
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 18 April 2023
Sec. Evolutionary and Population Genetics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1137618
This article is part of the Research Topic Genetic Basis of Adaptation to Extreme Environments Ⅱ View all 6 articles
 Jia-Rui Zhao1,2,3,4
Jia-Rui Zhao1,2,3,4 Shi-Yun Hu1,2,3,4
Shi-Yun Hu1,2,3,4 Li-Jun Zhang1,2,3,4
Li-Jun Zhang1,2,3,4 Li Zhang1,2,3,4
Li Zhang1,2,3,4 Xing-Zhuo Yang1,2,3,4
Xing-Zhuo Yang1,2,3,4 Ming-Long Yuan1,2,3,4*
Ming-Long Yuan1,2,3,4*Grassland caterpillars (Lepidoptera: Erebidae: Gynaephora) are important pests in alpine meadows of the Qinghai-Tibetan Plateau (QTP). These pests have morphological, behavioral, and genetic adaptations for survival in high-altitude environments. However, mechanisms underlying high-altitude adaptation in QTP Gynaephora species remain largely unknown. Here, we performed a comparative analysis of the head and thorax transcriptomes of G. aureata to explore the genetic basis of high-altitude adaptation. We detected 8,736 significantly differentially expressed genes (sDEGs) between the head and thorax, including genes related to carbohydrate metabolism, lipid metabolism, epidermal proteins, and detoxification. These sDEGs were significantly enriched in 312 Gene Ontology terms and 16 KEGG pathways. We identified 73 pigment-associated genes, including 8 rhodopsin-associated genes, 19 ommochrome-associated genes, 1 pteridine-associated gene, 37 melanin-associated genes, and 12 heme-associated genes. These pigment-associated genes were related to the formation of the red head and black thorax of G. aureata. A key gene, yellow-h, in the melanin pathway was significantly upregulated in the thorax, suggesting that it is related to the formation of the black body and contributed to the adaptation of G. aureata to low temperatures and high ultraviolet radiation in the QTP. Another key gene, cardinal, in the ommochrome pathway was significantly upregulated in the head and may be related to red warning color formation. We also identified 107 olfactory-related genes in G. aureata, including genes encoding 29 odorant-binding proteins, 16 chemosensory proteins, 22 odorant receptor proteins, 14 ionotropic receptors, 12 gustatory receptors, 12 odorant degrading enzymes, and 2 sensory neuron membrane proteins. Diversification of olfactory-related genes may be associated with the feeding habits of G. aureata, including larvae dispersal and searching for plant resources available in the QTP. These results provide new insights into high-altitude adaptation of Gynaephora in the QTP and may contribute to the development of new control strategies for these pests.
The genus Gynaephora (Lepidoptera: Erebidae: Lymantriinae) includes 15 species worldwide, 8 of which are endemic to the Qinghai-Tibetan Plateau (QTP) (Yan, 2006; Zhang and Yuan, 2013). Gynaephora species and the most destructive insect pests in the alpine meadows of the QTP and have caused serious feed shortages and grassland degradation (Yan, 2006; Zhang and Yuan, 2013). The QTP environment is characterized by hypoxia, low temperatures, and high ultraviolet (UV) radiation (Yu et al., 2016). The spread of QTP Gynaephora species in the harsh environment of the QTP can be explained by various morphological and behavioral adaptations (Yan, 2006; Zhang and Yuan, 2013; Barrio et al., 2014). In terms of morphology, black body hair can resist high UV radiation and a red head can protect against natural enemies in larvae of QTP Gynaephora species (Yan, 2006; Zhang and Yuan, 2013). The feeding habits of Gynaephora groenlandica and Gynaephora rossii in the Arctic differ from those of QTP Gynaephora species (Barrio et al., 2014; Cao, 2022). Our recent research on gut bacteria, transcriptome profiles, and mitochondrial genomes has contributed to our understanding of the high-altitude adaptation of QTP Gynaephora species (Zhang L. et al., 2017; Zhang Q. L. et al., 2017; Yuan et al., 2018; Cao et al., 2021). However, the molecular mechanisms underlying high-altitude adaptation in QTP Gynaephora species are still largely unknown.
QTP Gynaephora species are also known as red-headed black caterpillars. Different from other grassland caterpillars, the heads of QTP Gynaephora species are bright red, and the thorax and abdomen are covered with black body hair; these morphological features are associated with high-altitude adaptation. The skin and hair color of humans (such as Tibetans on the QTP), mammals (e.g., Psammodromus algirus and Sus scrofa domesticus), amphibians, and reptiles in high-altitude areas are significantly darker than the skin of those in low-altitude areas (Wang et al., 2000; Brenner and Hearing, 2008; Jablonski and Chaplin, 2010; Li et al., 2013; Reguera et al., 2014; Visscher, 2017; Gao et al., 2019; Zhang et al., 2021). Organisms at high-altitude areas show a deeper body color to resist high UV radiation, which reduces the damage to cells and DNA structure caused by the excessive production of reactive oxygen species (Rinnerthaler et al., 2015). In addition to protecting against UV radiation damage, a deeper body color also plays an important role in regulating the body temperature (DeJong et al., 1996; Carothers et al., 1998; Geen and Johnston, 2014; Gao et al., 2019). Black areas in the abdomens of Phrynocephalus in the QTP are significantly larger than those of individuals in low-altitude areas, and these areas are conducive to raising the body temperature (Jin and Liao, 2015; Gao et al., 2019). Dense black body hair on the thorax and abdomen of the larvae of QTP Gynaephora species may be an adaptation to the QTP environment (high UV levels and low temperatures). In many insects, bright colors, as warning colors, appear together with black, which has important ecological functions (Sun M. X. et al., 2020). The red heads of QTP Gynaephora species may be an adaptation to resist natural enemies. The formation of insect body color is closely related to pigment-related genes in the rhodopsin, ommochrome, pteridine, melanin, and heme biosynthesis pathways (Sugumaran et al., 1990; Ferreira, 1995; Ziegler et al., 2000; Tripoli et al., 2005; Di Pietro et al., 2006; Harris et al., 2011; Croucher et al., 2013; Sun M. X. et al., 2020). However, pigment pathway-associated genes and the molecular mechanism underlying the formation of body color in QTP Gynaephora species are still unknown.
QTP Gynaephora species are typical polyphagous pests. G. aureata, Gynaephora menyuanensis, and Gynaephora alpherakii can feed on 77 species of plants in 18 families, mainly herbaceous plants, such as Cyperaceae and Gramineae (Cao, 2022). Compared with G. groenlandica and G. rossii, which mainly feed on Arctic willow (Salix arctica) and avoid eat grasses and sedges (MacLean and Jensen, 1985; Barrio et al., 2014), the feeding habits of QTP Gynaephora species differ substantially. The change in feeding habits of QTP Gynaephora species is an adaptation to the abundant plant resources available in the QTP. Host finding relies on the olfactory system in insects to recognize environmental signals, such as volatiles from plants (Gols et al., 2012; Suh et al., 2014; Ballesteros et al., 2019). Insects can adapt to changes in host plants by adjusting their olfactory system (Linz et al., 2013; Claudianos et al., 2014; Ballesteros et al., 2019). Therefore, the change in the feeding habits of QTP Gynaephora is an olfactory system adaptation to the host plants. Insects have different types of olfactory proteins, with different roles in host preferences (Ballesteros et al., 2019). Odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) in the sensillar lymph of insect antennae can bind and transport lipophilic odorant molecules (Wanner et al., 2005; Pelosi et al., 2006; Guo et al., 2011; Dippel et al., 2014; Pelosi et al., 2014; Pelosi et al., 2018; Liu et al., 2020). Ionotropic receptors (IRs) play a key role in sensing different odors, such as acids, salts, ammonia, and aldehyde (Rytz et al., 2013; Chen et al., 2015; Enjin et al., 2016; Knecht et al., 2016; Frank et al., 2017; Zhang et al., 2019). IRs and odorant receptor proteins (ORs) expressed on the olfactory receptor neuron membrane detect transported odorant molecules (Benton et al., 2009; Abuin et al., 2011). Gustatory receptors (GRs) mainly detect sugar, bitterness, and pheromones and have important functions in finding hosts (Clyne et al., 2000; Park and Kwon, 2011; Mang et al., 2016). Odorant degrading enzymes (ODEs) are involved in the degradation of odorant molecules (Leal, 2013). Sensory neuron membrane protein genes (SNMPs) play an important role in the identification and detection of pheromones (Clyne et al., 1999; Mang et al., 2016). However, genes in the olfactory system of QTP Gynaephora species have not been characterized.
In this study, the first transcriptomic analyses of G. aureata were conducted via high-throughput sequencing. Significantly differentially expressed genes (sDEGs) were identified and analyzed via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. To explore the genetic basis of high-altitude adaptation in QTP Gynaephora species, 73 pigment-pathway associated and 107 olfactory-related genes in G. aureata were identified. The study results improve our understanding of high-altitude adaptation in QTP Gynaephora species and may guide the development of pest control strategies.
Specimens of 4th instar larvae (red head and black thorax) of G. aureata were collected from Azi Town, Maqu County, Gansu Province, China (3,400 m above sea level, 101°52′E, 33°40′N). Larvae were dissected under a dissecting microscope to collect the head and thorax into separate tubes. Six samples, including three biological replicates for the head (MQAZT1, MQAZT2, and MQAZT3) and thorax (MQAZX1, MQAZX2, and MQAZX3), were immediately frozen in liquid nitrogen and stored at −80°C before RNA extraction.
Total RNA was extracted using the RNA Extraction Kit RNeasy Mini Kit (Qiagen, Beijing, China) according to the manufacturer’s instructions. Residual genomic DNA was digested with RNase-free DNase (Qiagen, Hilden, Germany). The integrity and concentration of RNA were measured using 1.5% agarose gel electrophoresis and the NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, United States), respectively.
cDNA library construction was performed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, United States). Messenger RNAs (mRNAs) were enriched from total RNAs of G. aureata using oligo (dT) magnetic beads and then fragmented into short nucleotides using the fragmentation buffer solution. The cleaved mRNA was transcribed into first-strand cDNA using random hexamer primers and buffer solution. Second-strand cDNA was subsequently synthesized using buffer solution, deoxyribonucleotide triphosphate, DNA polymerase I, and Ribonuclease H. After end-repairing, dA-tailing, and adaptor ligation, the products were amplified by PCR and purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA, United States) to create the final sequencing library. Finally, the Illumina HiSeq2,500 platform (Illumina, San Diego, CA, United States) was used for sequencing (PE150). High-throughput sequencing of head and thorax RNA of G. aureata was completed by Annoroad Gene Technology Co., Ltd., Beijing, China.
To ensure analysis quality, the adapters, reads with >15% low-quality bases (Q value ≤ 19), and reads with unknown nucleotide (N) ratios greater than 5% were removed before de novo assembly. De novo transcriptome assembly was performed using Trinity (version 20140717) with default parameters (Grabherr et al., 2011), and unigene libraries of the high-quality head and thorax transcriptomes of G. aureata were obtained.
After assembly and evaluation, TransDecoder (version 20140717) was used to identify open reading frames (ORF). Trinotate (version 20140717) was used to predict protein signals and annotate unigenes in various databases, including the non-redundant (Nr) database at the National Center for Biotechnology Information (NCBI), the nucleotide (Nt) database at NCBI, protein family (Pfam) database, evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database, Gene Ontology (GO) database, Eukaryotic Orthologous Groups (KOG) database, and Kyoto Encyclopedia of Genes and Genomes (KEGG) database, with a cut-off E-value of 10–5 (Anderson and Brass, 1998).
Reads per kilobase million mapped reads (RPMK) were used to quantify gene expression levels (Mortazavi et al., 2008). DESeq2 (version 1.4.5) (Nunes et al., 2016) was adopted to analyze differentially expressed genes (DEGs). A gene with |Log2 Fold Change| ≥ 1 and FDR (corrected p-value) < 0.05 was considered DEGs and a gene with |Log2 Fold Change| ≥ 1 and FDR < 0.01 was considered an sDEG. The sDEGs were mapped to GO annotations using Blast2GO and to KEGG pathways using KOBAS. GO terms and pathways with Q ≤ 0.05 were regarded as significantly enriched in this study.
To verify the expression of head and thorax transcriptomes of G. aureata, quantitative real-time PCR (qRT-PCR) was performed for six DEGs. Primer Premier 5.0 was used to design qRT-PCR primers for DEGs (Supplementary Table S1). Elongation factor 1 alpha (EF-1α) was used as an internal reference gene. cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Kusatsu, Japan) according to the recommended protocol. All qRT-PCR analyses were performed in an ABI7500 real-time PCR system (Applied Biosystems, United States) using TB Green® Premix Ex TaqTM Ⅱ (Takara Bio Inc.), according to the recommended protocol. The PCR system and reaction conditions were conducted as described previously (Zhang L. et al., 2017). Relative gene expression levels were calculated using the 2−ΔΔCT method.
To identify candidate genes associated with the adaptation of Gynaephora, genes in major pigment pathways and olfactory-related genes were comprehensively screened. For pigment-pathway associated genes, referring to a previously described method, pigment-pathway associated proteins (n = 109) of Drosophila melanogaster were selected from the AmiGO database under the category “Pigment Metabolic Process” (GO: 0042440) (Croucher et al., 2013). The sequences (n = 109) of pigment-pathway associated proteins of D. melanogaster were downloaded from the NCBI database and searched by the BLASTP algorithm (E value < 10–5) against BLAST databases constructed from the head and thorax transcriptomes of G. aureata (Supplementary Table S2). Then, “significant” BLAST hits were extracted and compared by BLASTP (E value < 10–5) against the Nr database to obtain the pigment-pathway associated genes of G. aureata (Supplementary Table S2).
“OBP and odorant binding protein,” “CSP and chemosensory protein,” “OR and odorant receptor protein,” “IR and ionotropic receptor,” “GR and gustatory receptor,” “ODE and odorant degrading enzyme,” “CXE and carboxylesterase,” and “SNMP and sensory neuron membrane protein gene” were used as keywords to screen putative olfactory-related genes in the annotated head and thorax transcriptomes. All putative OBPs, CSPs, ORs, IRs, GRs, ODEs, and SNMPs were manually checked using the BLASTX program of NCBI with a cut-off E value of 10–5.
The putative N-terminal signal peptides and transmembrane domains (TMDs) of olfactory-related genes were predicted using SignalP 4.1 and TMHMM 2.0, respectively. Amino acid sequences were aligned using ClustalX (Thompson et al., 1994). MEME was used for motif pattern analyses of OBPs and CSPs to confirm the accuracy of annotations (Supplementary Table S3).
We obtained more than 4.9 Gb of raw reads from each sample in the head and thorax transcriptomes of G. aureata. The transcriptomes of three head samples of G. aureata yielded 45,053,582 (89.67%, MQAZT1), 47,478,452 (90.21%, MQAZT2), and 44,486,760 (96.29%, MQAZT3) clean reads. The transcriptomes of three thorax samples of G. aureata yielded 45,606,984 (89.71%, MQAZX1), 44,377,778 (89.7%, MQAZX2), and 45,345,264 (90.04%, MQAZX3) clean reads. The Q30 value in each sample was above 96.20% (Supplementary Table S4). After clean reads for six samples of head and thorax were assembled, a total of 132,598 unigenes were generated (Supplementary Table S5), with a maximum length of 31,778 bp, minimum length of 201 bp, and average length of 618 bp (Supplementary Figure S1). The raw reads have been deposited in the NCBI Sequence Read Archive (SRA) database under accession numbers SRR22475456-SRR22475461 (MQAZT1-3 and MQAZX1-3).
A total of 40,592 unigenes (30.61%) of G. aureata were successfully annotated to seven databases (Supplementary Table S6). The numbers of reference gene sequences annotated in Nr, KOG, Nt, GO, KEGG, eggNOG, and Pfam were 31,872 (24.04%), 8,801 (6.64%), 8,077 (6.09%), 19,385 (14.62%), 2,907 (2.19%), 12,153 (9.17%), and 16,531 (12.47%), respectively (Supplementary Table S6). Among the annotated unigenes, 50.56% had best matches to lepidopteran sequences, primarily Bombyx mori (26.72%), Plutella xylostella (12.41%), and Danaus plexippus (11.43%) (Supplementary Figure S2). GO annotation was used to classify the unigenes into 67 functional groups in three main categories. Of 40,592 unigenes of G. aureata, 19,385 (14.62%) were annotated (Supplementary Figure S3). Moreover, among the 40,592 unigenes, 8,801 unigenes (6.64%) were classified into 24 KOG categories (Supplementary Figure S4).
A total of 8,736 genes were significantly differential expressed between the head and thorax (Supplementary Table S7), 5,762 genes were upregulated and 2,974 genes were downregulated in the thorax compared with the head (Figure 1). Among sDEGs, 65 upregulated sDEGs and 14 downregulated were related to carbohydrate metabolism, 76 upregulated and 13 downregulated sDEGs were related to lipid metabolism, and 49 upregulated sDEGs and 5 downregulated sDEGs were related to detoxication. Among 66 genes encoding cuticular proteins, 14 were significantly upregulated and 52 were downregulated. We also detected three upregulated sDEGs related to the immune response, and two sDEGs (one upregulated and one downregulated) related to DNA repair (Supplementary Table S8). The results of high-throughput sequencing were verified by qRT-PCR for six DEGs (Supplementary Figure S5). GO functional analysis showed that 2,655 sDEGs (30.39% of 8,736 genes) were enriched for terms in the three GO categories: biological process (169 GO terms), cellular component (22 GO terms), and molecular function (121 GO terms) (Supplementary Table S9). In the cellular component category, the terms “regulation of peptidoglycan recognition protein signaling pathway” and “negative regulation of peptidoglycan recognition protein signaling pathway” were enriched (Supplementary Table S9; Supplementary Figure S6). “Myosin filament” was the most highly enriched in the molecular function domain (Supplementary Table S9; Supplementary Figure S6). In the biological process category, “l-threonine ammonia-lyase activity” and “amidase activity” were the enriched components (Supplementary Table S9; Supplementary Figure S6).
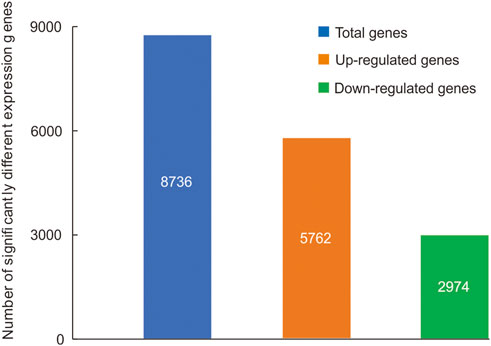
FIGURE 1. Significantly differentially expressed genes (sDEGs) between the head and thorax transcriptomes of Gynaephora aureata.
In total, 2,907 sDEGs (33.28% of 8,736 genes) were annotated to 335 KEGG pathways, including 16 significantly enriched pathways (Supplementary Figure S7). We found that seven pathways related to metabolic reactions were enriched, including two pathways related to carbohydrate metabolism (“Other glycan degradation” and “Galactose metabolism”), three pathways related to lipid metabolism (“Glycosphingolipid biosynthesis-globo series,” “Fatty acid elongation,” and “Fat digestion and absorption”), and two pathways related to amino acid metabolism (“Tryptophan metabolism” and “Glycine, serine and threonine metabolism”). sDEGs were also involved in two pathways (“Lysosome” and “Peroxisome”) related to antioxidant and detoxication (Supplementary Figure S7).
Pigment pathway-associated genes can be divided into five categories, namely, rhodopsin, ommochrome, pteridine, melanin, and heme. In this study, we searched the head and thorax transcriptomes of G. aureata for homologues corresponding to proteins obtained from the “Pigment Metabolic Process” GO term of D. melanogaster based on the AmiGO database. Of 109 D. melanogaster proteins, 99 proteins had putative homologues in G. aureata (Supplementary Table S2) and 73 proteins were identified as possible homologues by reciprocal best hits (RBH) (Supplementary Table S2; Figure 2).
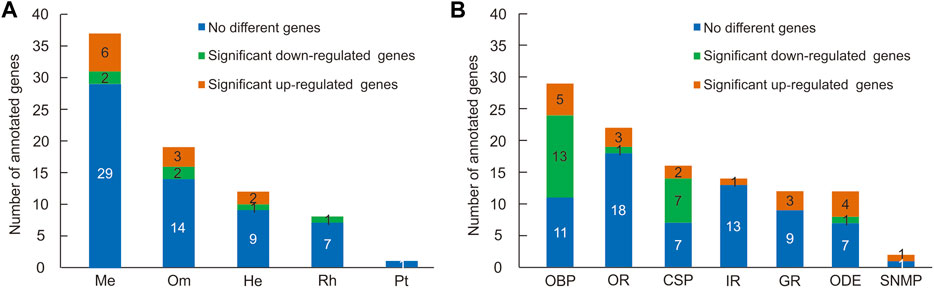
FIGURE 2. Numbers of pigment pathway and olfactory-related genes identified in the head and thorax transcriptomes of Gynaephora aureata. (A) Numbers of five kinds of pigment pathway genes. Pigment pathway genes are abbreviated as follows: melanin-associated genes (Me), ommochrome-associated genes (Om), heme-associated genes (He), rhodopsin-associated genes (Rh), and pteridine-associated genes (Pt). (B) Numbers of seven kinds of olfactory-related genes. Olfactory related genes are abbreviated as follows: odorant-binding protein (OBP), olfactory receptor (OR), chemosensory protein (CSP), ionotropic receptor (IR), gustatory receptor (GR), odorant degrading enzyme (ODE), and sensory neuron membrane protein (SNMP).
Eight of seventeen rhodopsin-associated genes were confirmed by RBH (Supplementary Table S2). The ninaG gene was significantly upregulated in the head (Supplementary Table S10; Figure 3). For ommochrome-associated genes, 19 out of 26 ommochrome-associated genes were confirmed by RBH (Supplementary Table S2). Among these, six ommochrome-associated genes, including Dihydropterin deaminase (DhpD), maroon-like (mal), Punch (Pu), sepia (se), claret (ca), and deep orange (dor), were not only related to the ommochrome biosynthesis pathway but also to the pteridine biosynthesis pathway. Two of these ommochrome-associated genes, cardinal (cd) and vermilion (v), were not only related to the ommochrome biosynthesis pathway but also to the heme biosynthesis pathway. The melanin and ommochrome biosynthesis pathways shared one gene (Rab32). Five key genes in the biosynthesis of ommochrome, namely, v, cinnabar (cn), cd, white (w), and scarlet (st), were identified by RBH (Supplementary Table S2). Three ommochrome-associated genes (v, Hn, and Rab32) were significantly upregulated in the thorax, and two ommochrome-associated genes, cd and brown (bw), were significantly upregulated in the head (Supplementary Table S10; Figure 3). One of seven pteridine-associated genes was confirmed by RBH (Supplementary Table S2). Additionally, 37 out of 50 melanin-associated genes were confirmed by RBH, including four key genes yellow-f, yellow-f2, yellow-h and ebony (e) involved in melanin synthesis (Supplementary Table S2). Six melanin-associated genes, including yellow-h, Rab32, Rac1, Integrin betanu subunit (Itgbn), Rho-like (RhoL), and atypical protein kinase C (aPKC), were significantly upregulated in the thorax, and two melanin-associated genes, e and Cyclin-dependent kinase 5 (Cdk5alpha), were significantly upregulated in the head (Supplementary Table S10; Figure 3). Furthermore, 12 out of 16 heme-associated genes were confirmed by RBH (Supplementary Table S2). Two heme-associated genes, Aminolevulinate synthase (Alas) and v, were significantly upregulated in the thorax, and one heme-associated gene (cd) was significantly upregulated in the head (Supplementary Table S10; Figure 3).

FIGURE 3. Significantly differentially expressed pigment pathway-associated genes between the head and thorax transcriptomes of Gynaephora aureata. Rhodopsin-associated genes, ommochrome-associated genes, melanin-associated genes, and heme-associated genes are shown in green, red, yellow, and blue boxes, respectively. NinaG, bw, cd, e, and Cdk5alpha are significantly upregulated in the head relative to the thorax of G. aureata. V, Hn, Rab32, yellow-h, Rac1, Itgbn, RhoL, aPKC, and Alas are significantly upregulated in the thorax relative to the head of G. aureata.
In the head and thorax transcriptomes of G. aureata, we identified 29 OBPs with ORFs ranging from 324 to 804 bp encoding 107–267 amino acids (Supplementary Tables S11, S12). Among 29 OBPs, 18 had complete ORFs. Based on a multiple sequence alignment, 22 OBPs belonged to the typical “Classic OBP” subfamily with six conserved cysteine residues. Seven OBPs were assigned to the “Minus-C OBP” subfamily with four conserved cysteine residues (Figure 4C). The homology of 23 OBP sequences from Lepidoptera orthologs ranged from 38.98% to 88.72% and the homology of 6 OBP sequences from Diptera orthologs ranged from 31.07% to 86.33% in the NCBI database (Supplementary Table S11). MEME results revealed 8 motifs in the identified OBPs (Figure 4A). Nine OBPs had the same motif pattern, 3-1-8-2. Six OBPs had the same motif pattern, 3-1-2 (Figure 4B). Five of the 29 OBPs were significantly upregulated in the thorax and 13 in the head (Table 1).
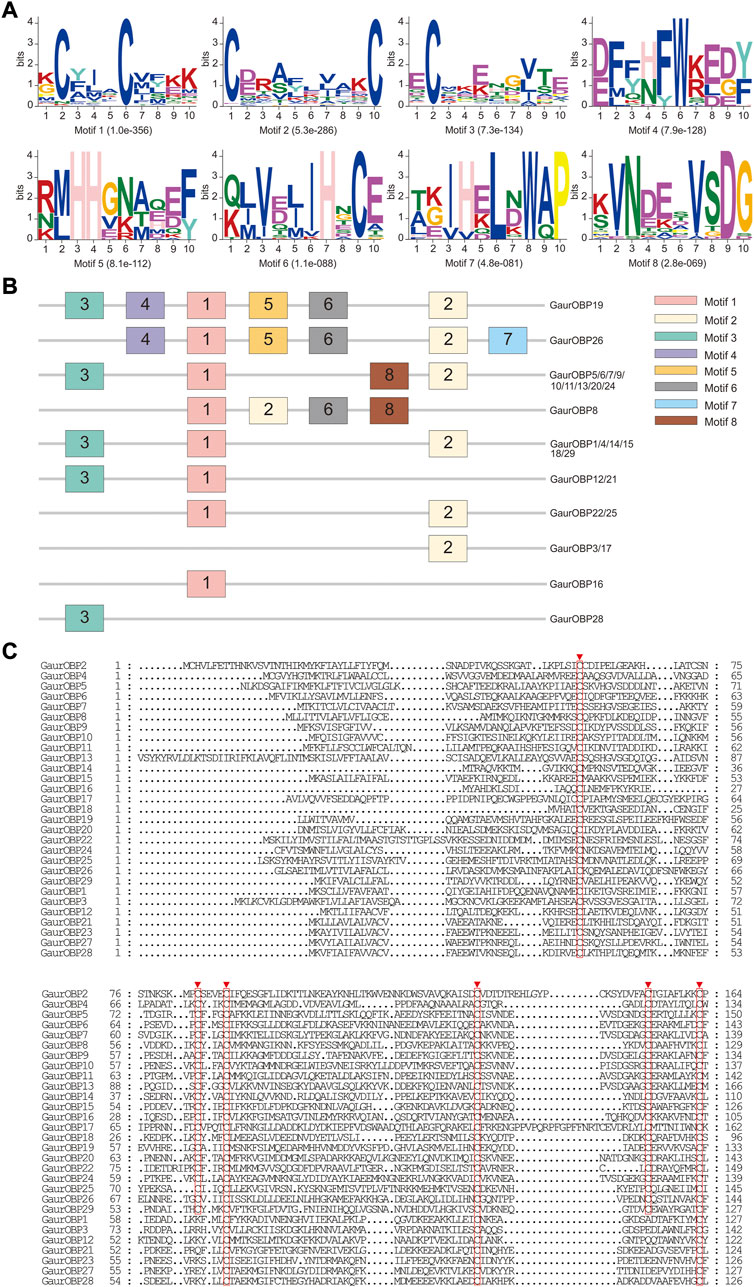
FIGURE 4. Motif pattern analysis and alignment of candidate odorant-binding proteins (OBPs) of Gynaephora aureata. (A) Eight motifs from G. aureata and other lepidopterans. The E-value for each motif is shown in parentheses. (B) Locations of each motif in the protein sequences. (C) Alignment of candidate OBPs of G. aureata. Highly conserved cysteine residues are marked with red borders.
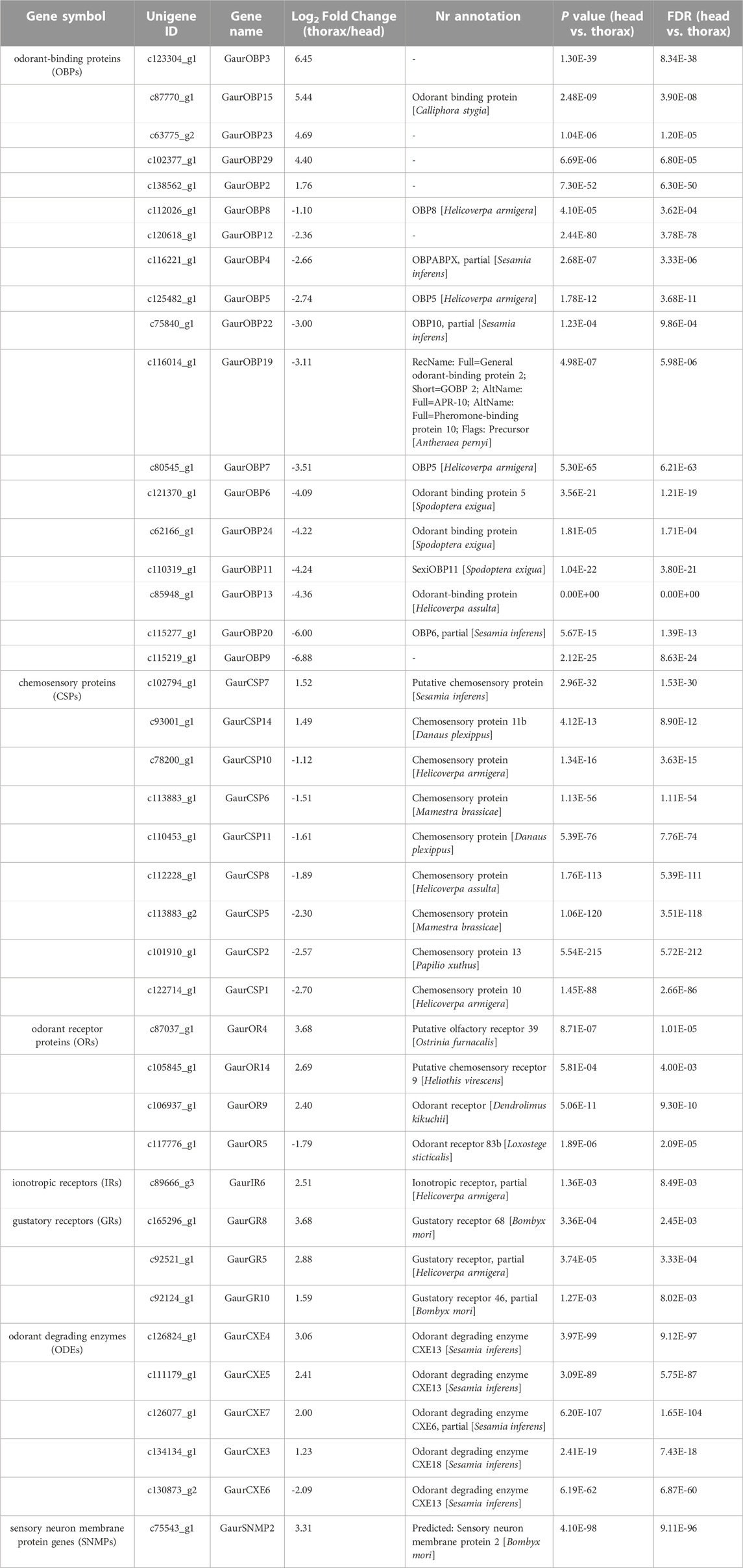
TABLE 1. Summary of all significantly differential expressed olfactory-related genes between the head and thorax transcriptomes of Gynaephora aureata. FDR represents the corrected p-value.
A total of 16 putative CSPs were identified from the head and thorax transcriptomes of G. aureata, of which 11 CSPs had complete ORFs (Supplementary Tables S11 and S12). The ORFs of 16 CSP genes ranged from 318 to 447 bp and encoded 105 to 148 amino acids (Supplementary Table S11). BLASTX results showed that in addition to the homology of GaurCSP14 (40.98%), the 15 CSPs had high homology (63.48%–92.73%) with lepidopteran orthologs (Supplementary Table S11). All CSPs except GaurCSP15 had signal peptides (Supplementary Table S11). All CSPs had four positionally conserved cysteine residues and the featured sequence C1-X6-7-C2-X18-C3-X2-C4 (where X indicates any amino acid) of typical insect CSPs (Figure 5C). MEME revealed 8 motifs in the identified CSPs (Figure 5A). Eleven CSPs had the same motif pattern, 7-3-6-1-5-2-8-4 (Figure 5B). Two CSPs had the same motif pattern, 7-3-6-1-5-2-4 (Figure 5B). The motif pattern of GaurCSP15 was 7-3-6-1-5-2-8 (Figure 5B). Two of the 16 CSPs were significantly upregulated in the thorax and seven were upregulated in the head (Table 1).
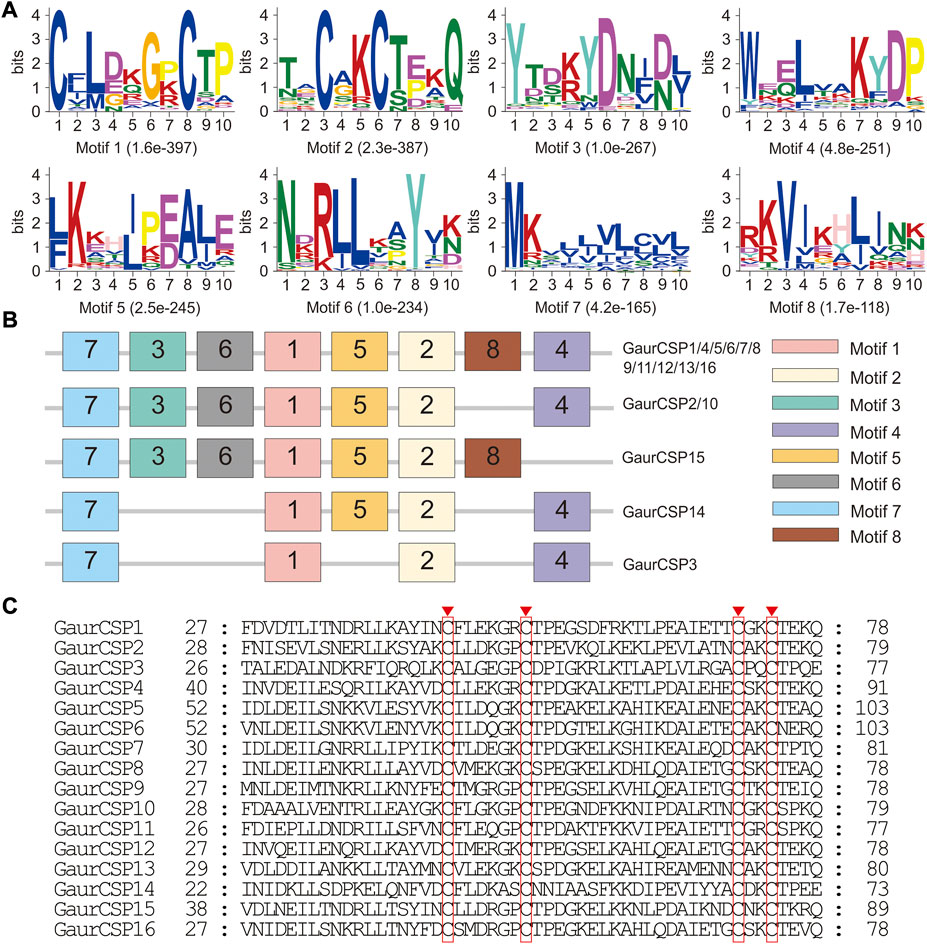
FIGURE 5. Motif pattern analysis and alignment of candidate chemosensory proteins (CSPs) of Gynaephora aureata. (A) Eight motifs from G. aureata and other lepidopterans. The E-value for each motif is shown in parentheses. (B) Locations of each motif in the protein sequences. (C) Alignment of candidate CSPs of G. aureata. The highly conserved cysteine residues are marked with red borders.
We identified 22 putative ORs in G. aureata based on sequence similarity to insect ORs (Supplementary Tables S11, S12). The genes of 22 ORs had ORFs encoding 105–336 amino acids and were predicted to have 0–5 TMDs, of which the genes encoding five ORs (GaurOR1, GaurOR2, GaurOR3, GaurOR5, and GaurOR12) had complete ORFs. The homology of OR sequences from lepidopteran orthologs ranged from 46.23% to 94.89% in the NCBI database (Supplementary Table S11 and S12). Three (GaurOR4, GaurOR9, and GaurOR14) of the 29 ORs were significantly upregulated in the thorax and one (GaurOR5) was upregulated in the head (Table 1).
The analysis of transcriptome data revealed 14 IRs (Supplementary Tables S11, S12). These IRs had ORFs ranging from 297 to 2,724 bp, encoded 98–907 amino acids (Supplementary Table S11), and had 0–4 predicted TMDs (Supplementary Table S11). Most best hit IRs shared more than 68% amino acid sequence identity with lepidopteran orthologs, whereas GaurIR6 and GaurIR8 shared 45.04% and 45.87% amino acid sequence identities, respectively (Supplementary Table S11). One (GaurIR6) of the 14 IRs was significantly upregulated in the thorax (Table 1).
We identified 12 putative GRs in G. aureata (Supplementary Tables S11, S12), containing 0–8 TMDs (Supplementary Table S11). The lengths of the deduced proteins for putative GR genes ranged from 106 to 455 amino acids. Of these, four of the GRs had complete ORFs (Supplementary Table S11). The BLASTX searches against the Nr database showed 32.56%–91.48% homology with sequences in other Lepidoptera (Supplementary Table S11). Three (GaurGR5, GaurGR8, and GaurGR10) of the 12 GRs were significantly upregulated in the thorax (Table 1).
We identified 12 putative ODEs in the head and thorax transcriptomes of G. aureata (Supplementary Tables S11, S12). The ORFs of 12 ODE genes ranged from 357 to 1785 bp and encoded 118–594 amino acids (Supplementary Table S11). Four of these ODEs had complete ORFs (Supplementary Table S11). The candidate ODEs shared 42.50%–75.47% amino acid sequence identities with other Lepidoptera ODEs in the NCBI database (Supplementary Table S11). Four (GaurCXE3, GaurCXE4, GaurCXE5, and GaurCXE7) of the 12 ODEs were significantly upregulated in the thorax and one (GaurCXE6) was upregulated in the head (Table 1).
We identified two putative SNMPs with complete ORFs, GaurSNMP1 and GaurSNMP2 (Supplementary Tables S11, S12). The ORFs of GaurSNMP1 and GaurSNMP2 were 1,560 and 1,521 bp and can encode 519 and 506 amino acids, respectively (Supplementary Table S11). The deduced GaurSNMP1 and GaurSNMP2 proteins contained two TMDs (Supplementary Table S11), conforming to the general characteristics of SNMPs. GaurSNMP1 and GaurSNMP2 had 78.65% and 60.67% homology with sequences of other Lepidoptera species (Supplementary Table S11). GaurSNMP2 was significantly upregulated in the thorax (Table 1).
The first transcriptomic analyses of G. aureata were conducted by high-throughput sequencing. Some sDEGs between the head and thorax of G. aureata were related to carbohydrate metabolism, lipid metabolism, epidermal protein, and detoxification. There were more upregulated sDEGs related to carbohydrate and lipid metabolism in the thorax than in the head, which may be related to the energy consumption caused by movement in the thorax. Genes encoding cuticle proteins had different expression patterns among tissues. The number of upregulated sDEGs encoding cuticle proteins was higher in the head than in the thorax, which may be due to the higher degree of ossification in head. The detoxification, immune response, and DNA repair functions of the QTP Gynaephora species may be related to high-altitude adaptation. Additionally, the detoxification function of the thorax may be stronger than that of the head in G. aureata (Supplementary Table S8).
Pigment pathway-associated genes are divided into five categories, namely, rhodopsin, ommochrome, pteridine, melanin, and heme. The upregulated pigment pathway-associated sDEGs in the head included ninaG, bw, cd, e, and Cdk5alpha. The rhodopsin-associated gene ninaG plays a role in the biochemical pathway of responsible for the conversion of retinal to the rhodopsin chromophore 3-hydroxyretinal (Sarfare et al., 2005). The upregulated pigment pathway-associated sDEGs in the thorax included v, Hn, Rab32, yellow-h, Rac1, Itgbn, RhoL, aPKC, and Alas. The heme-associated gene Alas is the first enzyme in the heme biosynthesis pathway. The melanin-associated gene Rab32 has a key role in the biogenesis of melanosomes and potentially other lysosome-related organelles (Wasmeier et al., 2006). There are few studies on the relationship between pigment-associated genes (Cdk5alpha, bw, Hn, Rac1, Itgbn, RhoL, and aPKC) and body color. Further functional experiments will be needed to link these pigment-associated genes to body color. QTP Gynaephora species show red head and black body hair, which are morphological features associated with high-altitude adaptation. The red and black color formation in G. aureata may be related to the melanin and ommochrome biosynthesis pathways. We focused on the melanin-associated genes e and yellow-h and ommochrome-associated genes cd and v. In the biosynthesis pathway of melanin, tyrosine hydroxylase encoded by pale hydroxylates the first tyrosine to dopa (Figure 6A). Under the action of dopamine decarboxylase (Ddc), dopamine is converted into dopamine (Barek et al., 2022). Dopamine under the action of NBAD synthetase (e) forms NBAD (N-β-alanyldopamine), which participates in epidermal fusion, making the epidermis yellow or light brown (Wittkopp et al., 2003a; Wittkopp et al., 2003b; Yang, 2021; Barek et al., 2022). In G. aureata, the e gene was significantly upregulated in the head (Figure 3), which may be related to the yellow appearance in the frons of the head. Phenoloxidase with wide specificity can oxidize dopa and dopamine, and convert them into quinones, which can rapidly cyclize to form amino pigments. Cuticular laccase oxidizes dopa and dopamine into corresponding quinones (Arakane et al., 2005), quinones rapidly cyclize to form amino chromes (Figure 6A) (Barek et al., 2022). In D. melanogaster, DCDT (dopachrome decarboxylase/tautomerases) encoded by yellow-f and yellow-f2 may convert dopachrome and dopaminechrome into the same end product, DHI (5,6-dihydroxyindole) (Figure 6A) (Barek et al., 2022). However, in other insects, DCDT is recalcitrant to dopachrome. Therefore, a separate DPT (dopaminechrome tautomerase) is required (Barek et al., 2022). Recent research has shown that DPT is the product of the yellow-h gene in D. melanogaster (Figure 6A) (Barek et al., 2022). In Papilio xuthus, yellow-h3 was detected at sites where melanin was deposited in the cuticle and may participate in the biosynthesis of melanin (Futahashi et al., 2011; Barek et al., 2022). In G. aureata, four yellow-related genes (yellow, yellow-f, yellow-f2, and yellow-h) were confirmed by RBH (Supplementary Table S2). Yellow-f and yellow-f2 may be involved in the conversion of dopa to DHI. According to a chemical analysis, melanin in insects mainly comes from dopamine, not dopa (Sugumaran and Barek, 2016; Barek et al., 2018; Barek et al., 2022). Therefore, gradual melanin formation via dopamine may be an important process. Yellow-h was significantly upregulated in the thorax of G. aureata (Supplementary Table S10; Figure 3). Yellow-h may be related to the transformation of dopamine into DHI, producing melanin to form the black body color of G. aureata. Therefore, yellow-h may be crucial in the formation of melanin in G. aureata; however, further studies are still needed to confirm this.
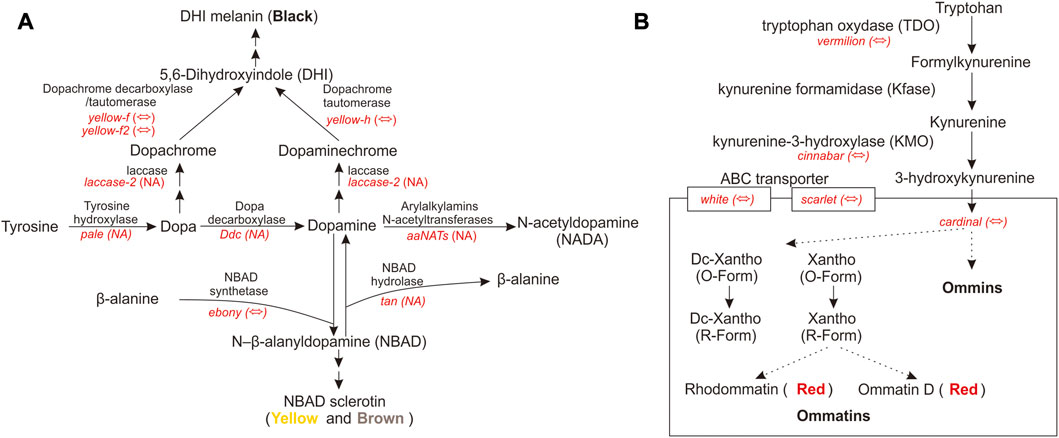
FIGURE 6. Melanin and ommochrome biosynthesis pathways (Sullivan and Sullivan, 1975; O'Hare et al., 1984; Searles and Voelker, 1986; ten Have et al., 1995; Yang, 2021; Barek et al., 2022). (A) Key gene products in the melanin biosynthesis pathway. (B) Key gene products in the ommochrome biosynthesis pathway. “⇔” indicates that the gene is supported by a reciprocal BLAST hit (RBH) analysis; “NA” indicates that the gene is either not supported by the RBH analysis or not part of the D. melanogaster AmiGO pigment gene set.
In the biosynthesis of ommochrome, tryptophan forms the precursor of the eye pigment 3-hydroxycaninurine under the action of tryptophan oxygenase (TDO), encoded by vermilion, kynurenine formamidase (Kfase), and kynurenine-3-hydroxylase (KMO) encoded by cinnabar, and it is transferred to pigment granules to form a heterodimer by the ABC transporter w and scarlet (st) (Figure 6B) (Sullivan and Sullivan, 1975; O'Hare et al., 1984; Searles and Voelker, 1986; ten Have et al., 1995; Yang, 2021). Ommochromes are divided into ommatins and ommins. The molecular mechanism underlying the transition from 3-hydroxykynurenine to ommochrome is largely unknown. Cd is important in the formation of eye pigments in B. mori (Osanai-Futahashi et al., 2016). In G. aureata, five key genes involved in the biosynthesis of ommochrome, namely, v, cn, cd, w, and st, were identified (Supplementary Table S2). Ommin A forms a purple phenotype, decarboxylated xanthommatin, decarboxylated dihydroxanthommatin, rhodommatin, and ommatin D form a red phenotype (Yang, 2021). In G. aureata, the cd gene was significantly upregulated in the head (Supplementary Table S10; Figure 3), which may be related to the red head of the species. The v gene was significantly upregulated in the thorax (Supplementary Table S10; Figure 3), suggesting that ommochromes formed in the thorax. The black phenotype of the thorax of G. aureata is the result of multiple pigments; however, further research is still needed to determine the detailed mechanisms.
In insects, olfaction plays an important physiological function by sensing chemical clues in the environment (Peng et al., 2022). Insects may adjust their olfactory system to adapt to changes in biotic (such as plant or host phenotype and genotype) and abiotic factors (Linz et al., 2013; Claudianos et al., 2014). The change in feeding habits of QTP Gynaephora species is an adaptation of the olfactory system to the environment of the QTP. In this study, 107 olfactory-related genes were identified in G. aureata (Supplementary Table S11; Figure 2), including OBPs, CSPs, ORs, IRs, GRs, ODEs, and SNMPs. As the first step in odor recognition, OBPs and CSPs can bind to and transport lipophilic odorant molecules (Wanner et al., 2005; Pelosi et al., 2006; Guo et al., 2011; Dippel et al., 2014; Pelosi et al., 2014; Pelosi et al., 2018; Liu et al., 2020). We identified 29 OBPs (Supplementary Table S11; Figure 2), which was fewer than the numbers in Spodoptera littoralis (49) and B. mori (46) (Gong et al., 2009; Vieira and Rozas, 2011; Walker et al., 2019) but similar to estimates in other species in Lepidoptera, such as Spodoptera frugiperda (25) and Chilo suppressalis (26) (Cao et al., 2014; Qiu et al., 2020). A total of 16 CSPs were identified, similar to corresponding counts in the lepidopterans Plodia interpunctella (15) and Galleria mellonella (18) (Jia et al., 2018; Jiang et al., 2021). The motif patterns of different OBPs and CSPs diff among insects. The most conserved motif pattern of OBPs was 3-1-8-2, and the most conserved motif pattern of CSPs was 7-3-6-1-5-2-8-4 in G. aureata. This showed that OBPs and CSPs may have conserved functions in odor recognition. Different motif patterns provide evidence for possible functional differentiation. Chemosensory receptor protein families are involved in the detection and transduction of odorant signals, mainly including ORs, IRs, and GRs. We identified 22 ORs (Supplementary Table S11; Figure 2), fewer than estimates in S. littoralis (60) (Walker et al., 2019), which may be related to the sequencing method and sequencing depth. We identified 12 GRs and 14 IRs (Supplementary Table S11; Figure 2). The number of GRs was similar to that in Peridroma saucia (10) (Sun Y. L. et al., 2020). The number of IRs was similar to counts in S. littoralis (17) and B. mori (18) (Croset et al., 2010; Walker et al., 2019). Carboxylesterases (CXEs) are among the most important ODEs. We identified 12 CXEs (Supplementary Table S11; Figure 2), which was fewer than the numbers in S. littoralis (20) (Durand et al., 2011; Liu et al., 2019), Spodoptera litura (24) (Zhang et al., 2016), and Semia inferens (20) (Zhang et al., 2014) but similar to the number in Cydia pomonella (12) (Huang et al., 2016; Liu et al., 2019). We identified two SNMPs (Supplementary Table S11; Figure 2), similar to count sin S. litura (2) (Zhang et al., 2014) and B. mori (Walker et al., 2019; Xu et al., 2021). Host finding relies on the olfactory system for the detection of environmental signals (Gols et al., 2012; Suh et al., 2014; Ballesteros et al., 2019). IRs play a key role in sensing different odors, such as acids, salts, ammonia, and aldehyde (Rytz et al., 2013; Chen et al., 2015; Enjin et al., 2016; Knecht et al., 2016; Frank et al., 2017; Zhang et al., 2019). GRs mainly detect sugar, bitterness, and pheromones, which have important functions in finding hosts (Clyne et al., 2000; Park and Kwon, 2011; Mang et al., 2016). OBPs and ORs play key roles in host preferences. Therefore, the differences in expression levels of different olfactory genes may be related to the recognition of different hosts; however, further research is still needed to verify this.
To explore the genetic basis of high-altitude adaptation in QTP Gynaephora species, we performed the sequencing, assembly and annotation of the head and thorax transcriptomes of G. aureata and identified 73 pigment pathway and 107 olfactory-related genes. Pigment pathway-associated genes are related to body color adaptation to high-altitude environments (low temperatures and high UV levels). Olfactory-related genes are related to the adaptation of G. aureata to the abundant plant resources in the QTP. Pigment pathway and olfactory-related genes provide insight into the adaptation of QTP Gynaephora species to high altitudes, including the genetic basis of body color and changes in food habits. A reference genome is not available for QTP Gynaephora species. All analyses in this study were based on the head and thorax transcriptomes of G. aureata. In the future, pigment pathway and olfactory-related genes should be identified by full-genome analyses. In addition, functional analyses of key melanin-associated yellow-h and ommochrome-associated cd genes are needed in the future.
In this study, transcriptomic analyses of G. aureata were conducted by high-throughput sequencing for the first time to clarify the molecular basis of key traits related to survival in the QTP. We detected sDEGs related to carbohydrate metabolism, lipid metabolism, epidermal proteins, and detoxification. Additionally, we identified 73 genes involved in pigment pathways and 107 olfactory-related genes in G. aureata. The melanin-associated gene yellow-h and the ommochrome-associated gene cd might play important roles in the formation of black and red body colors of G. aureata, respectively. However, further functional assays of yellow-h and cd are needed in the future. These results provide new insights into high-altitude adaptation of Gynaephora in the QTP, providing a basis for the development of new control strategies for these pests.
The original contributions presented in the study are publicly available. This data can be found here: SRR22475456-61.
M-LY conceived and designed the experiments. L-JZ, LZ, and X-ZY sampled G. aureata specimens. J-RZ, L-JZ, LZ, and X-ZY conducted experiments. J-RZ, L-JZ, and S-YH performed data analyses. J-RZ wrote the manuscript. M-LY revised the manuscript. All authors read and approved the final version of the manuscript.
This study was funded by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0302), the National Key Research and Development Program of China (2022YFD1401102) and the National Science & Technology Fundamental Resources Investigation Program of China (2019FY100404).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1137618/full#supplementary-material
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69 (1), 44–60. doi:10.1016/j.neuron.2010.11.042
Anderson, I., and Brass, A. (1998). Searching DNA databases for similarities to DNA sequences: When is a match significant? Bioinformatics 14, 349. doi:10.1093/bioinformatics/14.4.349
Arakane, Y., Muthukrishnan, S., Beeman, R. W., Kanost, M. R., and Kramer, K. J. (2005). Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. PNAS 102 (32), 11337–11342. doi:10.1073/pnas.0504982102
Ballesteros, G. I., Sepulveda, D. A., and Figueroa, C. C. (2019). Identification and expression profiling of peripheral olfactory genes in the parasitoid wasp aphidius ervi (hymenoptera: Braconidae) reared on different aphid hosts. Insects 10 (11), 397. doi:10.3390/insects10110397
Barek, H., Sugumaran, M., Ito, S., and Wakamatsu, K. (2018). Insect cuticular melanins are distinctly different from those of mammalian epidermal melanins. PIGM Cell Melanoma R. 31 (3), 384–392. doi:10.1111/pcmr.12672
Barek, H., Zhao, H., Heath, K., Veraksa, A., and Sugumaran, M. (2022). Drosophila yellow-H encodes dopaminechrome tautomerase: A new enzyme in the eumelanin biosynthetic pathway. PIGM Cell Melanoma R. 35 (1), 26–37. doi:10.1111/pcmr.13008
Barrio, I. C., Hik, D. S., and Liu, J. Y. (2014). Diet breadth of Gynaephora groenlandica (Lepidoptera: Erebidae): Is polyphagy greater in alpine versus arctic populations? Can. Entomol. 147 (2), 215–221. doi:10.4039/tce.2014.35
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136 (1), 149–162. doi:10.1016/j.cell.2008.12.001
Brenner, M., and Hearing, V. J. (2008). The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 84 (3), 539–549. doi:10.1111/j.1751-1097.2007.00226.x
Cao, D., Liu, Y., Wei, J., Liao, X., Walker, W. B., Li, J., et al. (2014). Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 10 (8), 846–860. doi:10.7150/ijbs.9297
Cao, Y. (2022). “Spatiotemporal dynamics of diet and gut bacteria of three Gynaephora species endemic to the Qinghai-Tibetan plateay,” (Lanzhou: Lanzhou University). (master).
Cao, Y., Yang, X.-Z., Zhang, L.-J., Li, M., and Yuan, M.-L. (2021). Gut bacteria communities differ between Gynaephora species endemic to different altitudes of the Tibetan Plateau. Sci. Total Environ. 777, 146115. doi:10.1016/j.scitotenv.2021.146115
Carothers, J. H., Marquet, P. A., and Jaksic, F. M. (1998). Thermal ecology of a Liolaemus lizard assemblage along an Andean altitudinal gradient in Chile. Rev. Chil. Hist. Nat. 71, 39–50.
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 527 (7579), 516–520. doi:10.1038/nature16148
Claudianos, C., Lim, J., Young, M., Yan, S., Cristino, A. S., Newcomb, R. D., et al. (2014). Odor memories regulate olfactory receptor expression in the sensory periphery. Eur. J. Neurosci. 39 (10), 1642–1654. doi:10.1111/ejn.12539
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287 (5459), 1830–1834. doi:10.1126/science.287.5459.1830
Clyne, P. J., Warr, C. G., Freeman, M. R., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi:10.1016/s0896-6273(00)81093-4
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6 (8), e1001064. doi:10.1371/journal.pgen.1001064
Croucher, P. J., Brewer, M. S., Winchell, C. J., Oxford, G. S., and Gillespie, R. G. (2013). De novo characterization of the gene-rich transcriptomes of two color-polymorphic spiders, Theridion grallator and T. californicum (Araneae: Theridiidae), with special reference to pigment genes. BMC Genom 14, 862. doi:10.1186/1471-2164-14-862
DeJong, P. W., Gussekloo, S. W. S., and Brakefield, P. M. (1996). Differences in thermal balance, body temperature and activity between non-melanic and melanic two-spot ladybird beetles (Adalia bipunctata) under controlled conditions. J. Exp. Biol. 199 (12), 2655–2666. doi:10.1242/jeb.199.12.2655
Di Pietro, S. M., Falcon-Perez, J. M., Tenza, D., Setty, S. R., Marks, M. S., Raposo, G., et al. (2006). BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell 17 (9), 4027–4038. doi:10.1091/mbc.e06-05-0379
Dippel, S., Oberhofer, G., Kahnt, J., Gerischer, L., Opitz, L., Schachtner, J., et al. (2014). Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant-binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genom 15, 1141. doi:10.1186/1471-2164-15-1141
Durand, N., Carot-Sans, G., Bozzolan, F., Rosell, G., Siaussat, D., Debernard, S., et al. (2011). Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS One 6 (12), e29147. doi:10.1371/journal.pone.0029147
Enjin, A., Zaharieva, E. E., Frank, D. D., Mansourian, S., Suh, G. S., Gallio, M., et al. (2016). Humidity sensing in Drosophila. Curr. Biol. 26 (10), 1352–1358. doi:10.1016/j.cub.2016.03.049
Ferreira, G. C. (1995). Heme biosynthesis: Biochemistry, molecular biology, and relationship to disease. J. Bioenergetics Biomembr. 27 (2), 147–150. doi:10.1007/BF02110029
Frank, D. D., Enjin, A., Jouandet, G. C., Zaharieva, E. E., Para, A., Stensmyr, M. C., et al. (2017). Early integration of temperature and humidity stimuli in the Drosophila brain. Curr. Biol. 27 (15), 2381–2388.e4. doi:10.1016/j.cub.2017.06.077
Futahashi, R., Tanaka, K., Matsuura, Y., Tanahashi, M., Kikuchi, Y., and Fukatsu, T. (2011). Laccase2 is required for cuticular pigmentation in stinkbugs. Insect biochem. Mol. Biol. 41 (3), 191–196. doi:10.1016/j.ibmb.2010.12.003
Gao, W., Fu, T., and Che, J. (2019). High-altitude adaptive evolution in amphibians and reptiles: Status and prospect. Sci. Sin. Vitae 49 (4), 345–360. doi:10.1360/n052018-00216
Geen, M. R., and Johnston, G. R. (2014). Coloration affects heating and cooling in three color morphs of the Australian bluetongue lizard, Tiliqua scincoides. J. Therm. Biol. 43, 54–60. doi:10.1016/j.jtherbio.2014.04.004
Gols, R., Veenemans, C., Potting, R. P. J., Smid, H. M., Dicke, M., Harvey, J. A., et al. (2012). Variation in the specificity of plant volatiles and their use by a specialist and a generalist parasitoid. Anim. Behav. 83 (5), 1231–1242. doi:10.1016/j.anbehav.2012.02.015
Gong, D. P., Zhang, H. J., Zhao, P., Xia, Q. Y., and Xiang, Z. H. (2009). The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genom 10, 332. doi:10.1186/1471-2164-10-332
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Guo, W., Wang, X., Ma, Z., Xue, L., Han, J., Yu, D., et al. (2011). CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLos Genet. 7 (2), e1001291. doi:10.1371/journal.pgen.1001291
Harris, D. A., Kim, K., Nakahara, K., Vasquez-Doorman, C., and Carthew, R. W. (2011). Cargo sorting to lysosome-related organelles regulates siRNA-mediated gene silencing. J. Cell Biol. 194 (1), 77–87. doi:10.1083/jcb.201102021
Huang, X., Liu, L., Su, X., and Feng, J. (2016). Identification of biotransformation enzymes in the antennae of codling moth Cydia pomonella. Gene 580 (1), 73–79. doi:10.1016/j.gene.2016.01.008
Jablonski, N. G., and Chaplin, G. (2010). Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. PNAS 107, 8962–8968. doi:10.1073/pnas.0914628107
Jia, X., Zhang, X., Liu, H., Wang, R., and Zhang, T. (2018). Identification of chemosensory genes from the antennal transcriptome of Indian meal moth Plodia interpunctella. PLoS One 13 (1), e0189889. doi:10.1371/journal.pone.0189889
Jiang, X. C., Liu, S., Jiang, X. Y., Wang, Z. W., Xiao, J. J., Gao, Q., et al. (2021). Identification of olfactory genes from the greater wax moth by antennal transcriptome analysis. Front. Physiol. 12, 663040. doi:10.3389/fphys.2021.663040
Jin, Y. T., and Liao, P. H. (2015). An elevational trend of body size variation in a cold-climate agamid lizard, Phrynocephalus theobaldi. Curr. Zool. 61 (3), 444–453. doi:10.1093/czoolo/61.3.444
Knecht, Z. A., Silbering, A. F., Ni, L., Klein, M., Budelli, G., Bell, R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife 5, e17879. doi:10.7554/eLife.17879
Leal, W. S. (2013). Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomology 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Li, M., Tian, S., Jin, L., Zhou, G., Li, Y., Zhang, Y., et al. (2013). Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45 (12), 1431–1438. doi:10.1038/ng.2811
Linz, J., Baschwitz, A., Strutz, A., Dweck, H. K., Sachse, S., Hansson, B. S., et al. (2013). Host plant-driven sensory specialization in Drosophila erecta. Proc. Biol. Sci. 280 (1760), 20130626. doi:10.1098/rspb.2013.0626
Liu, H., Lei, X., Du, L., Yin, J., Shi, H., Zhang, T., et al. (2019). Antennae-specific carboxylesterase genes from Indian meal moth: Identification, tissue distribution and the response to semiochemicals. J. Stored Prod. Res. 84, 101528. doi:10.1016/j.jspr.2019.101528
Liu, Z., Liang, X. F., Xu, L., Keesey, L. W., Lei, Z. R., Smagghe, G., et al. (2020). An antennae-specific odorant-binding protein is involved in bactrocera dorsalis olfaction. Front. Ecol. Evol. 8. doi:10.3389/fevo.2020.00063
MacLean, S. F., and Jensen, T. S. (1985). Food plant selection by insect herbivores in alaskan arctic tundra: The role of plant life form. Oikos 44 (1), 211–221. doi:10.2307/3544063
Mang, D., Shu, M., Endo, H., Yoshizawa, Y., Nagata, S., Kikuta, S., et al. (2016). Expression of a sugar clade gustatory receptor, BmGr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm Bombyx mori. Insect biochem. Mol. Biol. 70, 85–98. doi:10.1016/j.ibmb.2015.12.008
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5 (7), 621–628. doi:10.1038/nmeth.1226
Nunes, R. D., Romeiro, N. C., De Carvalho, H. T., Moreira, J. R., Sola-Penna, M., Silva-Neto, M. A., et al. (2016). Unique PFK regulatory property from some mosquito vectors of disease, and from Drosophila melanogaster. Parasites Vectors 9, 107. doi:10.1186/s13071-016-1391-y
O'Hare, K., Murphy, C., Levis, R., and Rubin, G. M. (1984). DNA sequence of the white locus of Drosophila melanogaster. J. Mol. Biol. 180 (3), 437–455. doi:10.1016/0022-2836(84)90021-4
Osanai-Futahashi, M., Tatematsu, K. I., Futahashi, R., Narukawa, J., Takasu, Y., Kayukawa, T., et al. (2016). Positional cloning of a Bombyx pink-eyed white egg locus reveals the major role of cardinal in ommochrome synthesis. Hered. (Edinb) 116 (2), 135–145. doi:10.1038/hdy.2015.74
Park, J. H., and Kwon, J. Y. (2011). Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS One 6 (12), e29022. doi:10.1371/journal.pone.0029022
Pelosi, P., Iovinella, I., Felicioli, A., and Dani, F. R. (2014). Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 5, 320. doi:10.3389/fphys.2014.00320
Pelosi, P., Iovinella, I., Zhu, J., Wang, G., and Dani, F. R. (2018). Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93 (1), 184–200. doi:10.1111/brv.12339
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63 (14), 1658–1676. doi:10.1007/s00018-005-5607-0
Peng, Z. L., Wu, W., Tang, C. Y., Ren, J. L., Jiang, D., and Li, J. T. (2022). Transcriptome analysis reveals olfactory system expression characteristics of aquatic snakes. Front. Genet. 13, 825974. doi:10.3389/fgene.2022.825974
Qiu, L., He, L., Tan, X., Zhang, Z., Wang, Y., Li, X., et al. (2020). Identification and phylogenetics of Spodoptera frugiperda chemosensory proteins based on antennal transcriptome data. Comp. Biochem. Physiol. D. Genomics Proteomics 34, 100680. doi:10.1016/j.cbd.2020.100680
Reguera, S., Zamora-Camacho, F. J., and Moreno-Rueda, G. (2014). The lizard Psammodromus algirus (Squamata: Lacertidae) is darker at high altitudes. Biol. J. Linn. Soc. 112 (1), 132–141. doi:10.1111/bij.12250
Rinnerthaler, M., Bischof, J., Streubel, M. K., Trost, A., and Richter, K. (2015). Oxidative stress in aging human skin. Biomolecules 5 (2), 545–589. doi:10.3390/biom5020545
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect biochem. Mol. Biol. 43 (9), 888–897. doi:10.1016/j.ibmb.2013.02.007
Sarfare, S., Ahmad, S. T., Joyce, M. V., Boggess, B., and O'Tousa, J. E. (2005). The Drosophila ninaG oxidoreductase acts in visual pigment chromophore production. J. Biochem. 280 (12), 11895–11901. doi:10.1074/jbc.M412236200
Searles, L. L., and Voelker, R. A. (1986). Molecular characterization of the Drosophila vermilion locus and its suppressible alleles. PNAS 83 (2), 404–408. doi:10.1073/pnas.83.2.404
Sugumaran, M., and Barek, H. (2016). Critical analysis of the melanogenic pathway in insects and higher animals. Int. J. Mol. Sci. 17 (10), 1753. doi:10.3390/ijms17101753
Sugumaran, M., Saul, S. J., and Dali, H. (1990). On the mechanism of side chain oxidation of N-β-Alanyldopamine by cuticular enzymes from Sarcophaga bullata. Arch. Insect Biochem. Physiol. 15, 255–269. doi:10.1002/arch.940150406
Suh, E., Bohbot, J., and Zwiebel, L. J. (2014). Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 6, 86–92. doi:10.1016/j.cois.2014.10.006
Sullivan, D. T., and Sullivan, M. C. (1975). Transport defects as the physiological basis for eye color mutants of Drosophila melanogaster. Biochem. Genet. 13 (9-10), 603–613. doi:10.1007/BF00484918
Sun, M. X., Yin, H. W., Wang, J. X., and Liang, A. P. (2020a). Progress in research on insect pigments. Chin. J. Appl. Entomology 57 (2), 298–309. doi:10.7679/j.issn.2095/1353.2020.033
Sun, Y. L., Dong, J. F., Gu, N., and Wang, S. L. (2020b). Identification of candidate chemosensory receptors in the antennae of the variegated cutworm, Peridroma saucia hubner, based on a transcriptome analysis. Front. Physiol. 11, 39. doi:10.3389/fphys.2020.00039
ten Have, J. F., Green, M. M., and Howells, A. J. (1995). Molecular characterization of spontaneous mutations at the scarlet locus of Drosophila melanogaster. MGG 249 (6), 673–681. doi:10.1007/BF00418037
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi:10.1093/nar/22.22.4673
Tripoli, G., D'Elia, D., Barsanti, P., and Caggese, C. (2005). Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Genome Biol. 6 (2), R11. doi:10.1186/gb-2005-6-2-r11
Vieira, F. G., and Rozas, J. (2011). Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490. doi:10.1093/gbe/evr033
Visscher, M. O. (2017). Skin color and pigmentation in ethnic skin. Facial Plastic Surg. Clin. N. Am. 25 (1), 119–125. doi:10.1016/j.fsc.2016.08.011
Walker, W. B., Roy, A., Anderson, P., Schlyter, F., Hansson, B. S., and Larsson, M. C. (2019). Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genom 20 (1), 428. doi:10.1186/s12864-019-5815-x
Wang, H., Wu, H., Liu, W., and Jin, L. (2000). Skin reflectance in the han and Tibetan nationality in China. Chin. J. Dermatology 33 (4), 257–258.
Wanner, K. W., Isman, M. B., Feng, Q., Plettner, E., and Theilmann, D. A. (2005). Developmental expression patterns of four chemosensory protein genes from the Eastern spruce budworm, Chroistoneura fumiferana. Insect Mol. Biol. 14 (3), 289–300. doi:10.1111/j.1365-2583.2005.00559.x
Wasmeier, C., Romao, M., Plowright, L., Bennett, D. C., Raposo, G., and Seabra, M. C. (2006). Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 175 (2), 271–281. doi:10.1083/jcb.200606050
Wittkopp, P. J., Carroll, S. B., and Kopp, A. (2003a). Evolution in black and white: Genetic control of pigment patterns in Drosophila. TRENDS Genet. 19 (9), 495–504. doi:10.1016/S0168-9525(03)00194-X
Wittkopp, P. J., Williams, B. L., Selegue, J. E., and Carroll, S. B. (2003b). Drosophila pigmentation evolution: Divergent genotypes underlying convergent phenotypes. PNAS 100 (4), 1808–1813. doi:10.1073/pnas.0336368100
Xu, W., Zhang, H., Liao, Y., and Papanicolaou, A. (2021). Characterization of sensory neuron membrane proteins (SNMPs) in cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae). Insect Sci. 28 (3), 769–779. doi:10.1111/1744-7917.12816
Yan, L. (2006). “Studies of taxonomy, geographic distribution in Gynaephora genus and life-history strategies on Gynaephora menyuanensis,” (Lanzhou: Lanzhou University). (doctor).
Yang, L. (2021). “Study on positional cloning and molecular mechanism of the fourth Brown egg (b-4) in the silkworm Bombyx mori,” (China: Southwest University Chongqing). (master).
Yu, L., Wang, G. D., Ruan, J., Chen, Y. B., Yang, C. P., Cao, X., et al. (2016). Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 48 (8), 947–952. doi:10.1038/ng.3615
Yuan, M. L., Zhang, Q. L., Zhang, L., Jia, C. L., Li, X. P., Yang, X. Z., et al. (2018). Mitochondrial phylogeny, divergence history and high-altitude adaptation of grassland caterpillars (Lepidoptera: Lymantriinae: Gynaephora) inhabiting the Tibetan Plateau. Mol. Phylogenet. Evol. 122, 116–124. doi:10.1016/j.ympev.2018.01.016
Zhang, J., Bisch-Knaden, S., Fandino, R. A., Yan, S., Obiero, G. F., Grosse-Wilde, E., et al. (2019). The olfactory coreceptor IR8a governs larval feces-mediated competition avoidance in a hawkmoth. PNAS 116 (43), 21828–21833. doi:10.1073/pnas.1913485116
Zhang, L., Zhang, Q. L., Wang, X. T., Yang, X. Z., Li, X. P., and Yuan, M. L. (2017a). Selection of reference genes for qRT-PCR and expression analysis of high-altitude-related genes in grassland caterpillars (Lepidoptera: Erebidae: Gynaephora) along an altitude gradient. Ecol. Evol. 7 (21), 9054–9065. doi:10.1002/ece3.3431
Zhang, Q. L., and Yuan, M. L. (2013). Research status and prospect of grassland caterpillars (Lepidoptera: Lymantriidae). Pratacultural Sci. 30, 638–646.
Zhang, Q. L., Zhang, L., Zhao, T. X., Wang, J., Zhu, Q. H., Chen, J. Y., et al. (2017b). Gene sequence variations and expression patterns of mitochondrial genes are associated with the adaptive evolution of two Gynaephora species (Lepidoptera: Lymantriinae) living in different high-elevation environments. Gene 610, 148–155. doi:10.1016/j.gene.2017.02.014
Zhang, Y. N., Li, J. B., He, P., Sun, L., Li, Z. Q., Fang, L. P., et al. (2016). Molecular identification and expression patterns of carboxylesterase genes based on transcriptome analysis of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). J. Asia-Pac. Entomol. 19 (4), 989–994. doi:10.1016/j.aspen.2016.07.020
Zhang, Y. N., Xia, Y. H., Zhu, J. Y., Li, S. Y., and Dong, S. L. (2014). Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker). J. Chem. Ecol. 40 (5), 439–451. doi:10.1007/s10886-014-0433-1
Zhang, Y. X., Wu, H., and Yu, L. (2021). Progress on coat color regulation mechanism and its association with the adaptive evolution in mammals. Hered. (Beijing) 43 (2), 118–133. doi:10.16288/j.yczz.20-390
Keywords: grassland caterpillars, Lymantriinae, RNA-Seq, Qinghai-Tibetan Plateau, high-altitude adaptation, pigment-associated genes, olfactory-related genes
Citation: Zhao J-R, Hu S-Y, Zhang L-J, Zhang L, Yang X-Z and Yuan M-L (2023) Differential gene expression patterns between the head and thorax of Gynaephora aureata are associated with high-altitude adaptation. Front. Genet. 14:1137618. doi: 10.3389/fgene.2023.1137618
Received: 04 January 2023; Accepted: 21 March 2023;
Published: 18 April 2023.
Edited by:
Chao Tong, University of Pennsylvania, United StatesCopyright © 2023 Zhao, Hu, Zhang, Zhang, Yang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming-Long Yuan, eXVhbm1sQGx6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.