually develop breast cancer, indicating that genetic factors play a key role. The genetic predisposition to breast cancer represents an important research focus in current studies. Classic BRCA1/2 germline mutations are associated with approximately 15%–20% of TNBC cases; nevertheless, the susceptibility genes and mutation sites have only been specified in few patients with TNBC (Pohl-Rescigno et al., 2020).
MYBL2 is an important regulatory gene in cell cycle progression and it affects cell proliferation and differentiation. Several studies have found an association of MYBL2 with malignant tumour development, such as breast, bladder, lung adenocarcinoma, and ovarian cancer (Liu Q et al., 2022; Inoue and Fry, 2016; Lee et al., 2022; Liu W et al., 2022). The MYBL2 S427G variant has been reported to be associated with an increased risk of basal-like breast cancer in the African American population; however, the specific mechanism has not been clarified (Thorner et al., 2009). Chen and Chen (2018) conducted a preliminary study on the mechanisms of MYBL2 in breast cancer development and found that miR-143-3p targets the MYBL2 3′-untranslated region, which inhibits MYBL2 expression and promotes proliferation and inhibits apoptosis of breast cancer cells. A MYBL2 splice variant lacking exon 9A has been found to function as a suppressor protein and counteract transactivation mediated by the whole exon-expressed MYBL2 (Horstmann et al., 2000). Although the study did not explore the effect of AS on cancer risk, several studies have shown that AS leads to an increase in cancer risk (Chan et al., 2022). Previous studies have explained part of the mechanism of MYBL2 in breast cancer risk; however, at present, no studies have systematically analysed the relationship between AS of MYBL2 and breast cancer risk (Bayley et al., 2020). Therefore, we performed bioinformatics analysis and found that two functional SNPs, rs285170 and rs405660, are likely to cause AS in MYBL2. These two SNPs are likely to influence the risk of TNBC by affecting AS of MYBL2. To our knowledge, this is the first study conducted worldwide on these two SNPs and the first systematic analysis of the association of MYBL2 AS-related genetic variants with AS risk.
We found that the C allele of rs285170 was associated with decreased susceptibility to TNBC in 217 of our cases, and the same results were confirmed in dominant and additive models. However, no significant difference was found in participants aged <50 years, and when this group of the cohort was excluded, the difference was more pronounced in the group aged ≧50 years. The G allele of rs405660 also exhibited a similar protective effect. A certain characteristic of the population aged ≧50 years is expected to play a role in this protective effect. Combined with the age characteristic analysis, we speculate that a joint effect relationship exists between menopausal status and mutations at this locus. After menopause, women experience certain changes in levels of hormones such as oestrogen and progesterone, and menopausal status is often found to be a risk factor for breast cancer (Howlader et al., 2014). ERβ expression is positively correlated with MYBL2 expression in endometrial cancer tissues, which supports a relationship between the increased risk of endometrial cancer and MYBL2 expression levels in premenopausal women (Luengo-Gil et al., 2019). Current studies have mostly focused on the relationship between hormone receptors and MYBL2 expression; however, ER, PR, and HER-2 are not expressed in TNBC cases. Therefore, to explain whether MYBL2 expression in TNBC is associated with hormone level changes, studies should confirm the existence of associations between hormone level changes and MYBL2 expression. In addition, unlike rs285170, where the homozygous rs405660 variant showed significant protection in the full-age analysis, the association of the homozygous rs285170 variant with TNBC risk was not significant. This may be explained by the low MAF of rs285170 with few homozygous mutant genotypes in our sample. Follow-up studies using larger sample sizes may obtain significant results. In addition, we found that rs405660 was significantly associated with a decreased risk of lymph node metastasis in TNBC, and this finding may provide a reference for subsequent clinicopathological studies on MYBL2 in association with TNBC.
On the basis of population association analyses, we performed biological functional analyses based on TCGA data. The results showed that both rs285170 and rs405660 were associated with MYBL2 exon 3 skipping. Whole exon spliceosomes significantly increase breast cancer susceptibility, but the exon 3-deleted spliceosome was not significantly different between breast cancer tissues and adjacent non-cancerous tissues. It reduced susceptibility to breast cancer compared with the whole exon spliceosome. This is consistent with the population association analysis results that MYBL2 AS-related genetic variants can reduce the risk of breast cancer.
In this study, we proposed a novel bioinformatic analysis strategy to search for genetic variants associated with AS, systematically analysed all SNPs potentially causing AS in MYBL2, and determined linkage loci in the SNPs most likely to exhibit AS function. Our findings may provide a reference for AS-related research. Our study had a few limitations. First, our cases were all recruited as inpatients in hospitals, and there may be a certain Berkson’s bias. Secondly, since the AS-related SNPs, splicing exons, splicing sites, and splicing patterns obtained in this study were all predicted by bioinformatics, we recommend that subsequent studies supplement biological functional verification, such as analysis of SNP association with MYBL2 splice isoform expression levels and minigene construct or subcutaneous tumour bearing experiments in nude mice.
To the best of our knowledge, this is the first study to systematically investigate the role of MYBL2 AS-related genetic variants in TNBC susceptibility in the Chinese population using bioinformatics analysis. We found an association between rs285170 and rs405660 and the development of TNBC. The SNPs rs285170 and rs405660 may produce different splicing isoforms by altering the transcription of MYBL2, which in turn affects the risk of TNBC. In addition, we found that rs405660 was associated with the risk of lymph node metastasis in TNBC. Our findings elucidate the mechanisms underlying TNBC development and cancer in general.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Ethics Committee of the Affiliated Hospital of Zunyi Medical University. The patients/participants provided their written informed consent to participate in this study.
XC designed the study and completed the writing of the article, JF was responsible for sample collection, YZ critically revised the article, XW confirmed the final manuscript, and the remaining authors participated in the experiments and data collection. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
This study was supported by grants from the National Natural Science Foundation of China (No. 82060620) and the Research Foundation Project for graduate students of Zunyi Medical University in China (No. ZYK88). Guizhou Science and Technology Program Project (No. Qiankehe Foundation - ZK[2023] General 502), the National Natural Science Foundation of China (Grant No. 31960209), the “Thousand” Level Project of Training High-Level Innovative Talents in Guizhou Province (Grant No. fzc20200612), Guizhou Science and Technology Fund Project (Grant No. (2020)1Y093), Zunyi Science and Technology Fund Project (Grant No. Zun Shi Ke He HZ Zi 2021-40), Future Eminent Clinician Plan of Zunyi Medical University (Grant No. 2022-02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1150976/full#supplementary-material
Aversa, R., Sorrentino, A., Esposito, R., Ambrosio, M. R., Amato, A., Zambelli, A., et al. (2016). Alternative splicing in adhesion- and motility-related genes in breast cancer. Int. J. Mol. Sci. 17 (1), 121. doi:10.3390/ijms17010121
Bayley, R., Ward, C., and Garcia, P. (2020). MYBL2 amplification in breast cancer: Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 1874 (2), 188407. doi:10.1016/j.bbcan.2020.188407
Berger, E. R., Park, T., Saridakis, A., Golshan, M., Greenup, R. A., and Ahuja, N. (2021). Immunotherapy treatment for triple negative breast cancer. Pharm. (Basel) 14 (8), 763. doi:10.3390/ph14080763
Bianchini, G., De Angelis, C., Licata, L., and Gianni, L. (2022). Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat. Rev. Clin. Oncol. 19 (2), 91–113. doi:10.1038/s41571-021-00565-2
Burguin, A., Diorio, C., and Durocher, F. (2021). Breast cancer treatments: Updates and new challenges. J. Pers. Med. 11 (8), 808. doi:10.3390/jpm11080808
Caswell, J. L., Camarda, R., Zhou, A. Y., Huntsman, S., Hu, D., Brenner, S. E., et al. (2015). Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum. Mol. Genet. 24 (25), 7421–7431. doi:10.1093/hmg/ddv432
Chan, J. J., Tabatabaeian, H., and Tay, Y. (2022). 3'UTR heterogeneity and cancer progression. Trends Cell Biol. S0962-8924 (22), 00232–X. doi:10.1016/j.tcb.2022.10.001
Chen, J., and Chen, X. (2018). MYBL2 is targeted by miR-143-3p and regulates breast cancer cell proliferation and apoptosis. Oncol. Res. 26 (6), 913–922. doi:10.3727/096504017X15135941182107
Desmet, F. O., Hamroun, D., Lalande, M., Collod-Béroud, G., Claustres, M., and Béroud, C. (2009). Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37 (9), e67. doi:10.1093/nar/gkp215
Dutertre, M., Vagner, S., and Auboeuf, D. (2010). Alternative splicing and breast cancer. RNA Biol. 7 (4), 403–411. doi:10.4161/rna.7.4.12152
Guo, H., Li, N., Sun, Y., Wu, C., Deng, H., Xu, L., et al. (2021). MYBL2 gene polymorphism is associated with acute lymphoblastic leukemia susceptibility in children. Front. Oncol. 11, 734588. doi:10.3389/fonc.2021.734588
Hong, R., and Xu, B. (2022). Breast cancer: An up-to-date review and future perspectives. Cancer Commun. (Lond). 42 (10), 913–936. doi:10.1002/cac2.12358
Horstmann, S., Ferrari, S., and Klempnauer, K. H. (2000). An alternatively spliced isoform of B-Myb is a transcriptional inhibitor. Oncogene 19 (48), 5428–5434. doi:10.1038/sj.onc.1203937
Howlader, N., Altekruse, S. F., Li, C. I., Chen, V. W., Clarke, C. A., Ries, L. A., et al. (2014). US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 106 (5), dju055. doi:10.1093/jnci/dju055
Inoue, K., and Fry, E. A. (2016). Novel molecular markers for breast cancer. Biomark. Cancer 8, 25–42. doi:10.4137/BIC.S38394
Kahles, A., Lehmann, K. V., Toussaint, N. C., Hüser, M., Stark, S. G., Sachsenberg, T., et al. (2018). Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34 (2), 211–224.e6. doi:10.1016/j.ccell.2018.07.001
Lauss, M., Kriegner, A., Vierlinger, K., Visne, I., Yildiz, A., Dilaveroglu, E., et al. (2008). Consensus genes of the literature to predict breast cancer recurrence. Breast Cancer Res. Treat. 110 (2), 235–244. doi:10.1007/s10549-007-9716-3
Lee, Y., Wu, Z., Yang, S., Schreiner, S. M., Gonzalez-Smith, L. D., and Rhie, S. K. (2022). Characterizing and targeting genes regulated by transcription factor MYBL2 in lung adenocarcinoma cells. Cancers (Basel) 14 (20), 4979. doi:10.3390/cancers14204979
Liu, Q., Liu, H., Huang, X., Fan, X., Xiao, Z., Yan, R., et al. (2022). A targetable MYBL2-ATAD2 axis governs cell proliferation in ovarian cancer. Cancer Gene Ther. 30 (1), 192–208. doi:10.1038/s41417-022-00538-2
Liu, W., Shen, D., Ju, L., Zhang, R., Du, W., Jin, W., et al. (2022). MYBL2 promotes proliferation and metastasis of bladder cancer through transactivation of CDCA3. Oncogene 41 (41), 4606–4617. doi:10.1038/s41388-022-02456-x
Luengo-Gil, G., García-Martínez, E., Chaves-Benito, A., Conesa-Zamora, P., Navarro-Manzano, E., González-Billalabeitia, E., et al. (2019). Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cell Oncol. (Dordr) 42 (5), 627–644. doi:10.1007/s13402-019-00450-2
Montgomery, S. B., Sammeth, M., Gutierrez-Arcelus, M., Lach, R. P., Ingle, C., Nisbett, J., et al. (2010). Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464 (7289), 773–777. doi:10.1038/nature08903
Musa, J., Aynaud, M. M., Mirabeau, O., Delattre, O., and Grünewald, T. G. (2017). MYBL2 (B-myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 8 (6), e2895. doi:10.1038/cddis.2017.244
Ness, S. A. (2003). Myb protein specificity: Evidence of a context-specific transcription factor code. Blood Cells Mol. Dis. 31 (2), 192–200. doi:10.1016/s1079-9796(03)00151-7
Nielsen, F. C., van Overeem Hansen, T., and Sørensen, C. S. (2016). Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer 16 (9), 599–612. doi:10.1038/nrc.2016.72
Pohl-Rescigno, E., Hauke, J., Loibl, S., Möbus, V., Denkert, C., Fasching, P. A., et al. (2020). Association of germline variant status with therapy response in high-risk early-stage breast cancer: A secondary analysis of the GeparOcto randomized clinical trial. JAMA Oncol. 6 (5), 744–748. doi:10.1001/jamaoncol.2020.0007
Reddel, R. R. (2000). The role of senescence and immortalization in carcinogenesis. Carcinogenesis 21 (3), 477–484. doi:10.1093/carcin/21.3.477
Ren, P., Lu, L., Cai, S., Chen, J., Lin, W., and Han, F. (2021). Alternative splicing: A new cause and potential therapeutic target in autoimmune disease. Front. Immunol. 12, 713540. doi:10.3389/fimmu.2021.713540
Schmid, B. C., Buluwela, L., Liu, Q., Fasching, B., Tong, D., Stimpfl, M., et al. (2002). Expression of MUCI splice variants correlates with invasive growth of breast cancer cell lines. Breast Cancer Res. Treat. 76 (3), 211–219. doi:10.1023/a:1020853900765
Shi, H., Bevier, M., Johansson, R., Grzybowska, E., Chen, B., Eyfjörd, J. E., et al. (2011). Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast Cancer Res. Treat. 130 (3), 905–916. doi:10.1007/s10549-011-1600-5
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Thorner, A. R., Hoadley, K. A., Parker, J. S., Winkel, S., Millikan, R. C., and Perou, C. M. (2009). In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 28 (5), 742–751. doi:10.1038/onc.2008.430
Tian, J., Wang, Z., Mei, S., Yang, N., Yang, Y., Ke, J., et al. (2019). CancerSplicingQTL: A database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res. 47 (D1), D909–d916. doi:10.1093/nar/gky954
Wang, Y., Bernhardy, A. J., Cruz, C., Krais, J. J., Nacson, J., Nicolas, E., et al. (2016). The BRCA1-d11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 76 (9), 2778–2790. doi:10.1158/0008-5472.CAN-16-0186
Yang, Q., Zhao, J., Zhang, W., Chen, D., and Wang, Y. (2019). Aberrant alternative splicing in breast cancer. J. Mol. Cell Biol. 11 (10), 920–929. doi:10.1093/jmcb/mjz033
Yin, L., Duan, J. J., Bian, X. W., and Yu, S. C. (2020). Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22 (1), 61. doi:10.1186/s13058-020-01296-5
Keywords: MYB proto-oncogene like 2, triple-negative breast cancer, alternative splicing, SNP, susceptibility
Citation: Chen X, Feng J, Zhang Y, Liu J, Zhang L, Zeng P, Wen L, Wang X and Zhang Y (2023) MYBL2 alternative splicing-related genetic variants reduce the risk of triple-negative breast cancer in the Chinese population. Front. Genet. 14:1150976. doi: 10.3389/fgene.2023.1150976
Received: 25 January 2023; Accepted: 06 April 2023;
Published: 18 April 2023.
Edited by:
Dharmendra Kumar Yadav, Gachon University, Republic of KoreaCopyright © 2023 Chen, Feng, Zhang, Liu, Zhang, Zeng, Wen, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wang, bGNod3hAYWxpeXVuLmNvbQ==; Yi Zhang, emhhbmd5aXptckAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.