- 1School of Life Sciences, Zhengzhou University, Zhengzhou, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
Hexokinase (HXK) is a bifunctional enzyme involved in carbohydrate metabolism and sugar signal sensing. HXK gene family has been extensively discussed in many species, while the detailed investigations of the family in Glycine max have yet to be reported. In this study, 17 GmHXK genes (GmHXKs) were identified in the G. max genome and the features of their encoded proteins, conserved domains, gene structures, and cis-acting elements were systematically characterized. The GmHXK2 gene isolated from G. max was firstly constructed into plant expression vector pMDC83 and then transformed with Agrobacterium tumefaciens into Arabidopsis thaliana. The expression of integrated protein was analyzed by Western Blotting. Subcellular localization analysis showed that the GmHXK2 was located on both vacuolar and cell membrane. Under salt stress, seedlings growth was significantly improved in Arabidopsis overexpressing GmHXK2 gene. Furthermore, physiological indicators and expression of salt stress responsive genes involved in K+ and Na+ homeostasis were significantly lower in GmHXK2-silenced soybean seedlings obtained by virus-induced gene silencing (VIGS) technique under salt stress compared with the control plants. Our study showed that GmHXK2 gene played an important role in resisting salt stress, which suggested potential value for the genetic improvement of abiotic resistant crops.
1 Introduction
Sugars play an important role during plant life, serving not only as the sources of energy and carbon structural components for growth and development but also as signaling molecules involved in many physiological processes such as germination (Kim et al., 2016), flowering (Zhang et al., 2020), senescence (Xiao et al., 2000), stomatal closure (Lugassi et al., 2015), and response to abiotic and biotic stresses (Sarowar et al., 2008; Granot et al., 2014; Lugassi et al., 2015). In higher plants, sucrose comprise the majority of carbohydrates produced by photosynthesis. Following synthesis, sucrose can then be directly stored and converted into hexose (glucose and fructose) by either invertase or sucrose synthase (Siemens et al., 2011; Granot et al., 2014). Afterwards, hexose is phosphorylated by hexokinase to produce hexose-6-P, which enters the glycolysis pathway and generates energy and intermediate metabolites involved in physiological activities of plants (David-Schwartz et al., 2013). HXK acts as a hexose sensor and signal in signaling networks to sense external nutrients, light, and hormones in addition to its role as a regulator of plant growth (Jang et al., 1997; Kim et al., 2013). Therefore, it has been recognized as a bifunctional enzyme and the key connecting element between sugar signaling and plant hormone signaling (Jang et al., 1997; Perata et al., 1997; Moore, 2004).
HXKs are encoded by a gene family in many plant species (Granot et al., 2014). AtHXKs genes were first isolated and identified from A. thaliana (Minet et al., 1992), revealing six members among which three proteins can phosphorylate hexoses, while the other three proteins lack catalytic activity (Karve et al., 2008). HXK gene family members have been systematically identified in various plant species. 10, 9, 4, 7, 11, 17 HXK genes members were identified respectively in Oryza sativa (Cho et al., 2006), Zea mays (Zhang et al., 2014), Lycopersicon esculentum (Kandel-Kfir et al., 2006), Manihot esculenta (Geng et al., 2017), Physcomitrella patens (Olsson et al., 2003) and Gossypium hirsutum L (Dou et al., 2022) in their genome.
Different HXKs are located in the membranes of various subcellular organelles such as the mitochondrion, chloroplast, cytoplasm, nucleus, Golgi, and vacuole (da-Silva et al., 2001; Olsson et al., 2003; Kandel-Kfir et al., 2006; Wang et al., 2014). Based on N-terminal amino acid sequences, plant HXK can be classified as Type A, Type B, or Type C (Giese et al., 2005; Karve et al., 2010). Type A HXKs has a plastid signal peptide structure containing a chloroplast transit peptide of about 30 amino acids, and it is mainly located in chloroplasts (Kandel-Kfir et al., 2006). Type B HXKs, which is mainly located in plastid (Olsson et al., 2003), contain an N-terminal hydrophobic membrane anchor domain of about 24 amino acids. Type C HXKs do not have signal peptides or a membrane anchor; they are cytosolic and seem to be only present in monocotyledonous plants and the moss P. patens (Karve et al., 2010; Cheng et al., 2011; Nilsson et al., 2011). As such, the diversity of HXK structure and subcellular localization lead to significant differences in their functions (Matschinsky et al., 2006; Riera et al., 2008).
Under salt stress, signaling substances such as ROS, ABA, Ca2+ in plants activate transcription factors through signal transduction, such as MYB, NAC, ERF, bZIP, etc., (Li et al., 2015; Wang et al., 2016; Baillo et al., 2019). The interaction between transcription factors and cis-regulatory elements promotes the expression of salt stress-responsive genes, inhibits the excessive accumulation of Na+ in plants and maintains the homeostasis of osmotic pressure. These include salt overly sensitive 1 (SOS1), high-affinity K+ transporter (HKT) and glutamate receptor-like channels (GLRs) located on the plasma membrane, sodium-hydrogen exchanger (NHX), cation/H+ exchanger (CHX and SALT3) and cyclic nucleotide-gated channels (CNGCs) located on the tonoplast and endomembranes, etc., (Plett et al., 2010; Guan et al., 2014; Ma et al., 2014). These genes are involved in the process of salt stress and regulate cation concentration to protect plants from damage caused by ion toxicity.
Soybean (Glycine. max L.) is the third most valuable plant crop worldwide as an oilseed crop (Bilal et al., 2020). However, the growth, production and quality of soybean are threatened by environment stress. With the increase in problems of soil salinity, salt stress has become one of the major stresses affecting growth and development of soybean (Leisner et al., 2017; Bilal et al., 2018). To date, little is known about any G. max HXK genes and their function in growth and development under abiotic salt stresses. In this study, we identified HXK gene family members of G. max based on the genome database. Next, we investigated the homology and phylogenetic relationship, gene structure, conserved motifs and cis-elements. The promoter sequences of most GmHXKs included many cis-elements including ABRE, ERE and LTR. It was supposed that GmHXKs were related with ABA signal pathway. Then, we assayed the expression pattern of GmHXKs under salt and drought stress, and found expression of GmHXK2 in roots was increased gradually after treatment with NaCl or drought stress for 72 h. GmHXK2 silenced plants obtained by VIGS technique were severely damaged after treatment with NaCl. We measured physiological indicators and the expression of some genes such as GmSOS1, GmSALT3, GmHKT1, GmbZIP44,etc., which were reported had a role in salt tolerance, in GmHXK2 soybean silenced plants under salt stress conditions. Moreover, the DNA sequence of GmHXK2 was cloned and constructed into the plant expression vector pMDC83, and it then transformed with Agrobacterium tumefaciens to A. thaliana. We analyzed the seed germination, plant growth and salt tolerance of homozygous T3 transgenic plants WT35 S:GmHXK2. Our study provided essential information concerning the physiological functions of GmHXK2 and an important theoretical basis for genetic engineering soybeans to better tolerate to salt stress.
2 Materials and methods
2.1 Identification of the HXK gene family members in G. max
HMMER 3.0 software was used to search the soybean genome database for genes containing the structural domain of hexokinase (PF03727 and PF00349) and 17 soybean hexokinase genes were retrieved finally. The HXK gene sequences of G. max was downloaded from Ensembl Plants (http://plants.ensembl.org/index.html). The physical and chemical parameters of the proteins, including molecular weight (MV) and theoretical isoelectric point (PI), were computed by ExPASy (http://web.expasy.org/protparam/). The presence of a chloroplast transit peptide (cTP) in the protein sequence was predicted using the ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/). The existence of transmembrane helices (TMHs) in the protein was predicted using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Chromosomal localizations of GmHXKs were mapped in Map Gene2Chromosome v2 (http://mg2c.iask.in/mg2c_v2.0/).
2.2 Multiple alignment, phylogenetic and expression pattern analysis
The sequences of the GmHXKs proteins obtained from the G. max genome were aligned using DNAMAN 7.0 software to search for conserved domains by inspection using sites present in AtHXK1 as a reference. To compare evolutionary relationships, the putative HXKs from G. max, A. thaliana, Solanum lycopersicum, O. sativa and Nicotiana tabacum were used to construct the phylogenetic tree using MEGA-X with the neighbor-joining (NJ) method and 1,000 bootstrap replicates (Kumar et al., 2018). Expression data on GmHXK gene family members at different developmental stages and in different tissues under normal conditions were downloaded from the Soybase (https://www.soybase.org/). Data on differential expression for only 14 members were eventually obtained and used for subsequent analysis.
To analyze expression pattern of soybean seedlings under salt stress, soybean seedlings were grown in a growth chamber under greenhouse conditions of 28°C under a16-h light/8-h dark cycle. Three-week-old seedlings were treated with 0.5% NaCl (salt stress) or drought treatment (10%PEG 6000). The root samples of the seedlings were collected after treatment for 2-h, 8-h, 24-h, and 72-h. Then, different samples were frozen quickly in liquid nitrogen, and stored at −80°C for RNA extraction and analysis. Total RNA was isolated using the Plant RNA Kit (CWBIO, Beijing, China), and its concentration and purity were determined by Nanodrop2000 nucleic acid analyzer (Thermo, America). First-strand cDNA was synthesized from 0.5 µg of total RNA using the HiFi-MMLV cDNA Kit (CWBIO, Beijing, China), and then used as a template for qRT-PCR analysis using gene-specific primers (Supplementary Table S1). Data analysis of RT-qPCR was performed using 2−ΔΔCT method (Livak and Schmittgen, 2001).
2.3 Gene structure, conserved motifs and cis-elements analysis
Gene structures were analyzed using Gene Structure Display Serve 2.0 (https://gsds.cbi.pku.edu. cn/) to investigate the exon-intron organizations of GmHXK genes based on their information given in the Ensembl Plants. The novel motifs of GmHXKs were searched using MEME (http://meme-suite. org/tools/meme) (Bailey et al., 2009). The parameters were set as follows: the site distribution was set to any number of repetitions (anr), the number of motifs was set to 10, and all other optional parameters remained default (He et al., 2019). The combination of gene structures, motifs, and phylogenetic tree was then generated using TBtools. In addition, the cis-acting regulatory elements in the 2000-bp genomic sequence upstream of the coding GmHXK gene sequences were investigated using the online PlantCARE databases. (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.4 Construction of plant expression vector and transformation of arabidopsis
GmHXK2 was obtained by PCR using the soybean genome DNA as a template with the primes, which are specific for GmHXK2 gene. The PCR was performed using standard conditions: initial denaturation at 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 1 min, followed by a final extension of 72°C for 5 min. The PCR products of GmHXK2 were gel purified using gel purification kit (TaKaRal MiniBEST Agarose Gel DNA Extraction Kit Ver.4.0) and used to construct the entry vector (Invitrogen, United States of America) by TOPO cloning reaction according to the manufacturer’s instructions. The entry vector containing the gene of the correct orientation and sequence was used to construct the target vector pMDC83 via the LR Clonase II enzyme mediated gateway cloning reaction according to manufacturer’s protocol. The recombinant plasmid pMDC83 with the hygomycin phosphotransferase gene under the regulation of double CaMV 35S promoter was generated, with GFP fused at the C-terminus of GmHXK2. Then, sequencing was performed to confirm the insertion into the correct gene in pMDC83. Next, the integrated vector was introduced into A. tumefaciens strains GV3101 by the liquid nitrogen freeze-thaw method. The A. tumefaciens transformation was transformed into WT Arabidopsis using the floral dipping technique. Transformed Arabidopsis plants were grown in a greenhouse under a 16/8 h light/dark cycle at 24/22°C with 70% relative humidity. Seeds harvested from the transformed plants (T0) were grown on 1/2 MS medium containing 20 mg L-1 hygromycin under the same growth conditions. Homozygous T3 progeny WT35S::GmHXK2 derived from T2 population were selected and confirmed by PCR for further salt tolerance analysis.
2.5 Molecular analysis of transgenic arabidopsis plant and analysis on subcellular localization of GmHXK2
Genomic DNA of from the putative T3 transgenic Arabidopsis was isolated and analyzed by PCR amplification using specific primers. The amplification fragments were monitored in transgenic Arabidopsis with 35S:GmHXK2-GFP in pMDC83. No transformed seedlings were used as control.
The roots from 4-week-old seedlings expressing GFP in the transgenic Arabidopsis transformed with 2 × 35S::GmHXK2-GFP in pMDC83 were monitored using confocal laser-scanning microscopes. Images were captured at an excitation of 480 nm and emission between 515 and 565 nm for GFP. The subcellular localization of GmHXK2-GFP fusion proteins was confirmed.
2.6 Western blot analysis of transgenic arabidopsis plants
WT and WT35S::GmHXK2 transgenic Arabidopsis plants were grown on 1/2 MS medium containing 20 mg L-1 hygromycin. After 15 days of cultivation, total proteins of the seedlings were extracted from WT and transgenic Arabidopsis seedlings with a buffer consisting of 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 0.2% (w/v) Triton X-100, 4% β-mercaptoethanol, 1 mM dithiothreitol (DTT), and 1% (v/v) protease inhibitor cocktail of which were then used for protein quantification using the BCA Protein Quantitative Kit (Boster). The protein samples (200 μg amounts) were electrophoresed in 8% SDS-PAGE and the gels were transferred to nitrocellulose membranes. The membranes were blocked with TBST buffer (10 mM Tris/HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween-20) supplemented with 5% non-fat milk for 2 h and incubated with primary antibodies (Anti-GFP antibody, abcam, diluted at 1:1,000) in TBST buffer with 5% BSA overnight at 4°C. Afterwards, the membranes were washed three times (10 min each) with TBST buffer and incubated with the secondary antibodies (Goat Anti-Mouse IgG H&L (HRP), abcam, dilution at 1:1,000) for 2 h. After washing three times with TBST buffer, the membranes were incubated with a chromogenic agent using the Enhanced HRP-DAB Chromogenic Substrate Kit (Boster).
2.7 Salt tolerance analysis of transgenic arabidopsis plants at the germination and seedling stage
Seeds of WT and WT35S::GmHXK2 transgenic Arabidopsis plants of homozygous T3 generation were sterilized using 75% ethanol and 5% sodium hypochlorite for 1 min and 10 min, respectively. The seeds were washed six to ten times in sterilized water and were sown on half-strength MS (1/2 MS) medium with 0 and 100 mM NaCl. They were then transferred to a culture room after 3 days of vernalization at 4°C. Finally, root lengths and fresh weight of the germinated seeds were measured after planting for 7 days to detect salt tolerance in transgenic Arabidopsis overexpressing GmHXK2.
To analyze salt tolerance of transgenic Arabidopsis plants at the seedlings stage, treatment of plant samples were as followed. After the seeds were sterilized, they were sown on MS medium after 3 days of vernalization at 4°C, then they were transferred to a culture room and grown for 9 days. Next, Arabidopsis seedlings were transplanted into 1/2 MS medium containing 0 mM, 100 mM and 150 mM NaCl with or without 100 mM glucose (Glc). Finally, fresh and dry weight, length of the roots, malondialdehyde, chlorophyll and proline contents of the seedlings were measured after 6 days.
2.8 Determination of chlorophyll content
Chlorophyll was extracted using ethanol as solvent (Matschinsky et al., 2006). 100 mg of leaves was pulverized with 2 mL 95% ethanol, and the sample supernatant was measured with a spectrophotometer UV-1800PC at 665 nm and 649 nm. The chlorophyll extraction solution was calculated using the following formula:
The chlorophyll content per unit fresh weight of leaf was calculated using the following formula: Chlorophyll content (mg g-1) = (CT×extraction solution volume×dilution ratio)/(fresh weight of leaves).
2.9 Determination of malondialdehyde (MDA) content
Fresh seedlings (0.1 g) were immediately homogenized with liquid nitrogen and were then mixed with 5 mL of 10% trichloroacetic acid (TCA) and centrifuged at 4,000 rpm for 10 min. The supernatant (2 mL) and 2 mL 0.6% thiobarbituric acid (TBA) were pipetted into a new tube. The mixture was incubated in a water bath at 100°C for 15 min and then cooled on ice and centrifuged at 5,000 rpm for 10 min. The absorbance of the supernatant was measured at 532 and 450 nm by spectrophotometer (Zhang et al., 2008). The MDA content was calculated using the formula: MDA (nmol g-1) = (6.45× 10−6×A532–0.56×10−6×A450)×V/W. V = volume of supernatant(L), W = weight of seedlings (g).
2.10 Determination of proline content
The proline content of Arabidopsis seedling was determined according to the method described by Jaemsaeng et al. (Zhang et al., 2008). Seedlings (0.05 g) were homogenized in 5 mL of sulphosalicylic acid (3%) and centrifuged for 10 min at 12,000 rpm. 2 mL of glacial acetic acid and 2 mL of acid ninhydrin were added to the supernatant (2.0 mL). The mixture was boiled in a water bath at 100°C for 30 min. After cooling, extraction was done with 4 mL of toluene. The absorbance was measured at 520 nm using toluene as a blank. The proline content (mg*g-1) = (Y × 5/2)/W. Y = content of proline in 2 mL supernatant. W = Weight of seedlings (g).
2.11 Determination of superoxide dismutase (SOD)
Seedings (0.1 g) were homogenized with 1 mL phosphate buffer (100 mM, ph = 7.8) and centrifuged at 12000 rpm for 30 min at 4°C. Then 0.1 mL of supernatant and phosphate buffer were added to the reaction solution (50 mM, ph = 7.8 Na3PO4,130 mM MET, 750uM NBT, 100uM EDTA, 20uM riboflavin) respectively. The reaction solution in which phosphate buffer was added served as the light control. The reaction was placed under fluorescent light for 15 min and then terminated in the dark. The absorbance value was measured at 560 nm. The SOD was calculated using the formula: SOD(U)=(ODL-ODS)*V/(0.5*ODL*VS*T*M). ODL = Absorbance value of light control. ODS = Absorbance value of samples. V = Total volume of sample extracts (ml). VS = Volume of sample extract for determination (ml). T = Light reaction time (min). M = Weight of seedings(g) (Donahue et al., 1997).
2.12 Determination of electrolyte leakage (EL)
The conductivity of the sample (0.2 g) immersed in 20 mL of deionized water for 2 h at room temperature was measured as C1. Then the sample was boiled in a water bath for 15 min and cooled to room temperature. The conductivity measured was C2. Conductivity of deionized water as a blank control (C0). The electrolyte leakage (%)=(C1-C0)/(C2-C0) (Jungklang et al., 2017).
2.13 Construction of VIGS vectors and sprout infiltration for silenced plants
The specific fragments from CDS of GmHXK2 and GmPDS were amplified using primers. The PCR products and the virus vector pTRV2 were digested with XbaI and BamHI(TAKARA),respectively. The products were ligated to obtain pTRV2-HXK2 and pTRV2-PDS vectors. For VIGS experiment, plasmids of pTRV1, pTRV2 and pTRV2 recombinant vectors (pTRV2-PDS and pTRV2-HXK2) were transformed into A. tumefaciens strain GV3101 cells by using the freeze-thaw method. The empty plasmid pTRV2 was used as control (TRV:00). The Agrobacterium strains were inoculated into 50 mL of LB medium as above on a shaker at 180 rpm at 28°C for 12–16 h to an OD600 of 1.2–1.5. The Agrobacterium cells were centrifuged at 4000 g for 10 min at room temperature and washed twice, resuspended with the infiltration solution (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone) to a final OD600 of 0.8–1.0 and placed at room temperature in darkness for 3 h. The infiltration solution of the Agrobacterium strain containing pTRV1 was mixed with the infiltration solution of pTRV2 or Agrobacterium carrying the constructs in a 1:1 ratio (v/v) and 20–40 washed soybean seeds were added respectively (Zhao et al., 2020b). Soybean seeds that had been soaked for 24 h in darkness at room temperature were removed and planted in nutrient soil. The silencing efficiency of soybean seedlings was determined by qRT-PCR when the first true leaf was fully expanded.
2.14 Statistical analysis
The results of root length, fresh weights, and dry weights were from five independent experiments, and results of Chlorophyll, MDA, proline contents, EL and SOD were from three independent experiments. GraphPad Prism 5 was utilized for all statistical analysis. Data significance analysis was performed using Student’s t-test (Yang et al., 2022). Data are presented as mean ± SE. Single, double and three asterisks denote significant differences compared with the values of WT at p < 0.05, p < 0.01 and p < 0.001, respectively.
3 Results
3.1 Identification of G. max HXK gene family
In total, 17 HXK genes were identified in the G. max genome, and they were designated as GmHXK1-17. The name, Ensembl Plants accession number, mRNA and CDS length of nucleotide sequence as well as the length of amino acid sequence, MW, PI, chromosome location, cTP, and number of TMHs, are summarized in Table 1. As depicted in Table 1, the length of the CDS varied from 471 to 1,812 bp, encoding 156 to 504 amino acids and corresponding to molecular weights ranging from 17.42 to 55.40 kDa. The theoretical PIs of these proteins ranged from 5.18 to 8.76. There were three genes on chromosomes 1, 3 genes on chromosomes 17, and 2 genes on chromosomes 5, 7, 8, and 11. GmHXK2, GmHXK10 and GmHXK4 were distributed on chromosomes 9, 12 and 14, respectively. GmHXK1, GmHXK2 and GmHXK11 contained no cTP or TMHs. GmHXK5, GmHXK6, GmHXK12–17 contained cTP and one TMH. GmHXK3 and GmHXK4 contained cTP but no TMHs.
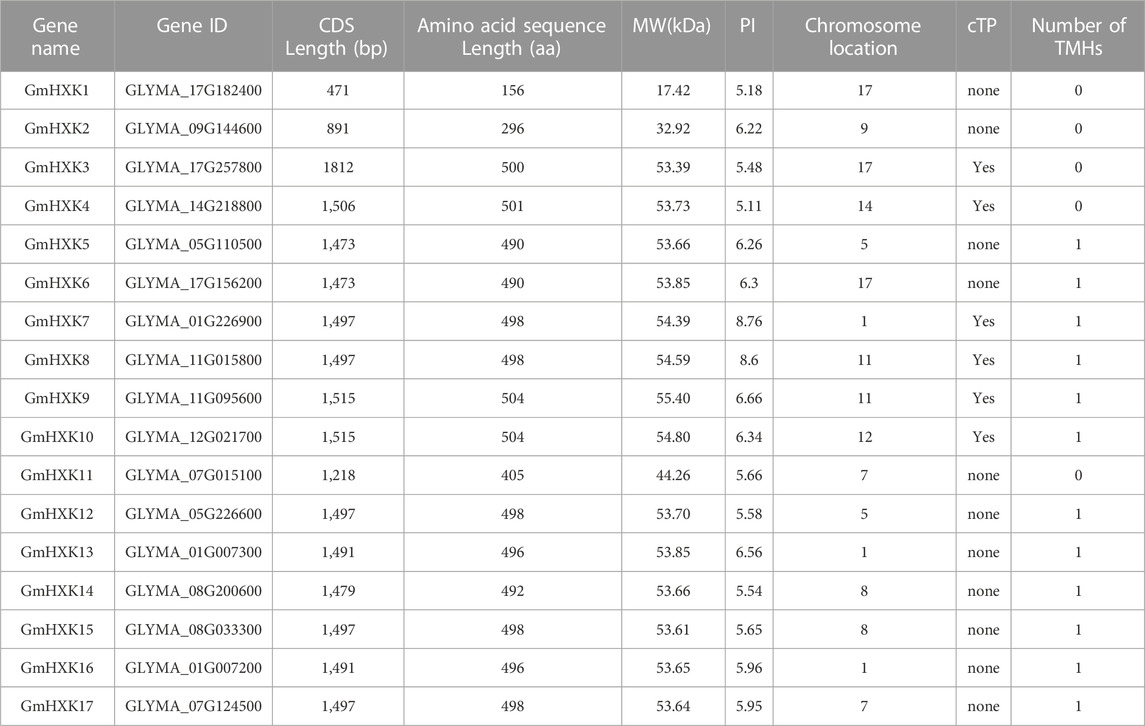
TABLE 1. Physicochemical property of HXK in G. max. CDS: coding sequences; MW (kDa): molecular weight (kilodaltons); PI: theoretical isoelectric point; cTP: chloroplast transit peptide; TMHs: transmembrane helices.
3.2 Multiple alignment and phylogenetic analysis of the GmHXK genes
To better study the structural difference between GmHXKs and their possible functional differences, the amino acid sequences of G. max were aligned and analyzed (Figure 1). Conserved domain analysis showed that most GmHXKs were multidomain proteins, which contained two phosphate sites (I and II), two connect sites (I and II), one α-helix site, one adenosine binding site, and one sugar binding site. However, GmHXK1 contained just three conserved domains, α-helix site, adenosine binding site, and sugar binding site. Additionally, GmHXK2 contained phosphate site II, two connect sites (I and II), one α-helix site, one adenosine binding site, and one sugar binding site.
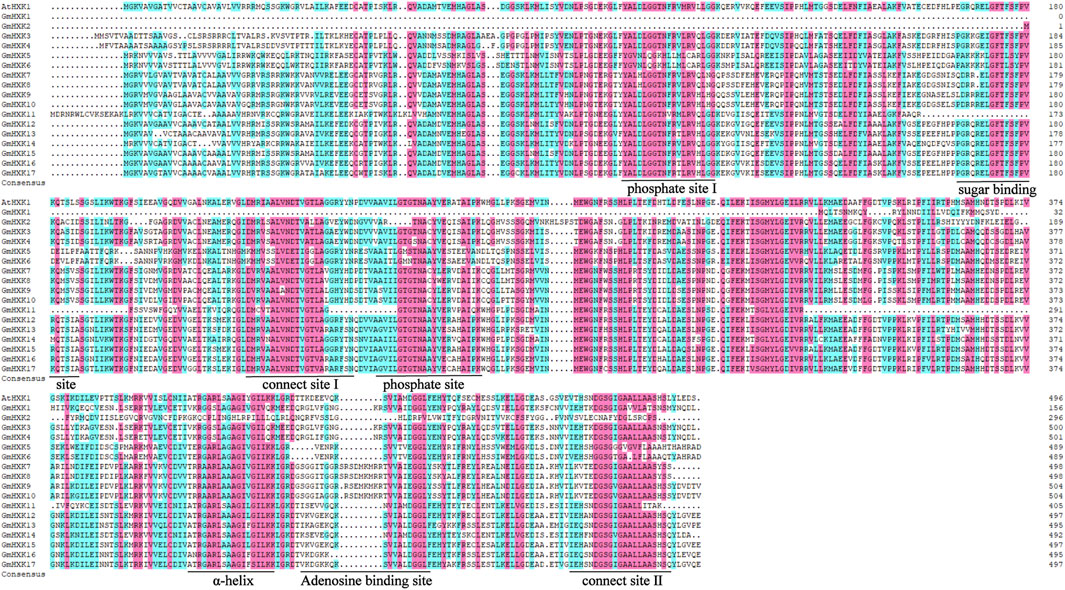
FIGURE 1. Multiple alignment of HXK amino acid sequences in G. max. Identical and similar amino acid residues are highlighted in black and pink shades, respectively. Core sugar binding site, phosphate sites (I and II), adenosine binding site, connection sites (I and II), and specific α-helix site are underlined.
To further reveal the evolutionary relationship of hexokinase family, the protein sequences of 56 hexokinases from five different plant species, including G. max, A. thaliana, S. lycopersicum, O. sativa and N. tobacum were used to construct phylogenetic tree using MEGA-X. As shown in Figure 2, all hexokinase genes were divided into three subfamilies (I–III). Most of the HXK genes of five different plant species were sorted into Cluster III. Cluster II contains fewer genes. It indicated the close relationship between the five plant species aforementioned. There were none GmHXKs in Cluster I, which contains only some hexokinase genes in S. lycopersicum.
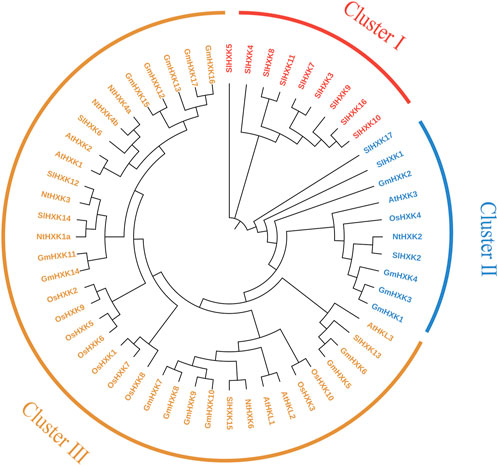
FIGURE 2. Phylogenetic analysis of the HXK gene family from G. max and other plant species. At, A. thaliana; Gm, G. max; Sl, S. lycopersicum; Os, O. sativa; Nt, N. tobacum. The amino acid sequences of HXK were aligned by Clustal X, and the tree was generated using the neighbor-joining method in MEGA-X with 1,000 bootstrap replicates.
3.3 Conserved motifs and gene structure analysis of GmHXKs
Generally speaking, the conserved regions of proteins frequently distinguish them from other proteins and determine the basis of their functions. To reveal the structural diversity and functional characteristics of the GmHXK family members, their conserved motifs were analyzed by MEME and mapped in the phylogenetic tree. In total, 10 conserved motifs were identified (Figure 3B). The detailed sequences and conserved motifs are shown in Supplementary Table S2. The identified motifs ranged from 29 to 50 amino acids in length. Among them, motifs 3, 6 and 8 were found to encode the hexokinase_1 domain, while motifs 1, 2, four and 5 encode the hexokinase_2 domain. Nevertheless, the functions of motif 7, 9 and 10 are unknown. GmHXK7-10, 12, 14–17 were found to have all 10 motifs, while other GmHXK family members had only portion motifs (Figure 3B). GmHXK3 and GmHXK4 contained all motifs except motif 2, and GmHXK5 and GmHXK6 contained all motifs except motif 9.
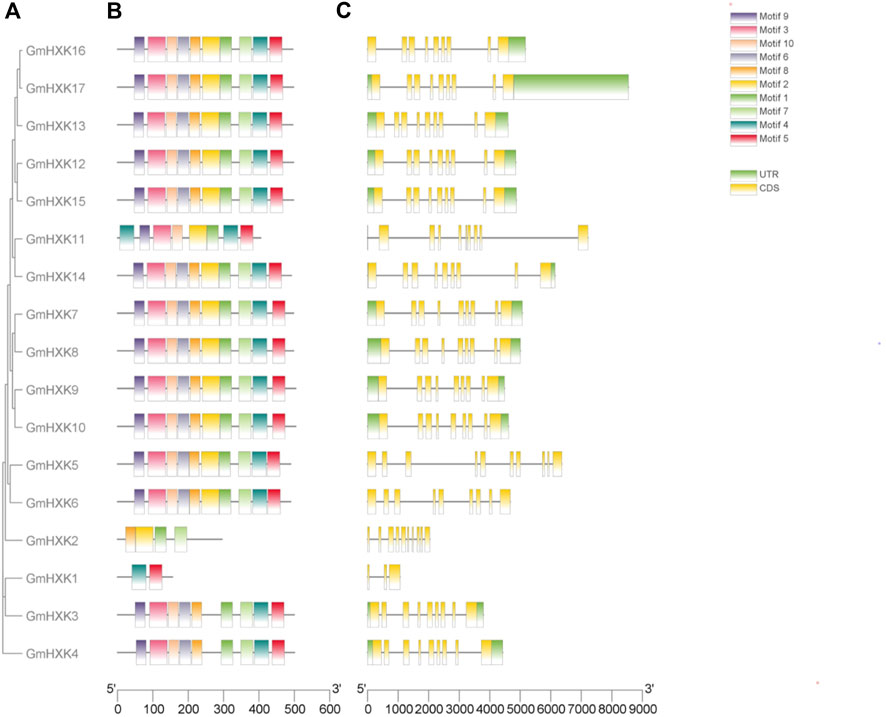
FIGURE 3. The conserved domain organization and exon-intron structure of GmHXKs genes. (A). Phylogenetic relationships of GmHXK genes. The GmHXK amino acid sequences were aligned by Clustal X and the tree was generated using the Neighbor-Joining method in MEGA-X with 1,000 bootstrap replicates. (B). The conserved domain analysis of GmHXK proteins using MEME. Different colors represent the different conserved motifs. (C). Exon–intron structures of GmHXK genes. Exons and introns are shown as yellow boxes and black lines, respectively.
Different combinations of exons and introns can lead to diverse gene function. To explore the structure diversity of GmHXK genes, Gene Structure Display Server 2.0 was employed to analyze the distribution of exon-intron structure based on the corresponding genome and coding sequences (Figure 3C). The results showed that most GmHXK genes contained nine exons and eight introns. GmHXK2 and GmHXK5 contained 10 exons and nine introns. GmHXK1, contained only three exons and two introns.
3.4 Cis-element analysis of the GmHXK genes
The identification of the cis-regulatory elements in the promoter part of the gene is important for functional and regulatory studies. To investigate the cis-regulatory elements of the 17 GmHXK genes, we analyzed the sequence 2,000 bp upstream of the start codon ATG. The cis-elements of the GmHXK genes were classified into three categories, involved in plant growth and development, stress responses and hormone-induced response (Supplementary Figure S1). The plant growth and development category contained seed-specific regulation (RY-element) and meristem expression (CAT-box) cis-elements. In the stress-responsive category, the elements included dehydration-responsive (DRE), anaerobic induction (ARE and GC-motif), low-temperature-responsive (LTR) and MYB-binding sites involved in drought inducibility (MBS), as well as defense and stress responsiveness (TCA). Seven types of phytohormone responsive cis-elements were detected, including auxin-responsive (AuxRR-core), abscisic acid-responsive (ABRE), methyl jasmonate-responsive (CGTCA-motif), ethylene-responsive (ERE), gibberellin responsive (GARE-motif), salicylic acid-responsive (TCA-element) and heat shock, osmotic stress, low pH, nutrient starvation-responsive (STRE). It is worth noting that the cis-elements MYB and MYC associated with phytohormone and abiotic stress were present in 17 GmHXK genes.
3.5 The expression pattern of GmHXK gene family
We analyzed the expression patterns of 14 GmHXK family members in different tissues including young-leaf, flower, pod, pod shell, seed, root and nodule using data in soybase (Figure 4A). It showed that GmHXK1,5,6,13,14 was expressed at a very low level or not expressed in seven tissues above. The level of expression of GmHXK4,7,8,9,17 was not obvious in majority of the plant tissues except for that in a certain tissue. GmHXK3,12,15,16 were expressed obviously in most tissues.
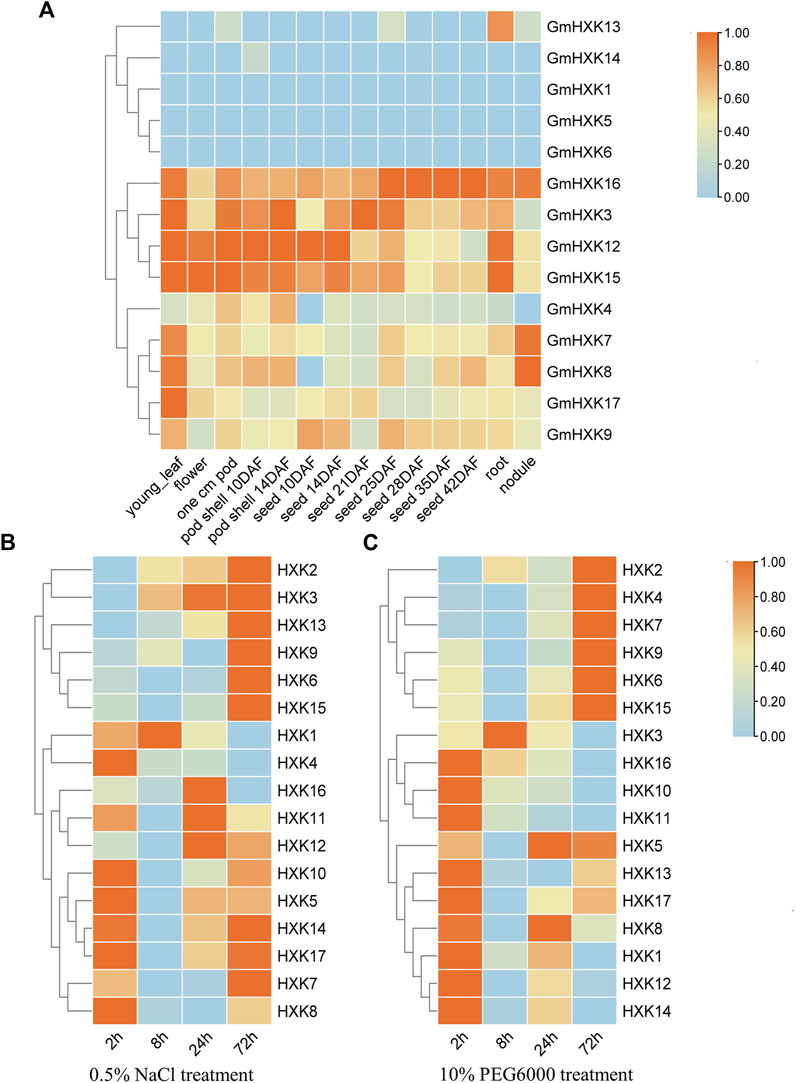
FIGURE 4. The expression pattern of GmHXK gene family. (A). Tissue-specific expression profile of GmHXK under normal condition. Expression of GmHXK in roots under salt stress (B) and drought stress (C). Data are presented as mean ± SE. Red indicates high expression levels blue indicates low expression levels.
And we analyzed also the expression of GmHXKs under salt and drought stress after treatments for different times by qRT-PCR. By analyzing expression data after treatment with NaCl or drought stress for 0, 2,8,24,72 h, we found that the expression of GmHXK 2,3,6,9,13,15 under treatment with NaCl (Figure 4B) and GmHXK2,4,6,7,9,15 under drought treatment with drought stress (Figure 4C) were increased after treatments within 72 h. The expression of other GmHXKs under salt or drought stress was obvious at the initial stage of treatment but decreased over time within 72 h. Owing to the increase in expression of GmHXK2,6,9,15 under both salt and drought stress, we selected GmHXK2 to study further its function under the salt stress.
3.6 Molecular characteristics of GmHXK2
GmHXK2 gene (Gene ID: GLYMA_09G144600) fragment containing introns without stop codon, was isolated by PCR with primers using genome DNA as a template. Fragments of size 2,033 bp were detected (Figure 5A). Then, the PCR product was inserted into a pMD18-T vector and sequenced. Sequence analysis suggested that the fragment was identical to the known GmHXK2 gene. The fragments were cloned into the entry vector TOPO by TOPO cloning reaction and subsequently into gateway destination vector pMDC83. Products amplified by PCR using genome DNA from T3 transgenic Arabidopsis WT35S::GmHXK2 seedlings of expected size 1,357 bp were detected (Figure 5B). Our results show that the GmHXK2 gene was integrated into the genome of WT.
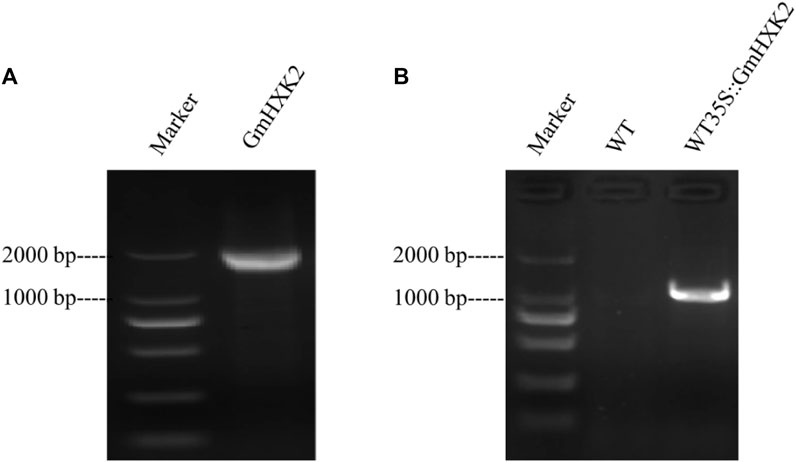
FIGURE 5. GmHXK2 gene amplification and identification of transgenic Arabidopsis seedlings. (A). PCR amplification of GmHXK2 gene. Marker: DL 2000; (B). PCR identification of transgenic Arabidopsis seedlings. Marker: DL 2000 (CWBIO, China).
3.7 Analysis on salt tolerance of GmHXK2-silenced plants
To further explore the role of GmHXK2 in plant salt tolerance, we constructed a TRV-VIGS vector by selecting specific sequence in the CDS region of the GmHXK2 gene and identified the efficiency in silencing of GmHXK2-silenced plants by qRT-PCR, which were 30%–50% of control plants (Supplementary Figure S2). The silenced plants showed different phenotype from the control. Margins of silenced plants in first true leaf were discolored and wilted (Figure 6B) compared with the control (TRV:00). The validity of silenced plants was further determined by the PDS phenotypes (Figure 6C). Finally, we selected the plants with the highest efficiency of silencing for the subsequent experiment on salt stress.
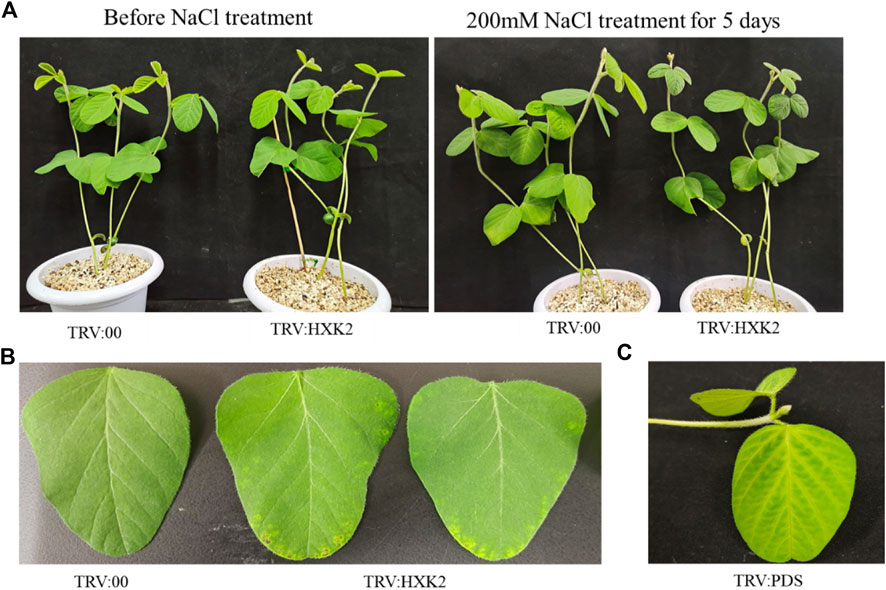
FIGURE 6. Phenotypic analysis of GmHXK2-silencing soybeans under salt stress. The seedings of TRV:00 and TRV:HXK2 divided into two groups (control and salt treatment) were grown for 7 days on pot under normal condition, then the plants were treated with 0 or 200 mM NaCl for 5 days, respectively. Salt treatment group phenotype as shown in figure (A–C) Phenotypes of TRV:00, TRV:HXK2 and TRV:PDS.
After treatment with salt stress, all leaves in soybean plants had been wilt, curled, and were yellow to some extent, and the leaves of GmHXK2-silenced plants were more severely damaged than control (Figure 6A).
To investigate the potential physiological mechanisms by which GmHXK2 enhances plant tolerance, the proline, chlorophyll content, EL and SOD in TRV:00 and TRV:HXK2 plants were measured under normal and salt stress conditions. Our results showed that the EL of TRV:HXK2 (26.18%) was significantly higher than that of TRV:00 (11.33%) (Figure 7B) under salt conditions. This indicated that TRV:HXK2 plants were damaged more seriously than the control plants. The content of proline, chlorophyll and SOD of TRV:HXK2 (0.60 mg/g, 3.06 mg/g and 12.61 U, respectively) were decreased compared with that of TRV:00 plants (1.11 mg/g, 4.31 mg/g and 16.80 U, respectively) (Figures 7A, C, D). These results suggested that capability of TRV:HXK2 plants in resisting salt stress and scavenging ROS generated intracellularly diminished compared with control plants.
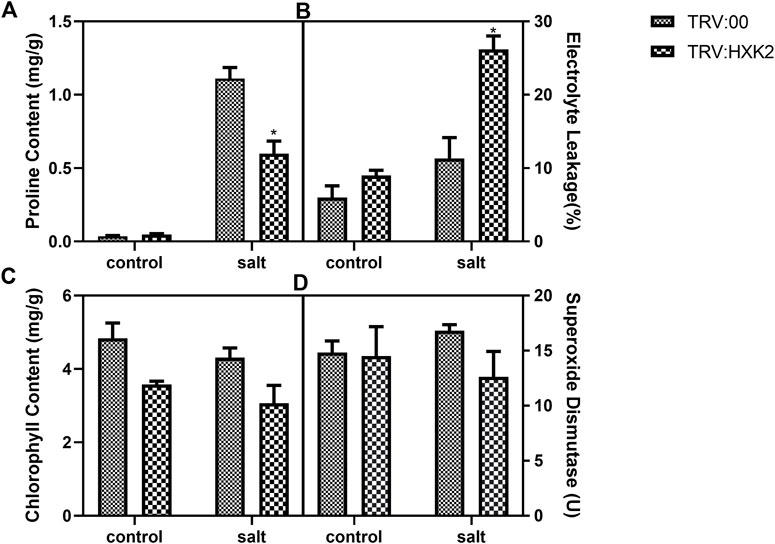
FIGURE 7. Physiological response of GmHXK2 silent plant under salt stress. Content of proline (A), EL (B), chlorophyll (C) and SOD (D) were measured in TRV:00 and TRV:HXK2 after treatment with 0 or 200 mM NaCl for 5-days. Data are presented as mean ± SE. Single asterisks denotes significant differences compared to the values of TRV:00 at p < 0.05; Student’s t-test.
3.8 Expression of Salt-Responsive genes in GmHXK2-silenced soybean plants
The plasma membrane Na+/H+ antiporters GmSOS1 (salt overly sensitive) (Ma et al., 2020), the high-affinity potassium transporters (GmHKTs) (Sun et al., 2021), GmbZIP44 (Zhao et al., 2020a), and GmSALT3 as one of cation/H exchangers, which play a vital role in response to environmental salinity (Xu et al., 2022a), were known to be involved in the regulation of salt tolerance in soybean. So we investigated the expression of these Salt-Responsive genes to ascertain the relationship of GmHXK2 and Na+ homeostasis.
Under control conditions, expressions of both GmSALT3 (Figure 8A) and GmSOS1 (Figure 8D) were decreased in silenced plants compared with WT, while expression of GmbZIP44 (Figure 8B) and GmHKT1 (Figure 8C) were not significantly different. However, the expression of all four genes in GmHXK2-silenced plants under salt stress was decreased significantly than WT (Figure 8). These results suggested that silence of the GmHXK2 gene downregulated the expression of salt stress-related genes involved in Na+ homeostasis, implying that GmHXK2 had an important role in remaining homeostasis of Na+ and K+.
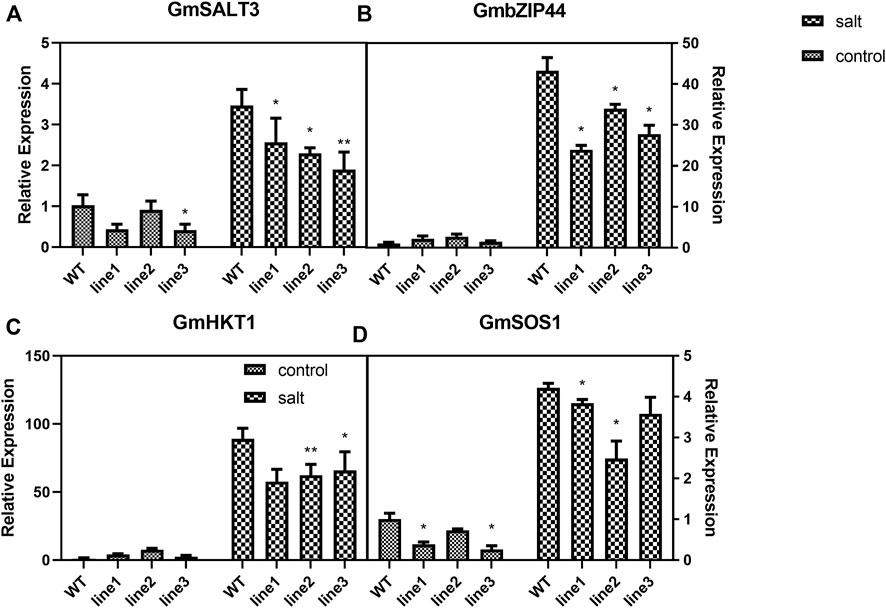
FIGURE 8. GmHXK2 affect the expression of Salt-Responsive genes. (A−D). Expression levels of salt stress-related genes. Data are presented as mean ± SE. Single and double asterisks denote significant differences compared to the values of WT at p < 0.05 and p < 0.01; Student’s t-test.
3.9 Subcellular localization and western blot analysis of transgenic arabidopsis plants
As shown in Figure 9A, signals of GmHXK2-GFP were distributed on cell wall and cell membrane, which revealed that GmHXK2 was predominantly localized on both vacuolar membrane and cell membrane. To examine GmHXK2 protein expression, transgenic Arabidopsis seedlings WT35S::GmHXK2 were further analyzed via immunoblotting with an antibody specific to GFP by Western blot. The results showed that T3-T6 transgenic lines expressed the integrated protein with an expected molecular mass of 59.92 kDa (Figure 9B), which corresponds to the fusion protein, while no proteins of this size were observed in that of WT seedlings.
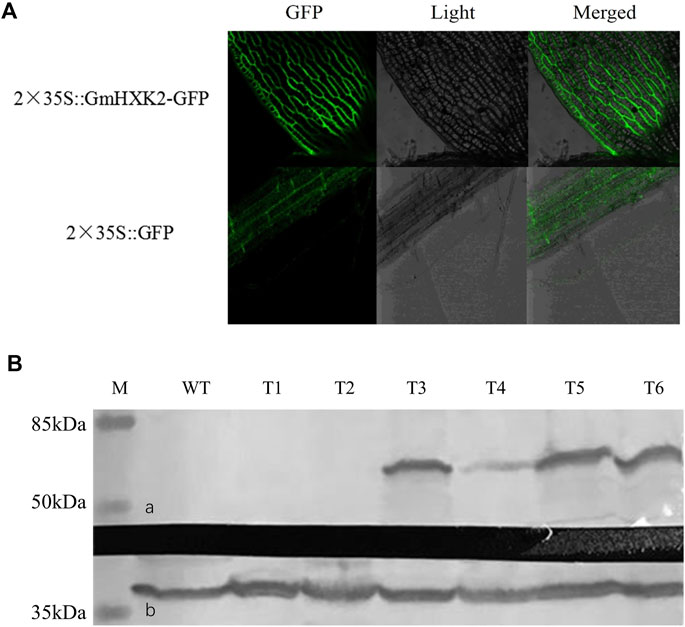
FIGURE 9. Subcellular localization and WB analysis of transgenic plants. (A). Subcellular localization of GmHXK2 in transgenic Arabidopsis root cells. (B). Expression of GmHXK2 in transgenic Arabidopsis detected by Western-blot analysis. (A). GmHXK2-GFP fusion protein; (B). GAPDH was used as internal control with molecular mass of 36 kDa (M: Marker; C: Wild type Arabidopsis; T1-T6: transgenic Arabidopsis plants.
3.10 Ectopic expression of GmHXK2 promote the growth of arabidopsis
As shown in Figure 10A, the WT and WT35S::GmHXK2 transgenic Arabidopsis grew well in the control group. After 100 mM NaCl treatment, the growth of the Arabidopsis was inhibited, but the growth of WT was more suppressed than that of WT35S::GmHXK2 transgenic Arabidopsis.
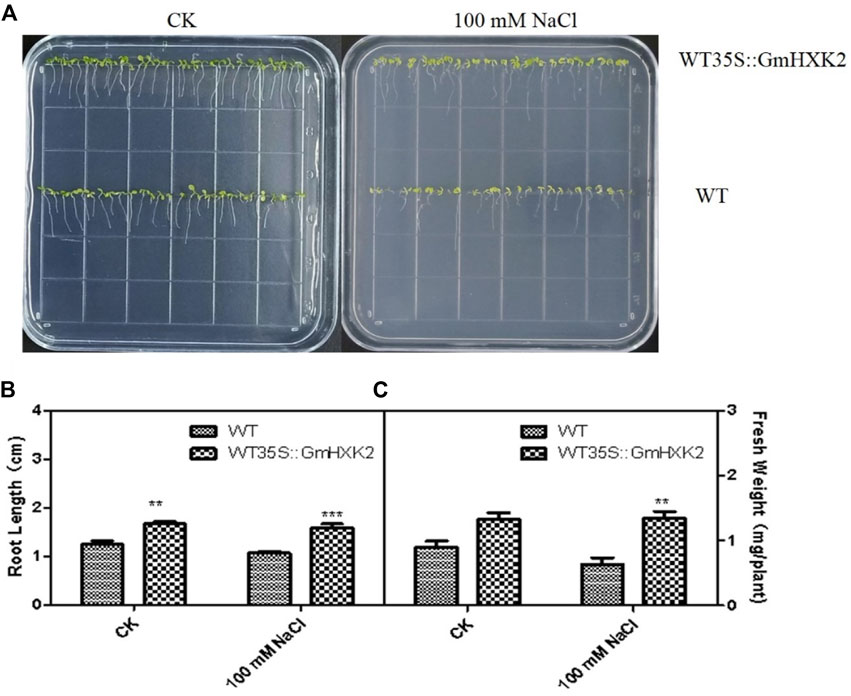
FIGURE 10. The phenotype (A), root length (B) and fresh weight (C) of the WT and WT35S::GmHXK2 transgenic Arabidopsis plants under salt stress at the germination stage. Seeds of WT and WT35S::GmHXK2 were planted in 1/2 MS medium with or without100 mM NaCl for 7 days. Data are presented as mean ± SE (n = 5, biological replicates). Single, double and three asterisks denote significant differences at p < 0.05, p < 0.01 and p < 0.001, respectively; Student’s t-test.
Under control condition, the root length of WT35S::GmHXK2 was significantly higher than that of WT, and fresh weight of WT35S::GmHXK2 were increased by 47% compared with WT. After NaCl treatment, the root length and fresh weight of Arabidopsis were decreased, and the root length and fresh weight of WT35S::GmHXK2 were significantly higher than that of WT. (Figures 10B, C). Hence, the expression of GmHXK2 promoted the growth of Arabidopsis.
3.11 Ectopic expression of GmHXK2 enhance the salt resistance of arabidopsis at the seedling stage
To verify whether GmHXK2-expressing Arabidopsis contributes to salt tolerance, we observed the phenotype of transgenic Arabidopsis and WT after 6 days of NaCl and glucose treatments and then measured the physiology and biochemistry indexes. Under salt stress, the growth of Arabidopsis seedlings were obviously inhibited, the leaves of all plants gradually became yellow and shriveled in addition to roots becoming shorter. In addition, the growth of WT was more suppressed than that of WT35S::GmHXK2. After adding 100 mM glucose, the growth condition of seedlings was much better compared to those at 0, 100 and 150 mM NaCl treatments. (Figure 11).
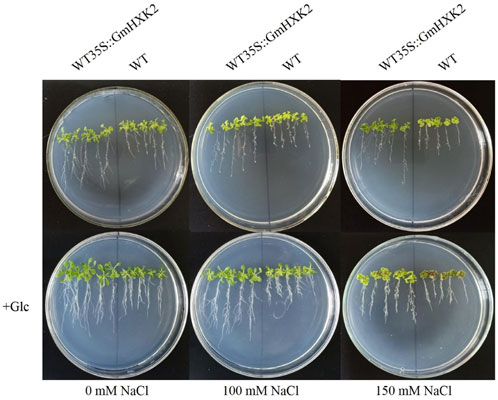
FIGURE 11. The phenotype of WT and WT35S::GmHXK2 transgenic Arabidopsis plants under salt stress. Glc represents 100 mM glucose treatment.
After NaCl treatment, the fresh and dry weight, root length and chlorophyll content decreased, though the MDA and proline contents increased (Figure 12). As shown in Figures 12A, B, the fresh weight and dry weight of WT35S::GmHXK2 transgenic Arabidopsis were higher than that of WT under 0, 100 and 150 mM NaCl treatment. After salt treatment, root length, chlorophyll and proline contents of WT35S::GmHXK2 seedlings were significantly increased compared to WT, but the MDA content of WT35S::GmHXK2 seedlings was much lower than that of WT (Figures 12C–F).
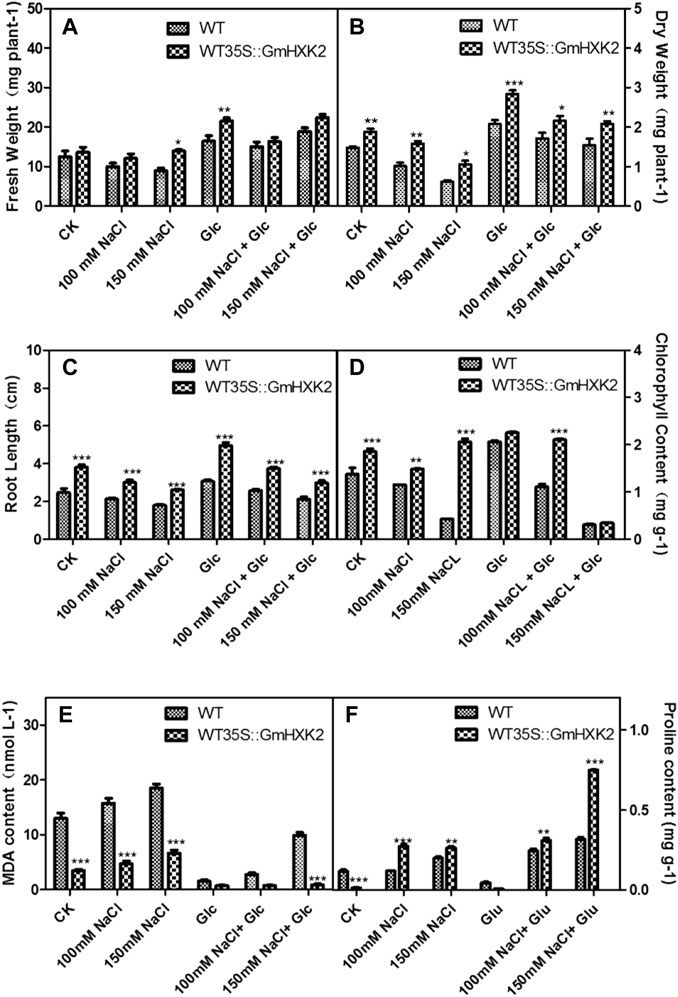
FIGURE 12. The fresh weight (A), dry weight (B), root length (C), chlorophyll content (D), MDA (E) and proline content (F) of the WT and WT35 S:GmHXK2 transgenic Arabidopsis plants under salt stress. Data are presented as mean ± SE. Single, double and three asterisks denote significant differences compared to the values of WT at p < 0.05, p < 0.01 and p < 0.001, respectively; Student’s t-test.
Exogenous glucose counteracted the effect on salt stress. The fresh weight and dry weight, root length, chlorophyll and proline contents of WT and WT35S::GmHXK2 were increased compared to those at the 0, 100 and 150 mM NaCl stress. And the application of glucose under salt treatment inhibited the increase of MDA content under salt stress. (Figure 12).
These results revealed the expression of GmHXK2 could enhanced the salt tolerance of plants during the seedling stage. In addition, 100 mM exogenous glucose alleviated the inhibition of salt stress on the growth of Arabidopsis plants.
4 Discussion
Hexokinases are very important enzymes in the growth and development of plants (Xiao et al., 2000). They not only play key roles in sugar signaling, but also involve in responding to abiotic stress in plants (Kim et al., 2013; Li et al., 2017). The OsHXK10 gene in rice can regulate plant reproduction (Xu et al., 2008), the prunus HXK3 gene can promote tolerance to drought and salt stress (Perez-Diaz et al., 2021), expression of Arabidopsis AtHXK1 in tobacco guard cells can attenuate transpiration in plants and improve salt and drought tolerance (Lugassi et al., 2019). In Malus domestica Borkh., MdHXK1 interacts with the salt tolerance gene MdNHX1 to improve the salt tolerance of plants (Sun et al., 2018). Though HXK genes have been widely studied in plants (Dai et al., 1999; Guglielminetti et al., 2000; Moore et al., 2003; Claeyssen et al., 2013; Li et al., 2017), HXK gene family members of G. max have not been characterized nor have their molecular properties under salt abiotic stresses been clarified until now.
Previous researches suggested that HXK proteins contain some conserved domains, phosphate 1 and 2, sugar binding site, and adenosine binding site, which are important to plant HXKs and essential for their enzymatic functions (Bork et al., 1993; Katz et al., 2000). In this study, a total of 17 HXK genes were identified in G. max. Except for GmHXK1, phosphate site, connect site, α-helix site, adenosine binding site, and sugar binding site, were well conserved in other GmHXKs (Figure 1). It suggested that they had the ability to phosphorylate hexoses. According to the characteristics of structure in Type A, B and C hexokinases and their distribution on subcellular organelle, we hypothesized that GmHXK3, GmHXK4 and GmHXK7-10 belong to Type A hexokinases and the other GmHXKs are Type B hexokinases using the data on the prediction of 17 hexokinase proteins and their subcellular localization in soybean.
ABA is a classical plant hormone whose level inside the plant is regulated by external environmental stress (Roychoudhury et al., 2013). Plants respond to environmental stress through interaction of transcription factors with a handful of cis-regulatory elements (Saidi and Hajibarat, 2019; Wang et al., 2021b). GmHXK2 exhibited abscisic acid-responsive (ABRE), ethylene-responsive (ERE), and MYB elements, etc. ABRE is an eight bp cis-acting sequence present in the promoter region of most ABA-inducible or ABA-responsive genes. It has been shown that bZIP transcription factor genes bind to ABRE cis-acting elements to regulate stress-induced expression of related genes in many plants such as A. thaliana and soybean (Kang et al., 2002; Liao et al., 2008). MYB transcription factor family genes are widely distributed in plants, which act on MYB cis-elements that involve in many important physiological and biochemical processes, such as cell development, signal transduction, and stress response (Ambawat et al., 2013; Li et al., 2015). Ethylene response factor (ERF) plays an important role in plant stress tolerance by regulating gene expression through binding to the ERE element of stress response genes (Wang et al., 2016). Transgenic plants expressing the ERF gene were shown to have significantly increased tolerance to abiotic stresses such as drought and salt stress (Zhang et al., 2009; Rong et al., 2014). GmHXKs not only contains Cis-acting regulatory element involved in seed-specific regulation (RY-element), Cis-acting regulatory element essential for meristem expression (CAT-box) but also ABRE, ERE, MYB and MYC elements related with response to abiotic environmental stress, suggesting GmHXKs might be induced by different signal such as ABA and ethylene to take part in regulation of development and abiotic tolerance to environmental stress through being activated by MYB or MYC transcription factor.
VIGS is a very effective method for gene function analysis (Ramegowda et al., 2014). We obtained GmHXK2-silenced soybean plants by this method and investigated the growth and physiological parameters under salt stress. During the growth of plants, many physiological indicators are often used to verify the tolerance of plants (Du et al., 2018; Sun et al., 2019; Wang et al., 2019; Leng et al., 2021; Li et al., 2021). It has been demonstrated that the accumulation of proline under adversity conditions can help plants to resist abiotic stresses (Lehmann et al., 2010; Xu et al., 2022b). To a certain extent chlorophyll content can indicate the degree of plant tolerance to stress. The reactive oxygen species (ROS) are by-products produced by various metabolic pathways within plant cells. Plants produce a range of defensive substances to maintain the dynamic intracellular equilibrium and prevent from causing damage to the organism due to accumulation of ROS. SOD is an enzyme that scavenges ROS in plants, and its level reflects the plant’s tolerance to stress (Apel and Hirt, 2004; Mittler et al., 2004; Wang et al., 2021a). EL can indicate the degree of plant cell membrane damage, higher EL suggests a greater degree of cell damage (Jungklang et al., 2017). Based on the data on phenotype (Figure 5) and physiological indicators that we obtained (Figure 6), silence of GmHXK2 decreased the salt tolerance of the plants, we deduced that the GmHXK2 gene is contributed to salt tolerance in soybean plants under salt stress.
Under salt stress, excessive sodium (Na+) inhibits enzyme activity and disrupts potassium (K+) uptake, leading to decrease in ration of K+/Na+. Plants have evolved multiple physiological mechanisms to cope with external stresses (Zhao et al., 2020a). GmHXKT1, GmSOS1, and GmSALT3 are genes that maintain ion homeostasis in cells in soybean plants. SOS1 is the key gene in the SOS pathway that regulates Na+ concentration and is believed to play an important role in osmoregulation (Shi et al., 2002; Olias et al., 2009; Ma et al., 2014). HKT is one of Na+/K+ transporters, which protects plants from Na+ accumulation in photosynthetic organs to improve the salt tolerance of plants by maintaining the Na+/K+ balance under salt stress (Berthomieu et al., 2003; Ren et al., 2005; Plett et al., 2010). GmSALT3 encodes a protein of the cation/H+ exchanger family which has a role in sensing or coping with salinity (Guan et al., 2014). GmbZIP44, as stress-responsive protein in the ABA pathway, can binds to the ABRE cis-regulatory element of target genes to regulate downstream gene expression (Kang et al., 2002; Liao et al., 2008). In our findings, expression of GmHXK2 gene were increased gradually in roots after treatment with NaCl for 2 h–72 h (Figures 4B, C). And the expression GmHXKT1, GmSOS1, and GmSALT3 were downregulated in GmHXK2-silenced soybean plants after treatment with NaCl compared with WT (Figure 8). These results confirmed that the silence of GmHXK2 in soybean plants was correlated significantly with decrease in expression of gene involving in uptake, transport and homeostasis of Na+, which reflected the important role of GmHXK2 through regulating homeostasis between K+ and Na+ to improve salt tolerance of soybean plants.
Plant transcription factors can bind to specific cis-element through their conserved domain. For example, overexpression of ThMYB8 decreased ROS levels and maintained K+/Na+ homeostasis (Liu et al., 2021). TaPIMP1 can acts by bind to a few of MYB-binding sites to regulate of genes related with abiotic stress in response to ABA or salicylic acid signal (Zhang et al., 2012). According to our results, we supposed that in response to salt stress, MYB or MYC transcription factor might be induced by ABA or ethylene, afterwards they might bind to GmHXK2 and triggered target genes downstream such as some genes related with homeostasis of Na+ to regulate the salt tolerance of soybean plants. The deduction on the molecular mechanisms of GmHXK2 gene in salt tolerance need to be further confirmed by extensive experiments. Furthermore, Arabidopsis plants of ectopic expressing GmHXK2 had the longer roots, lower levels of MDA contents, and higher levels of proline and chlorophyll content under salt stress, specially at conditions of exogenous application with glucose, indicating that GmHXK2 might also sense and respond to glucose signal to participate in salt tolerance under salt stress.
5 Conclusion
In this work, we not only identified HXK gene family members but also obtained GmHXK2 transgenic Arabidopsis lines and GmHXK2-silenced soybean plants. Subsequent analysis found that overexpressing GmHXK2 in Arabidopsis plants improved their tolerance to salt stresses. Moreover, 100 mM of exogenous glucose can alleviate plant growth inhibition under salt stress. Conversely, GmHXK2-silenced soybean plants reduced the expression of salt tolerance genes, which lead to less tolerant to salt stress. Our results not only provided the theoretical foundation for further research on HXK gene family in G. max, but also they could provide important information for breeding stress-resistant G. max cultivars.
Date availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YG designed the research. SC and ZT performed most of the experiments. All authors performed data analyses and took part in writing the manuscript.
Funding
This research was supported by the Scientific and Technological Projects in Henan province, China (172102110132).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1135290/full#supplementary-material
References
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: An overview. Physiology Mol. Biol. Plants 19 (3), 307–321. doi:10.1007/s12298-013-0179-1
Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi:10.1146/annurev.arplant.55.031903.141701
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. (Web Server issue). doi:10.1093/nar/gkp335
Baillo, E. H., Kimotho, R. N., Zhang, Z. B., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 10 (10), 771. doi:10.3390/genes10100771
Berthomieu, P., Conejero, G., Nublat, A., Brackenbury, W. J., Lambert, C., Savio, C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. Embo J. 22 (9), 2004–2014. doi:10.1093/emboj/cdg207
Bilal, S., Shahzad, R., Khan, A. L., Kang, S. M., Imran, Q. M., Al-Harrasi, A., et al. (2018). Endophytic microbial consortia of phytohormones-producing fungus paecilomyces formosus LHL10 and bacteria sphingomonas sp. LK11 to Glycine max L. Regulates physio-hormonal changes to attenuate aluminum and zinc stresses. Front. Plant Sci. 9, 1273. doi:10.3389/fpls.2018.01273
Bilal, S., Shahzad, R., Imran, M., Jan, R., Kim, K. M., and Lee, I. J. (2020). Synergistic association of endophytic fungi enhances Glycine max L. resilience to combined abiotic stresses: Heavy metals, high temperature and drought stress. Industrial Crops Prod. 143, 111931. doi:10.1016/j.indcrop.2019.111931
Bork, P., Sander, C., and Valencia, A. (1993). Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 2 (1), 31–40. doi:10.1002/pro.5560020104
Cheng, W., Zhang, H., Zhou, X., Liu, H., Liu, Y., Li, J., et al. (2011). Subcellular localization of rice hexokinase (OsHXK) family members in the mesophyll protoplasts of tobacco. Biol. Plant. 55 (1), 173–177. doi:10.1007/s10535-011-0025-7
Cho, J. I., Ryoo, N., Ko, S., Lee, S. K., Lee, J., Jung, K. H., et al. (2006). Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.) Planta 224 (3), 598–611. doi:10.1007/s00425-006-0251-y
Claeyssen, E., Dorion, S., Clendenning, A., He, J. Z., Wally, O., Chen, J., et al. (2013). The futile cycling of hexose phosphates could account for the fact that hexokinase exerts a high control on glucose phosphorylation but not on glycolytic rate in transgenic potato (Solanum tuberosum) roots. PLoS One 8 (1), e53898. doi:10.1371/journal.pone.0053898
da-Silva, W. S., Rezende, G. L., and Galina, A. (2001). Subcellular distribution and kinetic properties of cytosolic and non-cytosolic hexokinases in maize seedling roots: Implications for hexose phosphorylation. J. Exp. Bot. 52 (359), 1191–1201. doi:10.1093/jexbot/52.359.1191
Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., et al. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell. 11 (7), 1253–1266. doi:10.1105/tpc.11.7.1253
David-Schwartz, R., Weintraub, L., Vidavski, R., Zemach, H., Murakhovsky, L., Swartzberg, D., et al. (2013). The SlFRK4 promoter is active only during late stages of pollen and anther development. Plant Sci. 199-200, 61–70. doi:10.1016/j.plantsci.2012.09.016
Donahue, J. L., Okpodu, C. M., Cramer, C. L., Grabau, E. A., and Alscher, R. G. (1997). Responses of antioxidants to paraquat in pea leaves (relationships to resistance). Plant Physiol. 113 (1), 249–257. doi:10.1104/pp.113.1.249
Dou, L. L., Li, Z. H., Wang, H. Q., Li, H. Z., Xiao, G. H., and Zhang, X. L. (2022). The hexokinase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Front. Plant Sci. 13, 882587. doi:10.3389/fpls.2022.882587
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18 (1), 320. doi:10.1186/s12870-018-1551-7
Geng, M. T., Yao, Y., Wang, Y. L., Wu, X. H., Sun, C., Li, R. M., et al. (2017). Structure, expression, and functional analysis of the hexokinase gene family in cassava. Int. J. Mol. Sci. 18 (5), 1041. doi:10.3390/ijms18051041
Giese, J. O., Herbers, K., Hoffmann, M., Klösgen, R. B., and Sonnewald, U. (2005). Isolation and functional characterization of a novel plastidic hexokinase from Nicotiana tabacum. FEBS Lett. 579 (3), 827–831. doi:10.1016/j.febslet.2004.12.071
Granot, D., Kelly, G., Stein, O., and David-Schwartz, R. (2014). Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 65 (3), 809–819. doi:10.1093/jxb/ert400
Guan, R. X., Qu, Y., Guo, Y., Yu, L. L., Liu, Y., Jiang, J. H., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80 (6), 937–950. doi:10.1111/tpj.12695
Guglielminetti, L., Perata, P., Morita, A., Loreti, E., Yamaguchi, J., and Alpi, A. (2000). Characterization of isoforms of hexose kinases in rice embryo. Phytochemistry 53 (2), 195–200. doi:10.1016/s0031-9422(99)00541-5
He, X., Feng, T., Zhang, D., Zhuo, R., and Liu, M. (2019). Identification and comprehensive analysis of the characteristics and roles of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in Sedum alfredii Hance responding to cadmium stress. Ecotoxicol. Environ. Saf. 167, 95–106. doi:10.1016/j.ecoenv.2018.09.122
Jang, J. C., León, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell. 9 (1), 5–19. doi:10.1105/tpc.9.1.5
Jungklang, J., Saengnil, K., and Uthaibutra, J. (2017). Effects of water-deficit stress and paclobutrazol on growth, relative water content, electrolyte leakage, proline content and some antioxidant changes in Curcuma alismatifolia Gagnep. cv. Chiang Mai Pink. Saudi J. Biol. Sci. 24 (7), 1505–1512. doi:10.1016/j.sjbs.2015.09.017
Kandel-Kfir, M., Damari-Weissler, H., German, M. A., Gidoni, D., Mett, A., Belausov, E., et al. (2006). Two newly identified membrane-associated and plastidic tomato HXKs: Characteristics, predicted structure and intracellular localization. Planta 224 (6), 1341–1352. doi:10.1007/s00425-006-0318-9
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 14 (2), 343–357. doi:10.1105/tpc.010362
Karve, A., Rauh, B. L., Xia, X., Kandasamy, M., Meagher, R. B., Sheen, J., et al. (2008). Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228 (3), 411–425. doi:10.1007/s00425-008-0746-9
Karve, R., Lauria, M., Virnig, A., Xia, X., Rauh, B. L., and Moore, B. (2010). Evolutionary lineages and functional diversification of plant hexokinases. Mol. Plant 3 (2), 334–346. doi:10.1093/mp/ssq003
Katz, M. E., Masoumi, A., Burrows, S. R., Shirtliff, C. G., and Cheetham, B. F. (2000). The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of extracellular proteases. Genetics 156 (4), 1559–1571. doi:10.1093/genetics/156.4.1559
Kim, Y. M., Heinzel, N., Giese, J. O., Koeber, J., Melzer, M., Rutten, T., et al. (2013). A dual role of tobacco hexokinase 1 in primary metabolism and sugar sensing. Plant Cell. Environ. 36 (7), 1311–1327. doi:10.1111/pce.12060
Kim, H. B., Cho, J. I., Ryoo, N., Shin, D. H., Park, Y. I., Hwang, Y. S., et al. (2016). Role of rice cytosolic hexokinase OsHXK7 in sugar signaling and metabolism. J. Integr. Plant Biol. 58 (2), 127–135. doi:10.1111/jipb.12366
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Lehmann, S., Funck, D., Szabados, L., and Rentsch, D. (2010). Proline metabolism and transport in plant development. Amino Acids 39 (4), 949–962. doi:10.1007/s00726-010-0525-3
Leisner, C. P., Yendrek, C. R., and Ainsworth, E. A. (2017). Physiological and transcriptomic responses in the seed coat of field-grown soybean (Glycine max L. Merr.) to abiotic stress. BMC Plant Biol. 17 (1), 242. doi:10.1186/s12870-017-1188-y
Leng, Z. X., Liu, Y., Chen, Z. Y., Guo, J., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228-70 in response to drought and salt stresses. Front. Plant Sci. 12, 628299. doi:10.3389/fpls.2021.628299
Li, C. N., Ng, C. K. Y., and Fan, L. M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi:10.1016/j.envexpbot.2014.06.014
Li, N. N., Qian, W. J., Wang, L., Cao, H. L., Hao, X. Y., Yang, Y. J., et al. (2017). Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant (Camellia sinensis). J. Plant Physiol. 209, 95–104. doi:10.1016/j.jplph.2016.11.007
Li, M., Chen, R., Jiang, Q. Y., Sun, X. J., Zhang, H., and Hu, Z. (2021). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105 (3), 333–345. doi:10.1007/s11103-020-01091-y
Liao, Y., Zou, H. F., Wei, W., Hao, Y. J., Tian, A. G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228 (2), 225–240. doi:10.1007/s00425-008-0731-3
Liu, Z. Y., Li, X. P., Zhang, T. Q., Wang, Y. Y., Wang, C., and Gao, C. Q. (2021). Overexpression of ThMYB8 mediates salt stress tolerance by directly activating stress-responsive gene expression. Plant Sci. 302, 110668. doi:10.1016/j.plantsci.2020.110668
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lugassi, N., Kelly, G., Fidel, L., Yaniv, Y., Attia, Z., Levi, A., et al. (2015). Expression of Arabidopsis hexokinase in citrus guard cells controls stomatal aperture and reduces transpiration. Front. Plant Sci. 6, 1114. doi:10.3389/fpls.2015.01114
Lugassi, N., Yadav, B. S., Egbaria, A., Wolf, D., Kelly, G., Neuhaus, E., et al. (2019). Expression of Arabidopsis hexokinase in tobacco guard cells increases water-use efficiency and confers tolerance to drought and salt stress. Plants-Basel 8 (12), 613. doi:10.3390/plants8120613
Ma, D. M., Xu, W. R., Li, H. W., Jin, F. X., Guo, L. N., Wang, J., et al. (2014). Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.) Protoplasma 251 (1), 219–231. doi:10.1007/s00709-013-0540-9
Ma, X. J., Fu, J. D., Tang, Y. M., Yu, T. F., Yin, Z. G., Chen, J., et al. (2020). GmNFYA13 improves salt and drought tolerance in transgenic soybean plants. Front. Plant Sci. 11, 587244. doi:10.3389/fpls.2020.587244
Matschinsky, F. M., Magnuson, M. A., Zelent, D., Jetton, T. L., Doliba, N., Han, Y., et al. (2006). The network of glucokinase-expressing cells in glucose homeostasis and the potential of glucokinase activators for diabetes therapy. Diabetes 55 (1), 1–12. doi:10.2337/diabetes.55.01.06.db05-0926
Minet, M., Dufour, M. E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2 (3), 417–422. doi:10.1111/j.1365-313x.1992.00417.x
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 (10), 490–498. doi:10.1016/j.tplants.2004.08.009
Moore, B. (2004). Bifunctional and moonlighting enzymes: Lighting the way to regulatory control. Trends Plant Sci. 9 (5), 221–228. doi:10.1016/j.tplants.2004.03.005
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 (5617), 332–336. doi:10.1126/science.1080585
Nilsson, A., Olsson, T., Ulfstedt, M., Thelander, M., and Ronne, H. (2011). Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol. 11, 32. doi:10.1186/1471-2229-11-32
Olias, R., Eljakaoui, Z., Li, J., De Morales, P. A., Marin-Manzano, M. C., Pardo, J. M., et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell. Environ. 32 (7), 904–916. doi:10.1111/j.1365-3040.2009.01971.x
Olsson, T., Thelander, M., and Ronne, H. (2003). A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J. Biol. Chem. 278 (45), 44439–44447. doi:10.1074/jbc.M306265200
Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell. 9 (12), 2197–2208. doi:10.1105/tpc.9.12.2197
Perez-Diaz, J., Batista-Silva, W., Almada, R., Medeiros, D. B., Arrivault, S., Correa, F., et al. (2021). Prunus Hexokinase 3 genes alter primary C-metabolism and promote drought and salt stress tolerance in Arabidopsis transgenic plants. Sci. Rep. 11 (1), 7098. doi:10.1038/s41598-021-86535-1
Plett, D., Safwat, G., Gilliham, M., Moller, I. S., Roy, S., Shirley, N., et al. (2010). Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. Plos One 5 (9), e12571. doi:10.1371/journal.pone.0012571
Ramegowda, V., Mysore, K. S., and Senthil-Kumar, M. (2014). Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 5, 323. doi:10.3389/fpls.2014.00323
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 (10), 1141–1146. doi:10.1038/ng1643
Riera, A., Ahuatzi, D., Herrero, P., Garcia-Gimeno, M. A., Sanz, P., and Moreno, F. (2008). Human pancreatic beta-cell glucokinase: Subcellular localization and glucose repression signalling function in the yeast cell. Biochem. J. 415 (2), 233–239. doi:10.1042/bj20080797
Rong, W., Qi, L., Wang, A. Y., Ye, X. G., Du, L. P., Liang, H. X., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12 (4), 468–479. doi:10.1111/pbi.12153
Roychoudhury, A., Paul, S., and Basu, S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell. Rep. 32 (7), 985–1006. doi:10.1007/s00299-013-1414-5
Saidi, A., and Hajibarat, Z. (2019). Characterization of cis-elements in hormonal stress-responsive genes in Oryza sativa. Asia-Pacific J. Mol. Biol. Biotechnol. 27 (1), 95–102. doi:10.35118/apjmbb.2019.027.1.10
Sarowar, S., Lee, J. Y., Ahn, E. R., and Pai, H. S. (2008). A role of hexokinases in plant resistance to oxidative stress and pathogen infection. J. Plant Biol. 51 (5), 341–346. doi:10.1007/bf03036136
Shi, H. Z., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 14 (2), 465–477. doi:10.1105/tpc.010371
Siemens, J., González, M. C., Wolf, S., Hofmann, C., Greiner, S., Du, Y., et al. (2011). Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 12 (3), 247–262. doi:10.1111/j.1364-3703.2010.00667.x
Sun, M. H., Ma, Q. J., Hu, D. G., Zhu, X. P., You, C. X., Shu, H. R., et al. (2018). The glucose sensor MdHXK1 phosphorylates a tonoplast Na+/H+ exchanger to improve salt tolerance. Plant Physiol. 176 (4), 2977–2990. doi:10.1104/pp.17.01472
Sun, T. J., Fan, L., Yang, J., Cao, R. Z., Yang, C. Y., Zhang, J., et al. (2019). A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na(+) efflux rate and K(+)/Na(+) ratio in Arabidopsis. BMC Plant Biol. 19 (1), 469. doi:10.1186/s12870-019-2084-4
Sun, T. J., Ma, N., Wang, C. Q., Fan, H. F., Wang, M. X., Zhang, J., et al. (2021). A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 12, 638340. doi:10.3389/fpls.2021.638340
Wang, X. Q., Li, L. M., Yang, P. P., and Gong, C. L. (2014). The role of hexokinases from grape berries (Vitis vinifera L.) in regulating the expression of cell wall invertase and sucrose synthase genes. Plant Cell. Rep. 33 (2), 337–347. doi:10.1007/s00299-013-1533-z
Wang, H. Y., Wang, H. L., Shao, H. B., and Tang, X. L. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7, 67. doi:10.3389/fpls.2016.00067
Wang, D., Liu, Y. X., Yu, Q., Zhao, S. P., Zhao, J. Y., Ru, J. N., et al. (2019). Functional analysis of the soybean GmCDPK3 gene responding to drought and salt stresses. Int. J. Mol. Sci. 20 (23), 5909. doi:10.3390/ijms20235909
Wang, R., Zhang, Y., Wang, C., Wang, Y. C., and Wang, L. Q. (2021a). ThNAC12 from Tamarix hispida directly regulates ThPIP2;5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiology Biochem. 163, 27–35. doi:10.1016/j.plaphy.2021.03.042
Wang, X. P., Niu, Y. L., and Zheng, Y. (2021b). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22 (11), 6125. doi:10.3390/ijms22116125
Xiao, W., Sheen, J., and Jang, J. C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44 (4), 451–461. doi:10.1023/a:1026501430422
Xu, F. Q., Li, X. R., and Ruan, Y. L. (2008). RNAi-mediated suppression of hexokinase gene OsHXK10 in rice leads to non-dehiscent anther and reduction of pollen germination. Plant Sci. 175 (5), 674–684. doi:10.1016/j.plantsci.2008.07.002
Xu, C., Shan, J., Liu, T., Wang, Q., Ji, Y., Zhang, Y., et al. (2022a). CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant physiol., kiac573. doi:10.1093/plphys/kiac573
Xu, H. R., Liu, Y., Yu, T. F., Hou, Z. H., Zheng, J. C., Chen, J., et al. (2022b). Comprehensive profiling of tubby-like proteins in soybean and roles of the GmTLP8 gene in abiotic stress responses. Front. Plant Sci. 13, 844545. doi:10.3389/fpls.2022.844545
Yang, G., Chen, B. X., Chen, T., Chen, J. H., Lin, X. Y., Yue, X. L., et al. (2022). BYPASS1-LIKE regulates lateral root initiation via exocytic vesicular trafficking-mediated PIN recycling in Arabidopsis. J. Integr. Plant Biol. 64 (5), 965–978. doi:10.1111/jipb.13243
Zhang, G. W., Liu, Z. L., Zhou, J. G., and Zhu, Y. L. (2008). Effects of Ca(NO3)(2) stress on oxidative damage, antioxidant enzymes activities and polyamine contents in roots of grafted and non-grafted tomato plants. Plant Growth Regul. 56 (1), 7–19. doi:10.1007/s10725-008-9281-8
Zhang, G. Y., Chen, M., Li, L. C., Xu, Z. S., Chen, X. P., Guo, J. M., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60 (13), 3781–3796. doi:10.1093/jxb/erp214
Zhang, Z. Y., Liu, X., Wang, X. D., Zhou, M. P., Zhou, X. Y., Ye, X. G., et al. (2012). An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 196 (4), 1155–1170. doi:10.1111/j.1469-8137.2012.04353.x
Zhang, Z., Zhang, J., Chen, Y., Li, R., Wang, H., Ding, L., et al. (2014). Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 41 (9), 6157–6166. doi:10.1007/s11033-014-3495-9
Zhang, C., Zhang, L., Fu, J., and Dong, L. (2020). Isolation and characterization of hexokinase genes PsHXK1 and PsHXK2 from tree peony (Paeonia suffruticosa Andrews). Mol. Biol. Rep. 47 (1), 327–336. doi:10.1007/s11033-019-05135-5
Zhao, J. Y., Lu, Z. W., Sun, Y., Fang, Z. W., Chen, J., Zhou, Y. B., et al. (2020a). The ankyrin-repeat gene GmANK114 confers drought and salt tolerance in Arabidopsis and soybean. Front. Plant Sci. 11, 584167. doi:10.3389/fpls.2020.584167
Keywords: Glycine max, hexokinase, gene family, VIGS-induced, salt tolerance
Citation: Chen S, Tian Z and Guo Y (2023) Characterization of hexokinase gene family members in Glycine max and functional analysis of GmHXK2 under salt stress. Front. Genet. 14:1135290. doi: 10.3389/fgene.2023.1135290
Received: 31 December 2022; Accepted: 13 February 2023;
Published: 23 February 2023.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Chao Liu, Qujing Normal University, ChinaYalong Xu, Zhengzhou Tobacco Research Institute of CNTC, China
Copyright © 2023 Chen, Tian and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqi Guo, Z3VvX3l1cWkyMDIxQDEyNi5jb20=
†These authors have contributed equally to this work
 Shuai Chen1†
Shuai Chen1† Yuqi Guo
Yuqi Guo