- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Department of Anatomical Sciences, Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 3Clinical Research Development Unit of Tabriz Valiasr Hospital, Tabriz University of Medical Sciences, Tabriz, Iran
- 4Department of Clinical Analysis, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 5Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 6Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 7Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 8Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
SIRT1 is a member of the sirtuin family functioning in the process of removal of acetyl groups from different proteins. This protein has several biological functions and is involved in the pathogenesis of metabolic diseases, malignancy, aging, neurodegenerative disorders and inflammation. Several long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs) have been found to interact with SIRT1. These interactions have been assessed in the contexts of sepsis, cardiomyopathy, heart failure, non-alcoholic fatty liver disease, chronic hepatitis, cardiac fibrosis, myocardial ischemia/reperfusion injury, diabetes, ischemic stroke, immune-related disorders and cancers. Notably, SIRT1-interacting non-coding RNAs have been found to interact with each other. Several circRNA/miRNA and lncRNA/miRNA pairs that interact with SIRT1 have been identified. These axes are potential targets for design of novel therapies for different disorders. In the current review, we summarize the interactions between three classes of non-coding RNAs and SIRT1.
Introduction
As a member of the sirtuin family, Sirt1 has a function in removal of acetyl groups from different proteins. This nicotinamide adenosine dinucleotide (NAD)-dependent deacetylase has several biological functions and is involved in the pathogenesis of metabolic diseases, malignancy, aging, neurodegenerative disorders and inflammation (Rahman and Islam, 2011). SIRT1 has a lot of substrates including a number of transcription factors. These transcription factors include p53, FoxO family, HES1, HEY2, PPARγ, CTIP2, p300, PGC-1α, and NF-κB (Haigis and Guarente, 2006; Michan and Sinclair, 2007; Yamamoto et al., 2007; Pillarisetti, 2008). The enzymatic reaction catalyzed by SIRT1 leads to generation of nicotinamide and transfer of the acetyl group of the substrate to cleaved NAD, producing a distinctive metabolite, namely, O-acetyl-ADP ribose (Pillarisetti, 2008).
SIRT1 has an important role in the regulation of energy homeostasis in response to accessibility to nutrients. In the liver tissue, SIRT1 enhances expression of the nuclear receptor PPARα, thus regulating lipid homeostasis. Deletion of Sirt1 in this tissue has been shown to impair PPARα signaling and decrease ß-oxidation of fatty acids, resulting in the development of hepatic steatosis, induction of inflammatory responses in liver, and endoplasmic reticulum stress (Purushotham et al., 2009).
In addition to the regulation of metabolic pathways, SIRT1 is involved in the carcinogenic processes. Its expression has been found to be increased in both hematological malignancies (Bradbury et al., 2005) and solid tumors (Huffman et al., 2007; Stünkel et al., 2007). Possibly acting as an oncogene, SIRT1 interacts with p53 and induces its deacetylation at its C-terminal Lys382 residue (Vaziri et al., 2001), thus inactivating this tumor suppressor.
In fact, SIRT1 is involved in a variety of human disorders including malignant and nonmalignant conditions. Recently, researchers have focused on identification of the interaction between non-coding RNAs and SIRT1 in these disorders. These investigations have led to identification of a number of long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs) that regulate expression of SIRT1. In the current review, we provide an overview of these non-coding RNAs.
SIRT1-interacting miRNAs
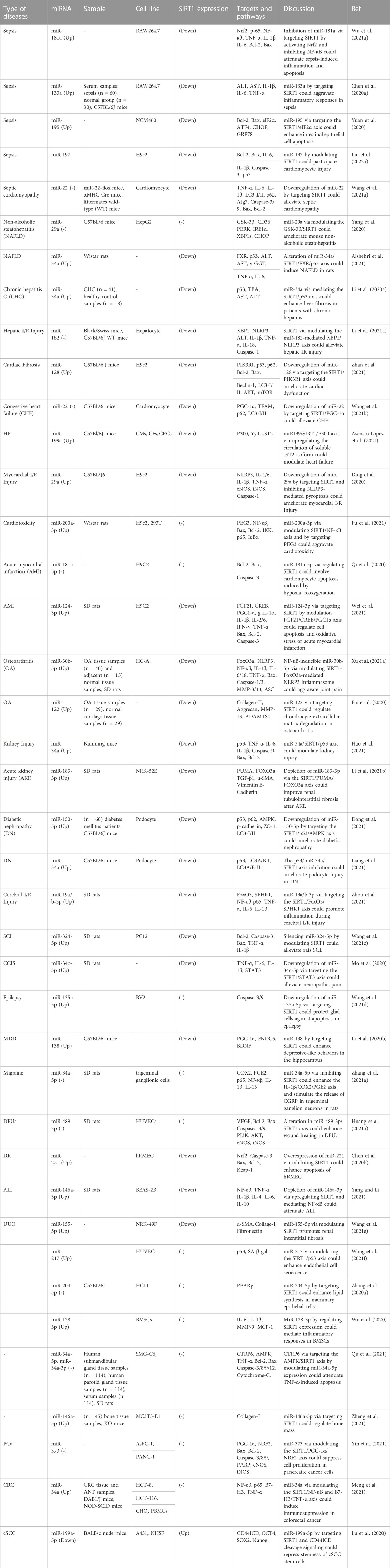
A class of non-coding RNAs known as miRNAs regulate gene expression by binding to specific target genes in distinct pathways, thereby modulating the expression of various genes (Ghafouri-Fard et al., 2021a; Hussen et al., 2021; Hussen et al., 2022a). Mature miRNAs are formed by further processing of pre-miRNAs, which are formed from the transcribed nucleic acids that make up primary miRNAs. Several miRNAs have been shown to target SIRT1, thus regulating its expression. Dysregulation of SIRT1-targeting miRNAs is involved in the pathogenesis of sepsis and its complications, non-alcoholic fatty liver disease (NAFLD), chronic hepatitis, hepatic and myocardial ischemia/reperfusion (I/R) injury, cardiac fibrosis, heart failure, myocardial infarction, osteoarthritis, kidney injury, diabetic nephropathy, cerebral I/R Injury, spinal cord injury, epilepsy and a number of malignant conditions (Table 1; Figure 1). In sepsis, upregulation of miR-181a (Wu Z. et al., 2021), miR-133a (Chen L. et al., 2020) and miR-195 (Yuan et al., 2020) has been shown to lead to downregulation of SIRT1 and aggravation of inflammatory responses. miR-29a, miR-34a and miR-182 are among SIRT1-interacting miRNAs being involved in the pathogenesis of hepatic disorders. For instance, miR-29a via modulating the GSK-3β/SIRT1 could ameliorate mouse non-alcoholic steatohepatitis (Yang et al., 2020). Alterations in the miR-34a/SIRT1/FXR/p53 axis have been found to induce NAFLD in rats (Alshehri et al., 2021). Moreover, miR-34a via mediating the SIRT1/p53 axis could enhance liver fibrosis in patients with chronic hepatitis (Li X. et al., 2020).
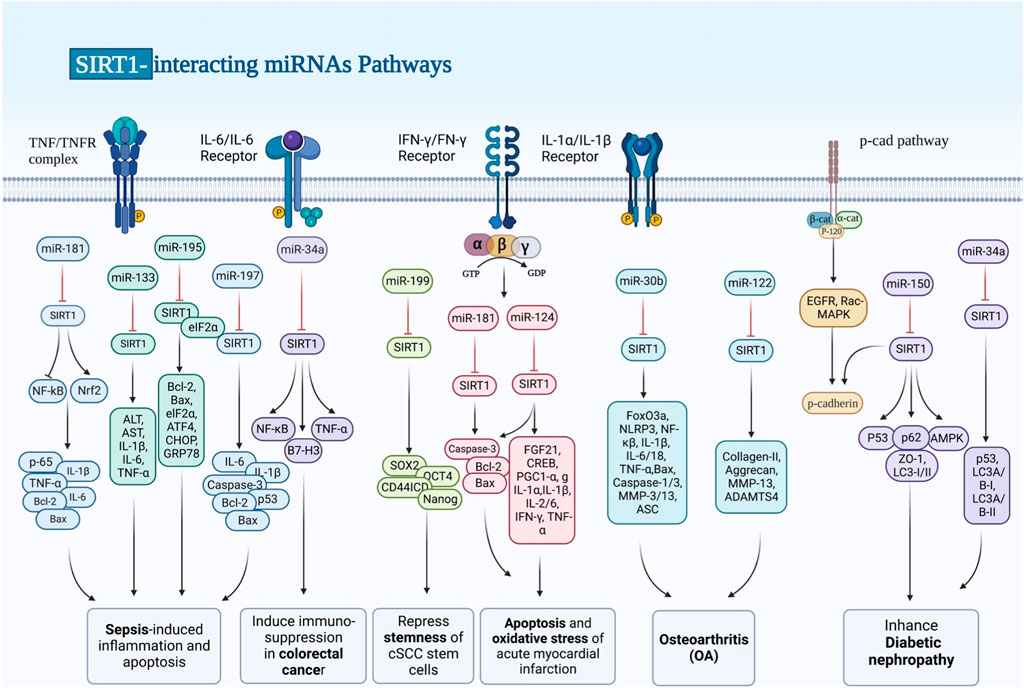
FIGURE 1. SIRT1 works with a lot of molecules, some of which are transcription factors. p53, the FoxO family, HES1, HEY2, PPAR, CTIP2, p300, PGC-1, and NF-B are all transcription factors. Dysregulation of SIRT1-targeting miRNAs plays a role in the pathogenesis of sepsis and its complications, chronic hepatitis, ischemia/reperfusion (I/R) injury to the liver and heart, cardiac fibrosis, myocardial infarction, osteoarthritis, diabetic nephropathy, and a number of malignant diseases like colorectal cancer.
miR-128 has been shown to be involved in the pathogenesis of chronic angiotensin II infusion-induced cardiac remodeling through modulation of SIRT1. Silencing this miRNA in the heart tissues of mice could ameliorate angiotensin II-induced cardiac dysfunction, hypertrophy, fibrosis and oxidative stress damage. Angiotensin II could induce upregulation of miR-128 in cell culture. Treatment of cells with miR-128 antagomir could attenuate angiotensin II -induced apoptosis and oxidative damage possibly through targeting the SIRT1/p53 pathway. Suppression of this miRNA could also activate PIK3R1/Akt/mTOR pathway, restrain angiotensin II-induced autophagy in cardiomyocytes, and mitigate oxidative stress and apoptosis (Zhan et al., 2021).
SIRT1-interacting miRNAs are also involved in the pathogenic processes in the acute myocardial infarction. Suppression of miR-29a has been shown to protect against myocardial I/R injury through influencing expression of SIRT1 and subduing oxidative stress and NLRP3-associated pyroptosis (Ding et al., 2020). In addition, miR-200a-3p has been found to aggravate doxorubicin-induced cardiotoxic effects through targeting PEG3 via SIRT1/NF-κB signaling pathway (Fu et al., 2021). miR-181a-5p is another miRNA which participates in the cardiomyocyte apoptosis induced by hypoxia–reoxygenation via regulation of SIRT1 (Qi et al., 2020). Moreover, an experiment in an animal model of acute myocardial infarction has shown that miR-124-3p targets SIRT1 to influence cell apoptosis, inflammatory responses, and oxidative stress through regulation of the FGF21/CREB/PGC1α axis (Wei et al., 2021). Besides, miRNAs that modulate expression of SIRT1 can affect pathogenesis of heart failure. For instance, downregulation of miR-22 by targeting SIRT1/PGC-1α could alleviate this disorder (Wang et al., 2021b). Finally, miR199/SIRT1/P300 axis has apotential function in the patheticlogy of this disorder (Asensio-Lopez et al., 2021).
Lastly, three SIRT1-interacting miRNAs have been revealed to participate in the carcinogenesis. miR-373 is a tumor suppressor miRNA that inhibits proliferation of pancreatic cancer cells through influencing activity of SIRT1/PGC-1α/NRF2 axis (Yin et al., 2021). On the other hand, miR-34a acts as an immunosuppressive miRNA in colorectal cancer via regulation of SIRT1/NF-κB/B7-H3/TNF-α axis (Meng et al., 2021). Lastly, miR-199a-5p has a role in repression of stemness of squamous cell carcinoma cells through influencing activity of SIRT1 and CD44ICD cleavage signaling (Lu et al., 2020).
SIRT1-interacting circRNAs
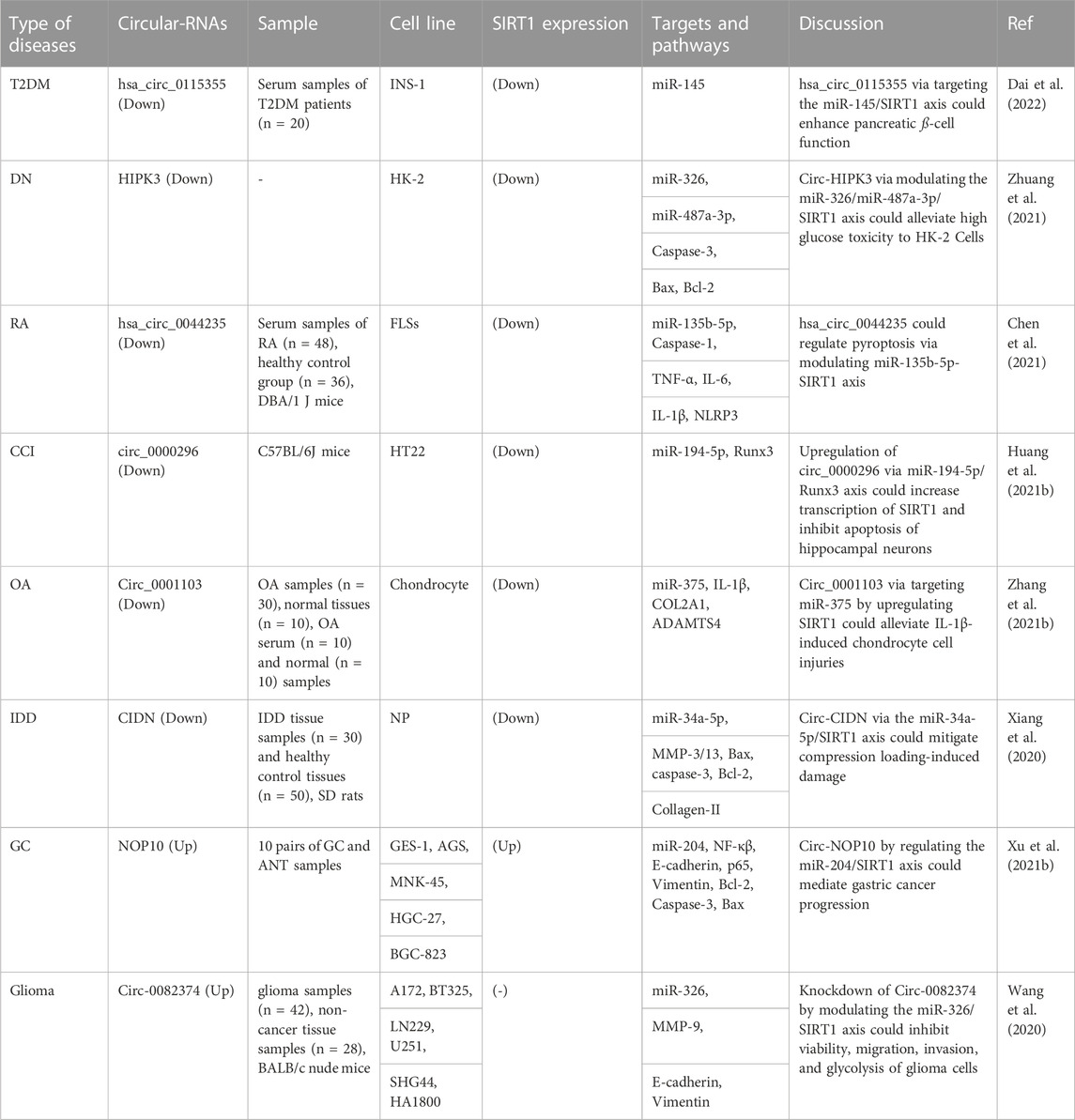
Circular RNAs (CircRNAs) are common in all animals, from viruses to mammals. They are single-stranded, endogenous covalently closed RNA molecules with highly stability. The biosynthesis, regulation, localization, destruction, and modification of circRNAs have all seen great progress (Sayad et al., 2022). CircRNAs play a role in a wide range of human disorders, particularly malignancies (Ghafouri-Fard et al., 2021b; Ghafouri-Fard et al., 2022). The impact of SIRT1-interacting circRNAs in the regulation of SIRT1 has been assessed in diabetes and its complications, rheumatoid arthritis, chronic cerebral ischemia, osteoarthritis, intervertebral disc degeneration as well as malignant disorders, particularly glioma (Table 2). All of these circRNAs have been shown to act as molecular sponges for miRNAs to subsequently affect expression of miRNAs targets (Figure 2). For instance, hsa_circ_0115355 has been found to regulate activity of miR-145/SIRT1 axis, thus enhancing function of pancreatic ß cells in patients with type 2 diabetes mellitus (Dai et al., 2022). CircHIPK3 is another circRNA which participates in the pathogenesis of diabetic complications. Expression of this circRNA has been significantly reduced in HK-2 cells following exposure with high glucose. Forced upregulation of circHIPK3 could reverse high glucose-induced pathologic events in HK-2 cells. SIRT1 has been found to be the target of miR-326 and miR-487a-3p, two downstream genes of circHIPK3. Silencing of these two miRNAs could induce proliferation and decrease apoptosis in high glucose-induced HK-2 cells. Taken together, upregulation of circHIPK3 can reduce the effects of high glucose in HK-2 cells via sponging miR-326 or miR-487a-3p and influencing expression of SIRT1 (Zhuang et al., 2021).
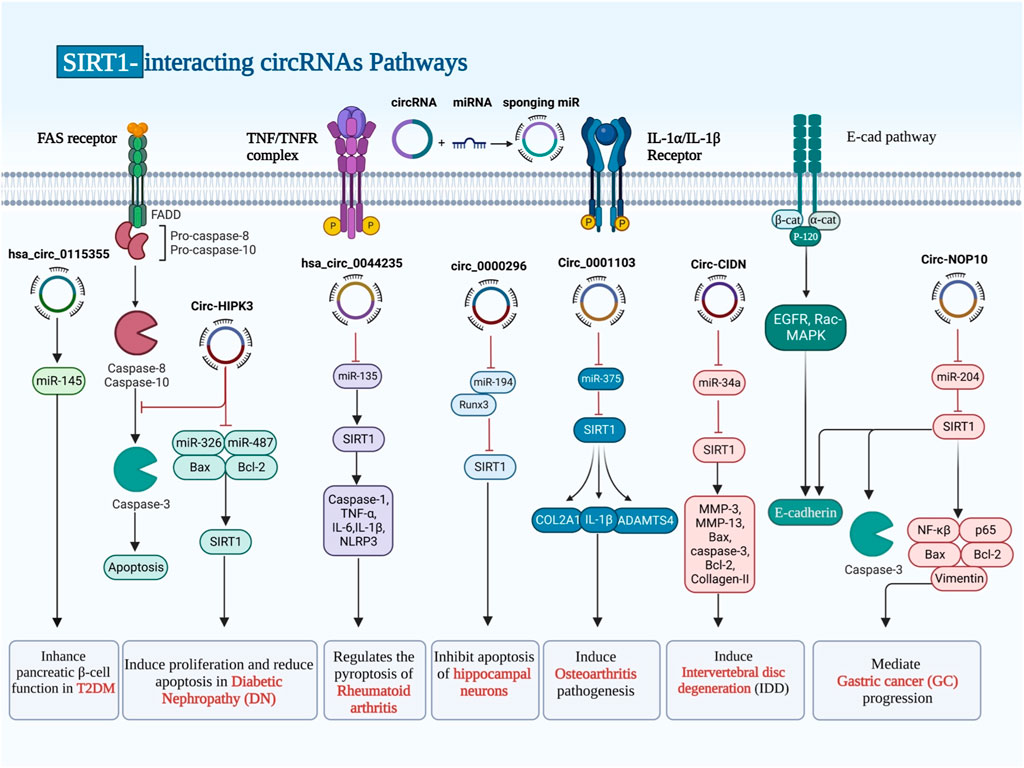
FIGURE 2. CircRNAs have been proven to serve as molecular sponges for miRNAs, thereby influencing the expression of miRNA targets. SIRT1 has been found to be the target of miRNA genes, which were already being sponged by different types of circRNAs that prevented or enhanced gene expression.
Hsa_circ_0044235 is another circRNA which has been shown to be downregulated in patients with rheumatoid arthritis (RA). Downregulation of this circRNA has been correlated with low levels of SIRT1 expression in these patients. Overexpression of hsa_circ_0044235 could attenuate joint inflammation, cell apoptosis, and joint injury, and reduce NLRP3-mediated pyroptosis but increasing SIRT1 expression. Upregulation of this circRNA could also inhibit caspase-1 content. Mechanistically, hsa_circ_0044235 increases expression of SIRT1 through sponging miR-135b-5p (Chen et al., 2021).
CircularNOP10 and circ0082374 are two putative oncogenic circRNAs that regulate expression of SIRT1. CircularNOP10 has a role in induction of progression of gastric cancer through regulation of miR-204/SIRT1 pathway (Xu J. et al., 2021). In glioma cells, circ0082374 has a role in induction of cell viability, migration, invasion and glycolysis through regulation of miR-326/SIRT1 axis (Wang et al., 2020).
SIRT1-interacting lncRNAs
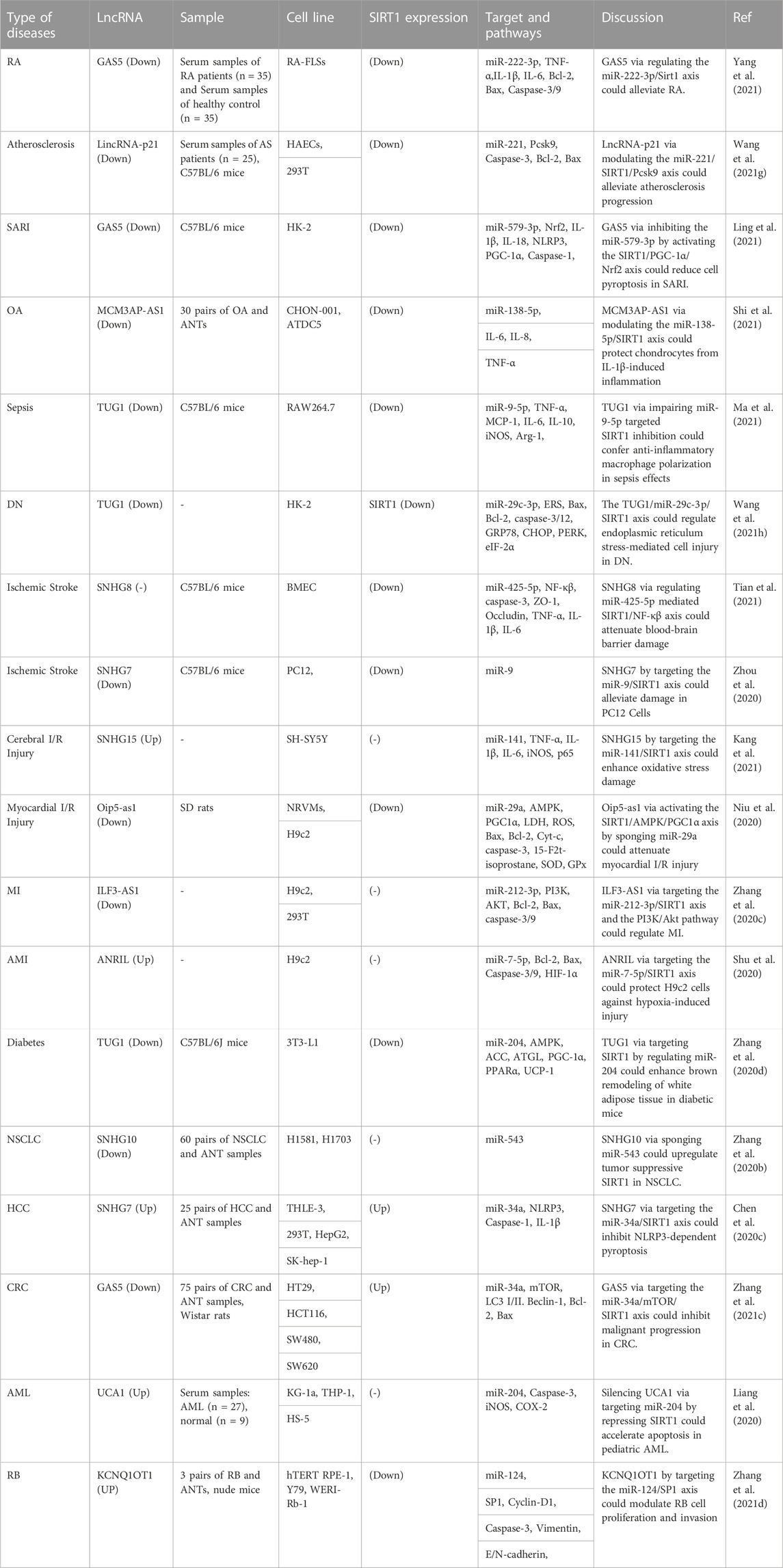
Transcripts larger than 200 nt are known as long non-coding RNAs (lnRNAs), which cannot code for proteins and may possess small open reading frames (ORFs). Because they interact with various proteins, mRNAs and DNA sequences, lncRNAs play significant roles in a number of disorders (Sabaie et al., 2021; Hussen et al., 2022b). GAS5, LincRNA-p21, MCM3AP-AS1, TUG1, SNHG7, SNHG8, SNHG10, SNHG15, Oip5-as1, ILF3-AS1, ANRIL, UCA1 and KCNQ1OT1 are examples of lncRNAs that regulate expression of SIRT1 through sponging miRNAs. These lncRNAs can affect pathogenesis of RA, atherosclerosis, sepsis-associated renal injury (SARI), diabetic nephropathy, ischemic stroke and a number of malignant conditions (Table 3). For instance, GAS5 via regulating the miR-222-3p/Sirt1 axis could alleviate RA (Yang et al., 2021). Moreover, GAS5 via inhibiting the miR-579-3p and activating the SIRT1/PGC-1α/Nrf2 axis could reduce cell pyroptosis in SARI (Ling et al., 2021). In the context of osteoarthritis, MCM3AP-AS1 via modulating the miR-138-5p/SIRT1 axis could protect chondrocytes from IL-1β-induced inflammation (Shi et al., 2021).
SIRT1-interacting lncRNAs have also been shown to affect pathogenesis of malignant conditions. For instance, SNHG10 has been found to sponge miR-543 in non small cell lung cancer (Zhang Z. et al., 2020). Moreover, SNHG7 has been demonstrated to inhibit NLRP3-associated pyroptosis through regulating miR-34a/SIRT1 axis in liver cancer (Chen Z. et al., 2020). GAS5 can inhibit malignant progression of colorecatl cancer cells through regulating macroautophagy and forming a negative feedback loop with the miR-34a/mTOR/SIRT1 axis (Zhang HG. et al., 2021). On the other hand, UCA1 has a role in induction of cell proliferation and suppression of apoptosis through affecting expression of SIRT1 and miR-204 in pediatric AML (Liang et al., 2020). The known interactions that SIRT1 has with a variety of lnRNAs are illustrated in Figure 3.
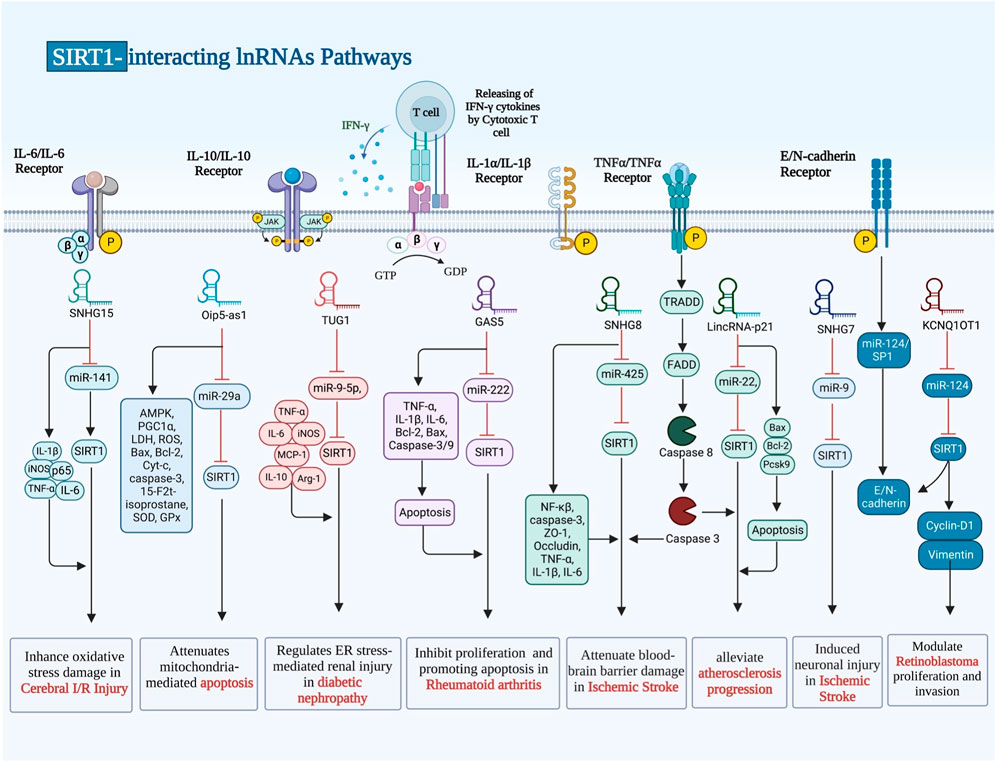
FIGURE 3. There are numerous ways in which SIRT1 and the other lnRNAs interact, and it has been demonstrated that these interactions have an impact on pathogenesis conditions.
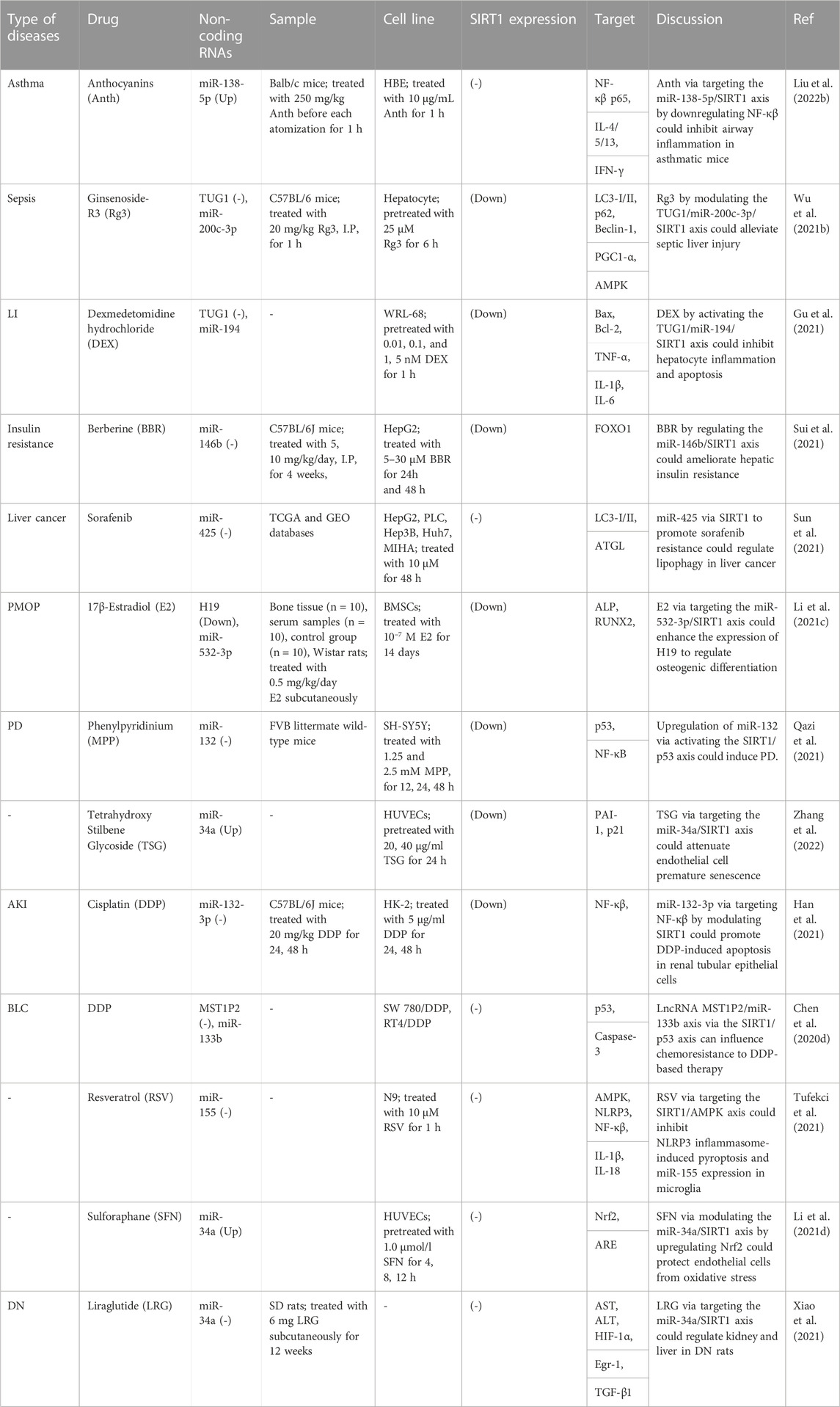
A number of therapeutic agents such as anthocyanins, ginsenoside-R3, dexmedetomidine hydrochloride, berberine, sorafenib, 17β-Estradiol, phenylpyridinium, tetrahydroxy stilbene glycoside, cisplatin, resveratrol, sulforaphane and liraglutide have been found to affect expression of non-coding RNAs/SIRT1 axes (Table 4). For instance, experiments in animal model of asthma have shown that anthocyanins suppresses inflammatory responses in airways through decreasing activity of NF-κB pathway via the miR-138-5p/SIRT1 axis (Liu Y. et al., 2022). Moreover, ginsenoside Rg3 can alleviate sepsis-related hepatic injury through modulation of TUG1/miR-200c-3p/SIRT1 axis (Wu P. et al., 2021). TUG1/miR-194/SIRT1 axis has been found to be targeted by dexmedetomidine hydrochloride to inhibit hepatocytes apoptosis and inflammatory responses (Gu et al., 2021). Additionally, the effects of berberine in amelioration of hepatic insulin resistance have been revealed to be mediated through regulation of miR-146b/SIRT1 axis (Sui et al., 2021).
Discussion
SIRT1 has a role as a deacetylase and is able to deacetylate a range of substrates. Thus, it participates in the regulation of a wide array of physiological processes such as gene expression, metabolic pathways and aging (Haigis and Guarente, 2006; Michan and Sinclair, 2007). This protein has functional interactions with lncRNAs, miRNAs and circRNAs. In fact, a complicated network exists between these non-coding RNAs and SIRT1. Hsa_circ_0115355/miR-326, hsa_circ_0115355/miR-487a-3p, HIPK3/miR-145, hsa_circ_0044235/miR-135b-5p, circ_0000296/miR-194-5p, circ_0001103/miR-375, CIDN/miR-34a-5p, NOP10/miR-204, circ-0082374/miR-324 are examples of circRNA/miRNA pairs that interact with SIRT1. Similarly, GAS5/miR-222-3p, GAS5/miR-579-3p, GAS5/miR-34a, MCM3AP-AS1/miR-138-5p, TUG1/miR-9-5p, TUG1/miR-29c-3p, TUG1/miR-204, SNHG8/miR-425-5p, SNHG7/miR-9, SNHG7/miR-34a, SNHG15/miR-141, SNHG10/miR-543, Oip5-as1/miR-29a, ILF3-AS1/miR-212-3p, ANRIL/miR-7-5p, UCA1/miR-204 and KCNQ1OT1/miR-124 are lncRNA/miRNA pairs that regulate expression of SIRT1 in different contexts. These interactions are possibly involved in the pathoetiology of a number of human disorders such as sepsis, cardiomyopathy, heart failure, non-alcoholic fatty liver disease, chronic hepatitis, cardiac fibrosis, myocardial ischemia/reperfusion injury, diabetes, ischemic stroke, immune-related disorders and cancers. In cancers, SIRT1-interacting non-coding RNAs not only affect cell proliferation but also regulate stemness and immunosuppressive responses in the tumor niche.
SIRT1 is a potential target for design of novel therapies. Most importantly, a number of drugs used for treatment of diverse asthma, sepsis, liver injury, insulin resistance, postmenopausal osteoporosis, Parkinson’s disease, diabetic nephropathy and cancers exert their effects through modulation of non-coding RNAs/SIRT1 axis. Thus, identification of the interactions between non-coding RNAs and SIRT1 has practical significance in design of novel therapeutic strategies for diverse disorders. Remarkably, non-coding RNAs that modulate expression of SIRT1 are putative modulators of the response of patients to different drugs.
Author contributions
SG-F wrote the manuscript and revised it. MT and GS supervised and designed the study. HS, YP, and BH collected the data and designed the figures and tables. All authors read and approved the submitted version.
Acknowledgments
The authors would like to thank the clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alshehri, A. S., El-Kott, A. F., El-Kenawy, A. E., Khalifa, H. S., and AlRamlawy, A. M. (2021). Cadmium chloride induces non-alcoholic fatty liver disease in rats by stimulating miR-34a/SIRT1/FXR/p53 axis. Sci. Total Environ. 784, 147182. doi:10.1016/j.scitotenv.2021.147182
Asensio-Lopez, M. C., Sassi, Y., Soler, F., Fernandez del Palacio, M. J., Pascual-Figal, D., and Lax, A. (2021). The miRNA199a/SIRT1/P300/Yy1/sST2 signaling axis regulates adverse cardiac remodeling following MI. Sci. Rep. 11 (1), 3915–4014. doi:10.1038/s41598-021-82745-9
Bai, Y., Chen, K., Zhan, J., and Wu, M. (2020). miR-122/SIRT1 axis regulates chondrocyte extracellular matrix degradation in osteoarthritis. Biosci. Rep. 40 (6), 20191908. doi:10.1042/BSR20191908
Bradbury, C., Khanim, F., Hayden, R., Bunce, C., White, D., Drayson, M., et al. (2005). Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 19 (10), 1751–1759. doi:10.1038/sj.leu.2403910
Chen, B., Wu, L., Cao, T., Zheng, H-M., and He, T. (2020b). MiR-221/SIRT1/Nrf2 signal axis regulates high glucose induced apoptosis in human retinal microvascular endothelial cells. BMC Ophthalmol. 20 (1), 300–310. doi:10.1186/s12886-020-01559-x
Chen, J., Li, Y., Li, Z., and Cao, L. (2020d). LncRNA MST1P2/miR-133b axis affects the chemoresistance of bladder cancer to cisplatin-based therapy via Sirt1/p53 signaling. J. Biochem. Mol. Toxicol. 34 (4), e22452. doi:10.1002/jbt.22452
Chen, L., Xie, W., Wang, L., Zhang, X., Liu, E., and Kou, Q. (2020a). MiRNA-133a aggravates inflammatory responses in sepsis by targeting SIRT1. Int. Immunopharmacol. 88, 106848. doi:10.1016/j.intimp.2020.106848
Chen, S., Luo, Z., and Chen, X. (2021). Hsa_circ_0044235 regulates the pyroptosis of rheumatoid arthritis via MiR-135b-5p-SIRT1 axis. Cell Cycle 20 (12), 1107–1121. doi:10.1080/15384101.2021.1916272
Chen, Z., He, M., Chen, J., Li, C., and Zhang, Q. (2020c). Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by targeting the miR-34a/SIRT1 axis in liver cancer. Oncol. Lett. 20 (1), 893–901. doi:10.3892/ol.2020.11635
Dai, Y., Ma, X., Zhang, J., Yu, S., Zhu, Y., and Wang, J. (2022). hsa_circ_0115355 promotes pancreatic β-cell function in patients with type 2 diabetes through the miR-145/SIRT1 axis. J. Clin. Laboratory Analysis 36, e24583. doi:10.1002/jcla.24583
Ding, S., Liu, D., Wang, L., Wang, G., and Zhu, Y. (2020). Inhibiting microRNA-29a protects myocardial ischemia-reperfusion injury by targeting SIRT1 and suppressing oxidative stress and NLRP3-mediated pyroptosis pathway. J. Pharmacol. Exp. Ther. 372 (1), 128–135. doi:10.1124/jpet.119.256982
Dong, W., Zhang, H., Zhao, C., Luo, Y., and Chen, Y. (2021). Silencing of miR-150-5p ameliorates diabetic nephropathy by targeting SIRT1/p53/AMPK pathway. Front. physiology 12, 624989. doi:10.3389/fphys.2021.624989
Fu, Q., Pan, H., Tang, Y., Rong, J., and Zheng, Z. (2021). MiR-200a-3p aggravates DOX-induced cardiotoxicity by targeting PEG3 through SIRT1/NF-κB signal pathway. Cardiovasc. Toxicol. 21 (4), 302–313. doi:10.1007/s12012-020-09620-3
Ghafouri-Fard, S., Hussen, B. M., Badrlou, E., Abak, A., and Taheri, M. (2021a). MicroRNAs as important contributors in the pathogenesis of colorectal cancer. Biomed. Pharmacother. 140, 111759. doi:10.1016/j.biopha.2021.111759
Ghafouri-Fard, S., Khoshbakht, T., Hussen, B. M., Sarfaraz, S., Taheri, M., and Ayatollahi, S. A. (2022). Circ_CDR1as: A circular RNA with roles in the carcinogenesis. Pathology - Res. Pract. 236, 153968. doi:10.1016/j.prp.2022.153968
Ghafouri-Fard, S., Taheri, M., Hussen, B. M., Vafaeimanesh, J., Abak, A., and Vafaee, R. (2021b). Function of circular RNAs in the pathogenesis of colorectal cancer. Biomed. Pharmacother. 140, 111721. doi:10.1016/j.biopha.2021.111721
Gu, X-X., Xu, X-X., Liao, H-H., Wu, R-N., Huang, W-M., Cheng, L-X., et al. (2021). Dexmedetomidine hydrochloride inhibits hepatocyte apoptosis and inflammation by activating the lncRNA TUG1/miR-194/SIRT1 signaling pathway. J. Inflamm. 18 (1), 20–12. doi:10.1186/s12950-021-00287-3
Haigis, M. C., and Guarente, L. P. (2006). Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes and Dev. 20 (21), 2913–2921. doi:10.1101/gad.1467506
Han, S., Lin, F., Ruan, Y., Zhao, S., Yuan, R., Ning, J., et al. (2021). miR-132-3p promotes the cisplatin-induced apoptosis and inflammatory response of renal tubular epithelial cells by targeting SIRT1 via the NF-κB pathway. Int. Immunopharmacol. 99, 108022. doi:10.1016/j.intimp.2021.108022
Hao, R., Song, X., Sun-Waterhouse, D., Tan, X., Li, F., and Li, D. (2021). Mir-34a/sirt1/p53 signaling pathway contributes to cadmium-induced nephrotoxicity: A preclinical study in mice. Environ. Pollut. 282, 117029. doi:10.1016/j.envpol.2021.117029
Huang, K., Yang, C., Zheng, J., Liu, X., Liu, J., Che, D., et al. (2021b). Effect of circular RNA, mmu_circ_0000296, on neuronal apoptosis in chronic cerebral ischaemia via the miR-194-5p/Runx3/Sirt1 axis. Cell Death Discov. 7 (1), 124–215. doi:10.1038/s41420-021-00507-y
Huang, L., Cai, H-A., Zhang, M-S., Liao, R-Y., Huang, X., and Hu, F-D. (2021a). Ginsenoside Rg1 promoted the wound healing in diabetic foot ulcers via miR-489–3p/Sirt1 axis. J. Pharmacol. Sci. 147 (3), 271–283. doi:10.1016/j.jphs.2021.07.008
Huffman, D. M., Grizzle, W. E., Bamman, M. M., Kim, J-S., Eltoum, I. A., Elgavish, A., et al. (2007). SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 67 (14), 6612–6618. doi:10.1158/0008-5472.CAN-07-0085
Hussen, B. M., Hidayat, H. J., and Ghafouri-Fard, S. (2022b). Identification of expression of CCND1-related lncRNAs in breast cancer. Pathology - Res. Pract. 236, 154009. doi:10.1016/j.prp.2022.154009
Hussen, B. M., Hidayat, H. J., Salihi, A., Sabir, D. K., Taheri, M., and Ghafouri-Fard, S. (2021). MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 138, 111528. doi:10.1016/j.biopha.2021.111528
Hussen, B. M., Salihi, A., Abdullah, S. T., Rasul, M. F., Hidayat, H. J., Hajiesmaeili, M., et al. (2022a). Signaling pathways modulated by miRNAs in breast cancer angiogenesis and new therapeutics. Pathology - Res. Pract. 230, 153764. doi:10.1016/j.prp.2022.153764
Kang, M., Ji, F., Sun, X., Liu, H., and Zhang, C. (2021). LncRNA SNHG15 promotes oxidative stress damage to regulate the occurrence and development of cerebral ischemia/reperfusion injury by targeting the miR-141/SIRT1 axis. J. Healthc. Eng. 2021, 6577799. doi:10.1155/2021/6577799
Li, C., Wang, F., Miao, P., Yan, L., Liu, S., Wang, X., et al. (2020b). miR-138 increases depressive-like behaviors by targeting SIRT1 in hippocampus. Neuropsychiatric Dis. Treat. 16, 949–957. doi:10.2147/NDT.S237558
Li, F., Zhang, L., Xue, H., Xuan, J., Rong, S., and Wang, K. (2021a). SIRT1 alleviates hepatic ischemia-reperfusion injury via the miR-182-mediated XBP1/NLRP3 pathway. Mol. Therapy-Nucleic Acids. 23, 1066–1077. doi:10.1016/j.omtn.2020.11.015
Li, H., Chou, P., Du, F., Sun, L., Liu, J., and Wang, W. (2021b). Depleting microRNA-183-3p improves renal tubulointerstitial fibrosis after acute kidney injury via SIRT1/PUMA/FOXO3a deacetylation. Life Sci. 269, 119017. doi:10.1016/j.lfs.2021.119017
Li, T., Jiang, H., Li, Y., Zhao, X., and Ding, H. (2021c). Estrogen promotes lncRNA H19 expression to regulate osteogenic differentiation of BMSCs and reduce osteoporosis via miR-532-3p/SIRT1 axis. Mol. Cell. Endocrinol. 527, 111171. doi:10.1016/j.mce.2021.111171
Li, T., Pang, Q., Liu, Y., Bai, M., Peng, Y., and Zhang, Z. (2021d). Sulforaphane protects human umbilical vein endothelial cells from oxidative stress via the miR-34a/SIRT1 axis by upregulating nuclear factor erythroid-2-related factor 2. Exp. Ther. Med. 21 (3), 186. doi:10.3892/etm.2021.9617
Li, X., Zhang, W., Xu, K., and Lu, J. (2020a). miR-34a promotes liver fibrosis in patients with chronic hepatitis via mediating Sirt1/p53 signaling pathway. Pathology-Research Pract. 216 (5), 152876. doi:10.1016/j.prp.2020.152876
Liang, Y., Li, E., Zhang, H., Zhang, L., Tang, Y., and Wanyan, Y. (2020). Silencing of lncRNA UCA1 curbs proliferation and accelerates apoptosis by repressing SIRT1 signals by targeting miR-204 in pediatric AML. J. Biochem. Mol. Toxicol. 34 (3), e22435. doi:10.1002/jbt.22435
Liang, Y., Liu, H., Zhu, J., Song, N., Lu, Z., Fang, Y., et al. (2021). Inhibition of p53/miR-34a/SIRT1 axis ameliorates podocyte injury in diabetic nephropathy. Biochem. Biophysical Res. Commun. 559, 48–55. doi:10.1016/j.bbrc.2021.04.025
Ling, H., Li, Q., Duan, Z-P., Wang, Y-J., Hu, B-Q., and Dai, X-G. (2021). LncRNA GAS5 inhibits miR-579-3p to activate SIRT1/PGC-1α/Nrf2 signaling pathway to reduce cell pyroptosis in sepsis-associated renal injury. Am. J. Physiology-Cell Physiology 321 (7), C117–C133. doi:10.1152/ajpcell.00394.2020
Liu, M., Zhang, Y., Cao, X., Shi, T., and Yan, Y. (2022a). miR-197 participates in lipopolysaccharide-induced cardiomyocyte injury by modulating SIRT1. Cardiol. Res. Pract. 2022, 7687154. doi:10.1155/2022/7687154
Liu, Y., Zhang, M., Zhang, H., Qian, X., Luo, L., and He, Z. (2022b). Cancer metastases from lung adenocarcinoma disappeared after molecular targeted therapy: A successfully clinical treatment experience. Int. Archives Allergy Immunol. 183 (5), 539–546. doi:10.2147/PGPM.S367978
Lu, R-H., Xiao, Z-Q., Zhou, J-D., Yin, C-Q., Chen, Z-Z., Tang, F-J., et al. (2020). MiR-199a-5p represses the stemness of cutaneous squamous cell carcinoma stem cells by targeting Sirt1 and CD44ICD cleavage signaling. Cell Cycle 19 (1), 1–14. doi:10.1080/15384101.2019.1689482
Ma, W., Zhang, W., Cui, B., Gao, J., Liu, Q., Yao, M., et al. (2021). Functional delivery of lncRNA TUG1 by endothelial progenitor cells derived extracellular vesicles confers anti-inflammatory macrophage polarization in sepsis via impairing miR-9-5p-targeted SIRT1 inhibition. Cell death Dis. 12 (11), 1056–1110. doi:10.1038/s41419-021-04117-5
Meng, F., Yang, M., Chen, Y., Chen, W., and Wang, W. (2021). miR-34a induces immunosuppression in colorectal carcinoma through modulating a SIRT1/NF-κB/B7-H3/TNF-α axis. Immunotherapy 70 (8), 2247–2259. doi:10.1007/s00262-021-02862-2
Michan, S., and Sinclair, D. (2007). Sirtuins in mammals: Insights into their biological function. Biochem. J. 404 (1), 1–13. doi:10.1042/BJ20070140
Mo, Y., Liu, B., Qiu, S., Wang, X., Zhong, L., Han, X., et al. (2020). Down-regulation of microRNA-34c-5p alleviates neuropathic pain via the SIRT1/STAT3 signaling pathway in rat models of chronic constriction injury of sciatic nerve. J. Neurochem. 154 (3), 301–315. doi:10.1111/jnc.14998
Niu, X., Pu, S., Ling, C., Xu, J., Wang, J., Sun, S., et al. (2020). lncRNA Oip5-as1 attenuates myocardial ischaemia/reperfusion injury by sponging miR-29a to activate the SIRT1/AMPK/PGC1α pathway. Cell Prolif. 53 (6), e12818. doi:10.1111/cpr.12818
Pillarisetti, S. (2008). A review of Sirt1 and Sirt1 modulators in cardiovascular and metabolic diseases. Recent Pat. Cardiovasc. Drug Discov. Discontin. 3 (3), 156–164. doi:10.2174/157489008786263989
Purushotham, A., Schug, T. T., Xu, Q., Surapureddi, S., Guo, X., and Li, X. (2009). Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell metab. 9 (4), 327–338. doi:10.1016/j.cmet.2009.02.006
Qazi, T. J., Lu, J., Duru, L., Zhao, J., and Qing, H. (2021). Upregulation of mir-132 induces dopaminergic neuronal death via activating SIRT1/P53 pathway. Neurosci. Lett. 740, 135465. doi:10.1016/j.neulet.2020.135465
Qi, M., He, L., Ma, X., and Li, Z. (2020). MiR-181a-5p is involved in the cardiomyocytes apoptosis induced by hypoxia–reoxygenation through regulating SIRT1. Biosci. Biotechnol. Biochem. 84 (7), 1353–1361. doi:10.1080/09168451.2020.1750943
Qu, L. H., Hong, X., Zhang, Y., Cong, X., Xiang, R. L., Mei, M., et al. (2021). C1q/tumor necrosis factor-related protein-6 attenuates TNF-α-induced apoptosis in salivary acinar cells via AMPK/SIRT1-modulated miR-34a-5p expression. J. Cell. Physiology 236 (8), 5785–5800. doi:10.1002/jcp.30262
Rahman, S., and Islam, R. (2011). Mammalian Sirt1: Insights on its biological functions. Cell Commun. Signal. 9 (1), 11–18. doi:10.1186/1478-811X-9-11
Sabaie, H., Salkhordeh, Z., Asadi, M. R., Ghafouri-Fard, S., Amirinejad, N., Askarinejad Behzadi, M., et al. (2021). Long non-coding RNA- associated competing endogenous RNA axes in T-cells in multiple sclerosis. Front. Immunol. 12, 770679. doi:10.3389/fimmu.2021.770679
Sayad, A., Najafi, S., Hussen, B. M., Jamali, E., Taheri, M., and Ghafouri-Fard, S. (2022). The role of circular RNAs in pancreatic cancer: New players in tumorigenesis and potential biomarkers. Pathology - Res. Pract. 232, 153833. doi:10.1016/j.prp.2022.153833
Shi, J., Cao, F., Chang, Y., Xin, C., Jiang, X., Xu, J., et al. (2021). Long non-coding RNA MCM3AP-AS1 protects chondrocytes ATDC5 and CHON-001 from IL-1β-induced inflammation via regulating miR-138-5p/SIRT1. Bioengineered 12 (1), 1445–1456. doi:10.1080/21655979.2021.1905247
Shu, L., Zhang, W., Huang, C., Huang, G., Su, G., and Xu, J. (2020). lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/SIRT1 axis. J. Cell. physiology 235 (2), 1175–1183. doi:10.1002/jcp.29031
Stünkel, W., Peh, B. K., Tan, Y. C., Nayagam, V. M., Wang, X., Salto-Tellez, M., et al. (2007). Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. Healthc. Nutr. Technol. 2 (11), 1360–1368. doi:10.1002/biot.200700087
Sui, M., Jiang, X., Sun, H., Liu, C., and Fan, Y. (2021). Berberine ameliorates hepatic insulin resistance by regulating microRNA-146b/SIRT1 pathway. Diabetes, metabolic syndrome Obes. targets Ther. 14, 2525–2537. doi:10.2147/DMSO.S313068
Sun, G., Yang, L., Wei, S., Jin, H., Li, B., and Li, H. (2021). miR-425 regulates lipophagy via SIRT1 to promote sorafenib resistance in liver cancer. Oncol. Lett. 22 (4), 695–710. doi:10.3892/ol.2021.12956
Tian, J., Liu, Y., Wang, Z., Zhang, S., Yang, Y., Zhu, Y., et al. (2021). LncRNA Snhg8 attenuates microglial inflammation response and blood–brain barrier damage in ischemic stroke through regulating miR-425-5p mediated SIRT1/NF-κB signaling. J. Biochem. Mol. Toxicol. 35 (5), e22724. doi:10.1002/jbt.22724
Tufekci, K. U., Eltutan, B. I., Isci, K. B., and Genc, S. (2021). Resveratrol inhibits NLRP3 inflammasome-induced pyroptosis and miR-155 expression in microglia through Sirt1/AMPK pathway. Neurotox. Res. 39 (6), 1812–1829. doi:10.1007/s12640-021-00435-w
Vaziri, H., Dessain, S. K., Eaton, E. N., Imai, S-I., Frye, R. A., Pandita, T. K., et al. (2001). hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell 107 (2), 149–159. doi:10.1016/s0092-8674(01)00527-x
Wang, B., Li, B., and Si, T. (2020). Knockdown of circ0082374 inhibits cell viability, migration, invasion and glycolysis in glioma cells by miR-326/SIRT1. Brain Res. 1748, 147108. doi:10.1016/j.brainres.2020.147108
Wang, C., Guo, X., Wang, Y., and Wang, H. (2021c). Silencing of miR-324-5p alleviates rat spinal cord injury by Sirt1. Neurosci. Res. 173, 34–43. doi:10.1016/j.neures.2021.05.010
Wang, H., He, F., Liang, B., Jing, Y., Zhang, P., Liu, W., et al. (2021g). LincRNA-p21 alleviates atherosclerosis progression through regulating the miR-221/SIRT1/Pcsk9 axis. J. Cell. Mol. Med. 25 (19), 9141–9153. doi:10.1111/jcmm.16771
Wang, R., Xu, Y., Niu, X., Fang, Y., Guo, D., Chen, J., et al. (2021b). miR-22 inhibition alleviates cardiac dysfunction in doxorubicin-induced cardiomyopathy by targeting the sirt1/PGC-1α pathway. Front. Physiology 12, 646903. doi:10.3389/fphys.2021.646903
Wang, R., Xu, Y., Zhang, W., Fang, Y., Yang, T., Zeng, D., et al. (2021a). Inhibiting miR-22 alleviates cardiac dysfunction by regulating Sirt1 in septic cardiomyopathy. Front. Cell Dev. Biol. 9, 650666. doi:10.3389/fcell.2021.650666
Wang, S., Yi, P., Wang, N., Song, M., Li, W., and Zheng, Y. (2021h). LncRNA TUG1/miR-29c-3p/SIRT1 axis regulates endoplasmic reticulum stress-mediated renal epithelial cells injury in diabetic nephropathy model in vitro. PloS one 16 (6), e0252761. doi:10.1371/journal.pone.0252761
Wang, Y., Yang, Z., Zhang, K., Wan, Y., Zhou, Y., and Yang, Z. (2021d). miR-135a-5p inhibitor protects glial cells against apoptosis via targeting SIRT1 in epilepsy. Exp. Ther. Med. 21 (5), 431–438. doi:10.3892/etm.2021.9848
Wang, Z., Chen, R., Xu, Z., Ru, W., Tian, H., Yang, F., et al. (2021e). MiR-155-5p promotes renal interstitial fibrosis in obstructive nephropathy via inhibiting SIRT1 signaling pathway. J. Recept. Signal Transduct. 41 (5), 466–475. doi:10.1080/10799893.2020.1825491
Wang, Z., Shi, D., Zhang, N., Yuan, T., and Tao, H. (2021f). MiR-217 promotes endothelial cell senescence through the SIRT1/p53 signaling pathway. J. Mol. Histology 52 (2), 257–267. doi:10.1007/s10735-020-09945-x
Wei, Y-J., Wang, J-F., Cheng, F., Xu, H-J., Chen, J-J., Xiong, J., et al. (2021). miR-124-3p targeted SIRT1 to regulate cell apoptosis, inflammatory response, and oxidative stress in acute myocardial infarction in rats via modulation of the FGF21/CREB/PGC1α pathway. J. physiology Biochem. 77 (4), 577–587. doi:10.1007/s13105-021-00822-z
Wu, L., Zhang, G., Guo, C., Zhao, X., Shen, D., and Yang, N. (2020). MiR-128-3p mediates TNF-α-induced inflammatory responses by regulating Sirt1 expression in bone marrow mesenchymal stem cells. Biochem. biophysical Res. Commun. 521 (1), 98–105. doi:10.1016/j.bbrc.2019.10.083
Wu, P., Yu, X., Peng, Y., Wang, Q-L., Deng, L-T., and Xing, W. (2021b). Ginsenoside Rg3 alleviates septic liver injury by regulating the lncRNA TUG1/miR-200c-3p/SIRT1 axis. J. Inflamm. 18 (1), 31–13. doi:10.1186/s12950-021-00296-2
Wu, Z., Chen, J., Zhao, W., Zhuo, C. H., and Chen, Q. (2021a). Inhibition of miR-181a attenuates sepsis-induced inflammation and apoptosis by activating Nrf2 and inhibiting NF-κB pathways via targeting SIRT1. Kaohsiung J. Med. Sci. 37 (3), 200–207. doi:10.1002/kjm2.12310
Xiang, Q., Kang, L., Wang, J., Liao, Z., Song, Y., Zhao, K., et al. (2020). CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine 53, 102679. doi:10.1016/j.ebiom.2020.102679
Xiao, S., Yang, Y., Liu, Y-T., and Zhu, J. (2021). Liraglutide regulates the kidney and liver in diabetic nephropathy rats through the miR-34a/SIRT1 pathway. J. Diabetes Res. 2021, 8873956. doi:10.1155/2021/8873956
Xu, H., Zhang, J., Shi, X., Li, X., and Zheng, C. (2021a). NF-κB inducible miR-30b-5p aggravates joint pain and loss of articular cartilage via targeting SIRT1-FoxO3a-mediated NLRP3 inflammasome. Aging (Albany NY) 13 (16), 20774–20792. doi:10.18632/aging.203466
Xu, J., Wang, X., Wang, W., Zhang, L., and Huang, P. (2021b). Candidate oncogene circularNOP10 mediates gastric cancer progression by regulating miR-204/SIRT1 pathway. J. Gastrointest. Oncol. 12 (4), 1428–1443. doi:10.21037/jgo-21-422
Yamamoto, H., Schoonjans, K., and Auwerx, J. (2007). Sirtuin functions in health and disease. Mol. Endocrinol. 21 (8), 1745–1755. doi:10.1210/me.2007-0079
Yang, Y., and Li, L. (2021). Depleting microRNA-146a-3p attenuates lipopolysaccharide-induced acute lung injury via up-regulating SIRT1 and mediating NF-κB pathway. J. Drug Target. 29 (4), 420–429. doi:10.1080/1061186X.2020.1850738
Yang, Y-L., Wang, P-W., Wang, F-S., Lin, H-Y., and Huang, Y-H. (2020). miR-29a modulates GSK3β/SIRT1-linked mitochondrial proteostatic stress to ameliorate mouse non-alcoholic steatohepatitis. Int. J. Mol. Sci. 21 (18), 6884. doi:10.3390/ijms21186884
Yang, Z., Lin, S-D., Zhan, F., Liu, Y., and Zhan, Y-W. (2021). LncRNA GAS5 alleviates rheumatoid arthritis through regulating miR-222-3p/Sirt1 signalling axis. Autoimmunity 54 (1), 13–22. doi:10.1080/08916934.2020.1846183
Yin, Q-H., Zhou, Y., and Li, Z-Y. (2021). miR-373 suppresses cell proliferation and apoptosis via regulation of SIRT1/PGC-1α/NRF2 axis in pancreatic cancer. Cell J. (Yakhteh) 23 (2), 199–210. doi:10.22074/cellj.2021.7038
Yuan, T., Zhang, L., Yao, S., Deng, S. Y., and Liu, J. Q. (2020). miR-195 promotes LPS-mediated intestinal epithelial cell apoptosis via targeting SIRT1/eIF2a. Int. J. Mol. Med. 45 (2), 510–518. doi:10.3892/ijmm.2019.4431
Zhan, H., Huang, F., Niu, Q., Jiao, M., Han, X., Zhang, K., et al. (2021). Downregulation of miR-128 ameliorates Ang II-induced cardiac remodeling via SIRT1/PIK3R1 multiple targets. Oxidative Med. Cell. Longev. 2021, 8889195. doi:10.1155/2021/8889195
Zhang, H., Yang, X., Xu, Y., and Li, H. (2021d). KCNQ1OT1 regulates the retinoblastoma cell proliferation, migration and SIRT1/JNK signaling pathway by targeting miR-124/SP1 axis. Biosci. Rep. 41 (1), 20201626. doi:10.1042/BSR20201626
Zhang, H., Zhang, X. M., Zong, Dd, Xy, Ji, Jiang, H., Zhang, F. Z., et al. (2021a). miR-34a-5p up-regulates the IL-1β/COX2/PGE2 inflammation pathway and induces the release of CGRP via inhibition of SIRT1 in rat trigeminal ganglion neurons. FEBS Open bio 11 (1), 300–311. doi:10.1002/2211-5463.13027
Zhang, H. G., Wang, F. J., Wang, Y., Zhao, Z. X., and Qiao, P. F. (2021c). lncRNA GAS5 inhibits malignant progression by regulating macroautophagy and forms a negative feedback regulatory loop with the miR-34a/mTOR/SIRT1 pathway in colorectal cancer. Oncol. Rep. 45 (1), 202–216. doi:10.3892/or.2020.7825
Zhang, J., Yang, Z., Fang, K., Shi, Z., Ren, D., and Sun, J. (2020c). Long noncoding RNA ILF3-AS1 regulates myocardial infarction via the miR-212-3p/SIRT1 axis and PI3K/Akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24 (5), 2647–2658. doi:10.26355/eurrev_202003_20534
Zhang, L., Guo, Y., Shi, S., Zhuge, Y., Chen, N., Ding, Z., et al. (2022). Tetrahydroxy stilbene glycoside attenuates endothelial cell premature senescence induced by H2O2 through the microRNA-34a/SIRT1 pathway. Sci. Rep. 12 (1), 1708–8. doi:10.1038/s41598-022-05804-9
Zhang, M., Cao, M., Kong, L., Liu, J., Wang, Y., Song, C., et al. (2020a). MiR-204-5p promotes lipid synthesis in mammary epithelial cells by targeting SIRT1. Biochem. Biophysical Res. Commun. 533 (4), 1490–1496. doi:10.1016/j.bbrc.2020.10.056
Zhang, M., Mou, L., Liu, S., Sun, F., and Gong, M. (2021b). Circ_0001103 alleviates IL-1β-induced chondrocyte cell injuries by upregulating SIRT1 via targeting miR-375. Clin. Immunol. 227, 108718. doi:10.1016/j.clim.2021.108718
Zhang, Y., Ma, Y., Gu, M., and Peng, Y. (2020d). lncRNA TUG1 promotes the Brown remodeling of white adipose tissue by regulating miR-204-targeted SIRT1 in diabetic mice. Int. J. Mol. Med. 46 (6), 2225–2234. doi:10.3892/ijmm.2020.4741
Zhang, Z., Nong, L., Chen, M-L., Gu, X-L., Zhao, W-W., Liu, M-H., et al. (2020b). Long noncoding RNA SNHG10 sponges miR-543 to upregulate tumor suppressive SIRT1 in nonsmall cell lung cancer. Cancer Biotherapy Radiopharm. 35 (10), 771–775. doi:10.1089/cbr.2019.3334
Zheng, M., Tan, J., Liu, X., Jin, F., Lai, R., and Wang, X. (2021). miR-146a-5p targets Sirt1 to regulate bone mass. Bone Rep. 14, 101013. doi:10.1016/j.bonr.2021.101013
Zhou, F., Wang, Y-K., Zhang, C-G., and Wu, B-Y. (2021). miR-19a/b-3p promotes inflammation during cerebral ischemia/reperfusion injury via SIRT1/FoxO3/SPHK1 pathway. J. neuroinflammation 18 (1), 122–213. doi:10.1186/s12974-021-02172-5
Zhou, T., Wang, S., Lu, K., and Yin, C. (2020). Long non-coding RNA SNHG7 alleviates oxygen and glucose deprivation/reoxygenation-induced neuronal injury by modulating miR-9/SIRT1 Axis in PC12 cells: Potential role in ischemic stroke. Neuropsychiatric Dis. Treat. 16, 2837–2848. doi:10.2147/NDT.S273421
Zhuang, L., Wang, Z., Hu, X., Yang, Q., Pei, X., and Jin, G. (2021). CircHIPK3 alleviates high glucose toxicity to human renal tubular epithelial HK-2 cells through regulation of miR-326/miR-487a-3p/SIRT1. Targets Ther. 14, 729–740. doi:10.2147/DMSO.S289624
Glossary
Keywords: SIRT1, lncRNA, miRNA, circRNA, biomar
Citation: Ghafouri-Fard S, Shoorei H, Hussen BM, Poornajaf Y, Taheri M and Sharifi G (2023) Interaction between SIRT1 and non-coding RNAs in different disorders. Front. Genet. 14:1121982. doi: 10.3389/fgene.2023.1121982
Received: 12 December 2022; Accepted: 16 June 2023;
Published: 27 June 2023.
Edited by:
Liqi Shu, Brown University, United StatesReviewed by:
Roopa Biswas, Uniformed Services University of the Health Sciences, United StatesAmin Safa, Complutense University of Madrid, Spain
Copyright © 2023 Ghafouri-Fard, Shoorei, Hussen, Poornajaf, Taheri and Sharifi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, TW9oYW1tYWQudGFoZXJpQHVuaS1qZW5hLmRl; Guive Sharifi, Z2libm93QHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Hamed Shoorei
Hamed Shoorei Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Yadollah Poornajaf
Yadollah Poornajaf Mohammad Taheri
Mohammad Taheri Guive Sharifi
Guive Sharifi