- 1Hainan Institute, Zhejiang University, Sanya, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
- 3State Key Lab of Rice Biology, Ministry of Agriculture Key Lab of Molecular Biology of Crop Pathogens and Insects, and Zhejiang Provincial Key Laboratory of Biology of Crop Pathogens and Insects, Zhejiang University, Hangzhou, China
- 4Institute of Insect Sciences, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 5Guizhou Province Tobacco Companies Qian xinan Municipal Tobacco Company, Xingyi, China
- 6Tobacco Leaf Purchase Center, Hunan China Tobacco Industry Co., Ltd., Changsha, China
Meteorus Haliday, 1835 is a cosmopolitan genus within Braconidae (Hymenoptera). They are koinobiont endoparasitoids of Coleoptera or Lepidoptera larvae. Only one mitogenome of this genus was available. Here, we sequenced and annotated three mitogenomes of Meteorus species, and found that the tRNA gene rearrangements in these mitogenomes were rich and diverse. Compared with the ancestral organization, only seven tRNAs (trnW, trnY, trnL2, trnH, trnT, trnP and trnV) were conserved and trnG had its own unique location in the four mitogenomes. This dramatic tRNA rearrangement was not observed in mitogenomes of other insect groups before. In addition, the tRNA cluster (trnA-trnR-trnN-trnS1-trnE-trnF) between nad3 and nad5 was rearranged into two patterns, i.e., trnE-trnA-trnR-trnN-trnS1 and trnA-trnR-trnS1-trnE-trnF-trnN. The phylogenetic results showed that the Meteorus species formed a clade within the subfamily Euphorinae, and were close to Zele (Hymenoptera, Braconidae, Euphorinae). In the Meteorus, two clades were reconstructed: M. sp. USNM and Meteorus pulchricornis forming one clade while the remaining two species forming another clade. This phylogenetic relationship also matched the tRNA rearrangement patterns. The diverse and phylogenetic signal of tRNA rearrangements within one genus provided insights into tRNA rearrangements of the mitochondrial genome at genus/species levels in insects.
1 Introduction
Braconidae is one of the most species-rich families of Hymenoptera, including 42 subfamilies represented by over 1,100 genera and more than 21,220 known species (Chen and van Achterberg, 2019). Meteorus Haliday, 1835 is a cosmopolitan genus within Euphorinae and more than 300 species have been described (Fujie et al., 2021). They are koinobiont endoparasitoids of Coleoptera or Lepidoptera larvae, and some of their hosts are considered pest insects, including some major pests such as Agrotis ipsilon, Helicoverpa armigera, Lymantria dispar and Spodoptera frugiperda (Yu et al., 2016). Several final-instar larvae of Meteorus can produce cocoons suspended by a common cable, and the cocoon architecture is one of the key characters for identifying these species (Fujie et al., 2021).
The mitochondrial genomes show extremely high rates of gene rearrangements in Hymenoptera compared with other orders in the Hexapoda (Wei et al., 2010; Li et al., 2016). Gene arrangements in the mitochondrial genome can be divided into two types: major rearrangements involving protein-coding genes (PCGs) and rRNAs and minor rearrangements involving tRNAs only based on gene type (Chen et al., 2016). The PCGs rearrangements have been found in Aculeata, Ceraphronoidea, Chalcidoidea, Cynipoidea, Gasteruptiidae, Ichneumonoidea and Trigonaloidea, while the tRNAs rearrangements occur in each family in Hymenoptera (Tang et al., 2019). In addition, rearrangements of the rRNAs have been found in Cynipoidea, Chrysidoidea and Megalyroidea (Tang et al., 2019; Zheng et al., 2021; Shu et al., 2022). In Braconidae, PCGs order is relatively conservative, while tRNA rearrangement patterns within subfamilies have a better taxon representation (Li et al., 2016; Jasso-Martinez et al., 2022a). Such tRNA rearrangement patterns are typically restricted to specific lineages, which can help with phylogenetic reconstruction in Braconidae at the subfamily level (Li et al., 2016).
Here, we re-sequenced the mitogenome of Meteorus pulchricornis and newly obtained two other Meteorus mitogenomes by next-generation sequencing. We further analyzed the main features of the three mitogenomes. Then we compared gene rearrangements of four known mitogenomes within the genus Meteorus. Finally, we confirmed the phylogenetic position of Meteorus within Braconidae based on mitogenome data.
2 Methods
2.1 Sample collection and DNA extraction
M. pulchricornis, M. sp. 1 and M. sp. 2 were all collected in the Chinese provinces of Zhejiang (Ningbo city), Guizhou (Guiyang city), and Hebei (Shijiazhuang city), respectively. The specimens were morphologically identified by Prof. Cornelis van Achterberg (Zhejiang University, China). All specimens were initially preserved in 100% ethanol and then stored at 4 °C before DNA extraction. Whole genomic DNA was extracted from every sample using the DNeasy tissue kit (Qiagen, Hilden, Germany).
2.2 High throughput sequencing and assembly
The libraries were prepared for each DNA sample using the VAHTS® Universal DNA Library Prep Kit. All constructed libraries were then sequenced as 150 bp paired-end on a full run (2 × 150 PE) using MGISEQ2000 platform. FastQC v0.11.9 (Andrews, 2015) was used to check the data quality, and fastp v0.23.1 (Chen et al., 2018) was used to trim adaptors and remove low quality reads with default parameters. More than 5 GB of clean data for each sample was used in de novo assembly. The mitogenomes were assembled using MitoZ v2.3 (Meng et al., 2019), IDBA v1.1.3 (Peng et al., 2012) and SPAdes v3.13.0 (Bankevich et al., 2012) with default parameters, respectively. Subsequently, to verify the accuracy of de novo assembly, one fragment in the cox1-cox2 junction was amplified with primers (C1-J-2195: 5’-TGATTTTTTGGGCATCCTGAAGT-3’, C2-N-3665: 5’-CCACAAATTTCAGAACATTGACC-3’) for three species by polymerase chain reaction (PCR) and was then Sanger-sequenced. Finally, all assemblies were then integrated with GENEIOUS v2020.0.5 (Biomatters Ltd. San Diego, CA, United States).
2.3 Mitochondrial genome annotation and analysis
Three assembled mitogenomes were annotated using MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py; accessed on 15 November 2022) (Bernt et al., 2013). The start and stop positions of 13 PCGs were manually adjusted and corrected by aligning published data of the Braconidae species in GenBank. tRNAScan-SE (Chan et al., 2021) was used to verify the results of putative tRNA genes. Nucleotide composition, codon usage, and relative synonymous codon usage (RSCU) values were estimated using MEGA 11 (Tamura et al., 2021). The bias of nucleotide composition was measured as AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) (Perna and Kocher, 1995). The gene rearrangements in Meteorus mitogenomes were analyzed by comparison with the ancestral insect mitogenomes (Boore, 1999).
2.4 Phylogenetic analysis
A total of 21 Braconidae mitogenomes were used for phylogenetic analyses, including three newly obtained mitogenomes. Two Ichneumonidae species, Euceros kiushuensis and Diadegma semiclausum, were used as outgroups (Supplementary Table S1). The PCGs were aligned using MAFFT v7.464 (Katoh and Standley, 2013) with the default algorithm, and the best partition schemes and substitution models (Supplementary Table S2) for the datasets were analyzed by PartitionFinder v1.1.1 (Lanfear et al., 2012). Bayesian inference (BI) and maximum likelihood (ML) were selected to reconstruct the phylogenetic trees. For BI analysis, MrBayes v3.2.7a (Ronquist and Huelsenbeck, 2003) was used to run four independent Markov chains for 100 million generations, with tree sampling occurring every 1,000 generations and a burn-in of 25% of the trees. The stationarity of the run was assessed by Tracer v1.7. (ESS values >200) (Rambaut et al., 2018). Maximum likelihood (ML) analysis was performed with RAxML-HPC2 v8.2.12 (Stamatakis, 2014) under the GTRGAMMA model. A total of 200 runs for different individual partitions were conducted with 1,000 bootstrap replicates.
3 Results and discussion
3.1 Mitochondrial genome organization
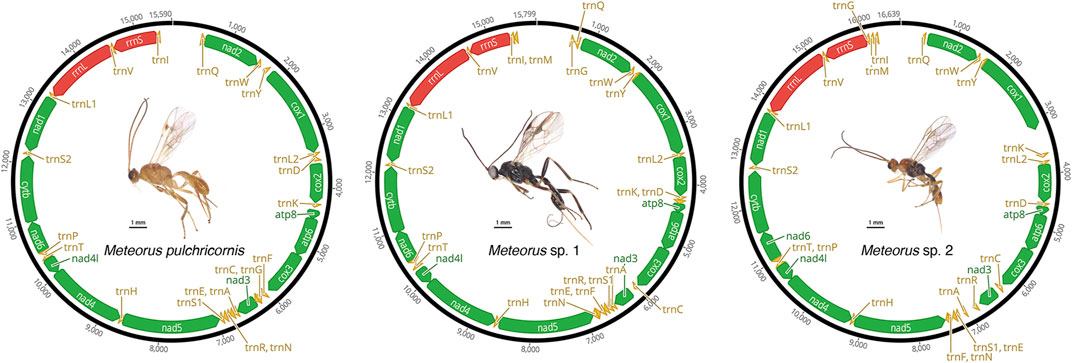
The mitogenomes of three Meteorus species were successfully obtained, and the newly sequenced mitogenomes were submitted to GenBank (accession numbers: OP832526 - OP832528) (Supplementary Table S1). All sequences were nearly complete mitogenomes, measuring 15,590 bp for M. pulchricornis, 15,799 bp for M. sp. 1, and 16,639 bp for M. sp. 2. In all assembled mitogenomes, all 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes were found (Figure 1). Non-etheless, the A-T control region was incomplete in all genomes due to the high A+ T content in the Hymenoptera mitogenomes (Zheng et al., 2021; Jasso-Martinez et al., 2022a).
The mitogenome of M. pulchricornis contained 19 non-coding regions ranging in size from 1 to 67 bp, with a total length of 407 bp. The nucleotides from 8 overlap regions were up to 22 bp in total. The maximum overlap length was 7 bp, located at two junctions (atp8-atp6, nad4-nad4l), while the minimum overlap length was 1 bp and occurred at four junctions (atp6-cox3, trnA-trnR-trnN, trnT-trnP). M. sp. 1 had 18 non-coding regions with lengths ranging from 1 to 83 bp, for a total of 303 bp. The nucleotides from 5 overlap regions amounted to 18 bp in total. Two junctions (atp8-atp6, nad4-nad4l) had overlaps of 7 bp in length. M. sp. 2 possessed 23 non-coding regions ranging from 1 to 465 bp in length and a total length of 1,138 bp, which had the longest intergenic nucleotides among the three Meteorus mitogenomes. It had 3 overlap regions with a total of 12 bp in length. Three junctions (nad4-nad4l, atp8-atp6, and trnE-trnF) had overlaps of 7 bp, 4 bp and 1 bp in length, respectively. The length of overlap (4 bp) at the junction atp8-atp6 in the mitogenomes of M. sp. 2 was unusually short compared to that of other wasps, which was typically 7 bp (Zhu et al., 2018; Tang et al., 2019).
3.2 Nucleotide composition
The A + T content for the sequenced region of the mitogenomes was 84.41% (M. pulchricornis), 82.70% (M. sp. 1) and 84.31% (M. sp.2) (Supplementary Table S3), which was similar to other wasp species (Tang et al., 2019). All 13 PCGs were detected in the newly generated mitogenomes, with sizes ranging from 11,063 bp (M. sp. 1) to 11,120 bp (M. sp. 2). The entire A + T content of all the PCGs was from 80.48% (M. sp. 1) to 83.37% (M. pulchricornis) (Supplementary Table S3). The size of the PCGs in the three mitochondrial genomes was similar to other wasps (Zheng et al., 2021; Shu et al., 2022; Zheng et al., 2022). The AT-skew in three Meteorus mitogenomes was negative (−0.1574 to −0.1479), and the GC-skew was positive (0.0662–0.1088) (Supplementary Table S3). All 22 typical tRNA genes were found in three mitogenomes. The size of all tRNA genes identified ranged from 57 bp to 72 bp (Supplementary Table S4). Two rRNA genes (rrnS and rrnL) were identified in all mitogenomes. The length of rrnS was 764 bp (M. pulchricornis), 735 bp (M. sp. 1) and 790 bp (M. sp. 2), and the size of rrnL was 1,253 bp, 1,325 bp and 1,347 bp, respectively (Supplementary Table S3).
3.3 Protein-coding genes and codon usage
The majority of the protein-coding genes used ATN (ATG, ATT, or ATA), except ATC, as an initiation codon. Although most PCGs terminated with the conventional TAA and TAG as stop codons, some of them have incomplete stop codons (T or TA) in some PCGs (Supplementary Table S5). The start and stop codons were typical of insect mitogenomes (Li et al., 2021; Ge et al., 2022). Relative synonymous codon usage (RSCU) values of three Meteorus species were analyzed (Figure 2; Supplementary Table S6). The total codon number of these three species was 3,689, 3,677 and 3,695, respectively. The codon GCG was not detected in all species, while codons UGC, CGC, AGC and GGC appeared in some species. Meanwhile, the five most frequently used codons UUA, AUU, UUU, AUA and AAU were observed in the mitogenomes due to the high A + T content in the mitogenomes. These results are consistent with other published wasp mitogenomes (Zhu et al., 2018; Zheng et al., 2022).
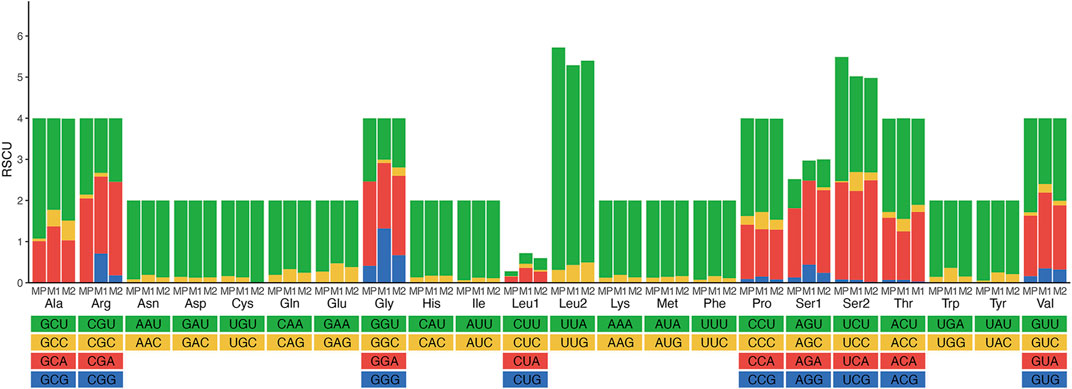
FIGURE 2. Relative synonymous codon usage (RSCU) of three Meteorus species. Codon families are provided on the X-axis along with the different combinations of synonymous codons that code for that amino acid. RSCU is defined on the Y-axis. MP, Meteorus pulchricornis; M1, Meteorus sp. 1; M2, Meteorus sp. 2.
3.4 Gene rearrangement
The order of PCGs was relatively conservative in the mitogenomes of Braconidae. So far, only Stenocorse bruchivora (Doryctinae), two Chelonus spp. (Cheloninae), and two Cotesia spp. (Microgastrinae) were found PCGs rearrangement in mitogenomes (Wei et al., 2010; Jasso-Martinez et al., 2022a; Yuan et al., 2022). In contrast, tRNA rearrangements occurred in all known Braconidae mitogenomes (Li et al., 2016; Jasso-Martinez et al., 2022a). PCG rearrangements did not occur in any of the four Meteorus mitogenomes studied, but tRNA rearrangements varied (Figure 3). Only seven tRNAs (trnW, trnY, trnL2, trnH, trnT, trnP and trnV) were conserved and trnG had different locations in each of the four mitogenomes. trnL1 and trnS2 were inverted and exchanged. Furthermore, trnC was translocated from nad2-cox1 junction to cox3-nad3 junction. The tRNA cluster (trnA-trnR-trnN-trnS1-trnE-trnF) between nad3 to nad5 exhibited two distinct patterns. In M. sp. USNM and M. pulchricornis, trnE was translocated upstream of trnA, and trnF was rearranged to cox3-nad3 junction, which reduced one tRNA in the tRNA cluster. In M. sp. 1 and M. sp. 2, the number of tRNAs between nad3 to nad5 remained at six, and trnN was translocated downstream of trnF. In addition, the positions of trnA and trnR were interchanged in M. sp. 2. Interestingly, trnI and trnM, which were usually conservative in Braconidae mitogenomes, were translocated from upstream of nad2 to the nad1-rrnL junction, and trnL2 was duplicated into the nad1-rrnL junction as well in M. sp. USNM. In summary, the pattern of tRNAs suggested that M. sp. USNM and M. pulchricornis were more closely related than the other two species.
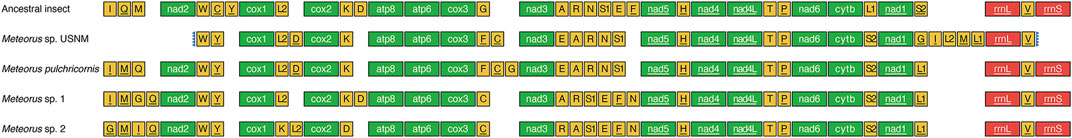
FIGURE 3. Mitochondrial gene order in Meteorus genus and ancestral insect. Genes are transcribed from left to right except those underlined, which have the opposite transcriptional orientation.
The rich and diverse rearrangements presented by the tRNAs in the genus Meteorus were the first to be found in the Braconidae. tRNA rearrangements are usually conserved in the same genus in Hymenoptera (Jasso-Martinez et al., 2022a). Although different rearrangement of tRNAs within the same genus in Hymenoptera has been reported in Aphelinidae, Chrysididae and Ichneumonidae (Zhu et al., 2018; Zheng et al., 2021; Zheng et al., 2022), the tRNA rearrangements within Meteorus in Braconidae were more dramatic than in the aforementioned families, which exhibited only one or two different tRNA rearrangements in the same genus. To our knowledge, the various tRNA rearrangement patterns within the same genus had not been found in the other insects.
3.5 Phylogenetic analysis
In this study, a phylogenetic analysis based on 13 PCGs was constructed using maximum likelihood and Bayesian methods, in order to validate the phylogenetic position of Meteorus within Braconidae (Figure 4). The consensus tree derived from the above two inference methods generated the congruent results. Most nodes had significantly supported bootstrap and posterior probability values. The tree recovered the well-accepted major lineages within the family and supported the division of Braconidae into cyclostomes and non-cyclostomes as generally accepted (Jasso-Martinez et al., 2022a; Jasso-Martinez et al., 2022b). Meanwhile, consistent with previous studies, within the non-cyclostomes, Euphorinae was recovered as a sister clade to the remaining non-cyclostomes (Jasso-Martinez et al., 2022b). The four Meteorus species formed a clade within Euphorinae, and were close to Zele chlorophthalmus rather than Dinocampus coccinellae. In the Meteorus, the species were clustered in two clades: one comprised of M. sp. USNM and M. pulchricornis while M. sp. 1 and M. sp. 2 grouped together in the second clade. This structure matched the tRNA rearrangement result presented above.
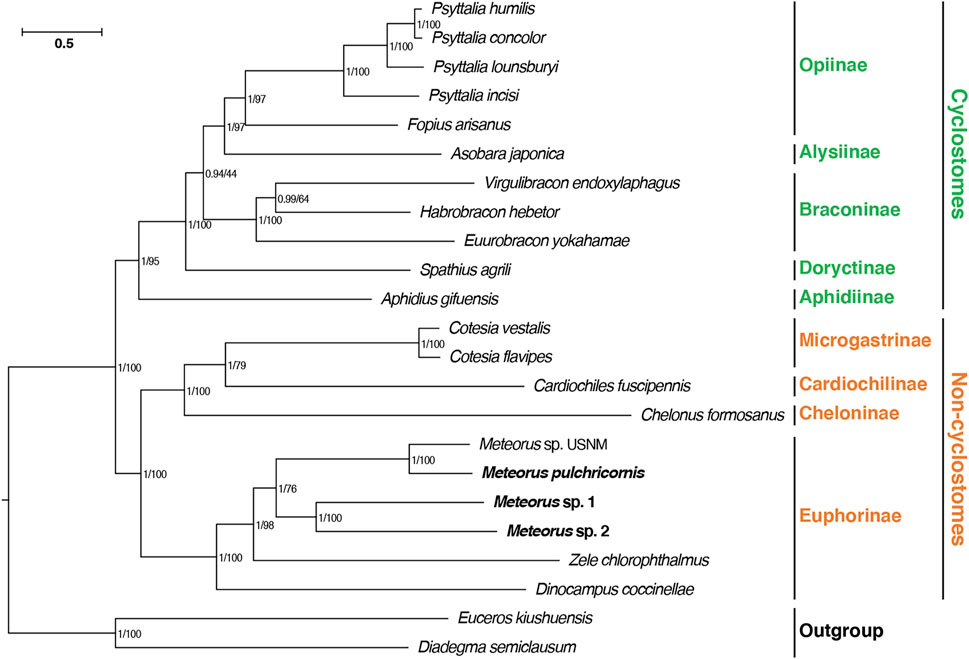
FIGURE 4. Phylogenetic analyses of Braconidae based on nucleotide datasets of 13 PCGs. The scale bar corresponds to the estimated number of substitutions per site. Numbers separated by a slash on the node are posterior probability (PP) and bootstrap value (BV).
4 Conclusion
In this study, three Meteorus mitogenomes were newly acquired using next-generation sequencing method, which expanded the data of mitochondrial genomes in the subfamily Euphorinae. Three mitogenomes of Meteorus spp. shared similar A + T content, AT- and GC-skew, and codon usage of PCGs, however, M. sp. 2 had longer intergenic nucleotides. Each mitogenome among the Meteorus spp. displayed dramatic divergent gene rearrangements within the same genus that had not previously been reported. The pattern of tRNAs between nad3 to nad5 had two types, trnE-trnA-trnR-trnN-trnS1 and trnA-trnR-trnS1-trnE-trnF-trnN. The BI and ML analyses showed consistent topology, Meteorus species formed a clade within Euphorinae, and were close to Zele. Two clades were reconstructed for the Meteorus, which matched the tRNA rearrangement patterns. In the future, more mitochondrial genomes from the same genus are needed to display further details of tRNA rearrangements. The diverse and phylogenetic signal of tRNA rearrangements within one genus may provide insights into the potential mechanism of tRNA rearrangements in the same genus and the phylogenetic relationships of taxa at the genus level.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OP832526—OP832528.
Author contributions
XC, QM, and PT contributed to conception and design of the study. XS, RY, ZX, GG, LY, and ZS performed and analyzed experiments. RY and XS contributed to all figures and tables. XS wrote the first draft of the manuscript. XC and PT reviewed and edited the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the Guangdong Laboratory of Lingnan Modern Agriculture Project (NT2021003), the Guizhou Tobacco Company of China National Tobacco Corporation (Science and Technology Major Project, No. 2022XM09), the Key International Joint Research Program of National Natural Science Foundation of China (31920103005), the General Program of National Natural Science Foundation of China (32070467), the Key R&D Program of Zhejiang Province (2020C02003), and the Fundamental Research Funds for the Central Universities (2021FZZX001-31). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We thank Prof. Cornelis van Achterberg for specimen identification and Xiqian Ye for providing access to the cluster computer system used for the phylogenetic analyses.
Conflict of interest
Author ZX, GG, LY, ZS, and QM were employed by Guizhou Province Tobacco Companies Qian xinan Municipal Tobacco Company. Author LY was also employed by Tobacco Leaf Purchase Center, Hunan China Tobacco Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1132606/full#supplementary-material
References
Andrews, S. (2015). FastQC. [Online]. Available: https://qubeshub.org/resources/fastqc (Accessed November 10, 2022).
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. doi:10.1089/cmb.2012.0021
Bernt, M., Donath, A., Juehling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69 (2), 313–319. doi:10.1016/j.ympev.2012.08.023
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27 (8), 1767–1780. doi:10.1093/nar/27.8.1767
Chan, P. P., Lin, B. Y., Mak, A. J., and Lowe, T. M. (2021). tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49 (16), 9077–9096. doi:10.1093/nar/gkab688
Chen, P. Y., Zheng, B. Y., Liu, J. X., and Wei, S. J. (2016). Next-generation sequencing of two mitochondrial genomes from family pompilidae (Hymenoptera: Vespoidea) reveal novel patterns of gene arrangement. Int. J. Mol. Sci. 17 (10), 1641. doi:10.3390/ijms17101641
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), 884–890. doi:10.1093/bioinformatics/bty560
Chen, X. X., and van Achterberg, C. (2019). Systematics, phylogeny, and evolution of braconid wasps: 30 Years of progress. Annu. Rev. Entomol. 64, 335–358. doi:10.1146/annurev-ento-011118-111856
Fujie, S., Shimizu, S., Tone, K., Matsuo, K., and Maeto, K. (2021). Stars in subtropical Japan: A new gregarious Meteorus species (Hymenoptera, Braconidae, Euphorinae) constructs enigmatic star-shaped pendulous communal cocoons. J. Hymenopt. Res. 86, 19–45. doi:10.3897/jhr.86.71225
Ge, X., Zang, H., Ye, X., Peng, L., Wang, B., Lian, G., et al. (2022). Comparative mitogenomic analyses of hydropsychidae revealing the novel rearrangement of protein-coding gene and tRNA (Trichoptera: Annulipalpia). Insects 13 (9), 759. doi:10.3390/insects13090759
Jasso-Martinez, J. M., Quicke, D. L. J., Belokobylskij, S. A., Santos, B. F., Fernandez-Triana, J. L., Kula, R. R., et al. (2022a). Mitochondrial phylogenomics and mitogenome organization in the parasitoid wasp family Braconidae (Hymenoptera: Ichneumonoidea). BMC Ecol. Evol. 22 (1), 46. doi:10.1186/s12862-022-01983-1
Jasso-Martinez, J. M., Santos, B. F., Zaldivar-Riveron, A., Fernandez-Triana, J. L., Sharanowski, B. J., Richter, R., et al. (2022b). Phylogenomics of braconid wasps (Hymenoptera, Braconidae) sheds light on classification and the evolution of parasitoid life history traits. Mol. Phylogenet. Evol. 173, 107452. doi:10.1016/j.ympev.2022.107452
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29 (6), 1695–1701. doi:10.1093/molbev/mss020
Li, Q., Wei, S. J., Tang, P., Wu, Q., Shi, M., Sharkey, M. J., et al. (2016). Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol. Evol. 8 (9), 2651–2662. doi:10.1093/gbe/evw184
Li, R., Lei, Z., Li, W., Zhang, W., and Zhou, C. (2021). Comparative mitogenomic analysis of heptageniid mayflies (insecta: Ephemeroptera): Conserved intergenic spacer and tRNA gene duplication. Insects 12 (2), 170. doi:10.3390/insects12020170
Meng, G. L., Li, Y. Y., Yang, C. T., and Liu, S. L. (2019). MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47 (11), e63. doi:10.1093/nar/gkz173
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2012). IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28 (11), 1420–1428. doi:10.1093/bioinformatics/bts174
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41 (3), 353–358. doi:10.1007/BF00186547
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67 (5), 901–904. doi:10.1093/sysbio/syy032
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 (12), 1572–1574. doi:10.1093/bioinformatics/btg180
Shu, X., Li, Z., Yuan, R., Tang, P., and Chen, X. (2022). Novel gene rearrangements in the mitochondrial genomes of cynipoid wasps (Hymenoptera: Cynipoidea). Genes. 13 (5), 914. doi:10.3390/genes13050914
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi:10.1093/bioinformatics/btu033
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Tang, P., Zhu, J. C., Zheng, B. Y., Wei, S. J., Sharkey, M., Chen, X. X., et al. (2019). Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenet. Evol. 131, 8–18. doi:10.1016/j.ympev.2018.10.040
Wei, S. J., Shi, M., Sharkey, M. J., van Achterberg, C., and Chen, X. X. (2010). Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 11, 371. doi:10.1186/1471-2164-11-371
Yuan, R. Z., Zhou, J. J., Shu, X. H., Ye, X. Q., Tang, P., and Chen, X. X. (2022). The mitochondrial genome of Chelonus formosanus (Hymenoptera: Braconidae) with novel gene orders and phylogenetic implications. Arch. Insect Biochem. Physiol. 111 (1), e21870. doi:10.1002/arch.21870
Zheng, B., Han, Y., Yuan, R., Liu, J., van Achterberg, C., Tang, P., et al. (2022). Comparative mitochondrial genomics of 104 Darwin wasps (Hymenoptera: Ichneumonidae) and its implication for phylogeny. Insects 13 (2), 124. doi:10.3390/insects13020124
Zheng, X. Y., Cao, L. J., Chen, P. Y., Chen, X. X., van Achterberg, K., Hoffmann, A. A., et al. (2021). Comparative mitogenomics and phylogenetics of the stinging wasps (Hymenoptera: Aculeata). Mol. Phylogenet. Evol. 159, 107119. doi:10.1016/j.ympev.2021.107119
Keywords: Braconidae, Meteorus, mitochondrial genome, tRNA gene, gene rearrangement
Citation: Shu X, Yuan R, Xia Z, Gao G, Yang L, Sun Z, Mu Q, Tang P and Chen X (2023) Comparative mitogenomes reveal diverse and novel gene rearrangements in the genus Meteorus (Hymenoptera: Braconidae). Front. Genet. 14:1132606. doi: 10.3389/fgene.2023.1132606
Received: 27 December 2022; Accepted: 03 February 2023;
Published: 13 February 2023.
Edited by:
Cristina P. Vieira, Universidade do Porto, PortugalReviewed by:
Elgion Lucio Silva Loreto, Federal University of Santa Maria, BrazilMaria Kamilari, Hellenic Agricultural Organization—ELGO, Greece
Copyright © 2023 Shu, Yuan, Xia, Gao, Yang, Sun, Mu, Tang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Mu, NDU1MTAxMjk4QHFxLmNvbQ==; Pu Tang, cHRhbmdAemp1LmVkdS5jbg==
 Xiaohan Shu
Xiaohan Shu Ruizhong Yuan2,3,4
Ruizhong Yuan2,3,4 Pu Tang
Pu Tang Xuexin Chen
Xuexin Chen