- 1Translational Oncology Laboratory, Department of Zoology, Hansraj College, Delhi University, New Delhi, India
- 2Radiation and Cancer Therapeutics Lab, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
- 3Immunology and Infectious Disease Biology Lab, CSIR-Institute of Genomics and Integrative Biology, New Delhi, India
- 4Department of Biotechnology, Maharishi Markandeshwar Engineering College, Maharishi Markandeshwar (Deemed to be University), Ambala, India
Breast cancer is the most commonly diagnosed cancer and a leading cause of death in women worldwide. It is a heterogeneous disease, as shown by the gene expression profiles of breast cancer samples. It begins in milk-producing ducts, with a high degree of diversity between and within tumors, as well as among cancer-bearing individuals. The enhanced prevalence of breast cancer is influenced by various hormonal, lifestyle, and environmental factors, and very early onset of the disease correlates strongly with the risk of local and distant recurrence. Many subtypes are difficult to treat with conventional therapeutic modalities, and therefore, optimal management and early diagnosis are the first steps to minimizing the mortality linked with breast cancer. The use of newer methods of nanotechnology extends beyond the concept of synthesizing drug delivery mechanisms into the creation of new therapeutics, such as delivering chemotherapeutics with nanomaterial properties. Exosomes, a class of nanovesicles, are emerging as novel tools for deciphering the patient-specific proteins and biomarkers across different disease models, including breast cancer. In this review, we address the role of exosomal miRNA in breast cancer diagnosis and treatment.
Highlights
➣ Exosomal miRNA role in the progression and diagnosis of breast cancer
➣ Exosomal miRNA role in chemoresistance of breast cancer
➣ Exosomal miRNA role in the different hallmarks of breast cancer
Introduction
New estimates suggest that about one in 300 women are diagnosed with an aggressive form of breast cancer before age 40 (DeSantis et al., 2019). Breast cancer is classified as either specific [20%–25%] or non-specific ductal carcinoma [60%–75%] subtypes, which include papillary, mucinous, lobular, and tubular tumors.
Its molecular classification is on the basis of the presence and absence of human epidermal growth factor 2 [HER2], estrogen or progesterone receptor [ERBB2] (hormone receptor negative/ERBB2− [70%], positive/ERBB2+ [15%–20%], and triple negative in which all the three markers are absent [15%]), basal-like [ER-/HER2−], HER2 enriched [HER2+], and combined luminal A and B [ER+/HER2−] (Johnson et al., 2020).
The triple-negative breast cancer (TNBC) subtype has a poor prognosis and a maximum rate of systemic recurrence. Various approaches have been applied to eradicate it, including modulation of the tumor microenvironment to increase CTL activity and other immunotherapy-based approaches like immune checkpoint inhibition by neutralizing antibodies and neoadjuvant-based immunotherapy (Jia et al., 2017).
These locally advanced and metastatic lesions show extensive nodal involvement and an inflammatory phenotype. The prognosis of these lesions is often unfavorable; despite an aggressive treatment regime, it eventually leads to an enhanced mortality rate (Hapach et al., 2021).
Newer innovations in systemic therapy, such as surgical procedures, radiotherapy, and the development of new advanced targeted agents, have improved the clinical outcomes of this malignancy. Therefore, optimal management and early diagnosis are the first key steps toward minimizing the mortality linked with this malignancy. Targeting molecular pathways and elucidating the molecular cascade and their relationship with other signaling molecules has led to the evolution of practical combination therapies that have been proven successful to a larger extent.
Currently, available cancer therapies are limited to surgery, radiation, and chemotherapy, with a high risk of bystander effect and damage to normal tissues. These conventional therapies have side effects and chemo/radioresistant and other toxicity-related issues. Nanotechnology-based methods selectively target cancerous cells, minimizing therapy side effects and enhancing the probability of survival. Exosomes, a class of nanovesicles that mediate cellular communications via delivering many types of biomolecules [oncogenes, protein DNA, and RNA, including different pharmacological compounds], are being explored to model the patient-specific proteins and biomarkers across different disease models, including breast cancer. An interesting aspect of these nanocarriers is that they can serve as informative sources of novel biomarkers, and their cargo can deliver therapeutic molecules across the target sites. Many studies have signified the role of exosomes in breast cancer biology, including the identification of signature molecules and regulation of the breast cancer tumor microenvironment. This comprehensive review addresses the role of exosomal miRNA as a diagnostic tool and treatment for breast cancer therapy.
The emerging role of liquid biopsy for breast cancer diagnosis
Identification of stage-specific cancer biomarkers is a prerequisite for early detection. Owing to the heterogeneous nature, small sample size, and varied genomic profiles of tumors, conventional biopsies often fail to reflect the whole nature of primary or secondary metastasis. Moreover, frequent tissue sampling from cancer patients for the identification of tumor-associated genetic changes, therapy responses, and investigating tumor dynamics is also a major concern. Therefore, there is an unmet need for novel low-cost and non-invasive sampling techniques and methods that could improve early detection and screening. In this context, identification of circulating tumor cells (CTCs) separated from the original tumor bulk has been considered a gold standard for identifying tumor-related evidence. However, due to the reduced availability of CTCs, isolation of genetic materials from the bloodstream or any other biological fluid is an alternate option that is equally convenient and minimally invasive.
Compared to direct tumor biopsies, collection from bioliquids seems an attractive alternative source for clinical application. In this context, the utility of exosomes has recently been reviewed in different tumor models. Exosome-based technologies offer several advantages over the existing traditional methods of biopsies as they show a universal presence across different biofluids, and the ease of isolation and characterization of their cargo makes them attractive tools (Wang et al., 2021).
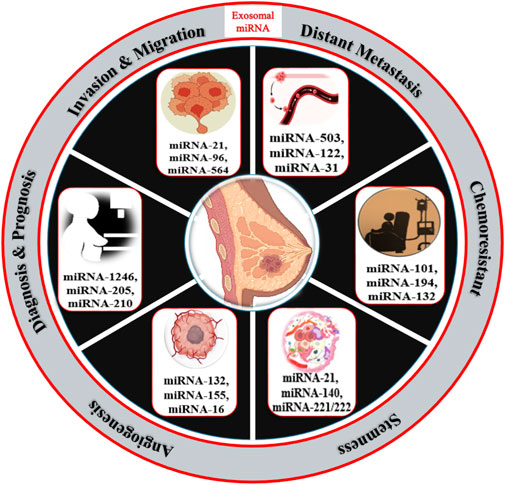
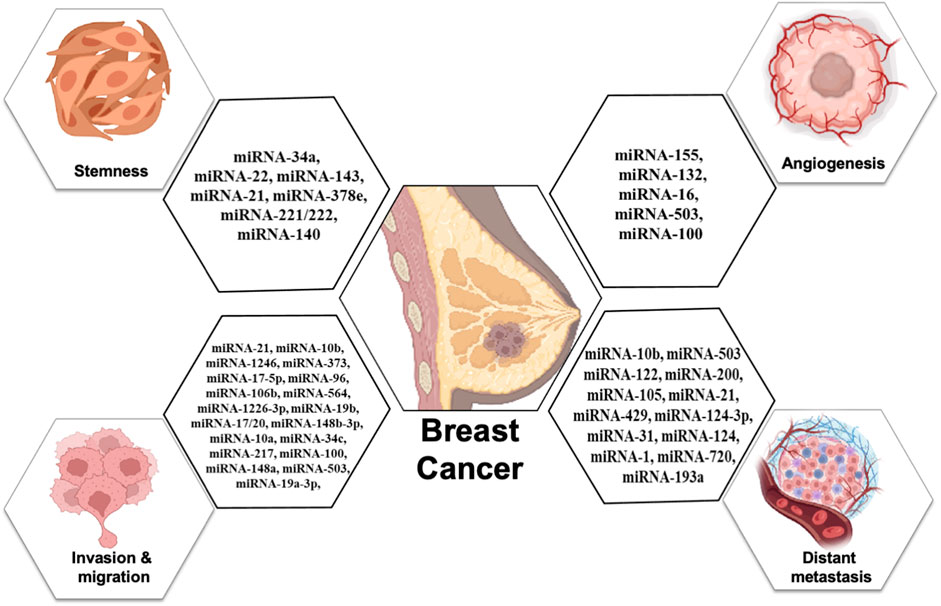
Breast cancer is one of the topmost threats to women’s health as diverse factors participate in tumorigenesis events. Among them, cell proliferation, stemness, metastasis, angiogenesis, epithelial-to-mesenchymal transition, and chemoresistance are major contributors to malignancy and reoccurrence. MicroRNAs that are specifically packed and secreted in exosomes are known as “exosomal microRNAs [miRNAs].” In contrast to different vesicular populations present in the biological fluids like apoptotic bodies and large vesicles, these exosomes more clearly represent the information present on the tumor. In this review, we summarize the utility and potential of small nanovesicles (exosomes) in different biological fluids, with a special focus on breast cancer (Figure 1), and show the different exosomal miRNAs in breast cancer progression.
Exosomal microRNAs in BC diagnosis
Cancer patients have more tumor-derived exosomes in circulation than healthy individuals (Tavoosidana et al., 2011). Exosomal miRNAs in blood have the potential to be new circulating biomarkers for early detection and diagnosis of many cancers, including breast cancer (Ogata-Kawata et al., 2014). Figure 1 shows some exosomal miRNAs in the diagnosis of breast cancer. Exosomal miR-1246 and miR-21 were reported in plasma as functional measures for breast cancer diagnosis (Hannafon et al., 2016; Singh et al., 2021). Li et al. (2018) reported an exosomal miR106a-363 cluster as a novel diagnostic biomarker in breast cancer (Li et al., 2018). Of them, four exosomal miRNAs are plasma-derived [miR-106a-5p, miR-92a-2-5p, miR-106a-3p, and miR-20b-5p], while four are serum-derived [miR-20b-5p, miR-106a-5p, miR-92a-2-5p, and miR-106a-3p] and showed higher expression in breast cancer patients compared with healthy controls (Stevic et al., 2018). The expression levels of exosomal miR-101 and miR-372 were higher in the serum of 50 breast cancer patients than in 12 healthy individuals (Eichelser et al., 2014). Of 435 breast cancer patients, 224 were TNBC patients, and 211 were HER-2 positive. Analysis of their plasma for exosomal miRNA revealed 13 miRNAs were lower and five were higher in HER-2 positive compared with the levels of TNBC patients (Stevic et al., 2018). The expression levels of miR-223-3p of invasive ductal carcinoma [IDC] patients were showing significantly many fold changes from ductal carcinoma in situ [DCIS] patients as well as from healthy controls (Yoshikawa et al., 2018). Ni et al. (2018) also reported when the expression of exosomal miR-30b, miR-16, and miR-93 were checked in 42 DCIS, 111 IDC patients, and 39 healthy individuals, out of them exosomal miR-16 show significant higher expression in plasma of BC patients as well as in DCIS patients than healthy control while exosomal miR-93 showed higher expression in DCIS patients as compared with IDC patients (Ni et al., 2018).
Exosomal microRNAs in BC for chemoresistance
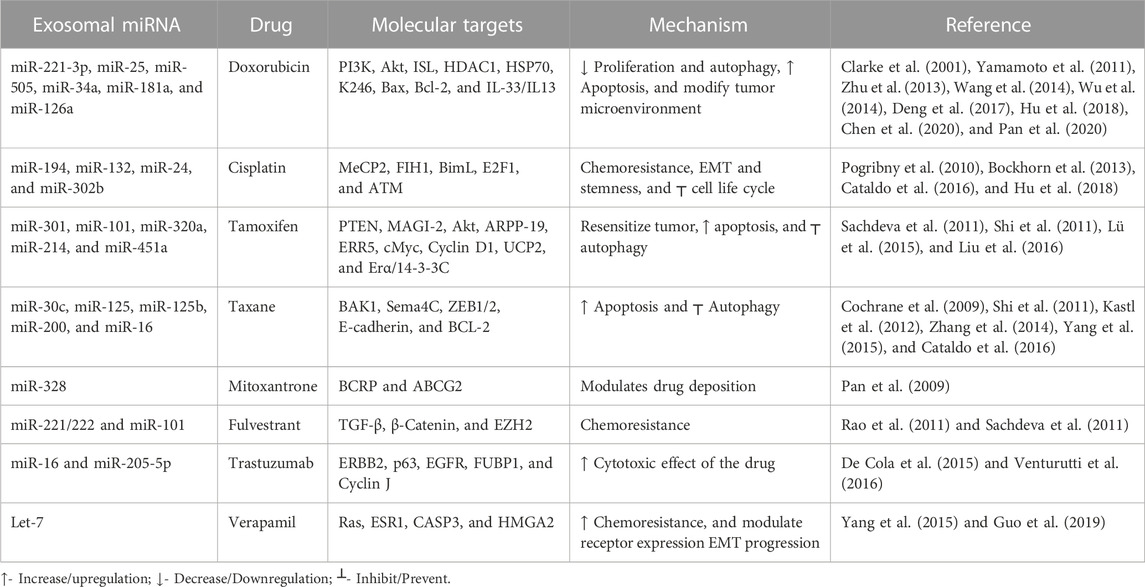
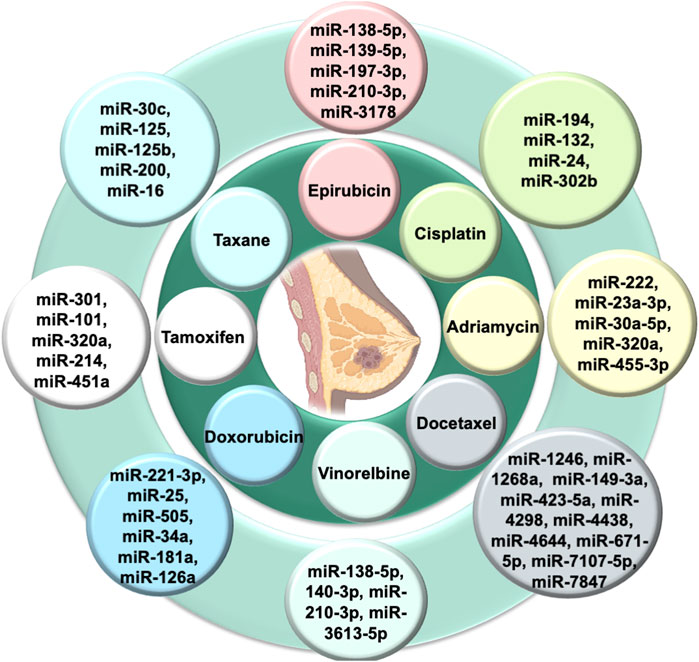
Almost 90% of chemotherapy failure occurs due to long-term and repetitive usage of similar types of drugs, leading to the chemoresistance that is a major hurdle for breast cancer treatment. There is a need to understand the molecular mechanism in chemoresistance to minimize the recurrence rate. Many studies showed the role of exosomal miRNA in the induction of chemoresistance in breast cancer (Tang et al., 2021). Here, we review some signaling pathways that are specifically targeted by exosomal miRNA to regulate the particular drug sensitivity of breast cancer (see Table 1).
Topoisomerase interactive agents
Doxorubicin is one of the commonly used drugs that intercalate into the DNA double helix, inhibit topoisomerase II enzyme activity, and attack mitochondrial and genomic DNA via ROS induction (van der Zanden et al., 2021). A previous study revealed a correlation between doxorubicin resistance and miRNA: around 309 miRNAs decreased, and 66 miRNAs increased (Chen et al., 2018). Akt is mainly involved in the signaling transduction pathway to modulate cell proliferation and DNA repair. It suppresses apoptosis, improves cell survival, and plays a significant role in chemoresistance (Yamamoto et al., 2011). Exosomal miR-221-3p regulates the PI3K/Akt/phosphoinositide-3-kinase regulatory subunit 1 [PIK3R1] to acquire the doxorubicin resistance (Pan et al., 2020). Meanwhile, exosomal miR-505 restores doxorubicin sensitivity by suppressing Akt3 activity and increasing apoptosis (Yamamoto et al., 2011). Mitoxantrone is also a commonly used topoisomerase II inhibitor, a chemotherapeutic drug that blocks the cell cycle (Evison et al., 2016). Increased expression of exosomal miR-328 was linked with increased sensitization of MCF-7/MX100 cells to mitoxantrone via downregulating the breast cancer resistance protein (BCRP/ABCG2) (Pan et al., 2009).
Platinum analogs: Cisplatin
Cisplatin causes double-strand DNA breaks by crosslinking via direct binding to DNA (Dickson et al., 2011). In cisplatin-resistant breast cancer cell lines, exosomal miR-194 and exosomal miR-132 inhibit the methyl-CpG-binding protein 2 [MECP2] (Cataldo et al., 2016). Some exosomal miRNAs downregulate chemoresistance (Hu et al., 2018). After cisplatin treatment, exosomal miRNA-302b causes cell cycle arrest and induces apoptosis (Bockhorn et al., 2013).
Antimicrotubule agents
Paclitaxel and docetaxel are tricyclic compounds under taxane that target the microtubules resulting in defects in cell division (Willson et al., 2019). Paclitaxel resistance occurs via exosomal miR-30c by modulating EMT-linked molecules, including twinfilin 1 and interleukin-11 (Kutanzi et al., 2011). Inversely, exosomal miR-125b inhibits the semaphorin 4C (inducer of EMT) and enhances paclitaxel sensitivity. BCL2 and BCL2 homologous killer/antagonist [BAK1] types of apoptosis-linked molecules are also involved in the exosomal microRNA-mediated chemoresistance. Exosomal miR-125 modulates BAK1 and downregulates the paclitaxel-induced apoptosis, and exosomal miR-16/BCL2 enhances docetaxel sensitivity and results in increased apoptosis (Kastl et al., 2012).
Hormonal agents
Tamoxifen inhibits estrogen-mediated growth (Cui et al., 2015). PTEN regulates the PI3K/Akt/mTOR signal transduction pathway. Loss of the function of PTEN (inhibitor of PI3K) promotes chemoresistance (Nagata et al., 2004). Exosomal miR-101 suppresses the PTEN, resulting in the activation of Akt by regulating membrane-linked guanylate kinase [MAGI-2] to impart tamoxifen resistance in breast cancer (Sachdeva et al., 2011). Autophagy is also a crucial factor in maintaining homeostasis and is linked to chemoresistance. Exosomal miR-214 inhibits uncoupling protein 2 [UCP2]-dependent autophagy and restores the tamoxifen response to cancer cells (Yu et al., 2015). Exosomal miR-221/222 promotes fulvestrant resistance in breast cancer cell lines by targeting β-catenin and TGF-β (Rao et al., 2011). Exosomal miR-205-5p sensitizes breast cells to trastuzumab by modulating ERBB2 and targeting the P63/EGFR axis (De Cola et al., 2015) Figure 3) shows the role of various exosomal miRNAs in breast cancer chemoresistance (Figure 2).
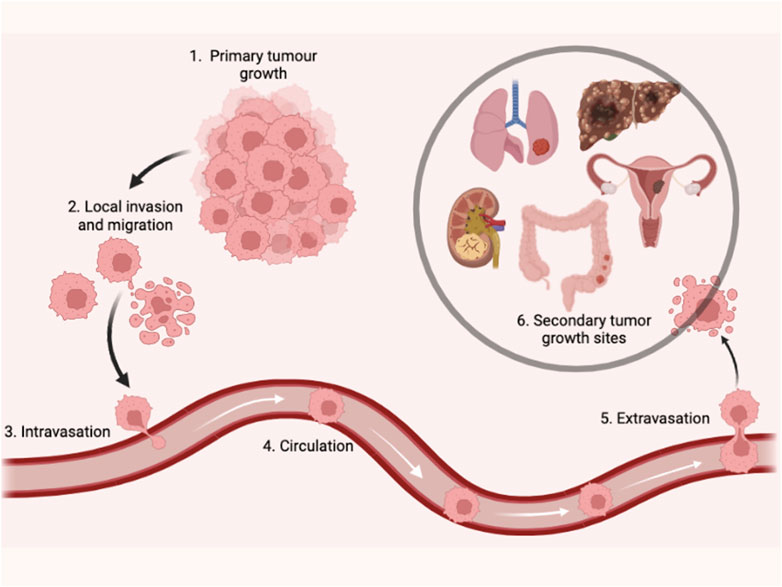
Exosomal miRNAs in BC invasion, migration, and metastasis
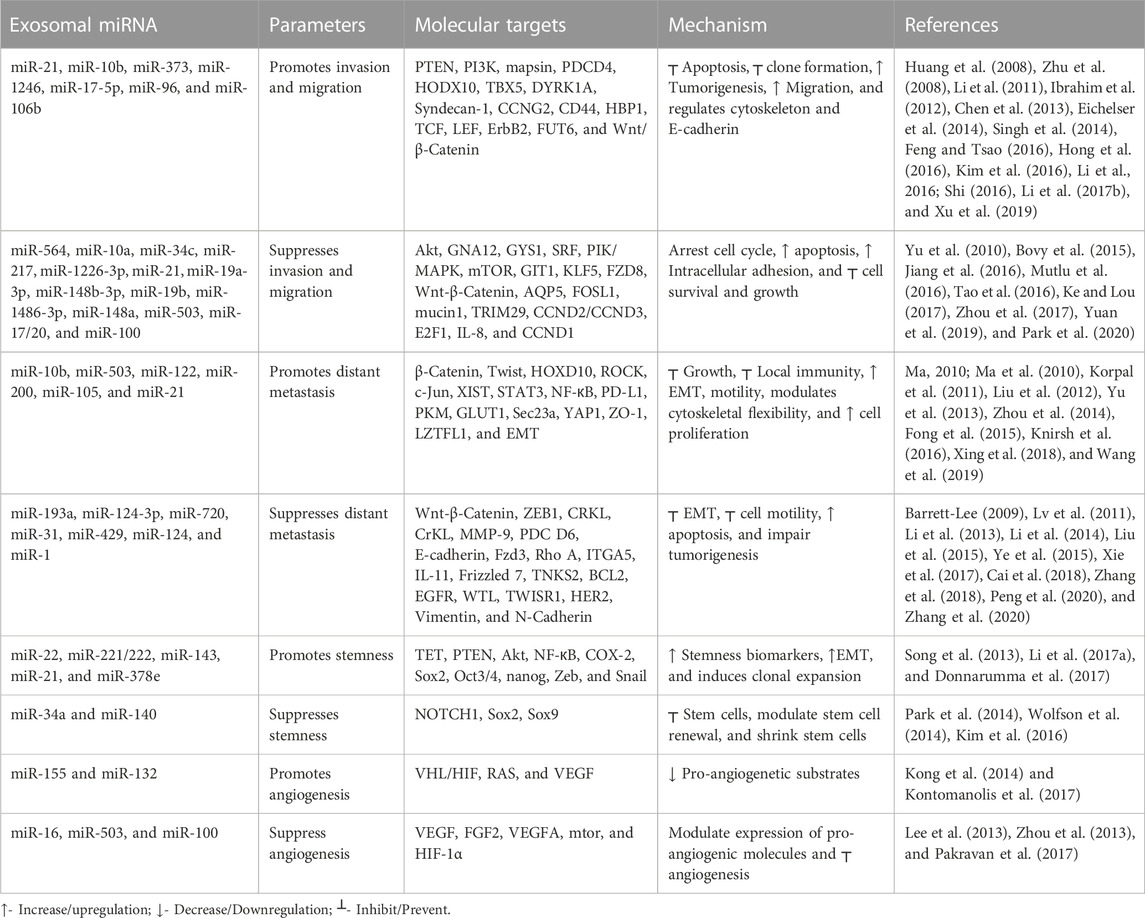
Metastasis is a pivotal factor for poor overall survival in breast cancer patients. It has been reported that exosomal miRNAs play a crucial role in almost every step of many biological processes in breast cancer (Liu et al., 2019; Figure 3). Many studies have demonstrated the dual role of exosomal miRNAs on breast cancer metastasis and related processes. In Table 2, we have mentioned some exosomal miRNAs along with their underlying mechanisms that are actively involved in metastasis, invasion, and migration. Table 2 shows the function of exosomal miRNAs in different hallmarks of BC, including invasion, migration, metastasis, stemness, and angiogenesis.
Exosomal microRNAs in the stemness of breast cancer cells
Cancer stem cells are groups of undifferentiated cells that have the potential to differentiate. Self-renewing populations of cells give rise to tumor heterogeneity, promoting metastasis, therapy resistance, and tumor recurrence (Vlashi and Pajonk, 2015). Breast cancer stem cells (BCSCs) mediate drug resistance and tumorigenesis because they are mostly found in the quiescent G0 phase, have high DNA repairability, and increase ABC transporter expression (Zhang et al., 2022). Exosomal miRNAs play a pivotal role in cancer stem cell maintenance. Exosomal miR-221/222 inhibits the PTEN activity that, in turn, activates the Akt/NF-κB/COX-2 signal transduction pathway and promotes stemness-like traits in breast cancer cells (Li B. et al., 2017). Exosomal miR-22 suppresses the TET/miR-200 axis via inhibiting TET [ten eleven translocation-DNA demethylase family], resulting in increased stemness and EMT (Song et al., 2013). Exosomal miRNAs target stemness by modulating the function and expression of breast cancer stemness-linked genes and their elements. Exosomal miR-34a downregulates the NOTCH1 expression in breast cancer, leading to decreased stemness in the mammosphere (Park et al., 2014). Sox9 is an oncogenic transcription factor that induces the transformation of mammary stem cells from differentiated mammary epithelial cells, which is crucial for breast cancer initiation and malignancy (Guo et al., 2012). Exosomal miR-140 decreases the expression of SOX2/SOX9 and reduces the stem cell population and disordered stem cell renewal (Wolfson et al., 2014).
Exosomal microRNAs in angiogenesis of breast cancer cells
Exosomal miRNAs play a significant role in angiogenesis to reduce the metastasis of breast cancer. Kong et al. (2014) reported that exosomal miR-155 modulates the Von Hippel–Lindau [VHL]/hypoxia-inducible factor and its downstream genes, including pyruvate kinase isozyme type M2, CD44, interleukin 6, and vascular endothelial growth factor in an in vivo mice model (Kong et al., 2014). Exosomal miR-132 enhances the sensitivity of endothelial cells to VEGF by decreasing the activity of the RAS suppressor, augmenting angiogenesis (Kontomanolis et al., 2017). Exosomal miR-16 halts the expression of VEGF by playing an anti-angiogenic role (Lee et al., 2013). Some exosomal miRNAs also target the tumor microenvironment and halt angiogenesis; for example, exosomal miR-503 regulates the expression of VEGFA and fibroblast growth factor 2 and hampers angiogenesis (Zhou et al., 2013). Exosomal miR-100 decreases the expression of human umbilical vein endothelial cells [HUVECs] and targets mTOR/[HIF-1α]/VEGF to decrease angiogenesis (Pakravan et al., 2017). Figure 4 shows the role of different exosomal miRNAs in different hallmarks (invasion and migration, distant metastasis, stemness, and angiogenesis) of breast cancer.
Conclusion and future perspective
Exosomal miRNAs play a significant role in almost all hallmarks of breast cancer, including chemoresistance, metastasis migration, invasion, stemness, and angiogenesis. Exosomal miRNAs show a dual mode of action w.r.t to chemoresistance and drug sensitivity. As breast cancer has many subtypes with different prognostic and clinicopathological features, it is necessary to examine the specific miRNA for multiple subtypes to evaluate the targeted therapeutic regime in breast cancer. The exosomal content w.r.t. miRNA increases to varying degrees that distinguish healthy controls from breast cancer patients and TNBC from other subtypes. Exosomal miRNAs may serve as an indication of the early stage of breast cancer; for example, exosomal miR-221/222 is found in almost every stage of breast cancer oncogenesis, and this exosomal miRNA-221 can predict the breast cancer origin, progress, and treatment effect. In stages I and II of breast cancer, miR-801, miR-127-3p, miR-148b, and miR-409-3p increased significantly, which is helpful for early detection.
The equilibrium types and numbers of exosomal miRNAs vary in pathological conditions; they serve as useful biomarkers for breast cancer prognosis and diagnosis. Exosomal miRNAs exert pleiotropic impacts on breast cancer hallmarks and clinical implications. (Luo et al. (2023) revealed the role of tumor-derived exosomes as a messenger and communicator between tumor cells and their microenvironment that can reshape and enhance tumor development and metastasis. They enhance the development of the pre-metastatic niche to promote tumor colonization and play a pivotal role in the early diagnosis and evaluation of future metastatic development (Luo et al., 2023). Exosomes are used as drug carriers (e.g., chemotherapy and miRNA) and as biomarkers because they exhibit antigen-presenting traits that have a crucial function in cancer immunotherapy (Hosseinikhah et al., 2023). Prakriti [phenotype-associated Ayurveda constitution] is linked with other fields like genomics, physiology, psychology, and therapeutics. It has a role in the evaluation of therapeutics such as early diagnosis, indicating disease susceptibility, prevention of diseases, drug design, and customization of therapy [lifestyle, drug, and diet]. This holds great potential for personalized medicine pharmacogenomics for predictive or preventive medicine (Sharma and Prajapati, 2020). This review gives a new insight that helps us examine prognostic, predictive, and diagnostic markers at the cellular and molecular levels to fill research gaps for novel therapeutic approaches and for future personalized medicine.
Author contributions
TS and MK: conceptualization, data curation, formal analysis, funding acquisition, investigation, project administration, resources, software, supervision, validation, visualization, and writing—original draft, and review and editing. LM, CB, VS, and HT: formal analysis and writing—review. All authors have read and approved the final manuscript.
Funding
The study was financially supported by ICMR-DHR, Government of India to TS (ICMR-DHR Young Scientist Fellowship, F.NO: R. 12014/29/2022/HR), and partly supported by college Research and Development Cell (RDC) Scheme (File no. HRC/RDC/2021/RP/13).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barrett-Lee, P. J. a. I. B. C. (2009). A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis, 6, 24–25.
Bockhorn, J., Dalton, R., Nwachukwu, C., Huang, S., Prat, A., Yee, K., et al. (2013). MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013 4, 1393. doi:10.1038/ncomms2393
Bovy, N., Blomme, B., Frères, P., Dederen, S., Nivelles, O., Lion, M., et al. (2015). Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 6, 10253, doi:10.18632/oncotarget.3520
Cai, W.-L., Huang, W.-D., Li, B., Chen, T.-R., Li, Z.-X., Zhao, C.-L., et al. (2018). microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol. Cancer 17, 9, doi:10.1186/s12943-017-0746-0
Cataldo, A., Cheung, D. G., Balsari, A., Tagliabue, E., Coppola, V., Iorio, M. V., et al. (2016). miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget 7, 786–797. doi:10.18632/oncotarget.6381
Chen, W.-X., Xu, L.-Y., Qian, Q., He, X., Peng, W.-T., Zhu, Y.-L., et al. (2018). Analysis of miRNA signature differentially expressed in exosomes from adriamycin-resistant and parental human breast cancer cells. Biosci. Rep. 38 (6), BSR20181090. doi:10.1042/BSR20181090
Chen, W., Cai, F., Zhang, B., Barekati, Z., and Zhong, X. Y. J. T. B. (2013). The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumour Biol. 34, 455–462. doi:10.1007/s13277-012-0570-5
Chen, Z., Pan, T., Jiang, D., Jin, L., Geng, Y., Feng, X., et al. (2020). The lncRNA-GAS5/miR-221-3p/DKK2 axis modulates ABCB1-mediated adriamycin resistance of breast cancer via the Wnt/β-catenin signaling pathway. Mol. Ther. Nucleic Acids 19, 1434–1448. doi:10.1016/j.omtn.2020.01.030
Clarke, R., Leonessa, F., Welch, J. N., and Skaar, T. C. J. P. R. (2001). Cellular and molecular pharmacology of antiestrogen action and resistance, Pharmacol. Rev. 53, 25–72.
Cochrane, D. R., Spoelstra, N. S., Howe, E. N., Nordeen, S. K., and Richer, J. K. J. M. C. T. (2009). MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 8, 1055–1066. doi:10.1158/1535-7163.MCT-08-1046
Cui, J., Yang, Y., Li, H., Leng, Y., Qian, K., Huang, Q., et al. (2015). MiR-873 regulates ERα transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene 34, 4018–3907. doi:10.1038/onc.2015.201
De Cola, A., Volpe, S., Budani, M., Ferracin, M., Lattanzio, R., Turdo, A., et al. (2015). miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 6 (7), e1823–e1823.
Deng, Z., Rong, Y., Teng, Y., Zhuang, X., Samykutty, A., Mu, J., et al. (2017). Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 36, 639, doi:10.1038/onc.2016.229
Desantis, C. E., Ma, J., Gaudet, M. M., Newman, L. A., Miller, K. D., Gooding Sauer, A., et al. (2019). Breast cancer statistics, 2019, Breast cancer stat. 2019 69, 438, doi:10.3322/caac.21583
Dickson, M. A., Carvajal, R. D., Merrill, A. H., Gonen, M., Cane, L. M., and Schwartz, G. K. J. C. C. R. (2011). A phase I clinical trial of safingol in combination with cisplatin in advanced solid tumors. Clin. Cancer Res. 17, 2484–2492. doi:10.1158/1078-0432.CCR-10-2323
Donnarumma, E., Fiore, D., Nappa, M., Roscigno, G., Adamo, A., Iaboni, M., et al. (2017). Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 8, 19592–19608. doi:10.18632/oncotarget.14752
Eichelser, C., Stückrath, I., Müller, V., Milde-Langosch, K., Wikman, H., Pantel, K., et al. (2014). Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 5, 9650–9663. doi:10.18632/oncotarget.2520
Evison, B. J., Sleebs, B. E., Watson, K. G., Phillips, D. R., and Cutts, S. M. J. M. R. R. (2016). Mitoxantrone, more than just another topoisomerase II poison. Med. Res. Rev. 36, 248–299. doi:10.1002/med.21364
Feng, Y.-H., and Tsao, C.-J. J. B. R. (2016). Emerging role of microRNA-21 in cancer. Biomed. Rep. 5, 395, doi:10.3892/br.2016.747
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194. doi:10.1038/ncb3094
Guo, L., Cheng, X., Chen, H., Chen, C., Xie, S., Zhao, M., et al. (2019). Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett. 452, 213–225. doi:10.1016/j.canlet.2019.03.032
Guo, W., Keckesova, Z., Donaher, J. L., Shibue, T., Tischler, V., Reinhardt, F., et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028. doi:10.1016/j.cell.2012.02.008
Hannafon, B. N., Trigoso, Y. D., Calloway, C. L., Zhao, Y. D., Lum, D. H., Welm, A. L., et al. (2016). Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 18, 90, doi:10.1186/s13058-016-0753-x
Hapach, L. A., Carey, S. P., Schwager, S. C., Taufalele, P. V., Wang, W., Mosier, J. A., et al. (2021). Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res. 81, 3649–3663. doi:10.1158/0008-5472.CAN-20-1799
Hong, Y., Liang, H., Wang, Y., Zhang, W., Zhou, Y., Chen, S. A., et al. (2016). miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 6, 37421, doi:10.1038/srep37421
Hosseinikhah, S. M., Gheybi, F., Moosavian, S. A., Shahbazi, M.-A., Jaafari, M. R., Sillanpää, M., et al. (2023). Role of exosomes in tumour growth, chemoresistance and immunity: State-of-the-art. State-of-the-art 31, 32–50. doi:10.1080/1061186X.2022.2114000
Hu, W., Tan, C., He, Y., Zhang, G., Xu, Y., Tang, J., et al. (2018). Functional miRNAs in breast cancer drug resistance. OncoTargets Ther., 1529–1541.
Huang, Q., Gumireddy, K., Schrier, M., Le Sage, C., Nagel, R., Nair, S., et al. (2008). The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 10, 202–210. doi:10.1038/ncb1681
Ibrahim, S. A., Yip, G. W., Stock, C., Pan, J. W., Neubauer, C., Poeter, M., et al. (2012). Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int. J. Cancer 131, E884–E896. doi:10.1002/ijc.27629
Jia, H., Truica, C. I., Wang, B., Wang, Y., Ren, X., Harvey, H. A., et al. (2017). Immunotherapy for triple-negative breast cancer: Existing challenges and exciting prospects. Drug resist. updat. 32, 1–15. doi:10.1016/j.drup.2017.07.002
Jiang, Q., He, M., Guan, S., Ma, M., Wu, H., Yu, Z., et al. (2016). MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumour Biol. 37, 5001–5011. doi:10.1007/s13277-015-4342-x
Johnson, K. S., Conant, E. F., and Soo, M. S. (2020). Molecular subtypes of breast cancer: A review for breast radiologists. J. Breast Imaging 3, 12–24. doi:10.1093/jbi/wbaa110
Kastl, L., Brown, I., and Schofield Andrew Craig, (2012). miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 131, 445–454.
Ke, K., and Lou, T. J. O. L. (2017). MicroRNA-10a suppresses breast cancer progression via PI3K/Akt/mTOR pathway. Oncol. Lett. 14, 5994–6000. doi:10.3892/ol.2017.6930
Kim, J., Siverly, A. N., Chen, D., Wang, M., Yuan, Y., Wang, Y., et al. (2016). Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Res. 76, 6424, doi:10.1158/0008-5472.CAN-16-1571
Knirsh, R., Ben-Dror, I., Modai, S., Shomron, N., and Vardimon, L. J. O. (2016). MicroRNA 10b promotes abnormal expression of the proto-oncogene c-Jun in metastatic breast cancer cells. Oncotarget 7, 59932–59944. doi:10.18632/oncotarget.11000
Kong, W., He, L., Richards, E., Challa, S., Xu, C., Permuth-Wey, J., et al. (2014). Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33, 679–689. doi:10.1038/onc.2012.636
Kontomanolis, E., Mitrakas, A., Giatromanolaki, A., Kareli, D., Panteliadou, M., Pouliliou, S., et al. (2017). A pilot study on plasma levels of micro-RNAs involved in angiogenesis and vascular maturation in patients with breast cancer. Med. Oncol. 34, 20, doi:10.1007/s12032-016-0881-2
Korpal, M., Ell, B. J., Buffa, F. M., Ibrahim, T., Blanco, M. A., Celià-Terrassa, T., et al. (2011). Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17, 1101–1108. doi:10.1038/nm.2401
Kutanzi, K. R., Yurchenko, O. V., Beland, F. A., Checkhun, V. F., and Pogribny, I. P. J. C. E. (2011). MicroRNA-mediated drug resistance in breast cancer. Clin. Epigenetics 2, 171–185. doi:10.1007/s13148-011-0040-8
Lee, J.-K., Park, S.-R., Jung, B.-K., Jeon, Y.-K., Lee, Y.-S., Kim, M.-K., et al. (2013). Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS One 8, e84256. doi:10.1371/journal.pone.0084256
Li, B., Lu, Y., Yu, L., Han, X., Wang, H., Mao, J., et al. (2017a). miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem. Biol. Interact., 277, 33, doi:10.1016/j.cbi.2017.08.014
Li, H., Bian, C., Liao, L., Li, J., and Zhao, R. C. (2011). miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res. Treat. 126, 565–575.
Li, L.-Z., Zhang, C. Z., Liu, L.-L., Yi, C., Lu, S.-X., Zhou, X., et al. (2014). miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis 35, 469–478. doi:10.1093/carcin/bgt330
Li, L., Luo, J., Wang, B., Wang, D., Xie, X., Yuan, L., et al. (2013). Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol. Cancer 12, 163, doi:10.1186/1476-4598-12-163
Li, M., Zhou, Y., Xia, T., Zhou, X., Huang, Z., Zhang, H., et al. (2018). Circulating microRNAs from the miR-106a–363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat., 170, 257, doi:10.1007/s10549-018-4757-3
Li, N., Liu, Y., Miao, Y., Zhao, L., Zhou, H., and Jia, L. J. I. L. (2016). MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life 68, 764–775. doi:10.1002/iub.1541
Li, X. J., Ren, Z. J., Tang, J. H., Yu, Q. J. C. P., and Biochemistry, (2017b). Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell. Physiol. Biochem. 44, 1741, doi:10.1159/000485780
Liu, Q., Peng, F., and Chen, J. J. I. J. O. M. S. (2019). The role of exosomal microRNAs in the tumor microenvironment of breast cancer. Int. J. Mol. Sci., 20, 3884, doi:10.3390/ijms20163884
Liu, T., Hu, K., Zhao, Z., Chen, G., Ou, X., Zhang, H., et al. (2015). MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin pathway. Oncotarget 6, 41638–41649. doi:10.18632/oncotarget.5873
Liu, Y., Zhao, J., Zhang, P.-Y., Zhang, Y., Sun, S.-Y., Yu, S.-Y., et al. (2012). MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med. Sci. Monit. 18, BR299–BR308. doi:10.12659/msm.883262
Liu, Z.-R., Song, Y., Wan, L.-H., Zhang, Y.-Y., and Zhou, L.-M. J. L. S. (2016). Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 149, 104–113. doi:10.1016/j.lfs.2016.02.059
Lü, M., Ding, K., Zhang, G., Yin, M., Yao, G., Tian, H., et al. (2015). MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRγ. Sci. Rep. 5, 8735.
Luo, X., Li, Y., Hua, Z., Xue, X., Wang, X., Pang, M., et al. (2023). Exosomes-mediated tumor metastasis through reshaping tumor microenvironment and distant niche. J. Control. Release 353, 327–336. doi:10.1016/j.jconrel.2022.11.050
Lv, X.-B., Jiao, Y., Qing, Y., Hu, H., Cui, X., Lin, T., et al. (2011). miR-124 suppresses multiple steps of breast cancer metastasis by targeting a cohort of pro-metastatic genes in vitro. Chin. J. Cancer 30, 821–830. doi:10.5732/cjc.011.10289
Ma, L. J. B. C. R. (2010). Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 12, 210–215. doi:10.1186/bcr2720
Ma, L., Reinhardt, F., Pan, E., Soutschek, J., Bhat, B., Marcusson, E. G., et al. (2010). Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 28, 341–347. doi:10.1038/nbt.1618
Mutlu, M., Saatci, Ö., Ansari, S. A., Yurdusev, E., Shehwana, H., Konu, Ö., et al. (2016). miR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Sci. Rep. 6, 32541.
Nagata, Y., Lan, K.-H., Zhou, X., Tan, M., Esteva, F. J., Sahin, A. A., et al. (2004). PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6, 117–127. doi:10.1016/j.ccr.2004.06.022
Ni, Q., Stevic, I., Pan, C., Müller, V., Oliveira-Ferrer, L., Pantel, K., et al. (2018). Different signatures of miR-16, miR-30b and miR-93 in exosomes from breast cancer and DCIS patients. Sci. Rep. 8, 12974, doi:10.1038/s41598-018-31108-y
Ogata-Kawata, H., Izumiya, M., Kurioka, D., Honma, Y., Yamada, Y., Furuta, K., et al. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9, e92921. doi:10.1371/journal.pone.0092921
Pakravan, K., Babashah, S., Sadeghizadeh, M., Mowla, S. J., Mossahebi-Mohammadi, M., Ataei, F., et al. (2017). MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1α/VEGF signaling axis in breast cancer cells. Cell. Oncol. 40, 457, doi:10.1007/s13402-017-0335-7
Pan, X., Hong, X., Lai, J., Cheng, L., Cheng, Y., Yao, M., et al. (2020). Exosomal MicroRNA-221-3p confers adriamycin resistance in breast cancer cells by targeting PIK3R1. Front. Oncol. 2020 10, 441. doi:10.3389/fonc.2020.00441
Pan, Y.-Z., Morris, M. E., and Yu, A.-M. J. M. P. (2009). MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol. Pharmacol. 75, 1374, doi:10.1124/mol.108.054163
Park, E. J., Jung, H. J., Choi, H. J., Jang, H. J., Park, H. J., Nejsum, L. N., et al. (2020). Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells 34, 3379, doi:10.1096/fj.201902434R
Park, E. Y., Chang, E., Lee, E. J., Lee, H.-W., Kang, H.-G., Chun, K.-H., et al. (2014). Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 74, 7573, doi:10.1158/0008-5472.CAN-14-1140
Peng, J., Yuan, C., Wu, Z., Wang, Y., Yin, W., Lin, Y., et al. (2020). Upregulation of microRNA-1 inhibits proliferation and metastasis of breast cancer. Mol. Med. Rep. 22, 454–464. doi:10.3892/mmr.2020.11111
Pogribny, I. P., Filkowski, J. N., Tryndyak, V. P., Golubov, A., Shpyleva, S. I., and Kovalchuk, O. J. I. J. O. C. (2010). Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 127, 1785–1794. doi:10.1002/ijc.25191
Rao, X., Di Leva, G., Li, M., Fang, F., Devlin, C., Hartman-Frey, C., et al. (2011). MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 30, 1082, doi:10.1038/onc.2010.487
Sachdeva, M., Wu, H., Ru, P., Hwang, L., Trieu, V., and Mo, Y. J. O. (2011). MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene 30, 822–831. doi:10.1038/onc.2010.463
Sharma, R., and Prajapati, P. K. J. C. P. R. (2020). Predictive, preventive and personalized medicine: Leads from ayurvedic concept of prakriti (human constitution). Curr. Pharmacol. Rep. 6, 441–450. doi:10.1007/s40495-020-00244-3
Shi, J. J. J. O. C. M. (2016). Considering exosomal miR-21 as a biomarker for cancer. J. Clin. Med. 5, 42. doi:10.3390/jcm5040042
Shi, W., Gerster, K., Alajez, N. M., Tsang, J., Waldron, L., Pintilie, M., et al. (2011). MicroRNA-301 mediates proliferation and invasion in human breast cancer, SKA2 71, 2926, doi:10.1158/0008-5472.CAN-10-3369
Singh, A., Singh, A. K., Giri, R., Kumar, D., Sharma, R., Valis, M., et al. (2021). The role of microRNA-21 in the onset and progression of cancer. Future Med. Chem. 13, 1885, doi:10.4155/fmc-2021-0096
Singh, R., Pochampally, R., Watabe, K., Lu, Z., and Mo, Y.-Y. J. M. C. (2014). Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 13, 256. doi:10.1186/1476-4598-13-256
Song, S. J., Poliseno, L., Song, M. S., Ala, U., Webster, K., Ng, C., et al. (2013). MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324. doi:10.1016/j.cell.2013.06.026
Stevic, I., Müller, V., Weber, K., Fasching, P. A., Karn, T., Marmé, F., et al. (2018). Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 16, 179, doi:10.1186/s12916-018-1163-y
Tang, L.-B., Ma, S.-X., Chen, Z.-H., Huang, Q.-Y., Wu, L.-Y., Wang, Y., et al. (2021). Exosomal microRNAs: Pleiotropic impacts on breast cancer metastasis and their clinical perspectives. Biol. (Basel) 10, 307. doi:10.3390/biology10040307
Tao, W.-Y., Wang, C.-Y., Sun, Y.-H., Su, Y.-H., Pang, D., and Zhang, G.-Q. J. J. O. C. (2016). MicroRNA-34c suppresses breast cancer migration and invasion by targeting GIT1. J. Cancer 7, 1653–1662. doi:10.7150/jca.14762
Tavoosidana, G., Ronquist, G., Darmanis, S., Yan, J., Carlsson, L., Wu, D., et al. (2011). Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 108, 8809–8814. doi:10.1073/pnas.1019330108
Van Der Zanden, S. Y., Qiao, X., and Neefjes, J. J. T. F. J. (2021). New insights into the activities and toxicities of the old anticancer drug doxorubicin 288, 6095, doi:10.1111/febs.15583
Venturutti, L., Cordo Russo, R. I., Rivas, M. A., Mercogliano, M. F., Izzo, F., Oakley, R., et al. (2016). MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 35, 6189, doi:10.1038/onc.2016.151
Vlashi, E., and Pajonk, F. (2015), “(Year). "Cancer stem cells, cancer cell plasticity and radiation therapy,” in Seminars in cancer biology (Elsevier), 28–35.
Wang, H., Tan, Z., Hu, H., Liu, H., Wu, T., Zheng, C., et al. (2019). microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer, 19, 738, doi:10.1186/s12885-019-5951-3
Wang, X., Sun, C., Huang, X., Li, J., Fu, Z., Li, W., et al. (2021). The advancing roles of exosomes in breast cancer, Front. Cell Dev. Biol. 9, 731062. 10.3389/fcell.2021.731062.
Wang, Z., Wang, N., Liu, P., Chen, Q., Situ, H., Xie, T., et al. (2014). MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 5, 7013–7026. doi:10.18632/oncotarget.2192
Willson, M. L., Burke, L., Ferguson, T., Ghersi, D., Nowak, A. K., and Wilcken, N. J. C. D. O. S. R. (2019). Taxanes for adjuvant treatment of early breast cancer, Cochrane Database Syst. Rev. 2019 (9), CD004421, doi:10.1002/14651858.CD004421.pub3
Wolfson, B., Eades, G., and Zhou, Q. J. W. J. O. S. C. (2014). Roles of microRNA-140 in stem cell-associated early stage breast cancer. World J. Stem Cells 6, 591–597. doi:10.4252/wjsc.v6.i5.591
Wu, M.-Y., Fu, J., Xiao, X., Wu, J., and Wu, R.-C. J. C. L. (2014). MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer Lett. 354, 311–319. doi:10.1016/j.canlet.2014.08.031
Xie, F., Hosany, S., Zhong, S., Jiang, Y., Zhang, F., Lin, L., et al. (2017). MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS One 12, e0185565. doi:10.1371/journal.pone.0185565
Xing, F., Liu, Y., Wu, S.-Y., Wu, K., Sharma, S., Mo, Y.-Y., et al. (2018). Loss of XIST in breast cancer activates MSN-c-met and reprograms microglia via exosomal miRNA to promote brain metastasis. Brain Metastasis 78, 4316–4330. doi:10.1158/0008-5472.CAN-18-1102
Xu, Y.-F., Hannafon, B. N., Khatri, U., Gin, A., and Ding, W.-Q. J. R. B. (2019). The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 16, 770–784. doi:10.1080/15476286.2019.1585738
Yamamoto, Y., Yoshioka, Y., Minoura, K., Takahashi, R.-U., Takeshita, F., Taya, T., et al. (2011). An integrative genomic analysis revealed the relevance of microRNA and gene expression for drug-resistance in human breast cancer cells. Mol. Cancer 10, 135. doi:10.1186/1476-4598-10-135
Yang, Q., Wang, Y., Lu, X., Zhao, Z., Zhu, L., Chen, S., et al. (2015). MiR-125b regulates epithelial-mesenchymal transition via targeting Sema4C in paclitaxel-resistant breast cancer cells. Oncotarget 6, 3268, doi:10.18632/oncotarget.3065
Ye, Z.-B., Ma, G., Zhao, Y.-H., Xiao, Y., Zhan, Y., Jing, C., et al. (2015). miR-429 inhibits migration and invasion of breast cancer cells in vitro. Int. J. Oncol. 46, 531–538. doi:10.3892/ijo.2014.2759
Yoshikawa, M., Iinuma, H., Umemoto, Y., Yanagisawa, T., Matsumoto, A., and Jinno, H. J. O. L. (2018). Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol. Lett. 15, 9584–9592. doi:10.3892/ol.2018.8457
Yu, S.-J., Hu, J.-Y., Kuang, X.-Y., Luo, J.-M., Hou, Y.-F., Di, G.-H., et al. (2013). MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin. Cancer Res. 19, 1389–1399. doi:10.1158/1078-0432.CCR-12-1959
Yu, X., Luo, A., Liu, Y., Wang, S., Li, Y., Shi, W., et al. (2015). MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 14, 208, doi:10.1186/s12943-015-0480-4
Yu, Z., Willmarth, N. E., Zhou, J., Katiyar, S., Wang, M., Liu, Y., et al. (2010). microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. U. S. A. 107, 8231–8236. doi:10.1073/pnas.1002080107
Yuan, L., Liu, Y., Qu, Y., Liu, L., and Li, H. J. F. I. O. (2019). Exosomes derived from MicroRNA-148b-3p-overexpressing human umbilical cord mesenchymal stem cells restrain breast cancer progression. Front. Oncol. 9, 1076, doi:10.3389/fonc.2019.01076
Zhang, J., Da Zhang, H., Chen, L., Fei Mao, C., Chen, W., Wu, J. Z., et al. (2014). β-elemene reverses chemoresistance of breast cancer via regulating MDR-related microRNA expression. Cell. Physiol. biochem. 34 (6), 2027–2037. doi:10.1159/000366398
Zhang, L., Chen, X., Liu, B., and Han, J. J. O. L. (2018). MicroRNA-124-3p directly targets PDCD6 to inhibit metastasis in breast cancer. Oncol. Lett. 15, 984, doi:10.3892/ol.2017.7358
Zhang, T., Zhou, H., Wang, K., Wang, X., Wang, M., Zhao, W., et al. (2022). Role, molecular mechanism and the potential target of breast cancer stem cells in breast cancer development 147, 112616.
Zhang, X., Yu, X., Zhao, Z., Yuan, Z., Ma, P., Ye, Z., et al. (2020). MicroRNA-429 inhibits bone metastasis in breast cancer by regulating CrkL and MMP-9. Bone 130, 115139. doi:10.1016/j.bone.2019.115139
Zhou, B., Ma, R., Si, W., Li, S., Xu, Y., Tu, X., et al. (2013). MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 333, 159–169. doi:10.1016/j.canlet.2013.01.028
Zhou, W., Fong, M. Y., Min, Y., Somlo, G., Liu, L., Palomares, M. R., et al. (2014). Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515. doi:10.1016/j.ccr.2014.03.007
Zhou, W., Song, F., Wu, Q., Liu, R., Wang, L., Liu, C., et al. (2017). miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One 12, e0176395. doi:10.1371/journal.pone.0176395
Zhu, S., Wu, H., Wu, F., Nie, D., Sheng, S., and Mo, Y.-Y. J. C. R. (2008). MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359. doi:10.1038/cr.2008.24
Keywords: breast cancer, exosome, miRNA, diagnosis, metastasis
Citation: Singh T, Kaushik M, Mishra LC, Behl C, Singh V and Tuli HS (2023) Exosomal miRNAs as novel avenues for breast cancer treatment. Front. Genet. 14:1134779. doi: 10.3389/fgene.2023.1134779
Received: 30 December 2022; Accepted: 27 February 2023;
Published: 22 March 2023.
Edited by:
Manoj Kumar Kashyap, Amity University Gurgaon, IndiaReviewed by:
Ankit Verma, Hebrew University of Jerusalem, IsraelNemat Ali, King Saud University, Saudi Arabia
Manohar Prasad Bhandari, University of Latvia, Latvia
Shoib Baba, Department of Education, India
Copyright © 2023 Singh, Kaushik, Mishra, Behl, Singh and Tuli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tejveer Singh, dGVqNjg3NUBnbWFpbC5jb20=, dGVqdmVlckBocmMuZHUuYWMuaW4=
 Tejveer Singh
Tejveer Singh Mahesh Kaushik2
Mahesh Kaushik2 Chesta Behl
Chesta Behl Vijay Singh
Vijay Singh Hardeep Singh Tuli
Hardeep Singh Tuli