
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 16 January 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1111318
This article is part of the Research TopicMolecular Dissection of the Genetic Basis Underlying Plant Complex TraitsView all 6 articles
Seed germination is vital for ensuring the continuity of life in spermatophyte. High-quality seed germination usually represents good seedling establishment and plant production. Here, we identified OsLTPL23, a putative rice non-specific lipid transport protein, as an important regulator responsible for seed germination. Subcellular localization analysis confirmed that OsLTPL23 is present in the plasma membrane and nucleus. The knockout mutants of OsLTPL23 were generated by CRISPR/Cas9-mediated genome editing, and osltpl23 lines significantly germinated slower and lower than the Nipponbare (NIP). Starch and soluble sugar contents measurement showed that OsLTPL23 may have alpha-amylase inhibitor activity, and high soluble sugar content may be a causal agent for the delayed seed germination of osltpl23 mutants. Transcript profiles in the germinating seeds exhibited that the abscisic acid (ABA)-responsive genes, OsABI3 and OsABI5, and biosynthesis genes, OsNCED1, OsNCED2, OsNCED3 and OsNCED4, are obviously upregulated in the osltpl23 mutants compared to NIP plants, conversely, ABA metabolism genes OsABA8ox1, OsABA8ox2 and OsABA8ox3 are stepwise decreased. Further investigations found that osltpl23 mutants displays weakened early seedling growth, with elevated gene expresssion of ABA catabolism genes and repressive transcription response of defence-related genes OsWRKY45, OsEiN3, OsPR1a, OsPR1b and OsNPR1. Integrated analysis indicated that OsLTPL23 may exert an favorable effect on rice seed germination and early seedling growth via modulating endogenous ABA homeostasis. Collectively, our study provides important insights into the roles of OsLTPL23-mediated carbohydrate conversion and endogenous ABA pathway on seed germination and early seedling growth, which contributes to high-vigor seed production in rice breeding.
Seed vigor in plant determines seed germination, seedling emergence and growth, and seed storage ability under favorable or adverse environmental conditions (Sun et al., 2007; Finch-Savage and Bassel, 2016). Seed germination, to some extent, is synonymous with seed vigor (Finch-Savage and Bassel, 2016; Leprince et al., 2017), in which the germination rate is usually used as morphological indicator of seed viability (Wang et al., 2010). High-quality seed germination results in robust and healthy post-germination seedling growth, with the ability to withstand stressful environment (Foolad et al., 2007). Therefore, identification of the genes responsible for seed germination will contribute to develop high-vigor plant varieties and crop production.
The phytohormone ABA acts an indispensable roles in inhibiting seed germination and promoting seed dormancy (Penfield, 2017). The degradation of ABA content in seeds alleviates the ABA signaling pathway during seed imbibition (Preston et al., 2009). The reports from independent groups have demonstrated that the ABA-responsive transcription factors, ABSCISIC ACID INSENSITIVE 3 (ABI3), ABSCISIC ACID INSENSITIVE 4 (ABI4) and ABSCISIC ACID INSENSITIVE 5 (ABI5), exert distinct roles in germinated seeds and early seedling growth. The loss-of-function alleles of ABI3 reduces the ABA-mediated inhibition during seed germination and post-germination growth (Ding et al., 2014). Salinity-induced ABI4 repress seed germination and post-germination growth by promoting ROS accumulation (Luo et al., 2021). ABI5 enhances exogenous ABA-mediated developmental arrest from seed germination to vegetative growth (Lopez-Molina et al., 2001). Soluble sugar is another important regulator in seed germination and post-germination seedling development in plants (Dekkers et al., 2004; Zhu et al., 2009; Zhao et al., 2018). The germination delay of plant seeds by exogenous glucose results from the suppressive ABA catabolism (Dekkers et al., 2004; Zhu et al., 2009), whereas exogenous ABA application inhibits seed germination through locally restricting glucose availability of the embryonic hypocotyl (Xue et al., 2021; Deng et al., 2020). In early seedling growth, the inhibitory effect of exogenous glucose is accomplished by through ABI4-mediated sugar-ABA signaling pathway (Arroyo et al., 2003; Bossi et al., 2009; Li et al., 2014a). However, the interaction of endogenous ABA and soluble sugar in plant seed germination and early seedling growth is still mysterious.
Plant non-specific lipid transfer proteins (nsLTPs) are small basic lipid-binding proteins, which precursors typically harbour an hydrophobic signal peptide in N-terminal and an internal hydrophobic molecules binding cavity forming by an conserved eight cysteine motif (8CM, C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C) (Kader, 1996; Salminen et al., 2016; Meng et al., 2018; Fleury et al., 2019). In rice, the nsLTPs, reportedly, reside in the plasma membrane (Li et al., 2020; Zhao et al., 2020; Chen et al., 2022), cytoplasm (Fujino et al., 2008; Li et al., 2020) and nucleolus (Fujino et al., 2008), with the function in abiotic stress (Guo et al., 2013; Zhao et al., 2020), pollen development (Zhang et al., 2010; Li et al., 2020; Tao et al., 2020; Chen et al., 2022), plant height (Li et al., 2014b; Ding et al., 2014), as well as embryo development and seed germination (Wang et al., 2015). There are only two investigations of nsLTPs on early seedling growth and seed germination in rice. The OsLTPL159 allele distributed in different rice groups confers a distinct seedling cold tolerance (Zhao et al., 2020), and OsLTPL36 contributes to rice embryo development, seed quality, seed germination and early seedling growth (Wang et al., 2015). Whereas, the association between ABA, soluble sugar, and nsLTPs-mediated seed germination and post-germination seedling growth awaits disclosure.
In this work, we characterized a rice plasma membrane- and nucleus-localized nsLTP, OsLTPL23. Site-specific mutation of OsLTPL23 resulted in an impeded seed germination and weakened seedling growth. Starch-sugar analysis implied that OsLTPL23 protein serves as an alpha-amylase inhibitor in seed. Prompted by the messenger RNA levels of OsABIs, OsNCEDs and OsABA8oxs, endogenous ABA contributed to OsLTPL23-dependent phenotype in seed germination and early seedling growth. Summarily, these findings indicated that OsLTPL23 promotes rice seed germination and post-germination seedling growth.
The germinated rice seeds were grown in a plant greenhouse at 30°C during 14 h daytime and 25°C with 10 h night under 70% humidity. All seeds were harvested at maturity stage, and stored at room temperature after drying at 42°C for 7 days.
The konck-out vector Cas9-OsLTPL23-gRNA was generated as previously reported (Xu et al., 2014). The specific sgRNA sequences targeted for OsLTPL23 were inserted into the pHUN4c12 vector digested with Bsa I-HF using T4 DNA ligase reaction (2011A; Takara, Japan). The resultant construct Cas9-OsLTPL23-gRNA was transformed into the japonica variety NIP through the Agrobacterium tumefaciens strain EH105-mediated stable transformation (Nishimura et al., 2006). Resistant rice calli were vigorously grown in hygromycin-containing medium, and finally transferred to regeneration medium to obtain transgenic plants.
To identify the osltpl23 mutants, the genomic DNA was extracted from the leaves of hygromycin-resistant plants using CTAB method, and then used as template to perform PCR amplification with OsLTPL23-Cas9-detection primers (Supplementary Table S4). All the DNA sequences of PCR products from the above plants were directly determinate to identify Cas9-editing events using Sanger sequencing techniques. Homozygous mutants and heterozygotes were indicated as normal sequencing chromatograms carrying simple indels and superimposed sequencing chromatograms, respectively.
To determine transgenic-free mutants, the T1 generation plants of Cas9-cutting lines were analyzed by PCR amplification with HPT-specific primers (Supplementary Table S4). The Cas9-OsLTPL23-gRNA vector and genomic DNA of NIP were chosen as positive control and negative control, repectively. The HPT-negative mutation lines were marked as transgenic-free mutants.
Fifty seeds per replicate of each osltpl23 mutants and NIP were germinated on Petri dishes containing moistened paper towels at 26°C for 6 days. The seed germination criterion and germination rate determination were as stated in Wang et al. (2021). The seeds germination assay was conducted in triplicate.
Total RNA was isolated from the germinating seeds (12, 24, 48, and 72 h post imbibition (hpi)) and various tissues at two developmental stages (root, leaf and sheath of five-leaf stage seedlings; stem, flag leaf and spike of booting stage plants) of NIP, and two-week-old seedlings of NIP, osltpl23-1 and osltpl23-2 mutants using the TRIzol™ reagent (15596-026; Invitrogen, Carlsbad, California, United States). 1 μg total RNA was used to first-strand cDNA synthesis by the ReverTra Ace qPCR RT Master Mix with gDNA Remover (FSQ-301; TOYOBO, Osaka, Japan). The qRT-PCR was performed using TransStart® Tip Green qPCR SuperMix (+Dye I) (AQ142-11; TransGen, Beijing, China) with corresponding primers on CFX96® Real-Time PCR system (Bio-Rad, Hercules, California, USA), and OsACTIN genes was used as the internal control for normalization. A complete list of primers is included in Supplementary Table S4. The relative transcript levels of indicated genes were quantified by 2−ΔΔCT method from the qRT-PCR data of three biological replicate experiments with three independent repeat (Schmittgen and Livak, 2008).
The protein-coding sequences of OsLTPL23 (without the termination codon) were cloned from the cDNA of 2-weeks old NIP seedlings, and inserted into the PMDC43 and the pSAT6-eYFP-N1 vector, respectively. Then, the plasma membrane marker AtPIP2A-mCherry (Cutler et al., 2000) or nucleus marker bZIP73-mCherry (Liu et al., 2019) were separately introduced into N. benthamiana epidermal cells with PMDC43-OsLTPL23 construct via Agrobacterium tumefaciens strain EH105-mediated transient transformation or infiltrated into rice protoplasts with pSAT6-OsLTPL23-eYFP-N1 construct using polyethylene glycol (PEG)-mediated transformation method, respectively (Chen et al., 2008). The fluorescence intensity of recombinant proteins was photographed at ZEISS LSM 710 NLO (Carl Zeiss, Oberkochen, Germany).
The primary sequences of OsLTPL23 were blasted and aligned to the plant homologues in NCBI and Phytozome database. A neighbour-joining phylogenetic tree between OsLTPL23 and its homologues was constructed by MEGA X with the Poisson correction model, pairwise deletion for gaps/missing data treatment and 1,000 of bootstrap replicates (1,000 replicates) (Kumar et al., 2016).
The dry rice seeds were successively ground into fine powder filtered with 100-, 200-, and 400-mesh sieves. The amount of starch and soluble sugar was separately quantified with 0.03 g and 0.2 g samples according to the manufacturer’s instructions (BC0705, BC0035; Solarbio, Beijing, China).
All experimental data were performed with GraphPad Pism 8 software and statistical analyses among samples were compared using Student’s t-test at the 5% and 1% levels of probability.
To reach a better understanding of nsLTP-based regulation of seed vigor and identify the nsLTPs responsible for seed germination in rice, we functionally analyzed the reported rice nsLTP members with gene expression in seed or embryo (Wang et al., 2012). Through phylogenetic relationship, we found that a total of 15 nsLTP-encoding proteins can be classified into five clade (Figure 1A), which agrees with previous investigation (Wang et al., 2012). Followed the tissue expression profiles from the Plant Regulomics database, the transcripts of the nsLTP genes, OsLTPL8, OsLTPL9, OsLTPL22 and OsLTPL23, in clade II were specifically enriched in the developing seed, with high expression in 5 days after pollination (DAP)-seed and -embryo and low accumulation in 5 DAP-endosperm, suggesting that this clade may be involved in carbohydrates accumulation during rice seed maturation (Figure 1B; Supplementary Table S1). The phytohormones ABA negatively regulates seed germination and maintains seed dormancy (Finkelstein et al., 2008; Rajjou et al., 2012; Penfield, 2017). To determine the candidate genes controlling seed germination, the responsive profiles of these nsLTP genes to ABA were downloaded from TENOR (https://tenor.dna.affrc.go.jp/) and investigated. The transcript abundance of OsLTPL5, OsLTPL8, OsLTPL9, OsLTPL11, OsLTPL22, OsLTPL23, OsLTPL26 and OsLTPL28 genes gradually decreased along ABA treatment, providing the possibility of OsLTPL8, OsLTPL9, OsLTPL22 and OsLTPL23 in regulating seed germination (Figure 1C; Supplementary Table S2). Further, we found that the transcription expression of three genes, OsLTPL16, OsLTPL23 and OsLTPL26, are induced in imbibed seeds, which reinforces the reliability of OsLTPL23 participating in seed germination (Figure 1D; Supplementary Table S3; Galland et al., 2014). Collectively, these data pointed to the idea that OsLTPL23 is the most candidate related to seed vigor and seed germination in these rice nsLTP members.
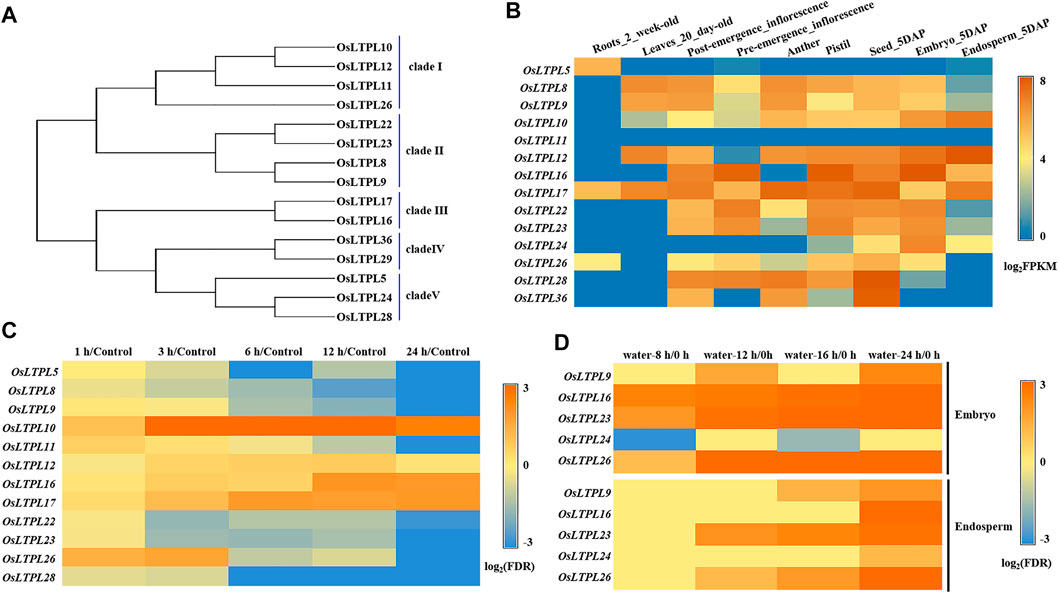
FIGURE 1. OsLTPL23 is specific expressed in seed development and germination process of rice. (A) Phylogenetic tree of the reported rice nsLTP genes expressed in seed or embryo. (B) Tissue expression profiles of rice nsLTP genes from Plant Regulomics. (C) Transcription patterns of rice nsLTP genes response to ABA treatments. Blue and orange red boxes represent decreased and enriched transcript abundance, respectively. The gene expression level at 0 h after ABA treatment serves as control. (D) Transcription patterns of rice nsLTP genes in the imbibed embryo and endosperm under water treatment, respectively. Blue and orange red boxes represent down-regulated and up-regulated expression level, respectively.
To ascertain the potential relationship between OsLTPL23 and seed germination, a phylogram of OsLTPL23 and its plant homologues was firstly generated through the amino acid (aa) sequence based neighbor-joining algorithm (Figures 2A, B). Homology analysis in 7 model plants displayed that the proteins high identity to OsLTPL23 are presented in gramineous C3 plants, including Triticum aestivum, Hordeum vulgare, Brachypodium distachyon. Unfortunately, there were no reports on the function of those proteins in seed germination. By the protein structure analysis, OsLTPL23 encodes a typical plant non-specific lipid transfer protein with 120 amino acids, including one 26 aa N-terminal hydrophobic signal peptide and the 8 CM in bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain (Figure 2A, C). Subsequently, we isolated total RNA from six tissue samples of NIP at vegetative growth and reproductive growth stages to verify the tissue specific expression pattern of OsLTPL23. The qRT-PCR analysis indicated that the highest gene expression level of OsLTPL23 is presented in rice spikes, followed by that in roots and leaves at five-leaf stage, and decreased in flag leaves, stem and sheathes at booting stage (Figure 3A). These results basically agreed with the data from the Plant Regulomics database, supporting the function of OsLTPL23 in seed development.
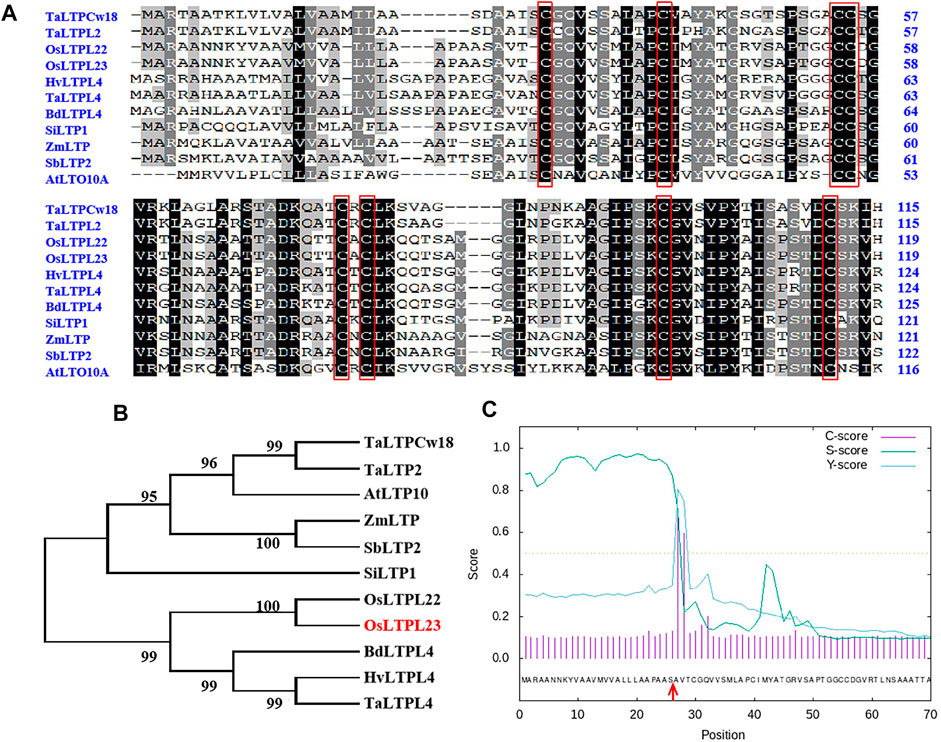
FIGURE 2. Sequence analysis of OsLTPL23 protein. (A) Primary sequence alignment of OsLTPL23 with homologues. Conserved eight-cysteine motif are represented by red rectangles. (B) Phylogenetic tree of OsLTPL23. Ta, Triticum aestivum; At, Arabidopsis thaliana; Zm, Zea mays; Sb, Sorghum bicolor; Si, Setaria italica; Os, Oryza sativa; Bd, Brachypodium distachyon; Hv, Hordeum vulgare. (C) Signal peptide prediction of OsLTPL23. The red arrows indicate the predicted signal peptide cleavage site using SignalP4.1 (https://services.healthtech.dtu.dk/service.php?SignalP-4.1).
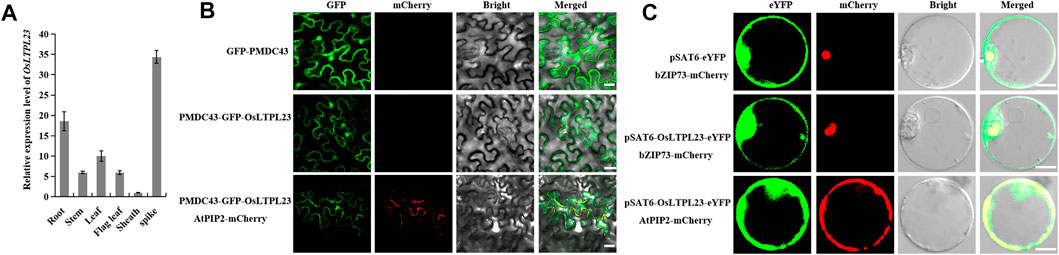
FIGURE 3. Tissue expression pattern and subcellular localization of OsLTPL23. (A) OsLTPL23 transcription expression in various tissues of NIP using qRT-PCR. (B) Subcellular localization of GFP-OsLTPL23 fusion protein in N. benthamiana epidermal cells. Plasmids PMDC43 and PMDC43-OsLTPL23 were introduced into tobacco leaf cells by Agrobacterium-mediated transformation, respectively. Scale bars, 20 μm. (C) Subcellular localization of OsLTPL23-eYFP fusion protein in rice protoplast. Plasmids pSAT6-eYFP-N1 and pSAT6-OsLTPL23-eYFP-N1 were introduced into rice protoplast by PEG-mediated transformation, respectively. Scale bar, 10 μm OsbZIP73 (bZIP transcription factor, nucleus marker, Os09g0474000); AtPIP2A (plasma membrane intrinsic protein 2A, At3g53420).
Several investigations showed that the rice nsLTP family members specifically localized to the plasma membrane, cytoplasm and nucleus to regulate the cold tolerance, fertility, seed development and low-temperature germinability, respectively (Fujino and Sekiguchi, 2011; Li et al., 2020; Zhao et al., 2020; Chen et al., 2022). To detect the functional compartment of OsLTPL23 at subcellular level, we monitored the fluorescence intensity of OsLTPL23 recombinant protein in different transient expression systems. The fluorescence signal in Nicotiana benthamiana exhibited that the fusion protein GFP-OsLTPL23 clearly localizes to the nucleus and colocalizes with the plasma membrane marker AtPIP2A-mCherry (Figure 3B). To further determine the subcellular localization of OsLTPL23, pSAT6-eYFP, OsbZIP73-mCherry, AtPIP2A-mCherry and pSAT6-OsLTPL23-eYFP constructs were also introduced into the rice protoplast. Laser scanning confocal microscope showed that the recombinant protein OsLTPL23-eYFP separately colocalizes with the plasma membrane marker AtPIP2A-mCherry and nucleus marker OsbZIP73-mCherry in rice (Figure 3C), implying that OsLTPL23 may be involved in transcriptional regulation, as well as plasma membrane biological function.
To explore the involvement of OsLTPL23 gene in rice seed germination, the generation of osltpl23 mutants in the NIP background was achieved through Cas9-induced gene editing (Figure 4A). Specifically, the target sequences were located at the position of 21–40 bp in the first exon of OsLTPL23 (Figure 4B). After the sequencing of site-specific PCR products, 12 osltpl23 mutants, including four homozygous and eight heterozygous mutations, were recovered from 21 T0 hygromycin-resistant transgenic plants (57.1%). To obtain more allele mutation types, the zygosity analysis of eight heterozygous mutants were carried out through the T vector sequencing of PCR products. Based on the results of zygosity analysis, 33.3%, 19.0% and 33.3% of the mutations were separately nucleotide insertions, deletions and substitutions, and others were characterized by the chimeric mutation (Figure 4C; Supplementary Figure S1).
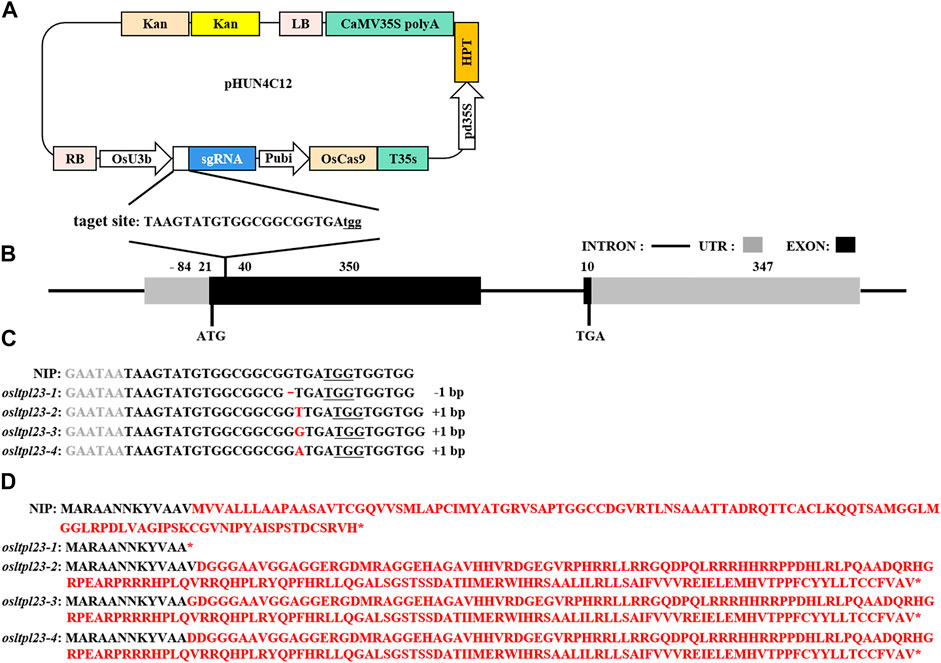
FIGURE 4. Cas9-mediated OsLTPL23 gene mutation in rice. (A) Target site sequence and CRISPR/Cas9 vector structure. Capital letters indicates the sgRNA sequences, and three underlined lowercase letters represent the protospacer adjacent motif (PAM). The expression of Cas9, sgRNA and hygromycin (HPT) is separately driven by the maize ubiquitin promoter (Pubi), rice U3b promoter (OsU3b) and 35S promoters (pd35S). (B) Schematic of the OsLTPL23 gene structure. Black rectangles, gray rectangles and black lines represent exon, untranslated region (UTR) and intron, respectively. (C) The gene editing events in the four rice homozygous mutants. The underlined nucleotide sequences are PAM. The red lines and capital letters represent deleted nucleotide and inserted nucleotide, respectively. (D) Predicted protein sequences encoded by mutational OsLTPL23 in osltpl23 mutants.
To obtain the transgene-free plants, all the T0 mutants were self-pollinated, and we isolated two T-DNA-free homozygous lines from T1 generation of osltpl23-1 and osltpl23-2 mutants. As shown in Figures 4C, D, the osltpl23-1 and osltpl23-2 mutants separately harbored a 1 bp deletion and insertion between nucleotide 37 and 39 of the OsLTPL23 coding region, which result in a truncated and recombinant variant of the OsLTPL23 respectively. Besides, we noticed that osltpl23-3 and osltpl23-4 mutants produce nearly identical protein variants with osltpl23-2 mutant. Given no differentially expressed OsLTPL23 in all the osltpl23-2, osltpl23-3, and osltpl23-4 mutants (Supplementary Figure S2), the osltpl23-1 and osltpl23-2 mutants were subjected to further investigation.
To validate the germination-responsiveness of OsLTPL23, seeds from wild type NIP, osltpl23-1 and osltpl23-2 mutants were harvested and stored. The seeds stored for 18 months post-harvest were soaked in distilled water to calculate the germination rate. The osltpl23 mutants supported distinctly slower germination before 24 hpi and lower germination percentage at 24 hpi to 96 hpi than NIP (Figures 5A–C). At 24 hpi, the germination percentage of NIP (11.83%) was clearly higher than that of osltpl23 (0%). Subsequently, there were a sharply augmented difference between NIP and osltpl23 mutants at 48 hpi and 72 hpi, which the germination percentages of NIP were 2.27- and 2.76-fold, and 1.73- and 1.86-fold that of osltpl23-1 and osltpl23-2 mutant, respectively. Eventually, the germination percentages of NIP and osltpl23 mutants at 96 h reached 98.20%、76.00% and 69.33%, respectively (Figure 5C). These data indicated that the dysfunction of OsLTPL23 delays and inhibits seed germination.
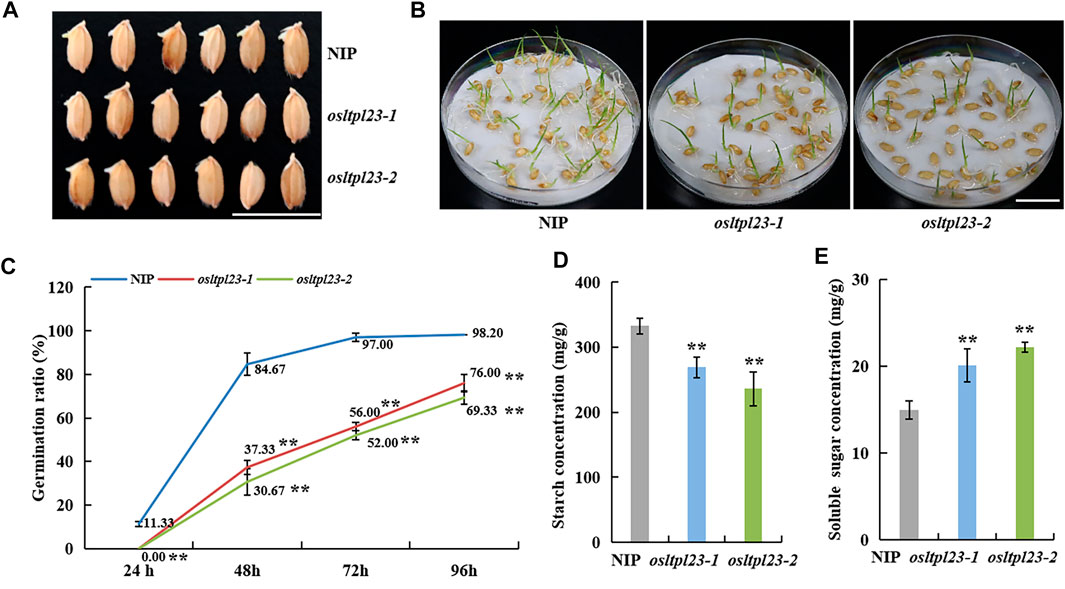
FIGURE 5. OsLTPL23 positively regulates rice seed germination. (A) Phenotypes of germinated seeds of NIP, osltpl23-1 and osltpl23-2 plants after 3 days. Scale bar, 2 cm. (B) Phenotypes of germinated seeds of NIP, osltpl23-1 and osltpl23-2 plants after 6 days. Scale bar, 2 cm. (C) Germination percentage of wild-type NIP, osltpl23-1 and osltpl23-2 mutant plants. (D) Starch content determination of dry seeds in NIP, osltpl23-1 and osltpl23-2 plants. (E) Soluble sugar content determination of dry seeds in wild-type NIP, osltpl23-1 and osltpl23-2 plants. Values are the mean ±standard deviations of three biological replicates. Significance were generated by Student’s t-test with *p < .05 and **p < .01.
In previous protein structure analysis, OsLTPL23 has a bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain. Further functional domain analysis showed that it may act as trypsin-alpha amylase inhibitor (https://www.ebi.ac.uk/interpro/entry/pfam/PF00234/). In rice, alpha-amylas isozymes are critical to convert the stored starch into soluble sugar for nourishing the seedling establishment (Damaris et al., 2019). To validate the role of OsLTPL23 in the conversion between starch and soluble sugar, we measured the starch and soluble sugar contents in the dry seeds of NIP, osltpl23-1 and osltpl23-2 mutants. The results uncovered that the starch contents of osltpl23-1 and osltpl23-2 mutants, with separately 268.90 mg/g and 236.17 mg/g, were significantly reduced compared with NIP (332.73 mg/g), whilst the soluble sugar contents were increased 34.11% and 48.19% relative to 14.98 mg/g in NIP (Figures 5D, E). There are reports that soluble sugar delays seed germination (Zhu et al., 2009), we speculated that the high soluble sugar contents may be a contributor responsible for the slower germination in osltpl23 mutants.
ABA is a central player in controlling seed dormancy and germination (Finkelstein et al., 2008; Rajjou et al., 2012; Penfield, 2017). To investigate whether ABA is involved OsLTPL23-trigger seed germination reduction, we firstly checked the expression level of ABA-responsive genes OsABI3, OsABI4 and OsABI5, which are major downstream components of ABA signalling pathway in seed dormancy and seed germination (Lopez-Molina et al., 2001; Liu et al., 2013; Luo et al., 2021; He et al., 2022). The transcripts of OsABI3 and OsABI5 in osltpl23 mutants obviously accumulated compared with those of NIP within 72 hpi, whereas the relative OsABI4 expression in osltpl23-1 mutant were only higher than NIP at 24 hpi (Figures 6A–C). Pioneer report has demonstrated that the ABA content of abi4 seeds was comparable with wild type after stratification (Shu et al., 2013). Therefore, we speculated that the amounts of ABA in osltpl23 mutants may be higher than that in NIP.
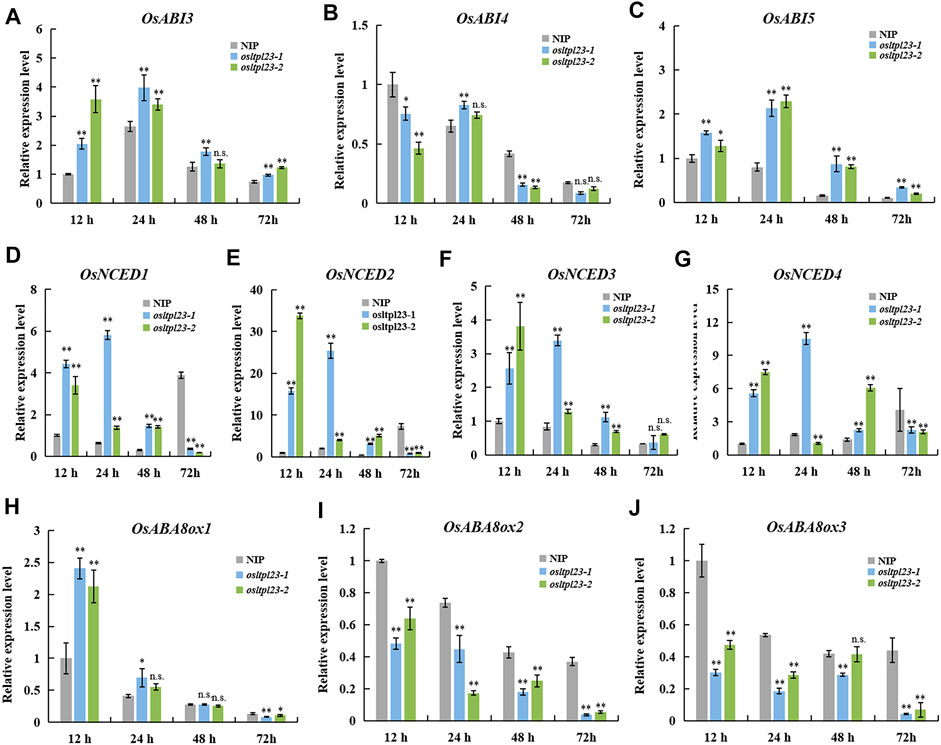
FIGURE 6. OsLTPL23 alters the transcriptional expression of the genes in ABA-responsive, -biosynthesis and -metabolism pathway during seed germination. (A–C) The OsABI3 (A), OsABI4 (B) and OsABI5 (C) expression in germinated seeds among NIP, osltpl23-1 and osltpl23-2 plants. (D–J) The relative gene expression levels of ABA biosynthesis genes OsNCED1 (D), OsNCED2 (E), OsNCED3 (F) and OsNCED4 (G), and ABA metabolism genes OsABA8ox1 (H), OsABA8ox2 (I), and OsABA8ox3 (J) in germinated seeds among NIP, osltpl23-1 and osltpl23-2 plants. The gene expression levels of NIP at 12 hpi was used as control. Values are the mean ±standard deviations of three biological replicates. Significance were generated by Student’s t-test with *p < .05 and **p < .01.
To investigate whether endogenous ABA is increased in osltpl23 mutants, the transcription levels of crucial ABA synthesis and metabolism genes (Figures 6D–J), including OsNCEDs (ABA biosynthesis) and OsABA8oxs (ABA degradation), were quantified among the imbibed seeds of NIP, osltpl23-1 and osltpl23-2 mutants. The gene transcription amounts of OsNCED1, OsNCED2, OsNCED3, and OsNCED4 were clearly upregulated in osltpl23 lines, compared with NIP within 12–48 hpi, whilst the increase trend of all the OsNCEDs were blocked at 72 hpi (Figures 6D–G). Concurrently, the mRNA amounts of OsABA8ox2 and OsABA8ox3 were markedly abolished in osltpl23 mutants relative to those in NIP at 12–72 hpi, whereas the transcription expression of OsABA8ox1 was gradually decreased within 3 days post imbibition (Figures 6H–J). Taken together, these data exposed that the disruption of OsLTPL23 may promote the endogenous ABA accumulation, thus leading to lower germination rate of osltpl23 mutants.
The transcript enrichment of OsABI3, OsABI5, OsNCEDs, and accumulative soluble sugar observed in osltpl23 mutants during seed germination provide a indicative cue that OsLTPL23 may regulate post-germination growth of rice. To verify the possibility, we observed the growth phenotype of 2-week old seedlings grown in greenhouse. Phenotypic investigation showed that the plants from osltpl23-1 and osltpl23-2 lines have significantly lower seedling height, with 30.74% less and 37.35% less, than the control plants, respectively (Figures 7A, B). To confirm whether the post-germination seedling growth inhibition of osltpl23 lines is ABA-dependent, we detected the mRNA levels of plant stress hormone-related genes in 2-week old seedlings between mutants and NIP. The qRT-PCR data displayed that all three genes, OsABA8ox1, OsABA8ox2 and OsABA8ox3 in ABA metabolic pathway are evidently increased in osltpl23 mutants relative to those of NIP, whereas ethylene signal transduction gene OsEiN3, salicylic acid signal pathway gene OsNPR1, and jasmonic acid-responsive transcription factor OsMYC2 have abortive or minor transcript enrichment in osltpl23 lines compared with those of NIP (Figure 7C). These data jointly pinpoint the idea that the ABA is the major limiting factor for early seedling growth in osltpl23 mutants. Meanwhile, we also checked the transcription levels of defence-related marker genes OsPR1a, OsPR1b, OsWRKY45 and OsPAD4 in all plants. All these biotic stress-responsive genes in the osltpl23 lines exhibited lower gene expression level compared with those in NIP (Figure 7D). All these results indicated that OsLTPL23 may positively regulate the rice early seedling growth.
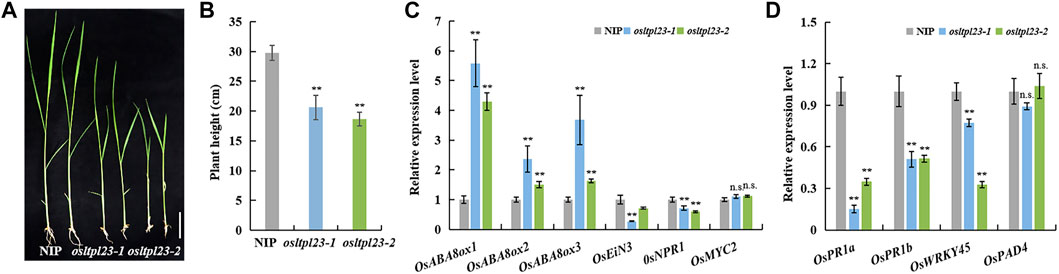
FIGURE 7. The osltpl23 mutants have weakened seedling growth. (A) Growth potential of NIP, osltpl23-1 and osltpl23-2 mutants after 14 days. Scale bar, 4 cm. (B) The plant height of 14 days old seedlings in NIP, osltpl23-1 and osltpl23-2 mutants. Scale bar, 4 cm. (C) Responsive expression of ABA-, ethylene-, JA- and SA-related genes of 14 days old seedlings in NIP, osltpl23-1 and osltpl23-2 mutants. (D) Expression level of defense-responsive genes of 14 days old seedlings in NIP, osltpl23-1 and osltpl23-2 mutants. Values are the mean ±standard deviations of three biological replicates. Significance were generated by Student’s t-test with *p < .05 and **p < .01.
The molecular dictation from a lower ABA/GA ratio in seeds determines the stage transition from dormancy to germination (Jacobsen et al., 2002; Piskurewicz et al., 2008; Shu et al., 2016; Yang et al., 2020). Sucrose is a rapidly consumed agent for growing embryonic axis in germinated seeds, which products, soluble sugar, is another factor influencing the germination process (Dekkers et al., 2004; Gibson, 2005; Zhu et al., 2009; Li et al., 2017; Matsukura et al., 2020). The effect and interplay between ABA and soluble sugar had been wildly investigated in the germinated seed (Dekkers et al., 2004; Zhu et al., 2009; Shu et al., 2013; Wang et al., 2021; Xue et al., 2021). In this present study, we provided evidences to depict the roles of lipid transfer protein OsLTPL23 in endogenous carbohydrates phase transition and seed germination. The abnormal starch-to-sugar conversion of the dry seed may lead to the delayed germination initiation in osltpl23 mutants, and endogenous ABA homeostasis is involved in OsLTPL23-controlled seed germination vigor. These results offered a new viewpoint regarding how nsLTPs act in the metabolism of carbohydrates and ABA during seed germination.
Plant nsLTPs are a class of small, secreted proteins, which localize in the cell wall (Thoma et al., 1993), plasma membrane (Li et al., 2020; Zhao et al., 2020; Chen et al., 2022), cytoplasm (Fujino et al., 2008; Li et al., 2020) and nucleus (Fujino et al., 2008). Accumulated evidences have shown that nsLTPs play important roles in wax assembly (Hollenbach et al., 1997), cell wall extension (Nieuwland et al., 2005), anther and pollen development (Chae et al., 2009; Zhang et al., 2010), seed development and quality (Wang et al., 2015) and plant-pathogen interaction (Segura et al., 1993; Molina and Garcia-Olmedo, 1997; Park et al., 2002; Gomes et al., 2003; Sarowar et al., 2009; Ahmed et al., 2017). Here, we identified a plasma membrane and nucleus colocalized nsLTP, OsLTPL23, which is distinct from starch-independent OsLTPL36 and ABA-dependent OsTPP1 in seed germination (Wang et al., 2015; Wang et al., 2021). We speculated that the putative alpha-amylase inhibitor activity and nucleus enrichment together issue in the phenotype of osltpl23 mutants, especially the nucleus localization representing the manipuility of ABA-related gene expression.
The stored starch in the endosperm is the major energy source during seed germination and seedling establishment, while the inhibitory effect on seed germination and post-germination seedling growth is largely dependent on the increase of ABA concentration (Damaris et al., 2019; Chen et al., 2020). In imbibed seed, the glucose, one hydrolysate of starch, treatment delays the germination process by alleviating endogenous ABA degradation, exogenous ABA, in turn, controls endogenous glucose partition to inhibit germination (Dekkers et al., 2004; Zhu et al., 2009; Xue et al., 2021). Initially, OsLTPL23 was identified as candidate positive regulator responsible for seed germination, with induced transcription in embryo and endosperm. Paradoxically, our carbohydrates determination result suggested that OsLTPL23 has alpha-amylase inhibitor activity in dry seeds, which must destroy the energy source of seed germination, seedling establishment and seedling growth through restraining starch-to-sugars conversion. There are two possible explanation for this discordance. One is that the microarray-based gene expression cannot reflect the natural transcription state of OsLTPL23. The other is that OsLTPL23 may not an key contributor in seed germination and post-germination seedling development. Here, we preferred the former, because osltpl23 mutants shows increased soluble sugar concentration in the dry seeds and the transcription feature of elevated ABA content in germinated seeds and early seedling growth. In the future, the molecular mechanism of endogenous ABA and soluble sugar interaction on seed germination should be emphasized. Overall, osltpl23 mutants provide an important resource to survey the seed germination, post-germination growth and seed development, as well as high vigor seed production in rice.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The work described in this paper was carried out with the cooperation of all authors. QL and YJ designed the experiments and managed the project. YJ and QL performed the experiments, analyzed the data and wrote the manuscript draft. JW and WZ reviewed the manuscript. All authors contributed to this article and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (32201777) and project funded by China Postdoctoral Science Foundation (2022M713356).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1111318/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Nucleotide sequences at the target site in eight heterozygous mutants. The recovered mutated alleles are shown below the NIP sequence. The target site nucleotides are indicated with black capital letters. The PAM site is underlined. The red dashes indicate deleted nucleotides. The red capital letters indicate inserted nucleotides.
SUPPLEMENTARY FIGURE S2 | OsLTPL23 expression detection in osltpl23 mutants. Each column presents the means±standard deviations of three biological replicates. Values are the mean±standard deviations of three biological replicates. Significance were generated by Student’s t-test with *p < .05 and **p < .01
Ahmed, S. M., Liu, P., Xue, Q., Ji, C., Qi, T., Guo, J., et al. (2017). TaDIR1-2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici. Front. Plant Sci. 8, 521. doi:10.3389/fpls.2017.00521
Arroyo, A., Bossi, F., Finkelstein, R. R., and Leon, P. (2003). Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 133, 231–242. doi:10.1104/pp.103.021089
Bossi, F., Cordoba, E., Dupre, P., Mendoza, M. S., Roman, C. S., and Leon, P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59, 359–374. doi:10.1111/j.1365-313X.2009.03877.x
Chae, K., Kieslich, C. A., Morikis, D., Kim, S. C., and Lord, E. M. (2009). A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21, 3902–3914. doi:10.1105/tpc.109.070854
Chen, H. M., Zou, Y., Shang, Y. L., Lin, H. Q., Wang, Y. J., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi:10.1104/pp.107.111740
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., and Zhao, Y. (2020). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi:10.1111/jipb.12899
Chen, L. B., Ji, C. H., Zhou, D. G., Gou, X., Tang, J. N., Jiang, Y. J., et al. (2022). OsLTP47 may function in a lipid transfer relay essential for pollen wall development in rice. J. Genet. Genomics 49, 481–491. doi:10.1016/j.jgg.2022.03.003
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP::DNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA. 97, 3718–3723. doi:10.1073/pnas.97.7.3718
Damaris, R. N., Lin, Z. Y., Yang, P. F., and He, D. L. (2019). The rice alpha-amylase, conserved regulator of seed maturation and germination. Int. J. Mol. Sci. 20, 450–466. doi:10.3390/ijms20020450
Dekkers, B. J., Schuurmans, J. A., and Smeekens, S. C. (2004). Glucose delays seed germination in Arabidopsis thaliana. Planta 218, 579–588. doi:10.1007/s00425-003-1154-9
Deng, W. J., Li, R. Q., Xu, Y. W., Mao, R. Y., Chen, S. F., Chen, L. B., et al. (2020). A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 71, 1294–1305. doi:10.1093/jxb/erz500
Ding, Z. J., Yan, J. Y., Li, G. X., Wu, Z. C., Zhang, S. Q., and Zheng, S. J. (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79, 810–823. doi:10.1111/tpj.12597
Finch-Savage, W. E., and Bassel, G. W. (2016). Seed vigour and crop establishment: Extending performance beyond adaptation. J. Exp. Bot. 67, 567–591. doi:10.1093/jxb/erv490
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi:10.1146/annurev.arplant.59.032607.092740
Fleury, C., Gracy, J., Gautier, M. F., Pons, J. L., Dufayard, J. F., Labesse, G., et al. (2019). Comprehensive classification of the plant non-specific lipid transfer protein superfamily towards its sequence-structure-function analysis. Peer J. 7, e7504. doi:10.7717/peerj.7504
Foolad, M. R., Subbiah, P., and Zhang, L. (2007). Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int. J. Plant Genom. 97386, 97386. doi:10.1155/2007/97386
Fujino, K., Sekiguchi, H., Matsuda, Y., Sugimoto, K., Ono, K., and Yano, M. (2008). Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA. 105, 12623–12628. doi:10.1073/pnas.0805303105
Fujino, K., and Sekiguchi, H. (2011). Origins of functional nucleotide polymorphisms in a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Plant Mol. Biol. 75, 1–10. doi:10.1007/s11103-010-9697-1
Galland, M., Boutet-Mercey, S., Lounifi, I., Godin, B., Balzergue, S., Grandjean, O., et al. (2014). Compartmentation and dynamics of flavone metabolism in dry and germinated rice seeds. Plant Cell Physiol. 55, 1646–1659. doi:10.1093/pcp/pcu095
Gibson, S. I. (2005). Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 8, 93–102. doi:10.1016/j.pbi.2004.11.003
Gomes, E., Sagot, E., Gaillard, C., Laquitaine, L., Poinssot, B., Sanejouand, Y. H., et al. (2003). Nonspecific lipid-transfer protein genes expression in grape (Vitis sp.) cells in response to fungal elicitor treatments. Mol. Plant Microbe Interact. 16, 456–464. doi:10.1094/MPMI.2003.16.5.456
Guo, C. K., Ge, X. C., and Ma, H. (2013). The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 82, 239–253. doi:10.1007/s11103-013-0057-9
He, Y., Chen, W. T., Tan, J. H., Luo, X. X., Zhou, T. J., Gong, X. T., et al. (2022). Rice CENTRORADIALIS 2 regulates seed germination and salt tolerance via ABA-mediated pathway. Theor Appl Genet 35, 4245–4259. doi:10.1007/s00122-022-04215-8
Hollenbach, B., Schreiber, L., Hartung, W., and Dietz, K. J. (1997). Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta 203, 9–19. doi:10.1007/s00050159
Jacobsen, J. V., Pearce, D. W., Poole, A. T., Pharis, R. P., and Mander, L. N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant 115, 428–441. doi:10.1034/j.1399-3054.2002.1150313.x
Kader, J. C. (1996). Lipid-transfer proteins in plants. Annu. Rev. Plant Physiology Plant Mol. Biol. 47, 627–654. doi:10.1146/annurev.arplant.47.1.627
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Leprince, O., Pellizzaro, A., Berriri, S., and Buitink, J. (2017). Late seed maturation: Drying without dying. J. Exp. Bot. 68, 827–841. doi:10.1093/jxb/erw363
Li, H. J., Kim, Y. J., Yang, L., Liu, Z., Zhang, J., Shi, H. T., et al. (2020). Grass-specific EPAD1 is essential for pollen exine patterning in rice. Plant Cell 32, 3961–3977. doi:10.1105/tpc.20.00551
Li, P., Zhou, H., Shi, X. L., Yu, B., Zhou, Y., Chen, S., et al. (2014a). The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 10, e1004213. doi:10.1371/journal.pgen.1004213
Li, R. Q., Xia, J. X., Xu, Y. W., Zhao, X. C., Liu, Y. G., and Chen, Y. L. (2014b). Characterization and genetic mapping of a Photoperiod-sensitive dwarf 1 locus in rice (Oryza sativa L.). Theor. Appl. Genet. 127, 241–250. doi:10.1007/s00122-013-2213-7
Li, T., Zhang, Y., Wang, D., Liu, Y., Dirk, L. M., Goodman, J., et al. (2017). Regulation of seed vigor by manipulation of raffinose family oligosaccharides in maize and Arabidopsis thaliana. Mol. Plant 10, 1540–1555. doi:10.1016/j.molp.2017.10.014
Liu, C. T., Schlappi, M. R., Mao, B. G., Wang, W., Wang, A. J., and Chu, C. C. (2019). The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 17, 1834–1849. doi:10.1111/pbi.13104
Liu, X. D., Zhang, H., Zhao, Y., Feng, Z. Y., Li, Q., Yang, H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 110, 15485–15490. doi:10.1073/pnas.1304651110
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A post germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA. 98, 4782–4787. doi:10.1073/pnas.081594298
Luo, X. F., Dai, Y. J., Zheng, C., Yang, Y. Z., Chen, W., Wang, Q. C., et al. (2021). The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 229, 950–962. doi:10.1111/nph.16921
Matsukura, C., Saitoh, T., Hirose, T., Ohsugi, R., Perata, P., and Yamaguchi, J. (2000). Sugar uptake and transport in rice embryo. expression of companion cell-specific sucrose transporter (OsSUT1) induced by sugar and light. Plant Physiol. 124, 85–93. doi:10.1104/pp.124.1.85
Meng, C., Yan, Y., Liu, Z., Chen, L., Zhang, Y., Li, X., et al. (2018). Systematic analysis of cotton non-specific lipid transfer protein family revealed a special group that is involved in fiber elongation. Front. Plant Sci. 9, 1285–1301. doi:10.3389/fpls.2018.01285
Molina, A., and Garcia-Olmedo, F. (1997). Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12, 669–675. doi:10.1046/j.1365-313x.1997.00669.x
Nieuwland, J., Feron, R., Huisman, B. A., Fasolino, A., Hilbers, C. W., Derksen, J., et al. (2005). Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17, 2009–2019. doi:10.1105/tpc.105.032094
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi:10.1038/nprot.2006.469
Park, C. J., Shin, R., Park, J. M., Lee, G. J., You, J. S., and Paek, K. H. (2002). Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol. Biol. 48, 243–254. doi:10.1023/a:1013383329361
Penfield, S. (2017). Seed dormancy and germination. Curr. Biol. 27, R874–R878. doi:10.1016/j.cub.2017.05.050
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi:10.1105/tpc.108.061515
Preston, J., Tatematsu, K., Kanno, Y., Hobo, T., Kimura, M., Jikumaru, Y., et al. (2009). Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 50, 1786–1800. doi:10.1093/pcp/pcp121
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi:10.1146/annurev-arplant-042811-105550
Salminen, T. A., Blomqvist, K., and Edqvist, J. (2016). Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 244, 971–997. doi:10.1007/s00425-016-2585-4
Sarowar, S., Kim, Y. J., Kim, K. D., Hwang, B. K., Ok, S. H., and Shin, J. S. (2009). Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 28, 419–427. doi:10.1007/s00299-008-0653-3
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Segura, A., Moreno, M., and Garcia-Olmedo, F. (1993). Purification and antipathogenic activity of lipid transfer proteins (LTPs) from the leaves of Arabidopsis and spinach.Arabiopsis and spinach. FEBS Lett. 332, 243–246. doi:10.1016/0014-5793(93)80641-7
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi:10.1016/j.molp.2015.08.010
Shu, K., Zhang, H. W., Wang, S. F., Chen, M. L., Wu, Y. R., Tang, S. Y., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9, e1003577. doi:10.1371/journal.pgen.1003577
Sun, Q., Wang, J. H., and Sun, B. Q. (2007). Advances on seed vigor physiological and genetic mechanisms. Agric. Sci. China 6, 1060–1066. doi:10.1016/S1671-2927(07)60147-3
Thoma, S., Kaneko, Y., and Somerville, C. (1993). A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J 3, 427–436. doi:10.1046/j.1365-313x.1993.t01-25-00999.x
Tao, Y., Zou, T., Zhang, X., Liu, R., Chen, H., Yuan, G. Q., et al. (2020). Secretory lipid transfer protein OsLTPL94 acts as a target of EAT1 and is required for rice pollen wall development. Plant J. 108, 358–377. doi:10.1111/tpj.15443
Wang, G. Q., Li, X. Z., Ye, N. H., Huang, M. K., Feng, L., Li, H. X., et al. (2021). OsTPP1 regulates seed germination through the crosstalk with abscisic acid in rice. New Phytol. 230, 1925–1939. doi:10.1111/nph.17300
Wang, H. W., Hwang, S. G., Karuppanapandian, T., Liu, A., Kim, W., and Jang, C. S. (2012). Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res. 19, 179–194. doi:10.1093/dnares/dss003
Wang, X., Wei, Z., Lu, Z. H., Ouyang, Y. D., Chol, S. O., and Yao, J. L. (2015). A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 239, 200–208. doi:10.1016/j.plantsci.2015.07.016
Wang, Z. F., Wang, J. F., Bao, Y. M., Wang, F. H., and Zhang, H. S. (2010). Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B 11, 958–964. doi:10.1631/jzus.B1000238
Xu, R. F., Li, H., Qin, R. Y., Wang, L., Li, L., Wei, P., et al. (2014). Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7, 5–8. doi:10.1186/s12284-014-0005-6
Xue, X., Yu, Y. C., Wu, Y., Xue, H., and Chen, L. Q. (2021). Locally restricted glucose availability in the embryonic hypocotyl determines seed germination under abscisic acid treatment. New Phytol. 231, 1832–1844. doi:10.1111/nph.17513
Yang, L. W., Liu, S. R., and Lin, R. C. (2020). The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 62, 1310–1326. doi:10.1111/jipb.13001
Zhang, D. S., Liang, W. Q., Yin, C. S., Zong, J., Gu, F. W., and Zhang, D. B. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162. doi:10.1104/pp.110.158865
Zhao, J., Wang, S., Qin, J., Sun, C., and Liu, F. (2020). The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnol. J. 18, 756–769. doi:10.1111/pbi.13243
Zhao, M., Zhang, H. X., Yan, H., Qiu, L., and Baskin, C. C. (2018). Mobilization and role of starch, protein, and fat reserves during seed germination of six wild grassland species. Front. Plant Sci. 9, 234. doi:10.3389/fpls.2018.00234
Keywords: seed germination, rice, OsLTPL23, soluble sugar, abscisic acid (ABA)
Citation: Li Q, Zhai W, Wei J and Jia Y (2023) Rice lipid transfer protein, OsLTPL23, controls seed germination by regulating starch-sugar conversion and ABA homeostasis. Front. Genet. 14:1111318. doi: 10.3389/fgene.2023.1111318
Received: 29 November 2022; Accepted: 02 January 2023;
Published: 16 January 2023.
Edited by:
Yuchen Yang, School of Ecology, Sun Yat-sen University, ChinaReviewed by:
Ping Hu, Henan Institute of Science and Technology, ChinaCopyright © 2023 Li, Zhai, Wei and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfeng Jia, eWZqaWFAZ2VuZXRpY3MuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.