
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 19 January 2023
Sec. Livestock Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1097825
This article is part of the Research TopicFisheries and Aquaculture GeneticsView all 18 articles
Members of the Doublesex and Mab-3-related transcription factor (Dmrt) gene family handle various vital functions in several biological processes, including sex determination/differentiation and gonad development. Dmrt1 and Sox9 (SoxE in invertebrates) exhibit a very conserved interaction function during testis formation in vertebrates. However, the dynamic expression pattern and functional roles of the Dmrt gene family and SoxE have not yet been identified in any echinoderm species. Herein, five members of the Dmrt gene family (Dmrt1, 2, 3a, 3b and 5) and the ancestor SoxE gene were identified from the genome of Apostichopus japonicus. Expression studies of Dmrt family genes and SoxE in different tissues of adult males and females revealed different expression patterns of each gene. Transcription of Dmrt2, Dmrt3a and Dmrt3b was higher expressed in the tube feet and coelomocytes instead of in gonadal tissues. The expression of Dmrt1 was found to be sustained throughout spermatogenesis. Knocking-down of Dmrt1 by means of RNA interference (RNAi) led to the downregulation of SoxE and upregulation of the ovarian regulator foxl2 in the testes. This indicates that Dmrt1 may be a positive regulator of SoxE and may play a role in the development of the testes in the sea cucumber. The expression level of SoxE was higher in the ovaries than in the testes, and knocking down of SoxE by RNAi reduced SoxE and Dmrt1 expression but conversely increased the expression of foxl2 in the testes. In summary, this study indicates that Dmrt1 and SoxE are indispensable for testicular differentiation, and SoxE might play a functional role during ovary differentiation in the sea cucumber.
The sea cucumber A. japonicus is an important nutritional seafood and is regarded as a precious traditional Chinese medicine (Yan J. et al., 2013; Yu Z. et al., 2014; Yang et al., 2015). The sea cucumber is widely distributed in the coastal regions of China, Far Eastern Russia, Japan, and Korea (Tian et al., 2017; Chen and Chang, 2015; Yan J. et al., 2013; Yu Z. et al., 2014), and its scale of breeding has continued to expand. Compared with the female sea cucumber, the male might have an advantage in terms of immunocompetence (Jiang et al., 2017; Jiang et al., 2019). Recently, several genes related to gonadal development have been identified, such as piwi (Sun et al., 2021), vasa (Yan M. et al., 2013) and foxl2 (Sun et al., 2022), and a hypothetical XX/XY sex determination system has been proposed (Wei et al., 2021). However, the mechanisms underlying sex determination and sex differentiation in the sea cucumber remain a mystery.
Identification of sex-related genes is an effective method to reveal the genetic basis of sex determination/differentiation in the sea cucumber. The Dmrt gene family encodes a large family of crucial transcription factors containing one or several conserved DM domains; this family was first identified in fruit fly (Drosophila melanogaster) (Arthropoda) (Burtis and Baker, 1989). The Dmrt gene family is involved in various biological processes, including sex determination, sex differentiation, testicular development, and embryo development (Dong et al., 2020). To date, the Dmrt gene family has been identified in diverse organisms and members of the Dmrt gene family exhibit substantial variation in different organisms. For instance, eight Dmrt genes (Dmrt1, Dmrt2, Dmrt3, Dmrt4, Dmrt5, Dmrt6, Dmrt7 and Dmrt8) have been identified in humans (Homo sapiens) (Ottolenghi et al., 2002) and mice (Mus musculus) (Kim et al., 2003). Six Dmrt genes, including Dmrt1, Dmrt2, Dmrt3, Dmrt4, Dmrt5 and Dmrt6, have been identified in tuatara (Sphenodon punctatus), and Dmrt1 is regarded as a crucial sex determination/differentiation gene in S. punctatus (Wang et al., 2006). Five Dmrt genes (Dmrt1, Dmrt2, Dmrt3, Dmrt5 and Dmrt6) have been identified in fish (Dong et al., 2020). In amphibians, five Dmrt genes (Dmrt1, Dmrt2, Dmrt3, Dmrt4 and Dmrt5) have been identified (Bewick et al., 2011; Watanabe et al., 2017). Due to teleost-specific whole-genome duplication (TGD) and rapid gene loss following TGD during evolution, the number of Dmrt genes is species-specific in teleost; there are five Dmrt genes in puffer fish (Takifugu rubripes) and zebrafish (Danio rerio) (Yamaguchi et al., 2006) but six Dmrt genes in Asian sea bass (Lates calcarifer), spotted gar (Lepisosteus oculatus) and channel catfish (Ictalurus punctatus) (Dong et al., 2020). Multiple Dmrt genes have also been identified from diverse invertebrate organisms. Seven Dmrt genes were identified both in Chinese mitten crab (Eriocheir sinensis) (Arthropoda), freshwater prawn (Macrobrachium rosenbergii) (Arthropoda) and mud crab (Scylla paramamosain) (Arthropoda) by screening transcriptome data using the bioinformatics method (Abayed et al., 2019; Du et al., 2019; Wan et al., 2021).
In addition to the identification of Dmrt gene families, several studies have focused on cloning, expression analysis and functional study of single Dmrt gene, especially the Dmrt1 gene. Dmrt1 is regarded as a conserved male-specific gene and plays a critical role in sex determination and sex differentiation in various species (Kopp, 2012). Sox9 (SRY-related HMG box gene 9) is a member of the SoxE subfamily and is another essential regulator of testis determination in many organisms (Hui et al., 2021). Both Dmrt1 and Sox9 lay downstream of the male sex differentiation pathway, and the expression of Dmrt1 was found to be upregulated before Sox9 during testis development (Elzaiat et al., 2014). Dmrt1 positively regulates the transcription of the Sox9b gene by directly binding to a specific cis-regulatory element (CRE) (Wei et al., 2019; Vining et al., 2021). Knockdown of the male sex-determining gene Dmrt1 leads to decreased Sox9 expression, while overexpression of Dmrt1 results in upregulation of Sox9 expression in red-eared slider turtle (Trachemys scripta) (Ge et al., 2017). In vertebrates, at least three members, including Sox8, Sox9 and Sox10, have been identified in the SoxE subfamily, but only one ancestral SoxE gene has been found in all studied invertebrate species (Heenan et al., 2016). The role of SoxE in the specification of the neural crest and the regulation of chondrogenesis has been revealed in invertebrates (McCauley, 2008); however, little is known about its function in reproduction and the interaction between Dmrt1 and SoxE.
In the current study, five members of the Dmrt gene family and a single SoxE gene were identified, and their molecular characteristics and phylogenetics were systemically analysed. Moreover, their dynamic expression patterns in different adult tissues and during different gonadal developmental stages were analysed using real-time quantitative PCR (RT-qPCR). Finally, the function and underlying interaction between Dmrt1 and SoxE in the process of gonadal development were explored using RNA interference (RNAi). These findings provide valuable information for understanding the sex determination and differentiation mechanisms of Echinodermata.
The published genome (taxid: 307972) data of the sea cucumber at the scaffold level (Zhang et al., 2017) was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). A BLASTP search was then performed using the common conserved domain protein sequence of the DM domain (XM_030995783.1) and the High Mobility Group box (HMG box) (XP_786809.2) to screen the homologous genes of the Dmrt family and SoxE, respectively. The E-value was set to ≤ e− 5. The suspected members of the Dmrt gene family and SoxE gene were validated by blasting against the NCBI database, in order to assess the reliability of the analysis.
The coding sequence of the Dmrt gene was predicted by ORF Finder online software (https://www.ncbi.nlm.nih.gov/orffinder). BioEdit software was used to perform multiple sequence alignments and visual analysis of the Dmrt proteins in the sea cucumber. The sequences used in this study were downloaded from the NCBI database (Supplementary Table S1). Phylogenetic analysis was performed using MEGA-7 software with bootstrapping (1,000 replicates) and the neighbour-joining (NJ) method.
Adult sea cucumbers were collected from the coastal areas of Dalian, China. No endangered or protected species were involved in this study. The expression patterns of the Dmrt genes and SoxE gene in different tissues were analysed by RT-qPCR. Briefly, total RNA from different tissues, including the tube feet, testes, intestines, ovaries, stomach, longitudinal muscle, coelomocytes and respiratory tree, were extracted using the Silica Membrane-based Vacuum Pump (SV) Total RNA Isolation System (Promega Z3100). The middle part of the intestine was used for gene expression analysis. According to previously reports (Sun et al., 2021), the gonads of A. japonicus are classified into four stages, including the early growing stage (Stage 1), growing stage (Stage 2), mature stage (Stage 3) and post-spawning stage (Stage 4). After gonadal histology analysis, gonadal tissues at different development stages were collected to examine the dynamic expression changes in Dmrt1 and SoxE. Gonadal tissues at Stage 2 were used to detect tissue expression specificity.
RT-qPCR experiments were performed in 20-μl reactions containing 10 µL of Fast Start Essential DNA Green Master (Roche, Mannheim, Germany), 6.4 µL of sterile water, 0.8 µL of each 10 mM primer, and 2 µL of cDNA. The protocol was as follows: 95°C (10 min) for heat denaturing; then, 40 cycles of 95°C (15 s) and 60°C (1 min). The efficiency of primers discovery through standard curve formulation reached 98%. The housekeeping gene NADH was selected as the internal reference gene (Sun et al., 2021). Data were performed from three independent experiments. Each sample was analyzed in triplicates, and the relative expression levels of target genes were calculated with the 2—ΔΔCt method. For statistical analysis, one-way ANOVA was calculated with SPSS software after the normal distribution and homogeneity of variance test (SPSS Inc.), and a probability (p) of ≤0.05 was considered statistically significant. The primers used in this study were designed using online software (http://biotools.nubic.northwestern.edu/OligoCalc.html) (Supplementary Table S2).
Gene-specific dsRNAs for the Dmrt1 and SoxE genes were designed using an online platform (https://www.dkfz.de/signaling/e-rnai3/). DsRNAs were synthesised using a T7 RiboMAX™ Express RNAi System (Promega), according to the manufacturer’s protocol. Three different dsRNAs were designed for each gene to ensure interference efficiency. The genetic sex of each sea cucumber was determined according to a previous report (Wei et al., 2021). In briefly, the tube feet tissues were used to extract genomic DNA by alkaline lysis method, then male-specific primers were used to amplify the male specific DNA marker, one specific band was amplified in males, but not in females. For the Dmrt1 RNAi experiment, 24 male sea cucumbers with an average weight of 100 ± 20 g were selected and were randomly divided into two groups: Dmrt1-knockdown (n = 12) and control (n = 12). For the SoxE RNAi experiment, 24 male sea cucumbers with an average weight of 60 ± 10 g were selected. The 24 males were divided into two groups: SoxE-knockdown (n = 12) and control group (n = 12). The injection experiments were performed as previously reported with slight modification (Sun et al., 2021). Specifically, before injecting, three gene-specific dsRNAs were mixed, then 100 μg of gene-specific dsRNA per 50 g body weight was injected into each sea cucumber in the knockdown group. Meanwhile, sea cucumbers in the control group were injected with GFP dsRNA at the same concentration. For the Dmrt1 RNAi experiment, three sea cucumbers from each group were removed randomly on the third day, seventh day and 10th day following the first injection. The gonads were surgically sampled for future research. In addition, repeat injections were performed on the third, sixth and ninth days following the first injection. For the SoxE RNAi experiment, three sea cucumbers from each group were removed randomly on the third, 7th and 23rd day following the first injection. The gonads were surgically sampled for future research. In addition, repeat injections were performed every 3 days after the first injection.
Gonadal tissues were surgically removed and fixed in 4% paraformaldehyde (PFA) at 4°C overnight. Then, they were washed three times in phosphate-buffered solution (PBS) and balanced in 30% sucrose at room temperature for 2 h. Next, the treated tissues were embedded in optimal cutting temperature (O.C.T) compound and frozen sections (5 μm) were cut and stained with haematoxylin/eosin. Digital photos were acquired under a Leica DM4B microscope
Genomic information for A. japonicus is available from NCBI (assembly ASM275485v1) (Zhang et al., 2017). To identify the Dmrt and SoxE genes in A. japonicus, the protein sequences of the conserved DM domain and HMG box of S. purpuratus (XM_030995783.1 and XP_786809.2) were used as the queries to perform a BLASTP search, respectively. Five predicted Dmrt genes (PIK43860.1, PIK34621.1, PIK44057.1, PIK33706.1 and PIK41536.1) were identified. Each Dmrt gene contained a complete coding DNA sequence (CDS) with lengths of 1,029, 651, 945, 507 and 849 bp, respectively. All identified Dmrt proteins had a DM domain (Supplementary Figure S1A), but had a different number of exons. PIK41536.1, PIK43860.1 and PIK34621.1 contained just a single intron, PIK44057.1 had two introns and PIK33706.1 had no introns (Supplementary Figure S1B). The identity between Dmrt cDNA sequences is about 32 %. Likewise, a predicted SoxE gene encoding a protein of 459 amino acids was identified AjSoxE. As expected, there was a 71-amino acid HMG box domain in the protein sequence (Supplementary Figure S1C).
To define these five Dmrt genes, 40 Dmrt genes from 17 species were downloaded from the NCBI database and multiple protein sequence alignments and phylogenetic analyses were performed. The protein sequences of the Dmrt genes exhibiting low similarity between species (4.94%–57.01%). However, the protein sequence of DM domain was highly conserved such that the similarity with other species was generally more than 60% (Supplementary Figures S2–S5). The phylogenetic analyses indicated that the Dmrt family genes can be classified into five branches (Dmrt1, -2, -3, -4 and -5). PIK41536.1 and PIK43860.1 were clustered into the Dmrt1 and Dmrt2 branches, and were named AjDmrt1 and AjDmrt2, respectively. PIK34621.1 and PIK44057.1 were both clustered with Dmrt3 homologs from other species, so were named AjDmrt3a and AjDmrt3b, respectively. The deduced amino acid sequences of PIK33706.1 were clustered into the Dmrt5 branch and were defined as AjDmrt5 (Figure 1A). Surprisingly, none of the AjDmrt genes clustered with Dmrt4 homologs from other species’ branches. The topology of clades of Dmrt1, Dmrt2, Dmrt3 and Dmrt5 were basically consistent with the known taxonomic relationships among these species. Moreover, the phylogenetic analyses showed that AjSoxE was firstly grouped with SoxE in invertebrate species, then clustered into Sox8, Sox9 and Sox10 branches in jawed vertebrates (Figure 1B). This phylogenetic tree further confirms that the ancestral SoxE gene is replicated in vertebrates and has produced at least three members (Sox8, Sox9, and Sox10).
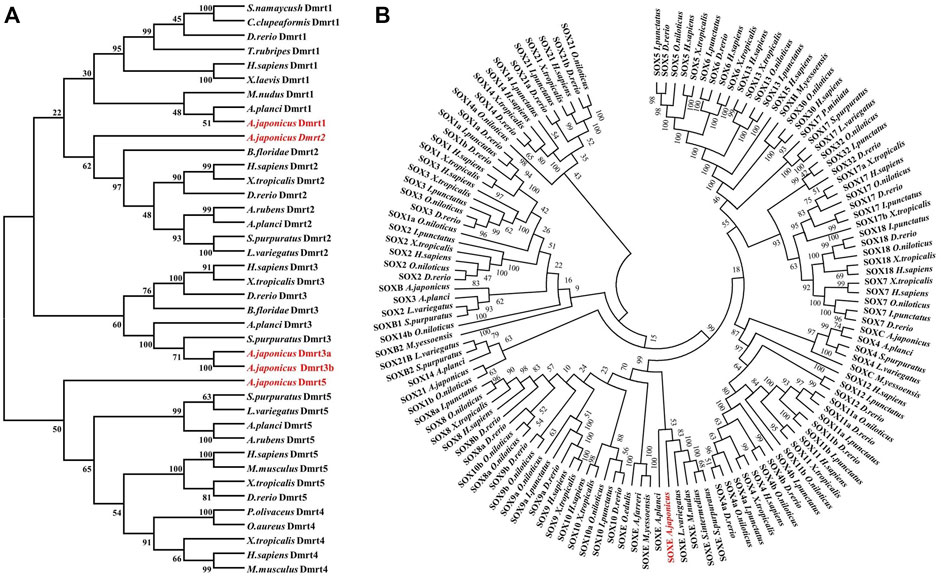
FIGURE 1. Phylogenetic analysis of the Dmrt and SOX protein family in Apostichopus japonicus (A) Dmrt.(B) SoxE. The phylogenetic tree was constructed with MEGA version 7.0 (1,000 replicates) by maximum likelihood analysis.
In order to reveal the expression differences, the transcripts of all five AjDmrt genes and the SoxE gene in adult tissues were examined by RT-qPCR. Specific primers of five AjDmrt genes were designed in the parts with the greatest difference in the nucleic acid sequence. AjDmrt1 was expressed predominantly in the testes, only low AjDmrt1 expression was detected in other tissues, including the ovaries, stomach, tube feet, longitudinal muscle, respiratory tree, and coelomocytes (Figure 2A). In contrast, AjDmrt2 was higher expressed in the tube feet (Figure 2B). AjDmrt3a and AjDmrt3b were expressed predominantly in the coelomocytes and tube feet, with low expression of AjDmrt3a and AjDmrt3b detected in other tissues, including the intestines, stomach, longitudinal muscle, ovaries, and respiratory tree (Figures 2C,D). The expression pattern of AjDmrt5 in adult tissue was similar to that of AjDmrt2, with predominant expression in the tube feet and slight expression in the other analysed tissues (Figure 2E). Strikingly, the SoxE gene had the highest expression in the tube feet. Its expression differed significantly between the male and female gonads, such that a large number of transcripts were detected in the ovaries while SoxE was nearly undetectable in the testes (Figure 2F).
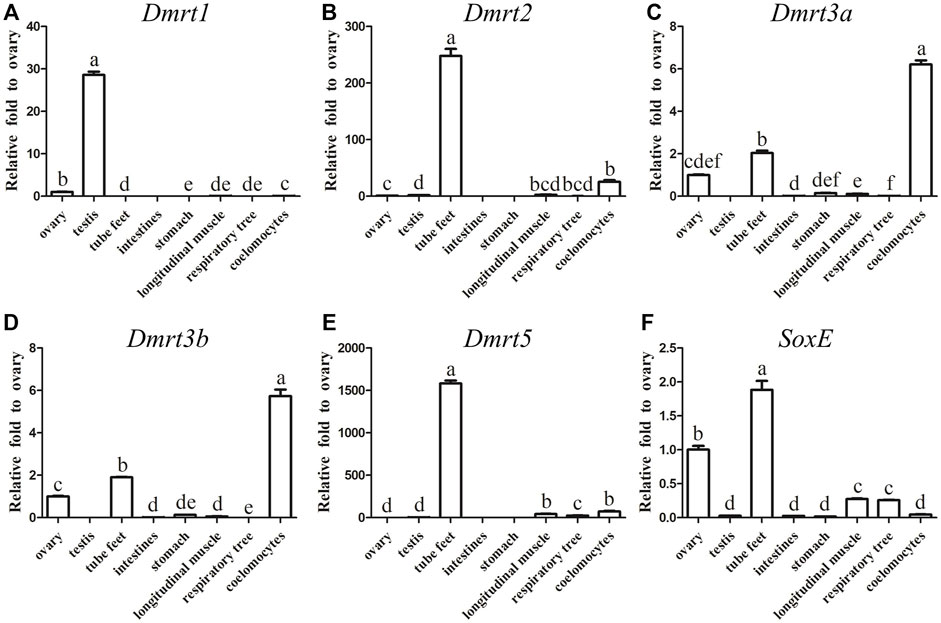
FIGURE 2. RT-qPCR analysis of Dmrts and SoxE Expression in adult tissues. NADH was used as internal control (A) Dmrt1. (B) Dmrt2. (C) Dmrt3a. (D) Dmrt3b. (E) Dmrt5. (F) SoxE. Data were performed from three independent experiments. Each bar represents mean ± SD. Different letters indicate significant differences between mean values (p ≤ 0.05), whilst shared letters indicate no significant difference.
Considering that the expression of Dmrt1 and SoxE differed significantly between the ovaries and testes in A. japonicus, gonadal tissues at different development stages were collected to examine the dynamic expression changes in Dmrt1 and SoxE. After gonadal histology analysis, gonadal tissues of four different development stages (i.e., the early growing stage (Stage 1), growing stage (Stage 2), mature stage (Stage 3) and post-spawning stage (Stage 4)) were obtained from male and female individuals, respectively. The expression of Dmrt1 was continuously maintained at a high level during the spermatogenesis process and was significantly downregulated during Stage 2 in the testes (Figure 3A). SoxE expression in the testes at the corresponding stage was much lower than that of Dmrt1, and there were no significant differences between Stage 1, Stage 2 and Stage 3. Along with the occurrence of spermatogenesis, the expression level of SoxE increased up to 6.3-fold during Stage 4 against to Stage1 (Figure 3B). Meanwhile, a large amount of SoxE transcript was detected in Stage 1 ovary tissue, but the relative expression level continuously decreased together with the oogenesis development process, reaching the lowest in Stage 4 (Figure 3C).
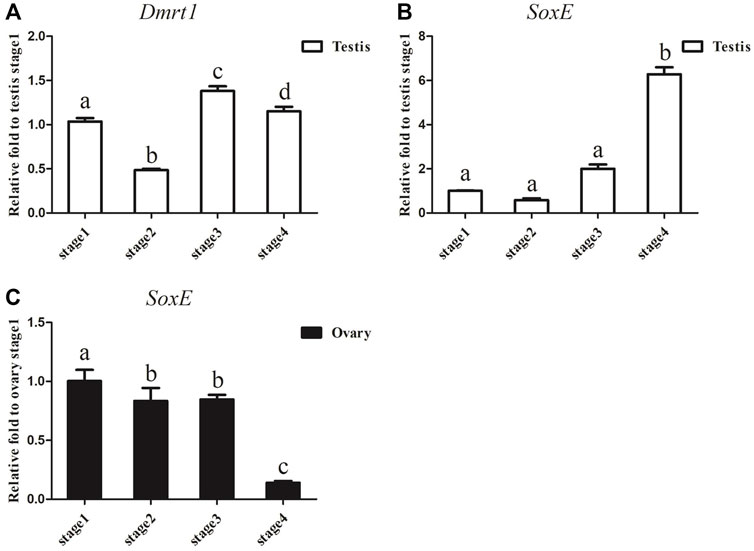
FIGURE 3. Dmrt1 and SoxE expression in gonad. (A) RT-qPCR analysis of Dmrt1 expression in testis at different development stages. (B) RT-qPCR analysis of SoxE expression in testis at different development stages. (C) RT-qPCR analysis of SoxE expression in ovary at different development stages. NADH was used as internal control. Data were performed from three independent experiments. Each bar represents mean ± SD. Different letters indicate significant differences between mean values (p ≤ 0.05), whilst shared letters indicate no significant difference.
RNAi was further utilised to examine the underlying function of Dmrt1 in testes development in male A. japonicus. As shown in Figure 4A, the expression levels of Dmrt1 significantly decreased, down 40.85% and 47.80% in the knockdown group compared to the control group at 3 days post-injection (dpi) and 7 dpi, respectively. On the 10 dpi, Dmrt1 expression was only 3.31% of that in the control group. Compared to the control group, the transcripts of SoxE were reduced in the knockdown group by 88.65% at 3 dpi, 45.52% at 7 dpi and 77.10% at 10 dpi (Figure 4B). Moreover, the dynamic expression changes in foxl2 and piwi, which have been reported to be involved in ovary differentiation and germ cell development, respectively (Ottolenghi et al., 2005; Gonzalez et al., 2021), were investigated. Interestingly, the expression level of foxl2 was significantly downregulated (about 46.17%) in the knockdown group at 3 dpi, but sharply increased 1.67–2.17-fold at 7 dpi and 10 dpi (Figure 4C). The expression level of piwi did not significantly differ between the knockdown and control groups at 3 dpi and 7 dpi, but was significantly reduced by 58.36% at 10 dpi in knockdown adults (Figure 4D). Although the expression levels of genes involved in testis differentiation, ovary differentiation and germ cell development showed dynamic changes after knockdown of Dmrt1 by RNAi, no obvious histological changes and no apoptosis cells were observed in the knockdown testes (Supplementary Figure S6).
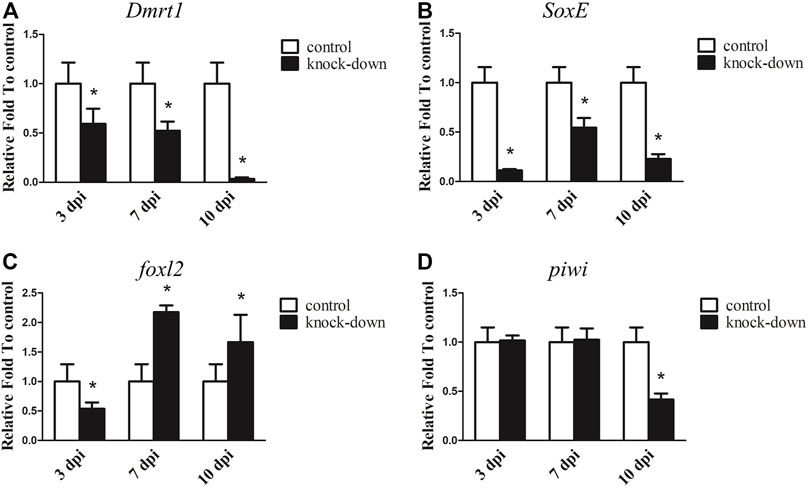
FIGURE 4. RNA interference (RNAi) of Dmrt1 in testis. RT-qPCR detected mRNA levels of Dmrt1 and other sex related genes after Dmrt1 RNAi. (A) Dmrt1. (B) SoxE. (C)foxl2. (D) piwi. NADH was used as internal control. Data were performed from three independent experiments. Each bar represents mean ± standard deviation (SD). Asterisks (*) indicate significant differences (p ≤ 0.05) between knock-down and control. dpi: days post-injection.
Since SoxE has been reported to play a conserved role in sexual differentiation, and it shows a sexually dimorphic expression pattern in the gonads of A. japonicus, its function during testis development was investigated by RNAi in an adult male individual. As shown in Figure 5A, the transcript level of SoxE was significantly decreased by 79.46% with SoxE knockdown compared to the control at 3 dpi. Its expression was continuously downregulated at 7 dpi and 23 dpi. In addition, the expression of Dmrt1 was sharply decreased to almost an undetectable level in the knockdown testes (Figure 5B). Conversely, the expression level of foxl2 was increased about 8.09-fold and 7.34-fold in the knockdown sample compared to the control sample at 3 dpi and 23 dpi, respectively. However, the expression level of foxl2 was not significantly different compared to the control at 7 dpi (Figure 5C). The transcripts of piwi were significantly decreased by 79.42%, 84.60% and 48.87% at 3 dpi, 7 dpi and 23 dpi, respectively (Figure 5D ). However, no obvious histological changes or apoptosis cells were observed in the knockdown testes (Supplementary Figure S7).
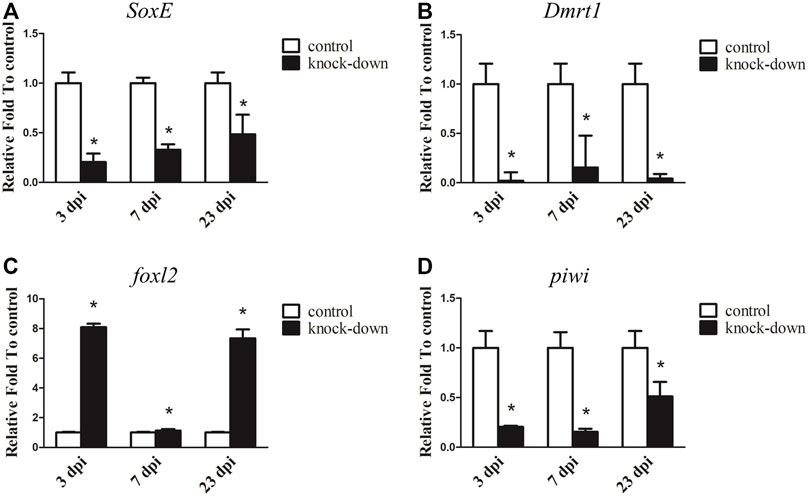
FIGURE 5. RNA interference (RNAi) of SoxE in testis. RT-qPCR detected mRNA levels of SoxE and other sex related genes after SoxE RNAi. (A) SoxE. (B) Dmrt1. (C)foxl2. (D) piwi. NADH was used as internal control. Data were performed from three independent experiments. Each bar represents mean ± standard deviation (SD). Asterisks (*) indicate significant differences (p ≤ 0.05) between knock-down and control. dpi: days post-injection.
With the development of sequencing technology, genome-wide investigations of gene families have become common, and several gene families, including the Dmrt gene family (Zafar et al., 2019), Sox gene family (Yang et al., 2020), DEAD-box helicase family (Xu et al., 2021), NRAMP gene family (Tian et al., 2021), TRAF gene family (Zhang et al., 2020), TGF-beta gene family (Cao et al., 2018) and TLR gene family (Habib et al., 2021) have been identified in various species. The origin of the Dmrt gene family can be traced back to coelenterates in the animal kingdom (Bellefroid et al., 2013; Wexler et al., 2014), and their composition, expression, and function show species-specificity. Here, Dmrt3 genes were duplicated in the A. japonicus genome and Dmrt3a and Dmrt3b were produced, whereas Dmrt4 was lost. In other species, Dmrt4 and Dmrt5 were clustered into a major branch (Wang et al., 2012; Dong et al., 2020), indicating that these genes possibly originated from a common ancestor of Dmrt. This loss or duplicated gene phenomenon possibly resulted from independent gene duplication (loss) during the evolution of the sea cucumber. In most vertebrates, the Dmrt1–Dmrt3–Dmrt2(2a) gene cluster is highly conserved and plays a key role in sex differentiation (Johnsen and Andersen., 2012; Dong et al., 2020). Since the existing sea cucumber genome is not assembled to the chromosome level (Zhang et al., 2017; Li et al., 2018), we failed to identify the Dmrt1–Dmrt3–Dmrt2(2a) conserved gene cluster (Supplementary Figure S8). Hence, a chromosome-level assembly of the sea cucumber (A. japonicus) should be sequenced to further elucidate the linkages among the conserved gene cluster and better understand the sex-determination mechanism.
Two different expression patterns of Dmrt1 have been identified, including testes-specific expression and sexually dimorphic expression. In most cases, Dmrt1 is specifically expressed in the testes and plays essential roles in male-sex determination and testicular differentiation. Knockdown or knockout of Dmrt1 leads to male-to-female sex reversal (Paul-Prasanth et al., 2006; Luo et al., 2015; Chakraborty et al., 2016). However, there are some situations in which Dmrt1 shows a sexually dimorphic expression pattern, with slight expression in the ovaries and abundant expression in the testes; for example, in red-eared slider turtle (Trachemys scripta) (Ge et al., 2017). In sea cucumber, Dmrt1 shares a similar expression pattern with the red-eared slider turtle, with higher expression in the testes than in the ovaries. In addition, its expression was found to be highly expressed throughout spermatogenesis, but declined at the growing stage (Figure 3A). Dmrt1 expression was decreased at spermiation stage in comparison with growing and maturing stages in Oncorhynchus mykiss and Odontesthes bonariensis (Marchand et al., 2000; Fernandino et al., 2006). These differences may be due to the different reproductive strategies and different testis structure between sea cucumber and fish, and should be investigated in the future. Curiously, there appears to be greater expression of SoxE in the ovaries than the testes at the same developmental stages. Both Dmrt1 and Sox9 are necessary for male sexual development in several species, knockdown (off) of SoxE (Sox9) and Dmrt1 leads to the disappearance of the male marker, ectopic expression of the ovarian regulator and the formation of an ovary-like structure (Barrionuevo et al., 2006; Kim et al., 2007; Krentz et al., 2009; Cui et al., 2017). In Nile tilapia knockdown of Dmrt1 led to the downregulation of Sox9b, and two potential cis-regulatory elements (CREs) for the Dmrt1 transcription factor were identified in the promoter of Sox9b (Wei et al., 2019). Loss of DMRT1 will inhibit SOX9 expression and activates FOXL2 transcription factor, even can reprogram granulosa cells into Sertoli cells in mice (Matson et al., 2011). In most species, sex is determined by male regulatory gene network in which Sry activates Sox9 and a female network involving WNT/b-catenin signalling (Matson et al., 2011), and Dmrt1 recruits Sox9 to reprogram sexual cell fate(Lindeman et al., 2021). Although no obvious histological changes were observed in SoxE and Dmrt1 knockdown testes in the current study, the expression level of the ovarian regulator foxl2 was significantly upregulated at 3dpi and 23dpi, indicating that SoxE and Dmrt1 may be closely related to the development and maintenance of the testes. Unexpectedly, foxl2 was significantly downregulated at 7dpi after SoxE-knockdown. Although RNAi provides a useful approach to study gene function in non-model species (Yang et al., 2017; Yu et al., 2014; Zhang et al., 2019), it is indeed a big challenge to ensure considerable knockdown efficiency among individuals especially in echinoderms with open-tube circulation. Hence, it is of great value to evaluate the functions of SoxE and Dmrt1 in testes differentiation through gene knockout biotechnologies.
Apart from Dmrt1, which is highly expressed in the testes, Dmrt2, 3a, 3b and 5, as well as SoxE, showed significantly higher expression in tube feet than in other adult tissues. Current studies have shown that the tube feet of the sea cucumber has a wide range of functions, including locomotion, feeding, chemoreception and light sensitivity respiration (Wang et al., 2014; Sun et al., 2013). According to previous reports, the expression pattern of Dmrt2 (2a, 2b) are species-specific, with expression primarily in both the ovaries and testes (Yamaguchi et al., 2006; Sheng et al., 2014; Su et al., 2015). In vertebrates, Dmrt3 shows high expression in the testes and nervous system; accordingly, it has been speculated to be involved in the development of nerves and germ cells (Yamaguchi et al., 2006; Li et al., 2008; Dong et al., 2010). In contrast, large transcripts of Dmrt3a and Dmrt3b have been detected in coelomocytes, indicating that they might contribute to the regulation of immune functions. Surprisingly, the expression of SoxE was higher in the ovaries than the testes. In vertebrates, the transcription factor Sox9 is highly expressed in developing male gonadal ridges and serves as the master regulator of Sertoli cell differentiation, playing a crucial role in sex determination (Kent et al., 1996; Jakob and Lovell-Badge., 2011). In invertebrates, SoxE has been detected in both the ovaries and testes and is mainly located in oocytes, spermatocytes and Sertoli cells (Santerre et al., 2014; Li et al., 2016; Yu et al., 2017). This indicates that SoxE may be involved in ovary development and differentiation as well as in the development of the testis in invertebrates.
In conclusion, this study identified and characterized the SoxE gene and Dmrt gene family in A.japonicus for the first time. A total of five Dmrt genes (Dmrt1, 2, 3a, 3b and 5) were identified and classified into four families based on phylogenetic analyses. Expression profile analyses demonstrated that the express patterns of Dmrt genes and the SoxE gene. Among them, Dmrt1 was higher expressed in the testes with low expression in the ovaries. Knocking-down of Dmrt1 led to the downregulation of SoxE expression and upregulation of the ovarian regulator foxl2 in the testes. This indicates that Dmrt1 may be a positive regulator of SoxE and may play a role in the development of the testes in the sea cucumber. In contrast, SoxE was more highly expressed in the ovaries than the testes, and the transcript level of Dmrt1 was significantly decreased while foxl2 expression was sharply increased in SoxE knockdown testes. This study provides a comprehensive overview of the Dmrt gene family and contributes to a better understanding of the role of the Dmrt1 and SoxE genes in the sex determination and differentiation mechanisms of the sea cucumber.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
“Conceptualization, Z-HS and Y-QC; methodology, B-ZL; investigation, B-ZL, J-JC and W-YS; resources, Z-HS and Y-QC; data curation Z-LH and B-ZL; writing—original draft preparation, Z-HS and B-ZL; writing—review and editing, Z-HS and B-ZL; funding acquisition, Z-HS and Y-QC; All authors have read and agreed to the published version of the manuscript.”
This work was supported by the National Key R&D Program of China (grant number 2018YFD0900201) and Liaoning Province “Xingliao Talents Plan” project (Grant no. XLYC2002107).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1097825/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Gene structure of Dmrt and SoxE. (A) Conserved DM domain diagram of Dmrts. (B) Schematic diagram of the gene structure of Dmrts. (C) Conserved HMG domain diagram of SoxE.
SUPPLEMENTARY FIGURE S2 | Amino acid alignments of Dmrt1 with the homologs from other species. (A) Full length of amino acid alignments of Dmrt1. (B) DM domain alignments of Dmrt1. The identities relative to are exhibited at the end of each sequence. The boule frame indicates the conserved position of the DM domain.
SUPPLEMENTARY FIGURE S3 | Amino acid alignments of Dmrt2 with the homologs from other species. (A) Full length of amino acid alignments of Dmrt2. (B) DM domain alignments of Dmrt2. The identities relative to are exhibited at the end of each sequence. The boule frame indicates the conserved position of the DM domain.
SUPPLEMENTARY FIGURE S4 | Amino acid alignments of Dmrt3 with the homologs from other species. (A) Full length of amino acid alignments of Dmrt3. (B) DM domain alignments of Dmrt3. The identities relative to are exhibited at the end of each sequence. The boule frame indicates the conserved position of the DM domain.
SUPPLEMENTARY FIGURE S5 | Amino acid alignments of Dmrt5 with the homologs from other species. (A) Full length of amino acid alignments of Dmrt5. (B) DM domain alignments of Dmrt5. The identities relative to are exhibited at the end of each sequence. The boule frame indicates the conserved position of the DM domain.
SUPPLEMENTARY FIGURE S6 | Histological examination of testes after RNAi of Dmrt1. Bars=50 μm.
SUPPLEMENTARY FIGURE S7 | Histological examination of testes after RNAi of soxE. Bars=50 μm.
SUPPLEMENTARY FIGURE S8 | Gene synteny of Dmrt3 with the homologues from other species. Conserved gene blocks are represented in matching colors, and transcription orientations are indicated by arrows.
Abayed, F. A. A., Manor, R., Aflalo, E. D., and Sagi, A. (2019). Screening for dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. General Comp. Endocrinol. 282, 113205. doi:10.1016/j.ygcen.2019.06.009
Barrionuevo, F., Bagheri-Fam, S., Klattig, J., Kist, R., Taketo, M. M., Englert, C., et al. (2006). Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod. 74 (1), 195–201. doi:10.1095/biolreprod.105.045930
Bellefroid, E. J., Leclère, L., Saulnier, A., Keruzore, M., Sirakov, M., Vervoort, M., et al. (2013). Expanding roles for the evolutionarily conserved dmrt sex transcriptional regulators during embryogenesis. Cell. Mol. life Sci. 70 (20), 3829–3845. doi:10.1007/s00018-013-1288-2
Bewick, A. J., Anderson, D. W., and Evans, B. J. (2011). Evolution of the closely related, sex-related genes DM-W and dmrt1 in African clawed frogs (Xenopus). Evol. Int. J. Org. Evol. 65 (3), 698–712. doi:10.1111/j.1558-5646.2010.01163.x
Burtis, K. C., and Baker, B. S. (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56 (6), 997–1010. doi:10.1016/0092-8674(89)90633-8
Cao, L., Xie, B. J., Cui, B. B., Chen, B. B., Ma, Z. S., Chen, X. B., et al. (2018). Genome-wide feature analysis of the sequence-specific recognition in intermolecular interaction between tgf-β pathway dna and meg3 lncrna in human cancer. J. Chemom. 32, e3042. doi:10.1002/cem.3042
Chakraborty, T., Zhou, L. Y., Chaudhari, A., Iguchi, T., and Nagahama, Y. (2016). Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 6 (1), 19480–19511. doi:10.1038/srep19480
Chen, J., and Chang, Y. (2015). Sea cucumber aquaculture in China. Echinoderm Aquac. 14, 317–330. doi:10.1002/9781119005810.ch14
Cui, Z., Liu, Y., Wang, W., Wang, Q., Zhang, N., Lin, F., et al. (2017). Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 7 (1), 42213–42310. doi:10.1038/srep42213
Dong, X. L., Chen, S. L., and Ji, X. S. (2010). Molecular cloning and expression analysis of Dmrt3 gene in half-smooth tongue sole (Cynoglossus semilaevis). J. Fish. China. 34, 829–835. doi:10.1007/s10695-018-0472-6
Dong, J. J., Li, J., Hu, J., Sun, C. F., Tian, Y. Y., Li, W. H., et al. (2020). Comparative genomics studies on the dmrt gene family in fish. Front. Genet. 11, 563947. doi:10.3389/fgene.2020.563947
Du, J., Liu, Y., Song, C. W., and Cui, Z. X. (2019). Discovery of sex-related genes from embryonic development stage based on transcriptome analysis in Eriocheir sinensis. Gene 710, 1–8. doi:10.1016/j.gene.2019.05.021
Elzaiat, M., Jouneau, L., Thépot, D., Klopp, C., Allais-Bonnet, A., Cabau, C., et al. (2014). High-throughput sequencing analyses of XX genital ridges lacking FOXL2 reveal DMRT1 up-regulation before SOX9 expression during the sex-reversal process in goats. Biol. Reprod. 91 (6), 153–161. doi:10.1095/biolreprod.114.122796
Fernandino, J. I., Guilgur, L. G., and Somoza, G. M. (2006). Dmrt1 expression analysis during spermatogenesis in pejerrey, Odontesthes bonariensis. Fish Physiol. Biochem. 32 (3), 231–240. doi:10.1007/s10695-006-9005-9
Ge, C. T., Ye, J., Zhang, H. Y., Zhang, Y., Sun, W., Sang, Y. P., et al. (2017). Dmrt1 induces the male pathway in a turtle species with temperature-dependent sex determination. Development 144 (12), 2222–2233. doi:10.1242/dev.152033
Gonzalez, L. E., Tang, X., and Lin, H. (2021). Maternal Piwi regulates primordial germ cell development to ensure the fertility of female progeny in Drosophila. Genetics 219 (1), iyab091. doi:10.1093/genetics/iyab091
Habib, Y. J., Wan, H. F., Sun, Y. L., Shi, J. L., Yao, C. J., Lin, J. M., et al. (2021). Genome-wide identification of toll-like receptors in Pacific white shrimp (litopenaeus vannamei) and expression analysis in response to vibrio parahaemolyticus invasion. Aquaculture 532, 735996. doi:10.1016/j.aquaculture.2020.735996
Heenan, P., Zondag, L., and Wilson, M. J. (2016). Evolution of the Sox gene family within the chordate phylum. Gene 575 (2), 385–392. doi:10.1016/j.gene.2015.09.013
Hui, H. B., Xiao, L., Sun, W., Zhou, Y. J., Zhang, H. Y., and Ge, C. T. (2021). Sox9 is indispensable for testis differentiation in the red-eared slider turtle, a reptile with temperature-dependent sex determination. Zool. Res. 42 (6), 721–725. doi:10.24272/j.issn.2095-8137.2021.136
Jakob, S., and Lovell-Badge, R. (2011). Sex determination and the control of Sox9 expression in mammals. FEBS J. 278 (7), 1002–1009. doi:10.1111/j.1742-4658.2011.08029.x
Jiang, J. W., Zhou, Z. C., Dong, Y., Gao, S., Sun, H. J., Chen, Z., et al. (2017). Comparative analysis of immunocompetence between females and males in the sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 63, 438–443. doi:10.1016/j.fsi.2017.02.038
Jiang, J. W., Zhao, Z. L., Pan, Y. J., Dong, Y., Gao, S., Li, S. L., et al. (2019). Gender specific differences of immune competence in the sea cucumber Apostichopus japonicus before and after spawning. Fish Shellfish Immunol. 90, 73–79. doi:10.1016/j.fsi.2019.04.051
Johnsen, H., and Andersen, Ø. (2012). Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. The fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505 (2), 221–232. doi:10.1016/j.gene.2012.06.021
Kent, J., Wheatley, S. C., Andrews, J. E., Sinclair, A. H., and Koopman, P. (1996). A male-specific role for SOX9 in vertebrate sex determination. Development 122 (9), 2813–2822. doi:10.1242/dev.122.9.2813
Kim, S., Kettlewell, J. R., Anderson, R. C., Bardwell, V. J., and Zarkower, D. (2003). Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 3 (1), 77–82. doi:10.1016/S1567-133X(02)00071-6
Kim, S., Bardwell, V. J., and Zarkower, D. (2007). Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol. 307 (2), 314–327. doi:10.1016/j.ydbio.2007.04.046
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28 (4), 175–184. doi:10.1016/j.tig.2012.02.002
Krentz, A. D., Murphy, M. W., Kim, S., Cook, M. S., Capel, B., Zhu, R., et al. (2009). The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc. Natl. Acad. Sci. 106 (52), 22323–22328. doi:10.1073/pnas.0905431106
Li, Q., Zhou, X., Guo, Y., Shang, X., Chen, H., Lu, H., et al. (2008). Nuclear localization, DNA binding and restricted expression in neural and germ cells of zebrafish Dmrt3. Biol. Cell. 100, 453–463. doi:10.1042/BC20070114
Li, Y., Zhang, L., Sun, Y., Ma, X., Wang, J., Li, R., et al. (2016). Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop Patinopecten yessoensis. Mar. Biotechnol. 18 (4), 453–465. doi:10.1007/s10126-016-9706-8
Li, Y., Wang, R., Xun, X., Wang, J., Bao, L., Thimmappa, R., et al. (2018). Sea cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discov. 4 (1), 29–17. doi:10.1038/s41421-018-0030-5
Lindeman, R. E., Murphy, M. W., Agrimson, K. S., Gewiss, R. L., Bardwell, V. J., Gearhart, M. D., et al. (2021). The conserved sex regulator DMRT1 recruits SOX9 in sexual cell fate reprogramming. Nucleic acids Res. 49 (11), 6144–6164. doi:10.1093/nar/gkab448
Luo, D., Liu, Y., Chen, J., Xia, X., Cao, M., Cheng, B., et al. (2015). Direct production of XYDMY− sex reversal female medaka (Oryzias latipes) by embryo microinjection of TALENs. Sci. Rep. 5 (1), 14057–14115. doi:10.1038/srep14057
Marchand, O., Govoroun, M., D’Cotta, H., McMeel, O., Lareyre, J. J., Bernot, A., et al. (2000). DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1493 (1-2), 180–187. doi:10.1016/s0167-4781(00)00186-x
Matson, C. K., Murphy, M. W., Sarver, A. L., Griswold, M. D., Bardwell, V. J., and Zarkower, D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476 (7358), 101–104. doi:10.1038/nature10239
McCauley, D. W. (2008). SoxE, Type II collagen, and evolution of the chondrogenic neural crest. Zool. Sci. 25 (10), 982–989. doi:10.2108/zsj.25.982
Ottolenghi, C., Fellous, M., Barbieri, M., and McElreavey, K. (2002). Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79, 333–343. doi:10.1006/geno.2002.6711
Ottolenghi, C., Omari, S., Garcia-Ortiz, J. E., Uda, M., Crisponi, L., Forabosco, A., et al. (2005). Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet. 14 (14), 2053–2062. doi:10.1093/hmg/ddi210
Paul-Prasanth, B., Matsuda, M., Lau, E. L., Suzuki, A., Sakai, F., Kobayashi, T., et al. (2006). Knock-down of DMY initiates female pathway in the genetic male medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 351 (4), 815–819. doi:10.1016/j.bbrc.2006.10.095
Santerre, C., Sourdaine, P., Adeline, B., and Martinez, A. S. (2014). Cg-SoxE and Cg-β-catenin, two new potential actors of the sex-determining pathway in a hermaphrodite lophotrochozoan, the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 167, 68–76. doi:10.1016/j.cbpa.2013.09.018
Sheng, Y., Chen, B., Zhang, L., Luo, M., Cheng, H., and Zhou, R. (2014). Identification of Dmrt genes and their up-regulation during gonad transformation in the swamp eel (Monopterus albus). Mol. Biol. Rep. 41 (3), 1237–1245. doi:10.1007/s11033-013-2968-6
Su, L., Zhou, F., Ding, Z., Gao, Z., Wen, J., Wei, W., et al. (2015). Transcriptional variants of Dmrt1 and expression of four Dmrt genes in the blunt snout bream, Megalobrama amblycephala. Gene 573 (2), 205–215. doi:10.1016/j.gene.2015.07.044
Sun, H., Zhou, Z., Dong, Y ., Yang, A., Jiang, B., Gao, S., et al. (2013). Identification and expression analysis of two Toll-like receptor genes from sea cucumber (Apostichopus japonicus). Fish shellfish Immunol. 34, 147–158. doi:10.1016/j.fsi.2012.10.014
Sun, Z. H., Wei, J. L., Cui, Z. P., Han, Y. L., Chang, Y. Q., Song, J., et al. (2021). Identification and functional characterization of piwi1 gene in sea cucumber, Apostichopus japonicus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 252, 110536. doi:10.1016/j.cbpb.2020.110536
Sun, J. J., Sun, Z. H., Wei, J. L., Ding, J., Song, J., and Chang, Y. Q. (2022). Identification and functional analysis of foxl2 and nodal in sea cucumber, Apostichopus japonicus. Gene Expr. Patterns 44, 119245. doi:10.1016/j.gep.2022.119245
Tian, Y., Jiang, Y., Shang, Y., Zhang, Y. P., Geng, C. F., Wang, L. Q., et al. (2017). Establishment of lysozyme gene RNA interference systemand its involvement in salinity tolerance in sea cucumber (Apostichopus japonicus). Fish Shellfish Immunol. 65, 71–79. doi:10.1016/j.fsi.2017.03.046
Tian, W. J., He, G. D., Qin, L. J., Li, D. D., Meng, L. L., Huang, Y., et al. (2021). Genome-wide analysis of the nramp gene family in potato (Solanum tuberosum): Identification, expression analysis and response to five heavy metals stress. Ecotoxicol. Environ. Saf. 208, 111661. doi:10.1016/j.ecoenv.2020.111661
Vining, B., Ming, Z., Bagheri-Fam, S., and Harley, V. (2021). Diverse regulation but conserved function: SOX9 in vertebrate sex determination. Genes 12 (4), 486. doi:10.3390/genes12040486
Wan, H., Zhong, J., Zhang, Z., Zou, P., and Wang, Y. (2021). Discovery of the dmrt gene family members based on transcriptome analysis in mud crab scylla paramamosain. Gene 145576, 145576. doi:10.1016/j.gene.2021.145576
Wang, Z., Miyake, T., Edwards, S. V., and Amemiya, C. T. (2006). Tuatara (Sphenodon) genomics: BAC library construction, sequence survey, and application to the dmrt gene family. J. Hered. 97 (6), 541–548. doi:10.1093/jhered/esl040
Wang, F., Yu, Y., Ji, D., and Li, H. (2012). The DMRT gene family in amphioxus. J. Biomol. Struct. Dyn. 30 (2), 191–200. doi:10.1080/07391102.2012.677770
Wang, H., Liu, S., Cui, J., Li, C., Qiu, X., Chang, Y., et al. (2014). Characterization and expression analysis of microRNAs in the tube foot of sea cucumber Apostichopus japonicus. PloS one 9 (11), e111820. doi:10.1371/journal.pone.0111820
Watanabe, M., Yasuoka, Y., Mawaribuchi, S., Kuretani, A., Ito, M., Kondo, M., et al. (2017). Conservatism and variability of gene expression profiles among homeologous transcription factors in xenopus laevis. Dev. Biol. 426, 301–324. doi:10.1016/j.ydbio.2016.09.017
Wei, L., Li, X., Li, M., Tang, Y., Wei, J., and Wang, D. (2019). Dmrt1 directly regulates the transcription of the testis-biased Sox9b gene in Nile tilapia (Oreochromis niloticus). Gene 687, 109–115. doi:10.1016/j.gene.2018.11.016
Wei, J. L., Cong, J. J., Sun, Z. H., Song, J., Zhao, C., and Chang, Y. Q. (2021). A rapid and reliable method for genetic sex identification in sea cucumber, Apostichopus japonicus. Aquaculture 543, 737021. doi:10.1016/j.aquaculture.2021.737021
Wexler, J. R., Plachetzki, D. C., and Kopp, A. (2014). Pan-metazoan phylogeny of the dmrt gene family: A framework for functional studies. Dev. Genes Evol. 224 (3), 175–181. doi:10.1007/s00427-014-0473-0
Xu, X. P., Chen, X. H., Shen, X., Chen, R. Z., Zhu, C., Zhang, Z. H., et al. (2021). Genome-wide identification and characterization of dead-box helicase family associated with early somatic embryogenesis in dimocarpus longan lour. J. Plant Physiol. 258-259, 153364. doi:10.1016/j.jplph.2021.153364
Yamaguchi, A., Lee, K. H., Fujimoto, H., Kadomura, K., Yasumoto, S., and Matsuyama, M. (2006). Expression of the dmrt gene and its roles in early gonadal development of the Japanese pufferfish Takifugu rubripes. Comp. Biochem. Physiol. Part D Genomics Proteomics 1 (1), 59–68. doi:10.1016/j.cbd.2005.08.003
Yan, J., Jing, J., Mu, X., Du, H., Tian, M., Wang, S., et al. (2013). A genetic linkage map of the sea cucumber (Apostichopus japonicus) based on microsatellites and SNPs. Aquaculture 404, 1–7. doi:10.1016/j.aquaculture.2013.04.011
Yan, M., Sui, J., Sheng, W., Shao, M., and Zhang, Z. (2013). Expression pattern of vasa in gonads of sea cucumber Apostichopus japonicus during gametogenesis and reproductive cycle. Gene Expr. Patterns 13 (5-6), 171–176. doi:10.1016/j.gep.2013.03.001
Yang, H., Hamel, J. F., and Mercier, A. (2015). The sea cucumber apostichopus japonicus: History, biology and aquaculture. Amsterdam: Elsevier: Academic Press.
Yang, D., Zhang, Z., Liang, S., Yang, Q., Wang, Y., and Qin, Z. (2017). A novel role of Krüppel-like factor 4 in Zhikong scallop Chlamys farreri during spermatogenesis. Plos one 12 (6), e0180351. doi:10.1371/journal.pone.0180351
Yang, J., Hu, Y., Han, J., Xiao, K., Liu, X., Tan, C., et al. (2020). Genome-wide analysis of the Chinese sturgeon sox gene family: Identification, characterisation and expression profiles of different tissues. J. Fish Biol. 96, 175–184. doi:10.1111/jfb.14199
Yu, J., Zhang, L., Li, Y., Li, R., Zhang, M., Li, W., et al. (2017). Genome-wide identification and expression profiling of the SOX gene family in a bivalve mollusc Patinopecten yessoensis. Gene 627, 530–537. doi:10.1016/j.gene.2017.07.013
Yu, Y. Q., Ma, W. M., Zeng, Q. G., Qian, Y. Q., Yang, J. S., and Yang, W. J. (2014). Molecular cloning and sexually dimorphic expression of two Dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agric. Res. 3 (2), 181–191. doi:10.1007/s40003-014-0106-x
Yu, Z., Zhou, Y., Yang, H., and Hu, C. (2014). Survival, growth, food availability and assimilation efficiency of the sea cucumber Apostichopus japonicus bottom-cultured under a fish farm in southern China. Aquaculture 426, 238–248. doi:10.1016/j.aquaculture.2014.02.013
Zafar, I., Rather, M. A., and Dhandare, B. C. (2019). Genome-wide identification of doublesex and Mab-3-Related transcription factor (dmrt) genes in nile tilapia (Oreochromis niloticus). Biotechnol. Rep. 24, e00398. doi:10.1016/j.btre.2019.e00398
Zhang, X. J., Sun, L. N., Yuan, J. B., Sun, Y. M., Yi, G., Zhang, L. B., et al. (2017). The sea cucumber genome provides insights into morphological evolution and visceral regeneration. Plos Biol. 15 (10), e2003790. doi:10.1371/journal.pbio.2003790
Zhang, J., Han, X., Wang, J., Liu, B. Z., Wei, J. L., Zhang, W. J., et al. (2019). Molecular cloning and sexually dimorphic expression analysis of nanos2 in the sea urchin, Mesocentrotus nudus. Int. J. Mol. Sci. 20 (11), 2705. doi:10.3390/ijms20112705
Keywords: RNAi, sea cucumber, gonad development, DMRT, SoxE
Citation: Liu B-Z, Cong J-J, Su W-Y, Hao Z-L, Sun Z-H and Chang Y-Q (2023) Identification and functional analysis of Dmrt1 gene and the SoxE gene in the sexual development of sea cucumber, Apostichopus japonicus. Front. Genet. 14:1097825. doi: 10.3389/fgene.2023.1097825
Received: 14 November 2022; Accepted: 06 January 2023;
Published: 19 January 2023.
Edited by:
Yuzine Esa, Putra Malaysia University, MalaysiaReviewed by:
Qian Wang, Yellow Sea Fisheries Research Institute (CAFS), ChinaCopyright © 2023 Liu, Cong, Su, Hao, Sun and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Hui Sun, c3VuemhpaHVpQGRsb3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.