- 1Laboratory of Endocrinology, Baoding No. 1 Central Hospital, Baoding, China
- 2Clinical Laboratory, The People’s Hospital of Baoding, Baoding, Hebei, China
- 3Infection Division, The People’s Hospital of Baoding, Baoding, Hebei, China
- 4Department of AIDS Research, Hebei Provincial Center for Disease Control and Prevention, Shijiazhuang, Hebei, China
Men who have sex with men (MSM) are the most frequent infection route of the human immunodeficiency virus (HIV) in Baoding, China, creating chances for the occurrence of unique recombinant forms (URFs) of the virus, i.e., recombination of different subtypes caused by co-circulation of multiple subtypes. In this report, two near-identical URFs (BDD002A and BDD069A) isolated from MSM in Baoding were identified. Phylogenetic tree analysis based on nearly full-length genomes (NFLGs) revealed that the two URFs formed a distinct monophyletic cluster with a bootstrap value of 100%. Recombinant breakpoints analysis identified that the NFLGs of BDD002A and BDD069A were both composed of CRF01_AE and subtype B, with six subtype B mosaic segments inserted into the CRF01_AE backbone. The CRF01_AE segments of the URFs clustered closely with the CRF01_AE reference sequences, and the B subregions clustered with the B reference sequences. The recombinant breakpoints of the two URFs were almost identical. These results suggest that effective interventions are urgently needed to prevent the formation of complex HIV-1 recombinant forms in Baoding, China.
Introduction
The human immunodeficiency virus-1 (HIV-1) possesses an extremely high mutation frequency, resulting in HIV-1-enriched gene polymorphisms (Yebra et al., 2018). During the spread of HIV, if more than two subtypes infect the same cell, their genomic information can exchange to generate recombinant virus genomes (Moore and Hu, 2009). A total of 132 circulating recombinant forms (CRFs) of HIV and many unique recombinant forms (URFs) have been reported worldwide (https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/crfs). In recent years, CRF01_AE and subtype B have become the two main HIV-1 genotypes prevalent in key populations of China, especially among men who have sex with men (MSM) (He et al., 2012). Hebei is a northern province of China with low HIV prevalence (Lu et al., 2017). By the end of October 2020, 15,178 individuals in the Hebei Province were diagnosed with HIV-1/AIDS, and the number of individuals infected with HIV-1 through MSM reached 77.5% (Lu et al., 2020; Wang, 2020). Baoding, which borders Beijing and Tianjin, was the second region severely affected by HIV-1 in Hebei Province, with MSM responsible for 84.8% of the HIV-1-infected population (Shi et al., 2021a). The prevalence of subtypes CRF01AE and B was similar to that reported in the whole province at 49.44% and 17.78%, respectively (Shi et al., 2021b). In this study, we identified two highly similar, nearly full-length genome (NFLG) sequences isolated from MSM in Baoding, Hebei Province. These two unique recombinant forms (URFs) were composed of subtypes CRF01_AE and B.
Case description
The two individuals, BDD002A and BDD069A, were a 35-year-old unmarried man and a 39-year-old married man, respectively. Moreover, their baseline CD4+ T-cell counts were 242 cells/ul and 128 cells/ul, respectively while the HIV-1 viral load were 1,140,000 copies/ml and 703,000copies/ml, respectively. They were infected through homosexual transmission. The study was approved by the Medical Ethics Committee of the Baoding People’s Hospital (protocol number: 2019-03). Written informed consent was obtained from the subjects prior to sample collection.
Diagnostic assessment
As described previously by us (Yang et al., 2022), HIV-1 RNA was extracted from 140 µL of the plasma samples of subjects BDD002A and BDD069A using a QIAamp Viral RNA Mini Accessory Set (QIAGEN, Hilden, Germany). PrimeScript IV 1st Strand cDNA Synthesis Mix (TaKaRa Biotechnology, Dalian, China) was used to reverse transcribe the RNA into 3′half-molecule cDNAs using the primers and reaction conditions listed in our previous report (Yang et al., 2022). Nested polymerase chain reactions (PCR) was performed using TaKaRa Premix Taq (TaKaRa Biotechnology, Dalian, China) to amplify 3′halfmolecule region of the NFLGs of BDD002A and BDD069A. HIV-1 near full-length pol and gag genes were amplified using Takara One-step RT-PCR Kit v2.0 (TaKaRa Biotechnology, Dalian, China). HIV-1 near full-length pol gene was amplified in two steps using the primers below:Pol-1e: TGGAAA TGTGGRAARGARGGAC (forward), Pol-x: CCTGTAATG CARMCCCCAATATG TT (reverse) and Pol-3: ACTGAGAGAC AGGCTAATTTTTTAGGGA (forward), Pol-4e:CTCCTAGTGG GGATRTGTACTTCTGARCTTA (reverse). HIV-1 near full-length gag gene was amplified in two steps using the primers below:gag-763:TGACTAGCGGAGGCTAGAAGG (forward), gag-5:TTCCYCC TATCATT TTTGGTTTCC (reverse) and gag-617: TGTGGAAAAT CTCTAGCAGTGG (forward), gag-6:TAATGCTTTTATT TTYTCYT CTGTCAATGGC (reverse). The reaction conditions used for amplification have been reported previously (Lu et al., 2017; Fan et al., 2022a). The positive PCR products were purified using 1.0% agarose gel electrophoresis and sequenced using the Sanger sequencing technology by Tianyi Huiyuan Bioscience & Technology Inc. (Beijing, China). Two NFLGs were obtained by assembling them with near full-length gag, pol and 3′half-genome gene sequences using Sequencher 5.4.6 (Gene Codes Corp., Ann Arbor, MI, United States of America).
The NFLG sequences were then submitted to the online tool HIV BLAST (https://www.hiv.lanl.gov/content/sequence/BASIC_BLAST/basic_blast.html) to determine whether the same recombinant sequences had been identified previously, but no sequences with high similarity (>95%) to BDD002A and BDD069A were found in the HIV database. In addition, phylogenetic tree and subregion phylogenetic trees were constructed using the neighbor-joining (N-J) method based on the Kimura two-parameter model with 1,000 bootstrap replications by Mega6.0. The recombination pattern was determined by Recombination Identification Program (https://www.hiv.lanl.gov/content/sequence/RIP/RIP.html) and jpHMM. Recombination breakpoints were identified by SimPlot (v3.5.1.0) and Bootscan analysis.
We acquired two NFLG sequences with 8,810 bp (HXB2: 761–9,613) and 8,944 bp (HXB2: 625–9,604) from BDD002A and BDD069A, respectively. The constructed NFLG N-J tree showed that both BDD002A and BDD069A formed a monophyletic branch with a bootstrap value of 100%, indicating that BDD002A and BDD069A are two novel recombinant forms (Figure 1). The recombinant breakpoints analysis revealed that BDD002A and BDD069A were composed of 12 interlaced mosaic gene segments, including six CRF01_AE subregions (I, III, V, VII, IX and XI) and 6 B regions (II, IV, VI, VIII, X and XII), with 11 recombinant breakpoints relative to the HXB2 coordinate (Figure 2; Figure 3; Figure 4). Subregion analysis (Figure 3) confirmed that the gene mosaic structure of the two NFLGs are: ICRF01_AE (HXB2, 790–1,172 nt), IIB (HXB2, 1,173–1,818 nt), IIICRF01_AE (HXB2, 1,819–2,098 nt), IVB (HXB2, 2,099–2,755 nt), VCRF01_AE (HXB2, 2,756–4,489 nt), VIB (HXB2, 4,490–4,881 nt), VIICRF01_AE (HXB2, 4,882–5,390 nt), VIIIB (HXB2, 5,391–5,617 nt), IXCRF01_AE (HXB2, 5,618–6,331 nt), XB (HXB2, 6,332–8,285 nt), XICRF01_AE (HXB2, 8,286–8,979 nt) and XIIB (HXB2, 8,980–9,411 nt) for BDD002A (Figure 3A); and ICRF01_AE (HXB2, 790–1,172 nt), IIB (HXB2, 1,173–1,818 nt), IIICRF01_AE (HXB2, 1,819–2,070 nt), IVB (HXB2, 2,071–2,726 nt), VCRF01_AE (HXB2, 2,727–4,489 nt), VIB (HXB2, 4,490–4,881 nt), VIICRF01_AE (HXB2, 4,882–5,390 nt), VIIIB (HXB2, 5,391–5,700 nt), IXCRF01_AE (HXB2, 5,701–6,331 nt), XB (HXB2, 6,332–8,285 nt), XI CRF01_AE (HXB2, 8,286–9,006 nt) and XIIB (HXB2, 9,007–9,411 nt) for BDD069A (Figure 3B). The above data revealed that both NFLGs shared seven almost identical breakpoints except a minor difference within the vif-vpr gene region (Figure 3). The parental origin of all fragments of the two NFLGs were analyzed and the CRF01_AE regions for both URFs were from the CRF01_AE cluster five lineage, which is circulating primarily among MSM in major northern cities of China (Figure 4A) (Feng et al., 2013). The subtype B regions for both URFs were clustered within the northern China subtype B lineage, which also circulates primarily among MSM in northern China (Figure 4B) (Li et al., 2011).
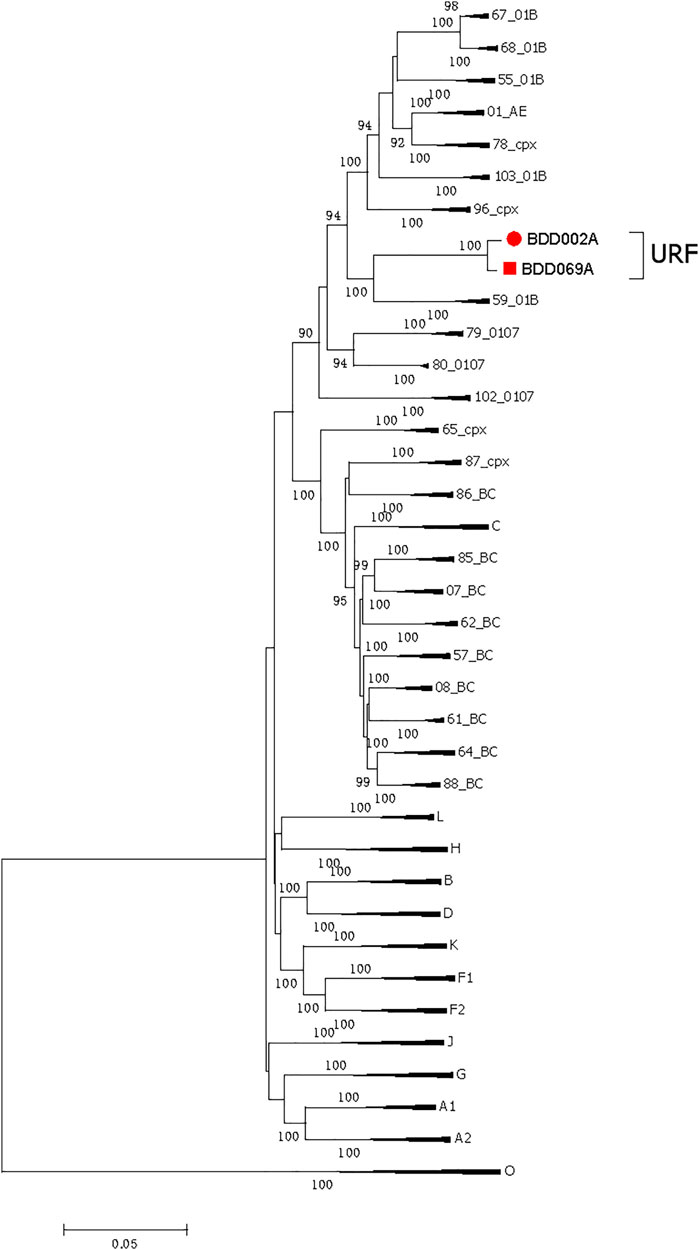
FIGURE 1. The phylogenetic tree is based on NFLG sequences BDD002A and BDD069A. The standard subtype references were downloaded from the Los Alamos National Laboratory HIV Database (www.hiv.lanl.gov). The neighbor-joining phylogenetic tree of BDD002A (8,810 bp, red-filled circle●) and BDD069A (8,944 bp, red-filled square■) was constructed based on the NFLG sequences using Mega6.0. The stability of each node was assessed by bootstrap tests with 1,000 replicates, and only bootstrap values ≥90% are shown at the corresponding nodes. The scale bar represents a 5% genetic distance. NFLG, near full-length genome.
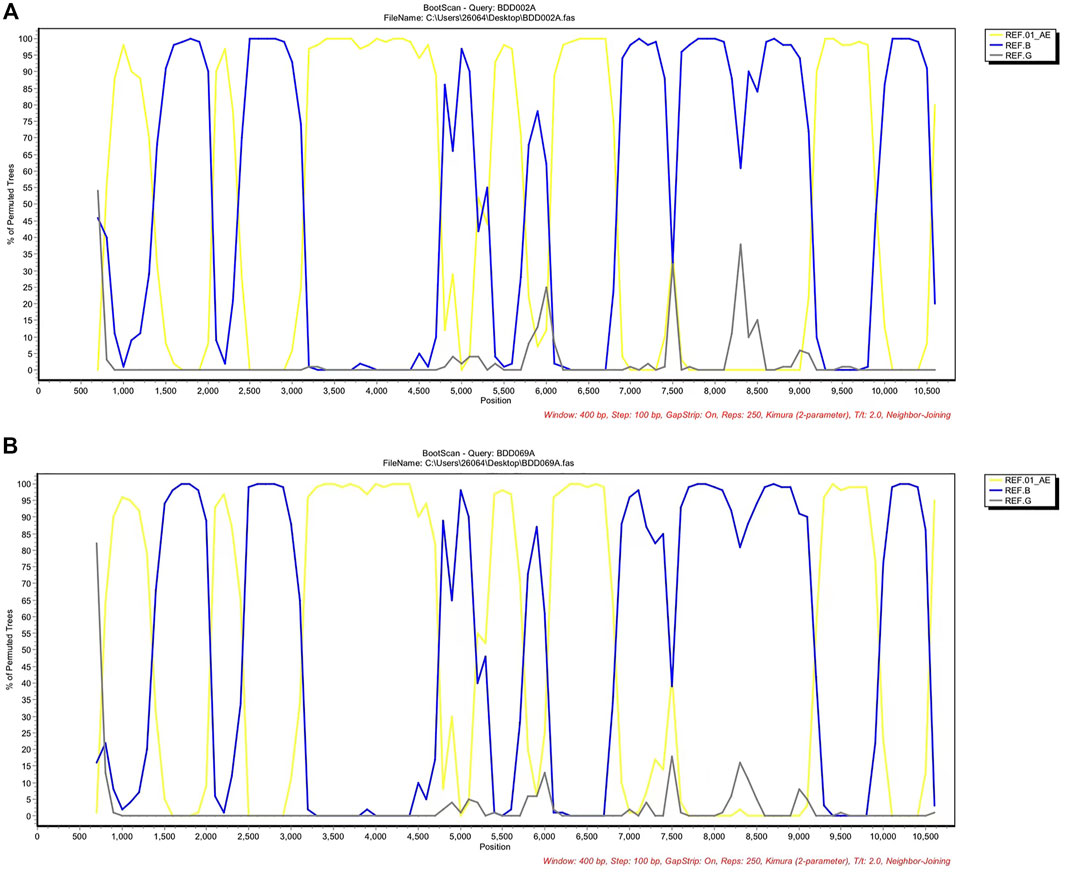
FIGURE 2. Bootscan results of the novel CRF01_AE/B identified. (A) Bootscan plots of BDD002A using CRF01_AE, subtype B and subtype G as references. The parameters of the bootscan analysis were a window size of 400 bp and a step size of 100 bp. (B) Bootscan plots of BDD069A using CRF01_AE and subtype B as putative parental reference sequences and subtype G as the outgroup. The parameters of the bootscan analysis were a window size of 400 bp and a step size of 100 bp. CRF, circulating recombinant form.
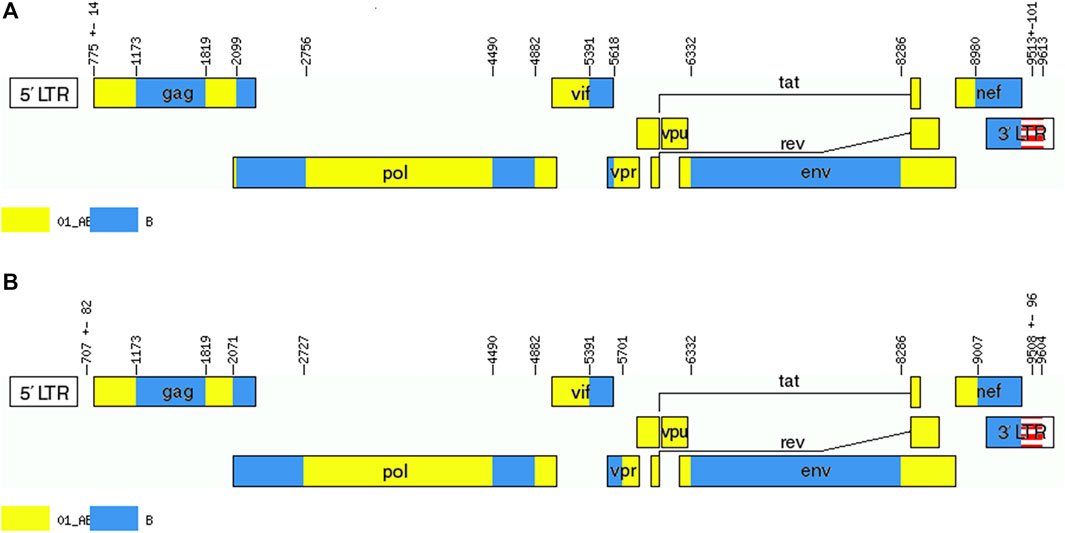
FIGURE 3. Recombinant maps of the novel CRF01_AE/B identified. The unique recombinant maps of (A) BDD002A and (B) BDD069A were drawn with the online Recombinant HIV-1 Drawing Tool (https://www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html). Six B segments were inserted into the CRF01_AE backbone, and 11 breakpoints divided the whole genome into 12 unique segments.
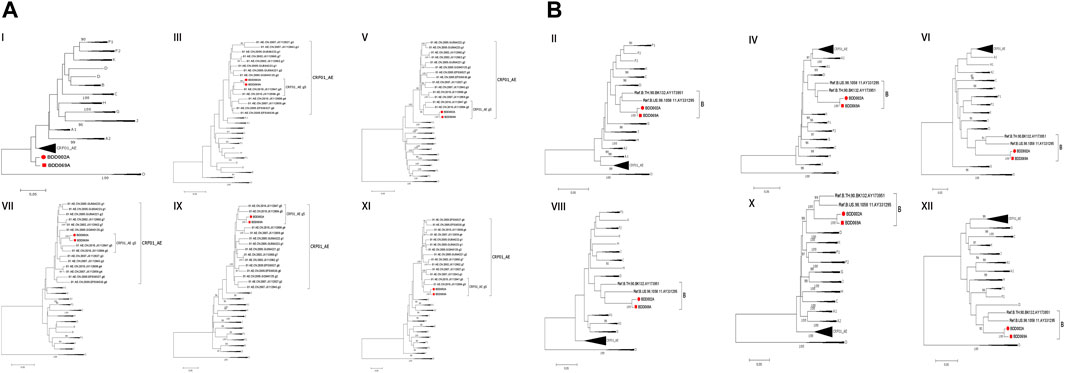
FIGURE 4. Subregional phylogenetic trees of the novel CRF01_AE/B identified. The trees were constructed by the neighbor-joining method with 1,000 bootstrap replications using Mega6.0. Bootstrap values ≥90% are shown at the corresponding nodes. The scale bars indicate a genetic distance of 5%. Each segment of BDD002A and BDD069A is marked by a red-filled circle and a red-filled square, respectively. (A) CRF01_AE regions for both URFs. (B) subtype B regions for both URFs.
Discussion
The diversity of HIV-1 is a significant challenge in preventing the global spread of HIV due to the different pathogenicity of subtypes. The co-circulation of multiple subtypes enables the occurrence of numerous URFs (Gao et al., 2021). In China, the prevalence of CRFs may be underestimated because of the high prevalence of URFs and the formation of many potential CRFs (Liu et al., 2019; Wang et al., 2021). URFs convert to CRFs if they spread widely and circulate in the population (Robertson et al., 2000; Hemelaar, 2012). In recent years, new CRFs such as CRF103_01 B and CRF112_01 B have been found because of the high occurrence of URFs among MSMs in Baoding (Zhou et al., 2020; Fan et al., 2022b; Wang et al., 2022). The continuous emergence of new recombinant forms has brought new barriers to the monitoring, treatment, vaccine development and prevention of HIV. In this study, we identified and characterized two novel recombinants derived from subtypes CRF01_AE and B, which were highly different from those reported previously in the cities of Langfang and Baoding and other Chinese provinces (Guo et al., 2014; Yan et al., 2015; Li et al., 2018; Huang et al., 2019; Li et al., 2019; Ou et al., 2019; Ge et al., 2020; Fan et al., 2022c). These two URFs were more complex than those reported previously. Although no new CRF has been formed, we infer that it may be a potential CRF. Currently, the sexual contact has been the main route of HIV-1 spread, it is key vital for us to carry out dynamic monitoring of new recombination forms in order to interdict HIV-1 spread.
In conclusion, we identified two almost identical HIV-1 URFs among MSMs in Baoding. The recombinant forms of CRF01_AE/B and CRF01_AE/CRF07_BC were found frequently in the MSM population of Baoding, indicating an increase in the genetic diversity of HIV-1 and the presence of many undiagnosed multiple infection cases in this region (Ou et al., 2019; Fan et al., 2022d; Yang et al., 2022), further bringing more barriers for HIV prevention and therapy. This study suggests that we should continuously monitor HIV-1 molecular epidemiology in order to provide effective suggestions to HIV-1 prevention and vaccine design.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP796643 https://www.ncbi.nlm.nih.gov/genbank/, OP796644.
Ethics statement
The studies involving human participants were reviewed and approved by The People’s Hospital of Baoding, Baoding Ethics committee. The patients/participants provided their written informed consent to participate in this study.
Author contributions
BZ, XL and WA designed the study. JM, MS and WF acquired the sequences. BZ, SC, WA and XL analyzed and interpreted the data. BZ, SC, JM and MS wrote the manuscript. All authors read and agreed to the published version of the manuscript.
Acknowledgments
The authors thank all staff involved in this study. We thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Fan, W., Liu, Y., Li, Y., Su, M., Meng, J., Lu, X., et al. (2022). Identification of three novel HIV-1 second-generation recombinant forms (CRF01_AE/CRF07_BC) among men who have sex with men in baoding, Hebei, China. AIDS Res. Hum. Retroviruses 38 (10), 812–816. doi:10.1089/AID.2022.0037
Fan, W., Su, M., Meng, J., Yang, X., Liu, Z., Wang, H., et al. (2022). Characterization of a new HIV-1 CRF01_AE/B recombinant virus form among men who have sex with men in baoding, Hebei, China. AIDS Res. Hum. Retroviruses 38 (3), 237–241. doi:10.1089/AID.2021.0209
Fan, W., Xing, Y., Han, L., Su, M., Meng, J., Dai, E., et al. (2022). HIV-1 genetic characteristics and pre-treatment drug resistance among newly diagnosed population in Baoding, Hebei Province [J]. Chin. J. Microbiol. Immunol. 42 (02), 88–93. doi:10.3760/cma.j.cn112309-20210814-00272
Fan, W., Yang, X., Liu, Z., Su, M., Meng, J., Lu, X., et al. (2022). Subtypes and transmission network analysis of HIV-1 among men who have sex with men in Baoding [J]. Int. J. Virology 29 (01), 30–34. doi:10.3760/cma.j.issn.1673-4092.2022.01.006
Feng, Y., He, X., His, J. H., Li, F., Li, X., Wang, Q., et al. (2013). The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS 27 (11), 1793–1802. doi:10.1097/QAD.0b013e328360db2d
Gao, Y., He, S., Tian, W., Li, D., An, M., Zhao, B., et al. (2021). First complete-genome documentation of HIV-1 intersubtype superinfection with transmissions of diverse recombinants over time to five recipients. PLoS Pathog. 17 (2), e1009258. doi:10.1371/journal.ppat.1009258
Ge, Z., Getaneh, Y., Liang, Y., Lv, B., Liu, Z., Li, K., et al. (2020). Identification of a novel HIV-1 second-generation (CRF01_AE/B) among men who have sex with men in Tianjin, China. AIDS Res. Hum. Retroviruses 36 (2), 138–142. doi:10.1089/AID.2019.0187
Guo, H., Hu, H., Zhou, Y., Yang, H., Huan, X., Qiu, T., et al. (2014). A Novel HIV-1 CRF01_AE/B recombinant among men who have sex with men in Jiangsu Province, China. AIDS Res. Hum. Retroviruses 30 (7), 706–710. doi:10.1089/AID.2014.0012
He, X., Xing, H., Ruan, Y., Hong, K., Cheng, C., Hu, Y., et al. (2012). A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 7 (10), e47289. doi:10.1371/journal.pone.0047289
Hemelaar, J. (2012). The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 18 (3), 182–192. doi:10.1016/j.molmed.2011.12.001
Huang, Q., Ou, W., Feng, Y., Li, F., Li, K., Sun, J., et al. (2019). Near full-length genomic characterization of HIV-1 CRF01_AE/B recombinant strains identified in Hebei, China. AIDS Res. Hum. Retroviruses 35 (2), 196–204. doi:10.1089/AID.2018.0130
Li, K., Liu, M., Li, J., Dong, A., Zhou, Y., Ding, Y., et al. (2019). Genomic characterization of a novel HIV-1 second-generation recombinant form (CRF01_AE/B) from men who have sex with men in guangxi Zhuang autonomous region, China. AIDS Res. Hum. Retroviruses 35 (10), 972–977. doi:10.1089/AID.2019.0149
Li, K., Ou, W., Feng, Y., Sun, J., Ge, Z., Xing, H., et al. (2018). Near full-length genomic characterization of a novel HIV type 1 recombinant form (CRF01_AE/B) identified from anhui, China. AIDS Res. Hum. Retroviruses 34 (12), 1100–1105. doi:10.1089/AID.2018.0144
Li, L., Lu, X., Li, H., Chen, L., Wang, Z., Liu, Y., et al. (2011). High genetic diversity of HIV-1 was found in men who have sex with men in Shijiazhuang, China. Infect. Genet. Evol. 11 (6), 1487–1492. doi:10.1016/j.meegid.2011.05.017
Liu, Y., Su, B., Zhang, Y., Jia, L., Li, H., Li, Z., et al. (2019). Brief report: Onward transmission of multiple HIV-1 unique recombinant forms among men who have sex with men in beijing, China. J. Acquir Immune Defic. Syndr. 81 (1), 1–4. doi:10.1097/QAI.0000000000001983
Lu, X., Kang, X., Liu, Y., Cui, Z., Guo, W., Zhao, C., et al. (2017). HIV-1 molecular epidemiology among newly diagnosed HIV-1 individuals in Hebei, a low HIV prevalence province in China. PLoS One 12 (2), e0171481. doi:10.1371/journal.pone.0171481
Lu, X., Zhang, J., Wang, Y., Liu, M., Li, Y., An, N., et al. (2020). Large transmission clusters of HIV-1 main genotypes among HIV-1 individuals before antiretroviral therapy in the Hebei province, China. AIDS Res. Hum. Retroviruses 36 (5), 427–433. doi:10.1089/AID.2019.0199
Moore, M. D., and Hu, W. S. (2009). HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 11 (2), 91–102.
Ou, W., Li, K., Feng, Y., Huang, Q., Ge, Z., Sun, J., et al. (2019). Characterization of a new HIV-1 CRF01_AE/B recombinant virus form among men who have sex with men in shanghai, China. AIDS Res. Hum. Retroviruses 35 (4), 414–418. doi:10.1089/AID.2018.0197
Robertson, D. L., Anderson, J. P., Bradac, J. A., Carr, J. K., Foley, B., Funkhouser, R. K., et al. (2000). HIV-1 nomenclature proposal. Science 288 (5463), 55–56. doi:10.1126/science.288.5463.55d
Shi, P., Chen, Z., Meng, J., Su, M., Yang, X., Fan, W., et al. (2021). Molecular transmission networks and pre-treatment drug resistance among individuals with acute HIV-1 infection in Baoding, China. PLoS One 16 (12), e0260670. doi:10.1371/journal.pone.0260670
Shi, P., Wang, X., Fan, W., Zhang, Z., Su, M., Meng, J., et al. (2021). Pre-treatment drug resistance analysis of HIV-1 infected patients in Baoding City, 2019-2020. Chin. J. Dermatovenereology 35 (09), 1012–1016+ 1020. doi:10.13735/j.cjdv.1001-7089.202103052
Wang, H., Zhao, X., Su, M., Meng, J., Fan, W., and Shi, P. (2022). Identification of a new HIV-1 circulating recombinant form CRF112_01B strain in baoding city, Hebei province, China. Curr. HIV Res. 20, 485–491. doi:10.2174/1570162X21666221027122528
Wang, T. X. (2020). A summary of HIV/AIDS epidemic situation across the country in 2020. Available at https://xw.qq.com/amphtml/20201207A0B1EE00 (accessed December 7, 2020).
Wang, X., Zhang, Y., Liu, Y., Li, H., Jia, L., Han, J., et al. (2021). Phylogenetic analysis of sequences in the HIV Database revealed multiple potential circulating recombinant forms in China. AIDS Res. Hum. Retroviruses 37 (9), 694–705. doi:10.1089/AID.2020.0190
Yan, J., Xin, R., Li, Z., Feng, Y., Lu, H., Liao, L., et al. (2015). CRF01_AE/B/C, a novel drug-resistant HIV-1 recombinant in men who have sex with men in beijing, China. AIDS Res. Hum. Retroviruses 31 (7), 745–748. doi:10.1089/AID.2015.0064
Yang, X., Zhu, H., An, W., Zhao, J., Lu, X., Sun, W., et al. (2022). Genetic characterizationof a novel HIV-1 CRF01_AE/CRF07_BC recombinant form found among men who have sex with men in Baoding City, Hebei Province, China. Arch. Virol. 167 (11), 2395–2402. doi:10.1007/s00705-022-05563-y
Yebra, G., Frampton, D., Gallo, C. T., Raffle, J., Hubb, J., Ferns, R. B., et al. (2018). A high HIV-1 strain variability in London, UK, revealed by full-genome analysis: Results from the ICONIC project. PLoS One 13 (2), e0192081. doi:10.1371/journal.pone.0192081
Keywords: HIV, near full-length genome, unique recombination forms, baoding, MSM
Citation: Zhang B, Chen S, Meng J, Su M, Fan W, An W and Lu X (2023) Identification of two near-identical novel HIV-1 unique recombinant forms (CRF01_AE/B) among men who have sex with men in baoding, hebei, China. Front. Genet. 14:1105739. doi: 10.3389/fgene.2023.1105739
Received: 23 November 2022; Accepted: 24 January 2023;
Published: 02 February 2023.
Edited by:
Hezhao Ji, Public Health Agency of Canada (PHAC), CanadaReviewed by:
Joan Fibla, Universitat de Lleida, SpainBinhua Liang, Public Health Agency of Canada (PHAC), Canada
Copyright © 2023 Zhang, Chen, Meng, Su, Fan, An and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weina An, YW53ZWluYTIwMjNAMTYzLmNvbQ==; Xinli Lu, bHhsaWkyMDA5QDE2My5jb20=
†These authors have contributed equally to this work
 Binbin Zhang1†
Binbin Zhang1† Weina An
Weina An Xinli Lu
Xinli Lu