- 1Department of Plant Pathology, The University of Agriculture Faisalabad, Faisalabad, Pakistan
- 2Centre of Agricultural Biochemistry and Biotechnology, The University of Agriculture Faisalabad, Faisalabad, Pakistan
- 3Institute of Horticultural Sciences, The University of Agriculture Faisalabad, Faisalabad, Pakistan
- 4Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Department of Plant Pathology, University of California, Davis, CA, United States
Dalbergia sissoo is one of the most economically important trees in forestry, agroforestry, and horticulture. This tree species is severely threatened by dieback. Widespread dieback outbreaks and infestations have drastically destroyed billions of D. sissoo trees. Hence, we attempted to resolve the dieback etiology through phylogenomics associated with D. sissoo mortality. The Ceratocystis species was evaluated using morphologically investigated fungal isolates collected from dieback-affected tissue plants. Based on the symptomatology, we have differentiated dieback from Fusarium wilt and concluded that the Ceratocystis fimbriata sensu lato complex is causing shisham dieback in Pakistan. As the Ceratocystis species complex is a cryptic species complex, we used genomics and phylogenetic analysis for deciphering its evolutionary hierarchical order. The pathogen’s operational taxonomy was unlocked with the help of phylogenomics, and it was discovered that isolates from D. sissoo represent a species distinct from the other species in the C. fimbriata sensu lato species complex. The name Ceratocystis dalbergicans sp. nov. has been given to the fungus causing dieback disease in D. sissoo.
1 Introduction
Dalbergia sissoo (Shisham) is a member of the eukaryotic kingdom Plantae, phylum Spermatophyta, subphylum Angiospermae, class Dicotyledonae, order Fabales, family Fabaceae, subfamily Faboideae, and genus Dalbergia (Ijaz et al., 2021a). It is a moisture-loving, fast-growing, medium-to large-sized deciduous tree that grows best on alluvial soils at a pH between 5.0 and 7.7 (Kumar and Khurana, 2016; Ijaz et al., 2021b). It is one of the trees with the greatest economic value in forestry, agroforestry, and horticulture (Ijaz et al., 2018). In addition to its commercial importance, D. sissoo exhibits medicinal and industrial uses: it is a source of fuel, alkaloids, fibers, neoflavonoids, resins, and tannins (Ijaz et al., 2021a), and is also used as a landscaping tree (Kumar and Khurana, 2016; Ijaz and Ul Haq, 2021).
Dieback is a commonly found disease in D. sissoo in Bangladesh, India, Nepal, and Pakistan (Ijaz et al., 2019; Naqvi et al., 2019; Ul Haq et al., 2021). In the past 100 years, widespread dieback outbreaks and infestations have drastically reduced D. sissoo densities and killed billions of D. sissoo trees (Webb and Hossain, 2005; Rehman et al., 2012; Naqvi et al., 2019; Ul Haq et al., 2021). Symptoms of D. sissoo dieback disease include yellowing, drying, and defoliation of leaves and branches; partial or complete wilting of the crown leading to yellowing of the whole plant; thinning of leaves and crown; internal stem and root browning; chlorosis; necrosis; top dieback; dieback of branches at initial and lateral stages; gummosis; vascular discoloration; and bark splitting (Poussio et al., 2010; Mukhtar et al., 2014b; Latif et al., 2021). Researchers have made several attempts to find out the pathological cause of the disease since its identification. They have documented many fungal pathogens that cause dieback in D. sissoo and proposed management strategies. Despite a long history of pathological research studies on D. sissoo dieback, the infection persists to this day in D. sissoo plantations with a varying frequency of incidence and severity. This indicates that further research is needed to explore the pathological cause of the disease by employing conventional and molecular pathological techniques (Ijaz et al., 2020). Numerous studies attempted to elucidate the biology of D. sissoo dieback (Hassan et al., 2021). Several fungal pathogens, including Botryodiplodia theobromae, Fusarium spp., Ganoderma lucidum, Phytophthora cinnamomi, and Ceratocystis, have been linked to dieback and mortality on D. sissoo plantations. Although some researchers have discussed managing D. sissoo dieback in the Asian subcontinent by using host resistance genes or chemical control, their work has focused primarily on F. solani as the causal agent (Dayaram et al., 2003; Javaid, 2008; Ul Haq and Ijaz, 2020). However, C. fimbriata was reported a decade ago as a leading cause of D. sissoo mortality in Pakistan (Al Adawi et al., 2009; Poussio et al., 2010; Ul Haq et al., 2019). There is a clear association between Ceratocystis and dieback in D. sissoo, although the specific pathogen that causes dieback in D. sissoo has not been determined. Identifying the etiology and overcoming the disease has proven challenging and needs to be addressed. Hence, we completed Koch’s postulates with isolates of Fusarium solani and the Ceratocystis fimbriata sensu lato species complex (Ul Haq et al., 2019) from symptomatic D. sissoo sampled from 117 sites across Pakistan. We observed the distinctive symptoms. Based on the symptomology, we have differentiated the dieback from Fusarium wilt and concluded that a species of Ceratocystis fimbriata sensu lato complex is causing shisham dieback in Pakistan.
This research provided a scientific understanding of D. sissoo dieback disease by focusing on the emerging pathogen, Ceratocystis, associated with D. sissoo mortality. This research study was conducted under CAS-PARB Project No. 952. In this study, the etiology of dieback was resolved through phylogenomics. Seven genetic loci were sequenced for phylogenetic analyses to decipher hierarchy, including the internal transcribed spacer (ITS), the translation elongation factor1-alpha (TEF1-α), the second-largest subunit of RNA polymerase II (RPBII), calmodulin (CAL), the guanine nucleotide-binding protein subunit beta-like protein (MS204), a subunit of mini-chromosome maintenance proteins (MCM7), and tubulin (β-tubulin). The Ceratocystis isolates were discovered by phylogenomics analysis to be a novel species in the Ceratocystis species complex that causes dieback disease in D. sissoo.
2 Materials and methods
2.1 Genomic analysis for unlocking the Ceratocystis taxonomy
2.1.1 DNA extraction
We isolated the DNA from mycelial cultures of Ceratocystis isolates using a genomic DNA extraction kit GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, USA) and monitored the integrity of the extracted DNA through agarose gel electrophoresis. The isolated DNA samples were quantified at an absorbance of 260/280 nm by UV-visible NANODROP (8,000 Spectrophotometer, Thermo Fisher Scientific).
2.1.2 PCR analysis
We explored the taxonomy of Ceratocystis isolates by unlocking the seven genetic loci: the internal transcribed spacer (ITS), the translation elongation factor 1-alpha (TEF1-α), the second-largest subunit of RNA polymerase II (RPBII), calmodulin (CAL), the guanine nucleotide-binding protein subunit beta-like protein (MS204), a subunit of minichromosome maintenance proteins (MCM7), and tubulin (β-tubulin) (Table 1). The partial region amplification of these loci using their respective sequence-specific primer pairs in the PCR analysis was carried out in the VeritiTM 96-Well Fast Thermal Cycler by Applied Biosystems. The PCR products were electrophoresed through high-resolution agarose (ACTGene, USA), eluted using the FavorPrep Gel Purification Kit (Favorgen Biotech Corporation, Taiwan), and then cloned into pTZ57R/T (InsTAcloneTM PCR cloning kit).
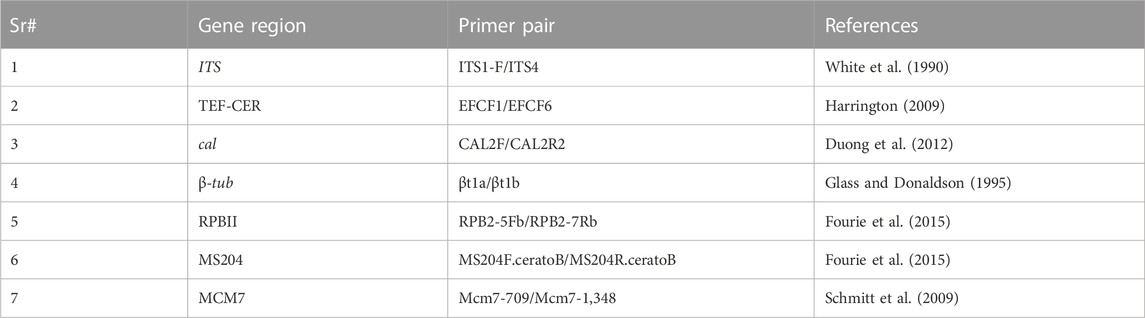
TABLE 1. Primers used for phylogenetic analysis at different loci (ITS, TEF1-α, RPBII, CAL, MS204, MCM7, and β-tubulin).
2.1.3 DNA sequencing
The sequencing of the cloned fragments was executed bidirectionally by Eurofins Genomics DNA sequencing services, United States. High-quality (HQ) sequences were obtained using the sequence alignment editor, BioEdit version 7.2.6. The DNASTAR Lasergene v. 7.1.0 SeqMan Pro (SeqManTMII) software package was used for generating consensus sequences before their deposit in GenBank to get accession numbers.
2.2 Phylogenomic studies
2.2.1 Phylogenetic analysis
For phylogenetic comparison, we supplemented the generated sequences of seven genic regions with available sequences from the NCBI database, based on their homology and literature (Supplementary Table S1). The ClustalW program was used to perform multiple sequence alignments (MSAs) of each individual dataset. The concatenated dataset from the aligned data of individual partitions was generated with Geneious software version 4.8.5, which was also used to check the quality of the alignments. Alignments were deposited in the nexus file in TreeBASE, which was generated using Mesquite version 3.51. In a phylogenetic analysis of a concatenated dataset of seven loci, Maximum Parsimony (MP) analysis using PAUP* version 4.0a161 software was employed for tree construction. Maximum-parsimony genealogies were constructed by selecting the bootstrap method in the heuristic search with 1,000 replicates, random stepwise addition for ten replicates, tree-bisection-reconstruction (TBR) as the branch-swapping algorithm with a reconnection limit (8), branches collapsing when the maximum branch length was zero, gaps being treated as a new state (5th base) with the “MulTrees” option in effect, and characters being of unord-type with equal weight. Different metrics, including tree length (TL), homoplasmy index, consistency index (CI), rescaled consistency index (RC), and retention index (RI), were also computed in the MP analysis.
2.2.2 Coalescent species tree method
For validating the concatenated dataset-based phylogenetic analysis, a coalescent species tree approach was implemented to construct a species tree by employing *BEAST software version 2.5.0. The BEAST (Bayesian Evolutionary Analysis for Sampling Tree) input files were generated through the BEAUti (Bayesian Evolutionary Analysis Utility) program under a non-standard template, StarBEAST, to set up an evolutionary model and an MCMC run to generate XML files importing aligned individual dataset files (input files) in NEX format. A linear and-constant root model was implemented as a multi-species coalescent model; however, Yule model selection as species tree priors was made to specify speciation time distribution. The maximum clade credibility species tree was constructed by TreeAnnotator version 2.3.0 with 10% burin and a 0.5 posterior probability limit to identify a tree that denotes the best posterior distribution. The Figtree software was used to visualize the species tree.
3 Results
We performed phylogenomic analysis on nine fungal isolates collected from diseased tissues of dieback-affected Dalbergia sissoo plants. These were selected for phylogenetic analysis using the findings of multilocus sequence typing (MLST) analysis of twenty-three isolates morphologically identified as the Ceratocystis fimbriata sensu lato species complex, a complex of cryptic species (Ul Haq et al., 2019). The generated sequences of seven loci were lodged in the NCBI database with GenBank accession numbers described in Supplementary Table S1.
A Maximum Parsimony (MP) analysis was executed on a concatenated dataset of seven loci for estimating species boundaries by generating a maximum parsimonious (MP) tree through PAUP* software. The majority rule bootstrap (50%) consensus tree was based on 3,590 parsimony-informative characters (PIC) with a tree length of 6,723. The computed matrices of this analysis were 0.2829 homoplasmy index (HI), 0.7289 consistency index (CI), 0.8910 rescaled consistency index (RC), and 0.6494 retention index (RI). The MP tree identified the FMB isolates (collected fungal isolates from the diseased tissues of dieback-affected D. sissoo plants) as a separate and distinctly diverged clade with 99.47% bootstrap support and a posterior probability (PP) support value of 0.99. A posterior probability value > 0.94 (94%) is considered to be strong support for separate taxa. The 94% speciation probability value is considered strong support for a speciation event (Figure 1). However, validating the MP analysis (Concatenation-based phylogenetic analysis), we employed the coalescent species tree method (Coalescent-based phylogenetic analysis) for the first time in Ceratocystis species delimitation studies. BEAST (Bayesian Evolutionary Analysis for Sampling Tree) analysis was made by executing *BEAST software for species tree generation. The species tree revealed strong concurrence with the most parsimonious phylogenetic tree and displayed clear divergence of these fungal isolates from other Ceratocystis species (Figure 2). Hence, considering the MP tree and species tree, we suggested these isolates as a new Ceratocystis species causing dieback in D. sissoo and named them Ceratocystis dalbergicans sp. nov.
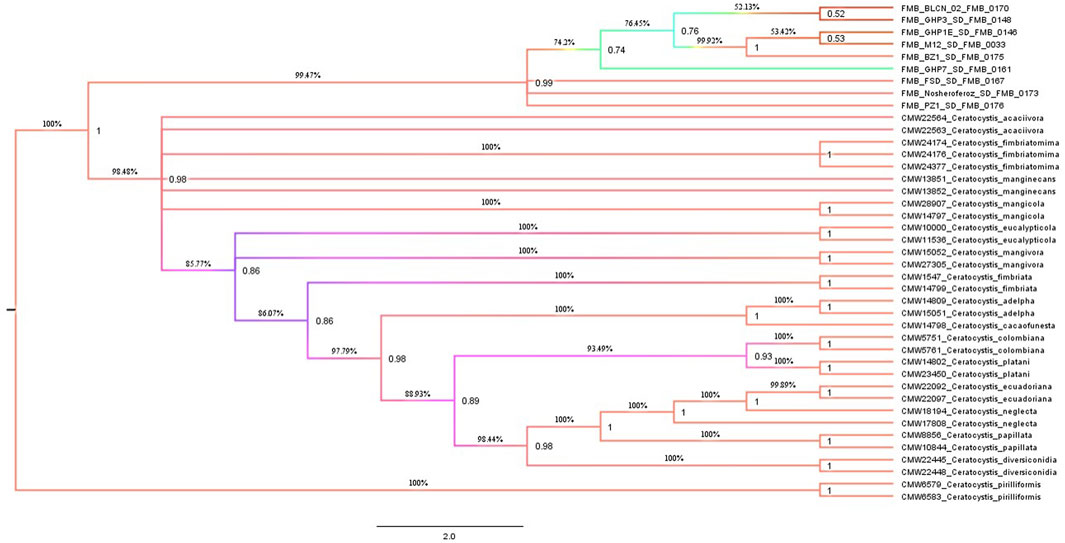
FIGURE 1. Maximum-parsimony genealogies were constructed by selecting the bootstrap method in heuristic search with 1,000 replicates, random stepwise addition for ten replicates, tree-bisection-reconstruction (TBR) as the branch swapping algorithm with a reconnection limit (8), branches collapsed when maximum branch length was zero, with the “MulTrees” option in effect.
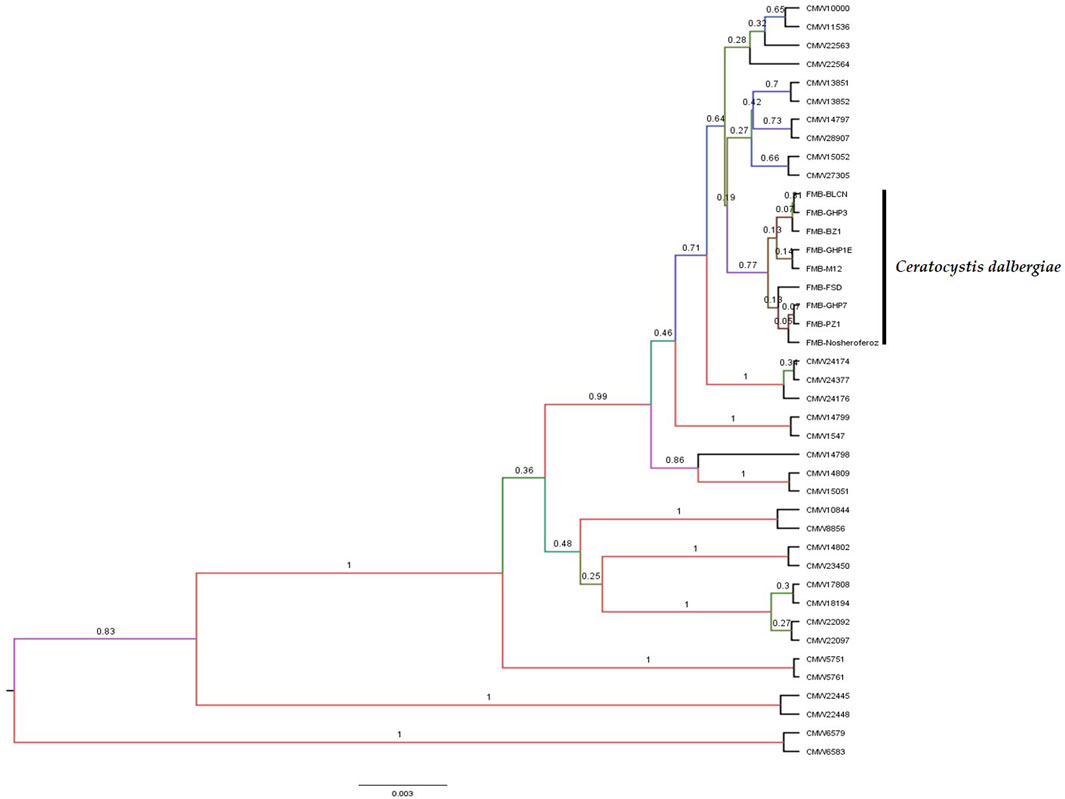
FIGURE 2. Species tree generated performing a BEAST analysis revealed that Ceratocystis isolates from Dalbergia sissoo dieback-affected tissues are more diverse than other Ceratocystis species and have been proven to be a new Ceratocystis species that causes dieback in Dalbergia sissoo. The scale bar depicts the expected number of changes per site.
4 Taxonomy
Ceratocystis dalbergicans I.U. Haq, S. Ijaz, I. A. Khan and M. Z. Latif, sp. nov; basionym: Ceratocystis dalbergiae; MycoBank 841380. NCBI:txid2870506.
Material examined: Pakistan, from living stems, and branches of the shisham tree, February and March 2017 and 2018, I.U. Haq (FMB H 13.1, Holotype, ex-type culture FMBCC 13.1).
Note: The strain is preserved in a metabolically inactive state.
Etymology: Named after the host (Shisham, in English; Dalbergia sissoo, botanical name) from which these were first isolated.
5 Discussion
Dalbergia sissoo dieback disease is a national issue in Pakistan and other South Asian countries (Dayaram et al., 2003; Webb and Hossain, 2005; Shakya and Lakhey, 2007; Mukhtar et al., 2014a). No data exist that concern the distribution, prevalence of dieback disease, and losses associated with Ceratocystis in Pakistan. However, reports are available on other fungal pathogens from limited areas of Punjab or Sindh province, which do not represent the overall situation of this issue (Bajwa et al., 2003; Khan et al., 2004; Rajput et al., 2010).
According to their morphology, the fungal isolates from dieback-affected D. sissoo trees were identified as possible Ceratocystis fimbriata sensu lato members (a cryptic species complex). Therefore, these isolates were characterized using primary and secondary DNA barcodes. The ITS region was used as the primary DNA barcode because this region has been used to distinguish species of the Ceratocystis genus (Van Wyk et al., 2009). However, two ITS types were identified Ceratocystis mangenicans suggested attentiveness while using this region for species delineation in the Ceratocystis species complex (Al Adawi et al., 2013), despite a phylogeny based on a combined dataset including TEF1-α and β tubulin strongly supported a speciation event (Tarigan et al., 2011; Al Adawi et al., 2013). Fourie et al. (2015) documented this combined tree gene phylogeny, which showed no well-supported species discrimination in the case of other Ceratocystis species and suggested βt1, MS204, and RPB11 as strong markers in combination to delineate the Ceratocystis species complex. Therefore, we used seven genetic loci: the internal transcribed spacer (ITS), the translation elongation factor1-alpha (TEF1-α), the second-largest subunit of RNA polymerase II (RPBII), calmodulin (CAL), guanine nucleotide-binding protein subunit beta-like protein (MS204), a subunit of mini-chromosome maintenance proteins (MCM7), and tubulin (β-tubulin). The combined seven gene phylogeny revealed support for species delineation for these Ceratocystis isolates as a new taxon of this complex.
Data presented in this study supports the hypothesis that Ceratocystis sp. is the primary fungal pathogen responsible for Pakistan’s widespread D. sissoo dieback disease. Our research findings are supported by other studies that conclude D. sissoo mortality in Pakistan is due to Ceratocystis sp. (Al Adawi et al., 2009; Poussio et al., 2010; Ul Haq et al., 2019). However, other scientists claimed that Botryodiplodia theobromae (Khan et al., 2004; Rehman et al., 2012), Fusarium spp. (Dayaram et al., 2003; Shakya and Lakhey, 2007; Rajput et al., 2008), Ganoderma lucidum (Parker, 1915; Troup, 1921; Sharma et al., 2000), and Phytophthora cinnamomi (Gill et al., 2002; Rehman et al., 2012) caused D. sissoo dieback. On the contrary, we have established that the novel species C. dalbergicans is the primary cause of dieback disease in D. sissoo in Pakistan.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceptualization, IH, SI, and IK; data curation, IH and SI; formal analysis, IH, SI, and ML; funding acquisition, SI; investigation, IH, SI, and ML; methodology, IH, SI, IK, and SK; project administration, SI and IK; resources, IK and HA; software, SI and ML; supervision, IH and SI; validation, IH, SI, and ML; writing–original draft, IH and SI; writing, review and editing, IH, SI, IK, HA, and SK.
Funding
Researchers Supporting Project number (RSP2023R123), King Saud University, Riyadh, Saudi Arabia.
Acknowledgments
Authors would like to extend their sincere appreciation to the Researchers Supporting Project number (RSP2023R123), King Saud University, Riyadh, Saudi Arabia
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1136688/full#supplementary-material
References
Al Adawi, A. O., Barnes, I., Al Jahwari, A. A., Deadman, M. L., Wingfield, B. D., and Wingfield, M. J. (2009). “Discovery of a Ceratocystis sp. associated with wilt disease of two native leguminous tree hosts in Oman and Pakistan,” in Australian Plant Pathology Society meeting (APPS 2009)'Plant health management: An integrated approach.
Al Adawi, A. O., Barnes, I., Khan, I. A., Al Subhi, A. M., Al Jahwari, A. A., Deadman, M. L., et al. (2013). Ceratocystis manginecans associated with a serious wilt disease of two native legume trees in Oman and Pakistan. Australas. Plant Pathol. 42 (2), 179–193.
Bajwa, R., Javaid, A., and Shah, M. B. (2003). Extent of shisham (dalbergia sissoo roxb.) decline in sialkot, gujranwala, lahore and sargodha districts. Mycopath 1 (1), 1–5.
Dayaram, M. K., Sharma, S., and Chaturvedi, O. P. (2003). Shisham mortality in Bihar extent and causes. Indian Phytopathol. 56 (4), 384–387.
Duong, T. A., De Beer, Z. W., Wingfield, B. D., and Wingfield, M. J. (2012). Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia 104 (3), 715–732. doi:10.3852/11-109
Fourie, A., Wingfield, M. J., Wingfield, B. D., and Barnes, I. (2015). Molecular markers delimit cryptic species in Ceratocystis sensu stricto. Mycol. Prog. 14 (1), 1020–1028. doi:10.1007/s11557-014-1020-0
Gill, M. A., Imtiaz, A., Khan, A. U., Muhamamd, A., Shaukat, A., Rafique, R. M., et al. (2002). “Phytophthora cinnamomi?-a cause of shisham decline in Punjab, Pakistan. InIntegrated plant disease management”. In Proceedings of 3rd National Conference of Plant Pathology, Islamabad, 1-3 Oct. 2002 (NARC), 33–37.
Glass, N. L., and Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61 (4), 1323–1330. doi:10.1128/aem.61.4.1323-1330.1995
Harrington, T. C. (2009). “The genus Ceratocystis. Where does the oak wilt fungus fit,” in Proceedings of the 2nd national Oak Wilt symposium 2009, Austin, Texas (USDA Forest Service, Forest Health Protection), 21–35.166.
Hassan, M., Ijaz, S., Sarwar, S., and Khan, A. H. (2021). “Genetics and breeding of dalbergia sissoo,” in Dalbergia sissoo: Biology, ecology and sustainable agroforestry (CRC Press), 103–114.
Ijaz, S., Babar, M., Razzaq, H. A., and Nasir, B. (2020). “Transgenic approaches in plants: Strategic control for disease management,” in Plant disease management strategies for sustainable agriculture through traditional and modern approaches (Springer Nature), 187–215.
Ijaz, S., Naheed, R., Razzaq, H. A., and Nasir, B. (2021a). “Physiological and biochemical dynamics of dalbergia sissoo,” in Dalbergia sissoo: Biology, ecology and sustainable agroforestry (CRC Press), 27–52.
Ijaz S., and Ul Haq I. (Editors) (2021). Dalbergia sissoo biology, ecology and sustainable agroforestry. 1st Ed. (CRC Press).
Ijaz, S., Ul Haq, I., and Babar, M. (2021b). “Reproductive biology, botany, and taxonomical description of dalbergia sissoo,” in Dalbergia sissoo: Biology, ecology and sustainable agroforestry (CRC Press), 3–12.
Ijaz, S., Ul Haq, I., Razzaq, H. A., and Babar, M. (2018). Assessment of population genetics of shisham (Dalbergia sissoo) based on genetic structure and diversity analyses. Int. J. Biosci. 13, 209–2022. doi:10.12692/ijb/13.3.209-222
S., Ijaz, I., Ul Haq, H. A., Razzaq, B., Nasir, and M., Babar (2019). ISSR-based population genetics study for tagging a diverse population of shisham (Dalbergia sissoo) in Pakistan. Appl. Ecol. Environ. Res. 17, 5851–5861. doi:10.15666/aeer/1703_58515861
Javaid, A. (2008). Research on shisham (Dalbergia sissoo Roxb.) decline in Pakistan-a review. Pak. J. Phytopathol. 20, 134–142.
Khan, S. H., Idrees, M., Muhammad, F. A., Mahmood, A. R., and Zaidi, S. H. (2004). Incidence of shisham (Dalbergia sissoo Roxb.) decline and in vitro response of isolated fungus spp. to various fungicides. Int. J. Agric. Biol. 6, 611–614.
Kumar, N., and Khurana, S. P. (2016). Biomanagement of wilting of a valuable timber and medicinal plant of Shisham (Dalbergia sissoo Roxb.)-A review. Int. J. Curr. Microbiol. Appl. Sci. 5(1), 32–54. doi:10.20546/ijcmas.2016.501.004
Latif, M. Z., Ul Haq, I., Ijaz, S., Habib, A., and Khan, I. A. (2021). Assessment of the distribution, incidence, and severity of Shisham (Dalbergia sissoo) dieback disease in Pakistan. Pak. J. Agri. Sci. 58, 1825–1532.
Mukhtar, I., Bajwa, R., Nasim, G., and Fy, H. (2014b). Evaluation of genetic diversity among phytopathogenic isolates of Fusarium solani complex causing shisham dieback disease in Pakistan. J. Anim. Plant. Sci. 1, 1728.
Mukhtar, I., Bajwa, R., and Nasim, G. (2014a). Trees survival exposed to dieback disease implies evolutionary modulation resistance in shisham (Dalbergia sissoo Roxb.) in various agro ecological zones of Punjab (Pakistan). Pak. J. Phytopathol. 26 (2), 289–300.
Naqvi, S. A., Mushtaq, S., Malik, M. T., ur Rehman, A., Fareed, S., Zulfiqar, M. A., et al. (2019). Factors leading towards dalbergia sissoo decline (syndrome) in Indian sub-continent: A critical review and future research agenda. Pak. J. Agric. Res. 32(2):302. doi:10.17582/journal.pjar/2019/32.2.302.316
Parker, R. N. (1915). A forest flora for the Punjab with Hazara and Delhi. Punjab: Superintendent Government Printing.
Poussio, G. B., Kazmi, M. R., Akem, C., and Fateh, F. S. (2010). First record of Ceratocystis fimbriata associated with shisham (Dalbergia sissoo) decline in Pakistan. Australas. Plant Dis. Notes. 5 (1), 63–65. doi:10.1071/DN10023
Rajput, N. A., Pathan, M. A., Jiskani, M. M., Rajput, A. Q., and Arain, R. R. (2008). Pathogenicity and host range of Fusarium solani (mart.) sacc., causing dieback of shisham (Dalbergia sissoo roxb.). Pak. J. Bot. 40 (6), 2631–2639.
Rajput, N. A., Pathan, M. A., Rajput, A. Q., Jiskani, M. M., Lodhi, A. M., Rajput, S. A., et al. (2010). Isolation of fungi associated with shisham trees and their effect on seed germination and seedling mortality. Pak. J. Bot. 42 (1), 369–374.
Rehman, A., Sahi, S. T., Khan, M. A., and Mehboob, S. (2012). Fungi associated with bark, twigs and roots of declined shisham (dalbergia sissoo roxb.) trees in Punjab, Pakistan. Pak. J. Phytopathol. 24 (2), 152–158.
Schmitt, I., Crespo, A., Divakar, P. K., Fankhauser, J. D., Herman-Sackett, E., Kalb, K., et al. (2009). New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia-Molecular Phylogeny Evol. Fungi 23 (1), 35–40. doi:10.3767/003158509X470602
Shakya, D. D., and Lakhey, P. B. (2007). Confirmation of Fusarium solani as the causal agent of dieback of Dalbergia sissoo in Nepal. Plant Pathol. 56 (6), 1041. doi:10.1111/j.1365-3059.2007.01637.x
Sharma, M. K., Singal, R. M., and Pokhriyal, T. C. (2000). Dalbergia sissoo in India. Dalbergia sissoo in India, 5–16.
Tarigan, M., Roux, J., Van Wyk, M., Tjahjono, B., and Wingfield, M. J. (2011). A new wilt and dieback disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South Afr. j.bot. 77 (2), 292–304. doi:10.1016/j.sajb.2010.08.006
Ul Haq, I., and Ijaz, S. (2020). “History and recent trends in plant disease control: An overview,” in Plant disease management strategies for sustainable agriculture through traditional and modern approaches (Springer Nature), 1–13.
Ul Haq, I., Ijaz, S., and Latif, M. Z. (2021). Diseases of dalbergia sissoo: Etiology and integrated management of economically important diseases. InDalbergia sissoo CRC Press, 63.
Ul Haq, I., Ijaz, S., and Latif, M. Z. (2019). Multilocus sequence typing (MLST) based genetic variation analysis of shisham dieback associated strains of Ceratocystis fimbriata sensu lato species complex in Pakistan. Appl. Ecol. Environ. Res. 17 (5), 12573–12582. doi:10.15666/aeer/1705_1257312582
Van Wyk, M., Wingfield, B. D., Clegg, P. A., and Wingfield, M. J. (2009). Ceratocystis larium sp. nov, a new species from Styrax benzoin wounds associated with incense harvesting in Indonesia. Persoonia-Molecular Phylogeny Evol. Fungi. 22 (1), 75–82. doi:10.3767/003158509X439076CRC Press
Webb, E. L., and Hossain, S. M. (2005). Dalbergia sissoo mortality in Bangladesh plantations: Correlations with environmental and management parameters. For. Ecol. Manag. 206(1-3), 61–69. doi:10.1016/j.foreco.2004.10.055
Keywords: novel species, fungi, dieback, Ceratocystis, Dalbergia sissoo, conservation
Citation: Haq IU, Ijaz S, Latif MZ, Khan IA, Ali HM and Kaur S (2023) Phylogenomics resolves the etiology of dieback disease and deciphers Ceratocystis dalbergicans sp.nov., causal agent of Dalbergia sissoo decline. Front. Genet. 14:1136688. doi: 10.3389/fgene.2023.1136688
Received: 03 January 2023; Accepted: 27 February 2023;
Published: 14 March 2023.
Edited by:
Feng Zhang, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Muhammad Imtiaz, National Institute for Biotechnology and Genetic Engineering, PakistanSajid Shokat, Nuclear Institute for Agriculture and Biology (NIAB), Pakistan
Copyright © 2023 Haq, Ijaz, Latif, Khan, Ali and Kaur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siddra Ijaz, c2lkZHJhaWphemtoYW5AeWFob28uY29t
 Imran Ul Haq
Imran Ul Haq Siddra Ijaz
Siddra Ijaz Muhammad Zunair Latif1
Muhammad Zunair Latif1 Hayssam M. Ali
Hayssam M. Ali