
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 05 April 2023
Sec. Statistical Genetics and Methodology
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1111144
This article is part of the Research Topic Causal Inference in Cancer-Omics Data View all 4 articles
Objectives: Correlations between obstructive sleep apnea (OSA) and gastroesophageal reflux disease (GERD) have been detected in previous observational studies. However, this association remains uncertain due to the potential presence of selection and confounding biases. Therefore, this bidirectional two-sample Mendelian randomization (MR) study was conducted to evaluate the causal relationship between OSA and GERD.
Methods: In this study, instrumental variables (IVs) for OSA were selected from publicly available genetic summary data (27,207 cases and 280,720 controls). Summary statistics for GERD were obtained from a genome-wide association study of 602,604 individuals. The inverse variance weighted (IVW) method was used as the main MR method. The MR-Egger intercept test, MR pleiotropy residual sum and outlier, and leave-one-out analysis were used to detect pleiotropy. Heterogeneity was detected by Cochran’s Q test.
Results: The IVW results revealed that OSA [odds ratio (OR): 1.19, 95% confidence interval (CI): 1.11–1.28, p = 8.88E-07] was causally associated with the incidence of GERD. Moreover, there was evidence of GERD leading to OSA in the IVW analysis (OR: 1.44, 95%CI: 1.33–1.57, p = 7.74E-19). No directional pleiotropy was detected by the MR-Egger intercept test (all p > 0.05).
Conclusion: This study found that OSA is linked to a higher incidence of GERD, and vice versa. This finding might be helpful for the screening and prevention of these two diseases.
Obstructive sleep apnea (OSA) is a prevalent disease characterised by disordered breathing during sleep. Specifically, in OSA, temporary upper airway stricture during sleep causes sleep deficiency, intermittent hypoxia and autonomic fluctuation (Patel, 2019). Epidemiological studies have estimated that approximately 22% of men and 17% of women have OSA (Franklin and Lindberg., 2015). Because of its troublesome symptoms and chronic course, OSA significantly reduces a patient’s health-related quality of life (Gottlieb and Punjabi., 2020). Gastroesophageal reflux disease (GERD) is a functional gastrointestinal disorder characterized by heartburn and regurgitation (Katz et al., 2022). According to current epidemiological research, the prevalence of GERD has increased by more than 77% over the past 10 years (Zhang D. et al., 2022). The increasing prevalence of GERD has resulted in increasingly high healthcare expenditure (Howden et al., 2021). Previous studies have identified a bidirectional relationship between OSA and GERD, where GERD appears to increase the risk of OSA and, on the other hand, OSA contributes to the development of GERD (Shibli et al., 2020).
A nationwide study reported that 12.21% of GERD patients have a concurrent diagnosis of OSA, with the prevalence being approximately triple that observed in the general population (Mahfouz et al., 2022). Similarly, compared to people without OSA, nocturnal gastroesophageal reflux is more common in OSA patients (Fass et al., 2005). In addition, continuous positive airway pressure treatment can reduce gastroesophageal reflux alongside improvements in OSA symptoms (Wang et al., 2020). A cross-sectional study observed a positive correlation between GERD and OSA, and this relationship was unlikely to be affected by potential confounders and different diagnostic criteria (Gilani et al., 2016). However, several studies have produced conflicting results. From example, Shepherd et al. found that obesity, rather than OSA per se, is the cause of GERD in OSA patients (Shepherd and Orr., 2016). Another study reported that the occurrence of GERD had no relation to the severity of OSA (Kuribayashi et al., 2010). Thus, the association between OSA and GERD remains controversial.
Mendelian randomization (MR) is a data analysis method that uses genotypes as instrumental variables (IVs) to infer causal relationships between features of interest (Xu et al., 2022). Mendel’s second law states that genotypes are allocated randomly at conception and are unlikely to be affected by environmental confounders. In addition, DNA sequences are invariant, and the flow of biological information is unidirectional. Therefore, MR can minimize reverse causation and avoid confusion (Swerdlow et al., 2016). Two-sample MR is an extension of the basic MR design. This MR analysis is conducted using the exposure and outcome data obtained from different or partially overlapping samples (Hartwig et al., 2016). It is widely applicable and can avoid pseudo-positive results. Therefore, this study implemented a two-sample MR analysis to reveal the causal relationship between OSA and GERD. The data for this analysis were obtained from published genome-wide association studies (GWAS).
Because the data were collected through publicly available online databases, ethical approval and informed consent were not applicable. The MR design flowchart of the current study is presented in Figure 1. For MR, the selected IVs must satisfy the following assumptions: 1) correlation: IVs must be strongly correlated with exposure; 2) exclusive: IVs should not directly affect the outcome; 3) independence: IVs should not affect the outcome through any confounding variables (Emdin et al., 2017). The “TwoSampleMR” package (version 0.5.6) and the “MRPRESSO” package (version 1.0) in R (version 4.2.1) were used to perform the statistical analyses.
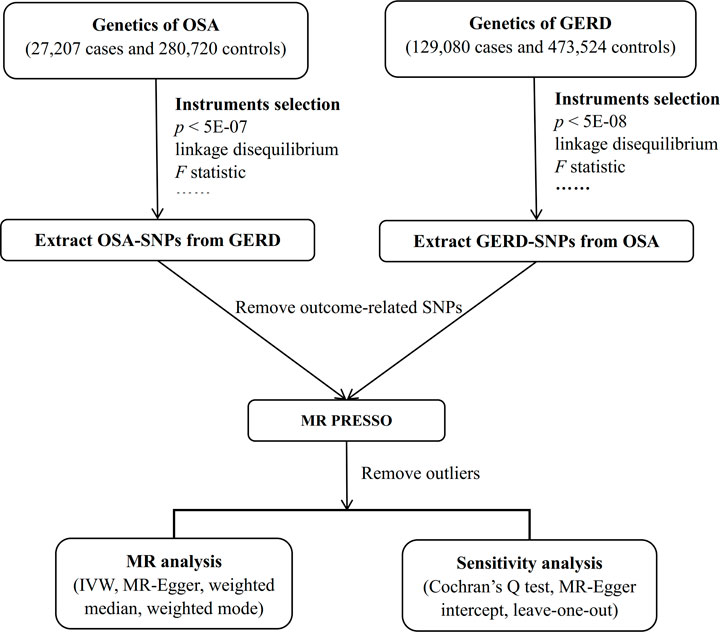
FIGURE 1. Workflow of the bidirectional two-sample MR study. Abbreviation: MR, Mendelian randomization; OSA, obstructive sleep apnea; GERD, gastroesophageal reflux disease; SNP, single nucleotide polymorphism; PRESSO, Pleiotropy REsidual Sum and Outlier; IVW, inverse variance weighted.
Candidate genetic instruments for OSA were extracted from the FinnGen database, which consists of 27,207 cases and 280,720 controls. These data can be found at https://r7.finngen.fi (Kurki et al., 2022). All subjects in the FinnGen data were of European ancestry.
Summary statistics for GERD phenotypes were obtained from the IEU GWAS database (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90000514/). This database comprises 602,604 European ancestry participants, including 129,080 GERD cases and 473,524 control cases (Ong et al., 2022) (Supplementary Table S1).
For the selection of eligible IVs, single nucleotide polymorphisms (SNPs) should meet various requirements. First, SNPs should be associated with the exposure at the genome-wide significance level (p < 5E-08). However, in this MR analysis, a more relaxed threshold (p < 5E-07) was used for OSA in order to include more SNPs; such a threshold has been used previously in many MR studies (Zhang Y. et al., 2022). Second, the PLINK algorithm was used to obtain SNPs that were not in linkage disequilibrium (r2 <0.001; clumping window size = 10,000 kb). Third, manual selection was performed in the PhenoScanner GWAS database (http://phenoscanner.medschl.cam.ac.uk) to exclude outcome trait-related IVs. Fourth, SNPs with an F statistic <10 were regarded as weak IVs and were excluded (Pierce et al., 2011). The calculation formula for the F statistic is F = R2(N-2)/(1-R2), where N is the sample size and R2 reflects the proportion of variance explained by the SNPs. R2 was calculated as 2 × MAF × (1-MAF) × beta2, where MAF is the minor allele frequency of the SNP and beta is the effect size of the SNP. Finally, palindromic SNPs and SNPs associated with the outcome at the genome-wide significance level were discarded.
In this study, inverse variance weighted (IVW), MR-Egger, weighted median and weighted mode were applied to investigate the causal relationship between OSA and GERD. IVW assumes that all SNPs are valid IVs. It has higher statistical power than other MR methods, and thus, IVW was used as the main analysis in this study (Lin et al., 2021). Unless otherwise specified, random-effects IVW was performed because the causal inference of this method is more conservative and takes into account uncertainty due to pleiotropy (Bowden et al., 2018). MR-Egger, weighted median and weighted mode were implemented as complementary methods to IVW; these have different underlying assumptions for horizontal pleiotropy and provide more robust MR estimates in broader scenarios. When at least half of the IVs are invalid, the causal relationship can also be estimated consistently by the weighted median (Bowden et al., 2016). MR-Egger considers the existence of an intercept term in weighted linear regression. It does not strictly require an absence of pleiotropy between genetic variants, which is in contrast to the IVW method (Bowden et al., 2015). The weighted mode method can evaluate the causal relationship between the outcome and a subset of SNPs by grouping the SNPs into subgroups based on similarities in their causal effects. However, compared to the IVW and weighted median methods, it is less able to detect a causal effect (Hartwig et al., 2017).
Sensitivity analyses were conducted to determine the robustness of the MR results. The heterogeneity test (Cochran’s Q test in the IVW method) was used to examine the differences between each IV; p < 0.05 was taken to represent the presence of heterogeneity. MR Pleiotropy RESidual Sum and Outlier (PRESSO) was used to detect horizontal pleiotropy and identify outlier variants. After removing SNP outliers, an outlier-corrected MR analysis was conducted to obtain an unbiased causal estimate (Verbanck et al., 2018). Directional pleiotropy was also appraised by the MR-Egger intercept test; a non-zero intercept suggests that the set of IVs suffers from directional pleiotropy (Burgess and Thompson., 2017). In addition, leave-one-out analysis was performed to evaluate whether the MR results were strongly driven by a specific SNP.
Initially, SNPs associated with OSA at the genome-wide significance level and those that were not in linkage disequilibrium were identified. After excluding palindromic SNPs and GERD-related SNPs, 21 SNPs were attained as IVs; these explain 3% of the phenotypic variation. The F statistic of each IV ranged from 325 to 492, indicating that all IVs were of good strength (all F >10) (Supplementary Tables S2). However, MR PRESSO indicated that rs10125995, rs10917318, rs11030323, rs2529273, rs76229479, and rs742760 were pleiotropic SNPs. After removing the SNP outliers, MR analysis was applied to evaluate the relationship between OSA and GERD. The IVW results [odds ratio (OR): 1.19, 95% confidence interval (CI): 1.11–1.28, p = 8.88E-07], weighted median results (OR: 1.24, 95% CI: 1.14–1.33, p = 5.65E-08) and weighted mode results (OR: 1.30, 95% CI: 1.14–1.48, p = 0.001) suggested that OSA was causally associated with the incidence of GERD. However, the MR-Egger results did not provide evidence of a causal association between OSA and GERD (OR: 1.04, 95% CI: 0.74–1.47, p = 0.826) (Figure 2A). This is probably because the MR-Egger method has lower causal estimate power than the other MR methods (Burgess and Thompson., 2017). The scatter plot and forest plot for these analyses are presented in Figures 3, 4. Heterogeneity was observed by Cochran’s Q test (Q = 25.75, p = 0.028). The MR-Egger intercept analysis results revealed no directional pleiotropy (p = 0.439) (Table 1). In the leave-one-out analysis, no single SNP strongly influenced the overall effect of OSA on GERD (Supplementary Figure S1).
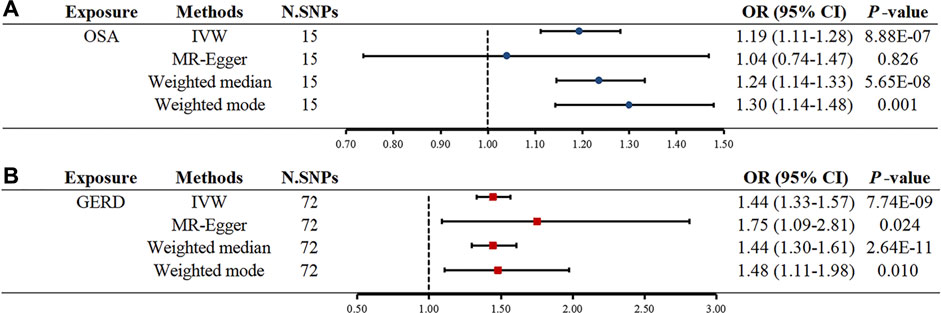
FIGURE 2. Two-sample MR estimates results of causal associations between OSA and GERD. (A) Causal estimates result for OSA on GERD. (B) Causal estimates result for GERD on OSA. Abbreviation: MR, Mendelian randomization; OSA, obstructive sleep apnea; GERD, gastroesophageal reflux disease; IVW, inverse variance weighted; N. SNPs is the number of SNPs after outliers removed; SNPs, single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval.
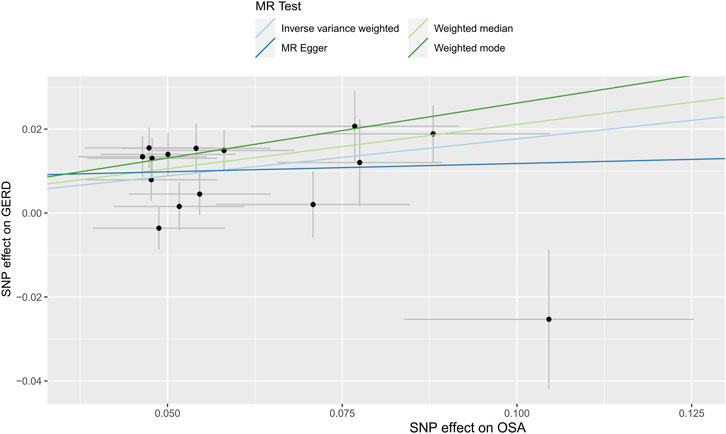
FIGURE 3. Scatter plot for the causal relationship of OSA on GERD. MR, Mendelian randomization; SNP, single nucleotide polymorphism; OSA, obstructive sleep apnea; GERD, gastroesophageal reflux disease.
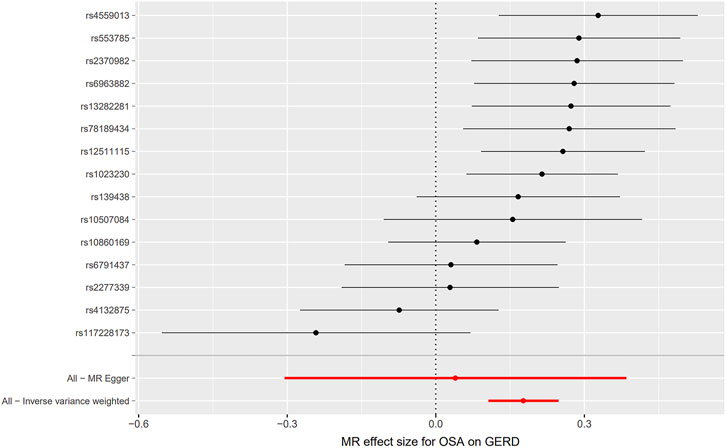
FIGURE 4. Forest plot for the causal relationship of OSA on GERD. MR, Mendelian randomization; OSA, obstructive sleep apnea; GERD, gastroesophageal reflux disease.
Seventy-four independent SNPs strongly associated with GERD were selected as IVs in the MR analysis. The F statistics for these IVs were all larger than 10 (ranging from 207 to 669), indicating that they were strong IVs (Supplementary Tables S3). These SNPs are responsible for 3% of the population-level variation in GERD. After removing two SNP outliers detected by MR PRESSO (rs2734839, rs7527682), MR analysis was performed. The IVW results (OR: 1.44, 95% CI: 1.33–1.57, p = 7.74E-19), MR-Egger results (OR: 1.75, 95% CI: 1.09–2.81, p = 0.024), weighted median results (OR: 1.44, 95% CI: 1.30–1.61, p = 2.64E-11) and weighted mode results (OR: 1.48, 95% CI: 1.11–1.98, p = 0.010) all indicated that GERD was significantly associated with an increased risk of OSA (Figure 2B). The scatter plot and forest plot are shown in Figures 5, 6. Cochran’s Q test detected slight heterogeneity (Q = 91.75, p = 0.049). The MR-Egger intercept analysis did not reveal directional pleiotropy (p = 0.423) (Table 1). Moreover, the leave-one-out analysis indicated that no single SNP drove the results (Supplementary Figure S2).
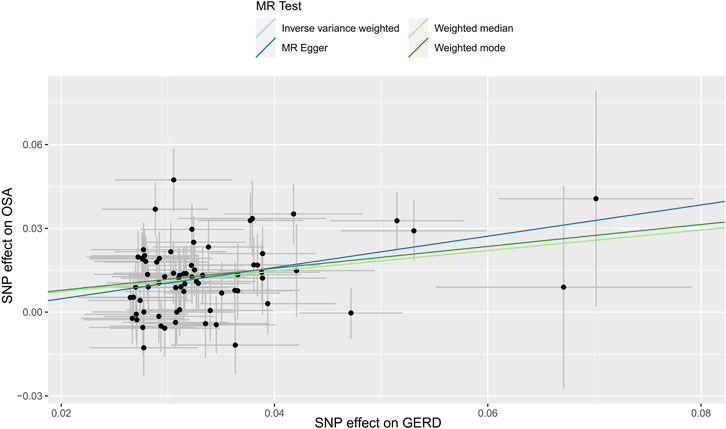
FIGURE 5. Scatter plot for the causal relationship of GERD on OSA. Abbreviation: MR, Mendelian randomization; SNP, single nucleotide polymorphism; GERD, gastroesophageal reflux disease; OSA, obstructive sleep apnea.
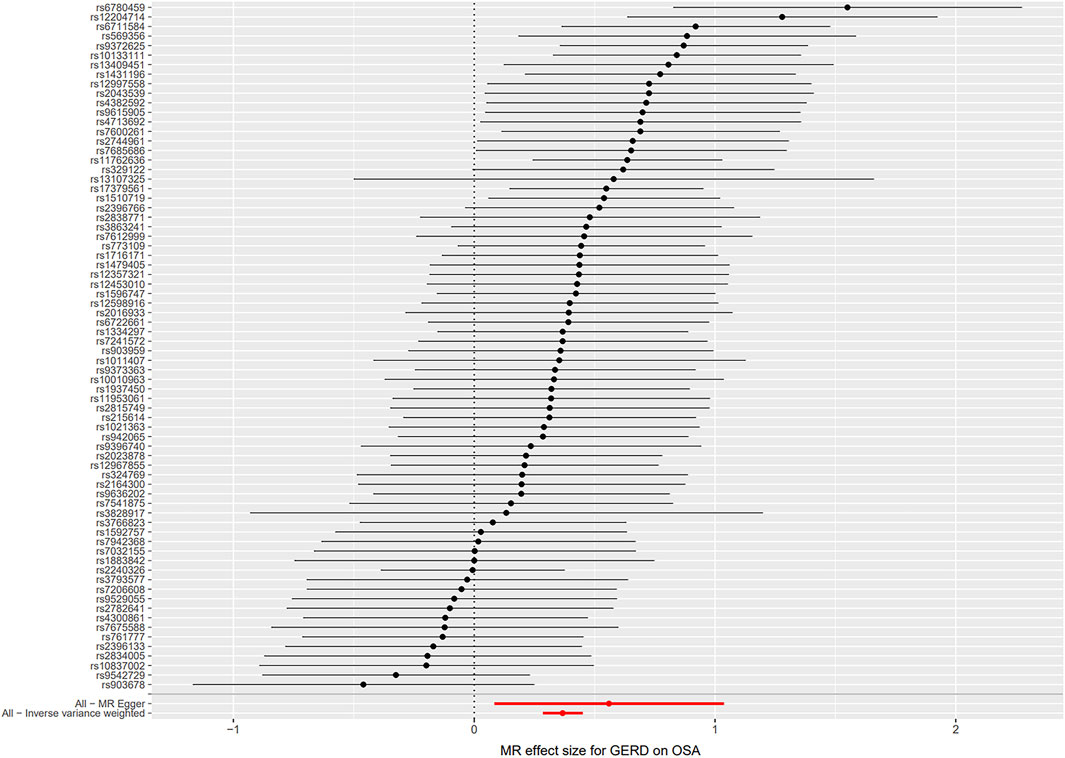
FIGURE 6. Forest plot for the causal relationship of GERD on OSA. Abbreviation: MR, Mendelian randomization; GERD, gastroesophageal reflux disease; OSA, obstructive sleep apnea.
To the best of our knowledge, this is the first two-sample MR study to explore the causal association between OSA and GERD. According to our findings, OSA is causally related to GERD in individuals of European descent. Moreover, GERD also causally increases the risk of OSA. In view of the increasing health burden caused by OSA and GERD, this study provides new insight into the diagnosis and treatment of these two diseases.
Several case-control studies have failed to reveal a causal relationship between OSA and GERD due to the potential influence of confounders such as gender, age, obesity, physical activity, and alcohol use (Shibli et al., 2020). MR analysis can effectively avoid confounders and reverse causation bias in observational studies. It can also overcome the high cost and low practicability of randomized controlled trials. Thus, MR analysis is a powerful tool that can provide strong evidence that OSA is causally associated with GERD and that GERD is causally associated with OSA.
Results from a meta-analysis indicated that OSA and GERD are closely related, with a pooled OR of 1.75 (Wu et al., 2019). It is generally believed that OSA results in sleep fragmentation and sleep deprivation from awakenings (Yeghiazarians et al., 2021). A randomized controlled trial reported that sleep deficiency can increase oesophageal acid exposure, and even in healthy subjects, a lack of sleep can contribute to abnormal pH testing (Yamasaki et al., 2019). This might be the reason why OSA increases the incidence of GERD. An alternative hypothesis is that OSA can increase the respiratory workload and raise the abdominal-gastric pressure, which leads to lower oesophageal sphincter relaxations, and then reflux occurring (Tamanna et al., 2016; Ozcelik et al., 2017; Gleeson and McNicholas., 2022).
Previous cohort studies have revealed that more severe OSA symptoms are frequently encountered in patients with GERD, as compared to the healthy population (Emilsson et al., 2013; Kim et al., 2018). In addition, oesophagogastroduodenoscopy findings have demonstrated that GERD can cause OSA (Kim et al., 2018). These results illustrated that GERD might be associated with a higher risk of OSA, which is consistent with the current MR estimates. Thus, GERD may be a novel predictor of OSA diagnosis. The possible mechanism involves activation of the oesophageal-bronchial neuronal pathway due to reflux of the stomach contents, leading to bronchoconstriction (Houghton et al., 2016). However, the complex pathophysiology underlying the causal relationship between GERD and OSA is not completely understood. More research is required to further clarify this mechanism.
In the current MR analysis, the exposure and outcome genetic data were obtained from different European countries, which minimizes population stratification and sample overlap. Further, the GWAS summary data were obtained from large case-control samples (307,927 for OSA; 602,604 for GERD). Hence, this MR analysis is powered to detect genetic effects on outcome risks. Although a more liberal threshold (p < 5E-07) was used to select OSA-related IVs, the included genetic IVs were all strong IVs, with a minimum F statistic of 207. Non-etheless, this study has several limitations that should be acknowledged. First, the results of this study are based on data from individuals of European ancestry; thus, the findings may not apply to other ethnicities. Second, the p values for the Cochran Q test were less than 0.05, suggesting the presence of some heterogeneity in the effect estimates. However, this did not invalidate the random-effects IVW estimates, and the MR-Egger intercept analysis did not detect any directional pleiotropy, which provides validation of the robustness of the MR results.
Even though the mechanism underlying the association between OSA and GERD is not fully clear, the current MR analysis results suggest that OSA plays an important role in the pathogenesis of GERD, and vice versa. These findings have implications for reducing the risks of OSA and GERD. When managing and treating OSA patients, we must pay attention to their gastrointestinal symptoms. If we suspect that a patient has GERD symptoms (such as chest pain, chronic cough, laryngitis) (Gyawali et al., 2018), 24-h pH monitoring or upper gastrointestinal endoscopy is recommended. Non-pharmacologic and behavioural interventions for GERD patients (such as dietary control, weight reduction, and cutting alcohol consumption) may also be effective for OSA patients. In addition, patients with GERD can have latent OSA, so including questions about GERD when taking the patient’s clinical history may be significant in the screening and prevention of OSA.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
QZ conceived the idea and drafted the manuscript. QZ, LC, DD, and JX contributed to designing the research and visualized the results. LH and TM assisted in performing the computations and supervised the method. CS revised the final manuscript. All authors read and approved the final manuscript.
We want to acknowledge the participants and investigators in the original research of this MR study. We thank the FinnGen and IEU GWAS for sharing these data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1111144/full#supplementary-material
Bowden, J., Davey, S. G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: Effect estimation and bias detection through egger regression. Int. J. Epidemiol. 44 (2), 512–525. doi:10.1093/ije/dyv080
Bowden, J., Davey, S. G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40 (4), 304–314. doi:10.1002/gepi.21965
Bowden, J., Spiller, W., Del, G. M. F., Sheehan, N., Thompson, J., Minelli, C., et al. (2018). Improving the visualization, interpretation and analysis of two-sample summary data mendelian randomization via the radial plot and radial regression. Int. J. Epidemiol. 47 (4), 1264–1278. doi:10.1093/ije/dyy101
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32 (5), 377–389. doi:10.1007/s10654-017-0255-x
Emdin, C. A., Khera, A. V., and Kathiresan, S. (2017). Mendelian randomization. JAMA 318 (19), 1925–1926. doi:10.1001/jama.2017.17219
Emilsson, Ö. I., Bengtsson, A., Franklin, K. A., Torén, K., Benediktsdóttir, B., Farkhooy, A., et al. (2013). Nocturnal gastro-oesophageal reflux, asthma and symptoms of OSA: A longitudinal, general population study. Eur. Respir. J. 41 (6), 1347–1354. doi:10.1183/09031936.00052512
Fass, R., Quan, S. F., O'Connor, G. T., Ervin, A., and Iber, C. (2005). Predictors of heartburn during sleep in a large prospective cohort study. Chest 127 (5), 1658–1666. doi:10.1378/chest.127.5.1658
Franklin, K. A., and Lindberg, E. (2015). Obstructive sleep apnea is a common disorder in the population-a review on the epidemiology of sleep apnea. J. Thorac. Dis. 7 (8), 1311–1322. doi:10.3978/j.issn.2072-1439.2015.06.11
Gilani, S., Quan, S. F., Pynnonen, M. A., and Shin, J. J. (2016). Obstructive sleep apnea and gastroesophageal reflux: A multivariate population-level analysis. Otolaryngol. Head. Neck. Surg. 154 (2), 390–395. doi:10.1177/0194599815621557
Gleeson, M., and McNicholas, W. T. (2022). Bidirectional relationships of comorbidity with obstructive sleep apnoea. Eur. Respir. Rev. 31 (164), 210256. doi:10.1183/16000617.0256-2021
Gottlieb, D. J., and Punjabi, N. M. (2020). Diagnosis and management of obstructive sleep apnea: A review. JAMA 323 (14), 1389–1400. doi:10.1001/jama.2020.3514
Gyawali, C. P., Kahrilas, P. J., Savarino, E., Zerbib, F., Mion, F., Smout, A., et al. (2018). Modern diagnosis of GERD: The lyon consensus. Gut 67 (7), 1351–1362. doi:10.1136/gutjnl-2017-314722
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46 (6), 1985–1998. doi:10.1093/ije/dyx102
Hartwig, F. P., Davies, N. M., Hemani, G., and Davey, S. G. (2016). Two-sample mendelian randomization: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int. J. Epidemiol. 45 (6), 1717–1726. doi:10.1093/ije/dyx028
Houghton, L. A., Lee, A. S., Badri, H., DeVault, K. R., and Smith, J. A. (2016). Respiratory disease and the oesophagus: Reflux, reflexes and microaspiration. Nat. Rev. Gastroenterol. Hepatol. 13 (8), 445–460. doi:10.1038/nrgastro.2016.91
Howden, C. W., Manuel, M., Taylor, D., Jariwala-Parikh, K., and Tkacz, J. (2021). Estimate of refractory reflux disease in the United States: Economic burden and associated clinical characteristics. Gastroenterol. 55 (10), 842–850. doi:10.1097/MCG.0000000000001518
Katz, P. O., Dunbar, K. B., Schnoll-Sussman, F. H., Greer, K. B., Yadlapati, R., and Spechler, S. J. (2022). ACG clinical guideline for the diagnosis and management of gastroesophageal reflux disease. Am. J. Gastroenterol. 117 (1), 27–56. doi:10.14309/ajg.0000000000001538
Kim, Y., Lee, Y. J., Park, J. S., Cho, Y. J., Yoon, H. I., Lee, J. H., et al. (2018). Associations between obstructive sleep apnea severity and endoscopically proven gastroesophageal reflux disease. Sleep. Breath. 22 (1), 85–90. doi:10.1007/s11325-017-1533-2
Kuribayashi, S., Kusano, M., Kawamura, O., Shimoyama, Y., Maeda, M., Hisada, T., et al. (2010). Mechanism of gastroesophageal reflux in patients with obstructive sleep apnea syndrome. Neurogastroenterol. Motil. 22 (6), 611–e172. doi:10.1111/j.1365-2982.2010.01485.x
Kurki, M. I., Karjalainen, J., Palta, P., Sipilä, T. P., Kristiansson, K., Donner, K., et al. (2022). “Finngen: Unique genetic insights from combining isolated population and national health register data,”. medRxiv.
Lin, Z., Deng, Y., and Pan, W. (2021). Combining the strengths of inverse-variance weighting and Egger regression in mendelian randomization using a mixture of regressions model. PLoS Genet. 17 (11), e1009922. doi:10.1371/journal.pgen.1009922
Mahfouz, R., Barchuk, A., Obeidat, A. E., Mansour, M. M., Hernandez, D., Darweesh, M., et al. (2022). The relationship between obstructive sleep apnea (OSA) and gastroesophageal reflux disease (GERD) in inpatient settings: A nationwide study. Cureus 14 (3), e22810. doi:10.7759/cureus.22810
Ong, J. S., An, J., Han, X., Law, M. H., Nandakumar, P., Schumacher, J., et al. (2022). Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett’s oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut 71 (6), 1053–1061. doi:10.1136/gutjnl-2020-323906
Ozcelik, H., Kayar, Y., Danalioglu, A., Arabaci, E., Uysal, O., Yakar, F., et al. (2017). Does CPAP treatment lead to gastroesophageal reflux in patients with moderate and severe OSA? Eur. Arch. Otorhinolaryngol. 274 (3), 1223–1229. doi:10.1007/s00405-016-4116-0
Patel, S. R. (2019). Obstructive sleep apnea. Ann. Intern. Med. 171 (11), C81–C96. doi:10.7326/AITC201912030
Pierce, B. L., Ahsan, H., and Vanderweele, T. J. (2011). Power and instrument strength requirements for mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40 (3), 740–752. doi:10.1093/ije/dyq151
Shepherd, K., and Orr, W. (2016). Mechanism of gastroesophageal reflux in obstructive sleep apnea: Airway obstruction or obesity? J. Clin. Sleep. Med. 12 (1), 87–94. doi:10.5664/jcsm.5402
Shibli, F., Skeans, J., Yamasaki, T., and Fass, R. (2020). Nocturnal gastroesophageal reflux disease (GERD) and sleep: An important relationship that is commonly overlooked. J. Clin. Gastroenterol. 54 (8), 663–674. doi:10.1097/MCG.0000000000001382
Swerdlow, D. I., Kuchenbaecker, K. B., Shah, S., Sofat, R., Holmes, M. V., White, J., et al. (2016). Selecting instruments for mendelian randomization in the wake of genome-wide association studies. Int. J. Epidemiol. 45 (5), 1600–1616. doi:10.1093/ije/dyw088
Tamanna, S., Campbell, D., Warren, R., and Ullah, M. I. (2016). Effect of CPAP therapy on symptoms of nocturnal gastroesophageal reflux among patients with obstructive sleep apnea. J. Clin. Sleep. Med. 12 (9), 1257–1261. doi:10.5664/jcsm.6126
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet. 50 (5), 693–698. doi:10.1038/s41588-018-0099-7
Wang, L., Han, H., Wang, G., Liu, H., Sun, Z., Li, B., et al. (2020). Relationship between reflux diseases and obstructive sleep apnea together with continuous positive airway pressure treatment efficiency analysis. Sleep. Med. 75, 151–155. doi:10.1016/j.sleep.2020.07.024
Wu, Z. H., Yang, X. P., Niu, X., Xiao, X. Y., and Chen, X. (2019). The relationship between obstructive sleep apnea hypopnea syndrome and gastroesophageal reflux disease: A meta-analysis. Sleep. Breath. 23 (2), 389–397. doi:10.1007/s11325-018-1691-x
Xu, J., Li, M., Gao, Y., Liu, M., Shi, S., Shi, J., et al. (2022). Using mendelian randomization as the cornerstone for causal inference in epidemiology. Environ. Sci. Pollut. Res. Int. 29 (4), 5827–5839. doi:10.1007/s11356-021-15939-3
Yamasaki, T., Quan, S. F., and Fass, R. (2019). The effect of sleep deficiency on esophageal acid exposure of healthy controls and patients with gastroesophageal reflux disease. Neurogastroenterol. Motil. 31 (12), e13705. doi:10.1111/nmo.13705
Yeghiazarians, Y., Jneid, H., Tietjens, J. R., Redline, S., Brown, D. L., El-Sherif, N., et al. (2021). Obstructive sleep apnea and cardiovascular disease: A scientific statement from the American heart association. Circulation 144 (3), e56–e67. doi:10.1161/CIR.0000000000000988
Zhang, D., Liu, S., Li, Z., and Wang, R. (2022a). Global, regional and national burden of gastroesophageal reflux disease, 1990-2019: Update from the GBD 2019 study. Ann. Med. 54 (1), 1372–1384. doi:10.1080/07853890.2022.2074535
Keywords: Mendelian randomization, obstructive sleep apnea, gastroesophageal reflux disease, bidirectional, causal
Citation: Zhu Q, Hua L, Chen L, Mu T, Dong D, Xu J and Shen C (2023) Causal association between obstructive sleep apnea and gastroesophageal reflux disease: A bidirectional two-sample Mendelian randomization study. Front. Genet. 14:1111144. doi: 10.3389/fgene.2023.1111144
Received: 29 November 2022; Accepted: 28 March 2023;
Published: 05 April 2023.
Edited by:
Michelle Lacey, Tulane University, United StatesReviewed by:
Yunping Tang, Zhejiang Ocean University, ChinaCopyright © 2023 Zhu, Hua, Chen, Mu, Dong, Xu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuizhen Shen, c2hlbmN1aXpoZW5AMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.