- 1Department of Medical Oncology, Qiantang Campus of Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
- 2Department of Medical Oncology, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
- 3Department of Pathology, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
- 4Department of Radiology, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
- 5Key Laboratory of Digital Technology in Medical Diagnostics of Zhejiang Province, Dian Diagnostics Group Co., Ltd., Hangzhou, Zhejiang, China
Background: Cancer of unknown primary (CUP) is a class of metastatic malignant tumors whose primary location cannot be determined. The diagnosis and treatment of CUP are a considerable challenge for clinicians. Herein, we report a CUP case whose corresponding primary tumor sites were successfully identified, and the patient received proper treatment.
Case report: In February 2022, a 74-year-old woman was admitted to the Medical Oncology Department at Sir Run Run Shaw Hospital for new lung and intestinal tumors after more than 9 years of breast cancer surgery. After laparoscopically assisted right hemicolectomy, pathology revealed mucinous adenocarcinoma; the pathological stage was pT2N0M0. Results from needle biopsies of lung masses suggested poorly differentiated cancer, ER (-), PR (-), and HER2 (-), which combined with the clinical history, did not rule out metastatic breast cancer. A surgical pathology sample was needed to determine the origin of the tumor tissue, but the patient’s chest structure showed no indications for surgery. Analysis of the tumor’s traceable gene expression profile prompted breast cancer, and analysis of next-generation amplification sequencing (NGS) did not obtain a potential drug target. We developed a treatment plan based on comprehensive immunohistochemistry, a gene expression profile, and NGS analysis. The treatment plan was formulated using paclitaxel albumin and capecitabine in combination with radiotherapy. The efficacy evaluation was the partial response (PR) after four cycles of chemotherapy and two cycles combined with radiotherapy.
Conclusion: This case highlighted the importance of identifying accurate primary tumor location for patients to benefit from treatment, which will provide a reference for the treatment decisions of CUP tumors in the future.
Introduction
Cancer of unknown primary (CUP) is a malignant tumor whose primary focus cannot be determined after detailed examination and evaluation. It accounts for 1–5% of all malignancies (Fizazi et al., 2015). A small number of CUP patients (15–20%) can be inferred from primary tissue according to clinical and histopathological results, and the prognosis is usually better with corresponding tumor-type-specific treatment (Li et al., 2019; Rassy and Pavlidis, 2020). However, the majority of CUP patients cannot identify the primary tumor. The median survival time of these patients is only 4.5 months, the 1-year survival rate is 20%, and the 5-year survival rate is only 4.7% (Richardson et al., 2015; Pu et al., 2021). The current standard treatment for CUPs is based on the primary site, so they can only receive empirical chemotherapy and have a poor prognosis (Pavlidis, 2007; Daud, 2013; Kim et al., 2018a). In recent years, molecular detection in tumor tissue tracing has shown that gene expression profiling can be used to trace the location of primary tumors by detecting the expression level of specific genes in tumor tissue (Dermawan and Rubin, 2021; Liu et al., 2021; Chen et al., 2022). Here, we report a CUP patient who sank into an unfavorable subset with lung and intestinal tumors. The location of primary tumor tissue was inferred based on molecular expression profile analysis. Then, the treatment plan was formulated using immunohistochemical (IHC) analysis and NGS analysis. The patient obtained clinical benefits from the combination of chemotherapy and radiotherapy.
Case presentation
In February 2022, a 74-year-old woman was presented with a mass shadow in the lower lobe of her left lung during a periodic review after a breast cancer operation. (Figure 1A). This patient underwent a left mastectomy in September 2012 due to a malignant tumor in the left breast in another hospital. Postoperative pathology showed that the malignant tumor in the left breast was an invasive lobular carcinoma. Metastatic cancer was not found in lymph nodes and did not metastasize in pectoralis fibro-fatty tissue. Tumor cells appeared as cords and trabeculae with fibrous tissue hyperplasia (Supplementary Figure S1). A series of IHC results suggested ER (+), PR (-), HER2 (-), Ki-67 (+, 5%), E-cadherin (-), P120 (+), EGFR (-), CK5/6 (-), P63 (-), calponin (-), CK7 (+), CK20 (-), LCA (-), TTF-1 (-), HEP (-), GCDFP-15 (-), PAS (+), and AB (-). The patient was treated with six cycles of anthracycline drugs after surgery, and exemestane was prescribed until December 2021, during which the periodic review showed no abnormalities. The CT scan found a pulmonary nodule in the left lung until February 2022. The abnormal focus of the lung was on the left side of the body, which was the same as the breast lesions 9 years ago. PET-CT showed that the nodules in the left lower lobe of the lung were well demarcated and had morphological rules that were not typical of lung cancer (Figure 1B). On 4 March 2022, the left lower lobe tumor biopsy of the patient’s lung showed poorly differentiated carcinoma (Figure 1C). The combined results of differential IHC detection for lung adenocarcinoma and squamous cell carcinoma suggested TTF-1 (-), napsin A (-), CK7 (+), p40 (-), and p63 (-). The results of IHC detection in the left lower lobe of the lung to analyze whether it originated from breast cancer suggested ER (-), PR (-), HER2 (-), Ki-67 (+, 15%), GCDFP-15 (-), and GATA-3 (-). The results of IHC detection in the left lower lobe of the lung to analyze whether it originated from intestinal cancer suggested CK20 (-), SATB2 (-), and CDX-2 (-). PD-L1 (22C3) showed a TPS<1%. Tumor markers were all in the normal range, including CA125, CEA, SCC, NSE, ProGRP, and CYFRA21-1. Practitioners considered not to exclude metastatic breast cancer through a combination of clinical history, IHC analysis, and imaging manifestations. Furthermore, PET-CT was implemented to understand the patient’s general condition. Results showed that a malignant tumor is considered in the lower lobe of the left lung, and lymph node metastasis should be considered under the left pulmonary hilum (Figure 1B). There were no space-occupied lesions and abnormal metabolism of FDG in the postoperative area of left breast cancer. It was worth noting that PET-CT found a malignant space-occupying lesion in the initial segment of the ascending colon (Figure 2A). The subsequent colonoscopic biopsy revealed mucinous adenocarcinoma. IHC results suggested GATA-3 (-), mammaglobin (-), CDX-2 (+), SATB2 (+, individual cells are positive), GCDFP-15 (-), and HER2 (-). Pathological analysis after laparoscopically assisted right hemicolectomy confirmed mucinous adenocarcinoma of the colon again (Figure 2B). The pathological stage was pT2N0M0, which was different from the pathology of the left inferior pulmonary lobe. At this point, the origin of the malignant lung mass in this patient had not been determined after a pathological assessment of needle biopsy and imaging evaluation. A surgical pathology sample was needed to determine the origin of the tumor tissue, but the patient’s chest structure showed no indications for surgery. We used two simultaneous approaches to identify the source of malignant tumors in the lower lobe of the left lung. The tissue traceability expression profile was initially detected in punctured tissue, and the details are provided in Supplementary Table S1. The results showed that the malignant tumor in the lower lobe of the left lung originated from breast cancer (Figure 3). The maximum tumor tissue similarity score was 100, and the results suggested that lung and colorectal cancer similarity scores were deficient, at only 0.7 and 1.9, respectively. It indicated that the lesion in the lower lobe of the left lung was not a primary pulmonary lesion and was not metastatic from the ascending colon. This was consistent with the pathological analysis results. The left lower lobe nodule was a poorly differentiated carcinoma, and the ascending colon nodule was a mucinous adenocarcinoma. There were no apparent standard features between the two lesions. The tissue traceability expression profile results exhibited essential reference values for subsequent diagnosis and treatment. Then, more than 600 hot spot exon regions and some intron regions of tumorigenesis and developmental genes were analyzed by NGS. DNA was extracted from plasma. The Illumina NextSeq 500 System (Illumina, San Diego, USA) was used for NGS. The average sequencing depth was at least ×1,000. Unfortunately, no tumor-driving gene was found in this test. Only a somatic point mutation FAT1 p.G4570S was obtained, which has not been found in other studies and has not been recorded in COSMIC or other databases. Based on the comprehensive evaluation results, a malignant lung tumor in the left lower lobe was initially diagnosed as stage IV triple-negative cancer. After a multidisciplinary treatment (MDT) discussion, the patient was treated with C1–C4 paclitaxel albumin and capecitabine on 2022-4-7, 2022-4-29, 2022-5-24, and 2022-6-20. The specific regimen was planned with 1,500 mg orally twice daily on days 1–14 and 200 mg intravenous infusion of paclitaxel albumin on days 1 and 8. The radiotherapy plan was added on 22 July 2022, including 10 MV-X SAD 100 DT 214 cGy/F/d in the intensity-modulated field of the chest tumor area (including lesions in the lower left lung and mediastinal metastatic lymph nodes) and 10 MV-X SAD 100 DT 180 cGy/F/d in the intensity-modulated field of the high-risk chest area. She was scheduled to finish radiotherapy 28 times. Combined with the chemotherapy regimen, the specific regimen was provided with 1,500 mg orally twice daily and 190 mg intravenous infusion of paclitaxel albumin on days 1 and 8 (q3w). The longest diameter of the left lower pulmonary nodule narrowed from 29 mm to 14 mm in September 2022. A partial response (PR) was achieved, with lesion diameters decreasing by 51.72% (Figure 4). She is currently in stable condition.
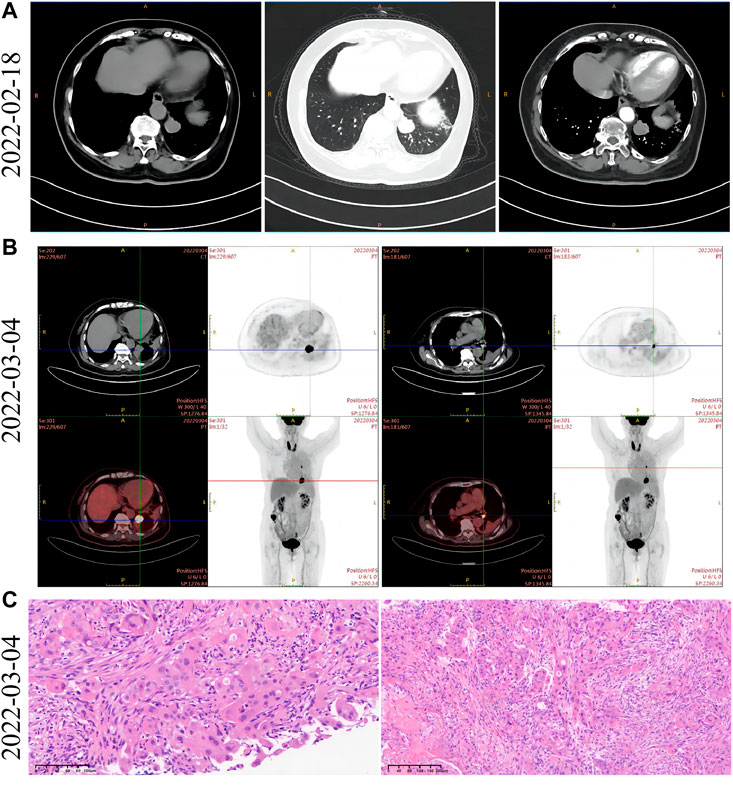
FIGURE 1. Imaging and pathological data of lung lesions at initial diagnosis. (A) Left side: the contrast-enhanced mediastinal chest CT window showed a mass of high-density shadow in the left lower lobe of the lung. Middle: the pulmonary window in the arterial phase of chest contrast-enhanced CT revealed a shallow 25-mm lobular mass in the left lower lobe. The boundary was clear and regular. Right side: the mediastinal arterial window of chest contrast-enhanced CT revealed a mass of high-density shadow in the left lower lobe that exhibited mild-to-moderate progressive enhancement and noticeable marginal enhancement. (B) Left side: PET-CT showed a soft tissue mass in the left lower lobe of the lung. It increased FDG uptake, and the maximum SUV is 15.57. Right side: an enlarged lymph node with increased FDG metabolism is observed below the left pulmonary hilum. The diameter is about 1 cm, and the maximum SUV is 4.38. (C) Pathological analysis of tumor biopsies in the left lower lobe of the lung. The morphology of tumor cells was observed after staining with H&E.

FIGURE 2. Imaging and pathological data of ascending colon lesions. (A) PET-CT of ascending colon lesions. There was more content in the ascending colon, and the local nodular FDG metabolism was increased. SUVmax = 12.61 for early imaging, and SUVmax = 14.11 for delayed imaging. (B) Pathological analysis of tumor biopsy in ascending colon lesions. The morphology of tumor cells was observed after staining with H&E.
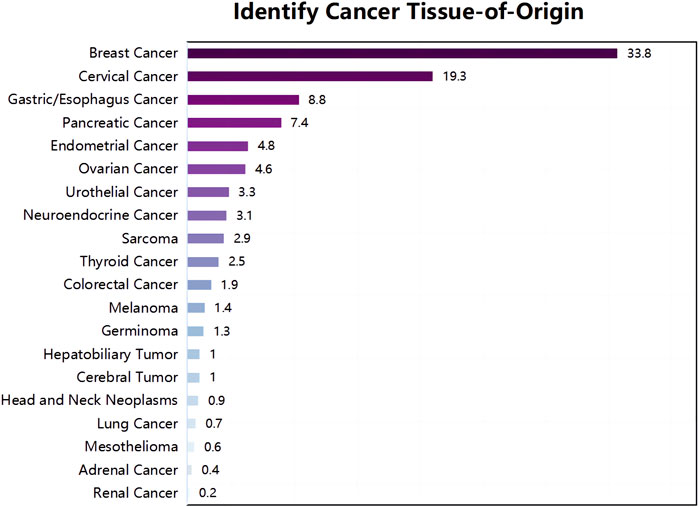
FIGURE 3. Tumor tissue traceability expression assay results. The maximum similarity score for tumor tissue traceability was 100.
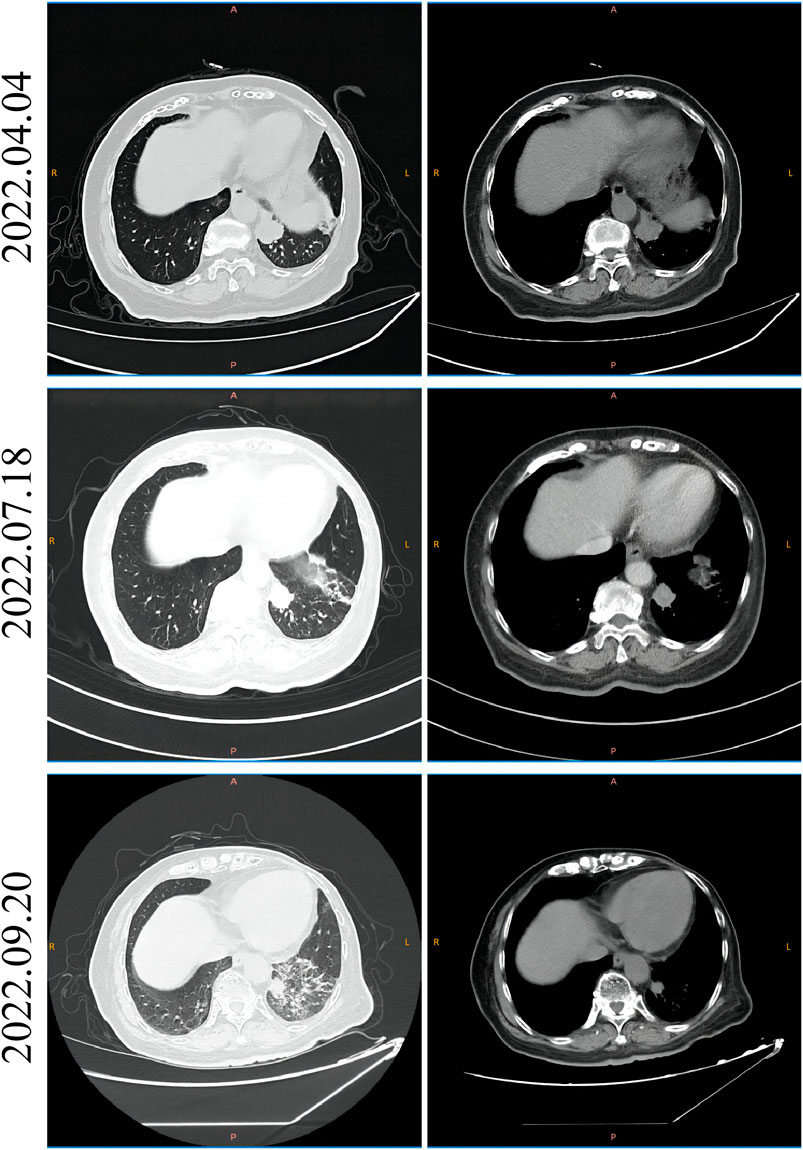
FIGURE 4. CT imaging of the patient before and after the initial chemotherapy and radiotherapy. The left side is the lung CT scan window, and the right side is the mediastinum CT scan window. The CT image of the patient before receiving chemotherapy on 4 April 2022; the CT image of the patient receiving chemotherapy but not radiotherapy on 18 July 2022; and the CT image of the patient receiving chemotherapy and radiotherapy on 20 September 2022.
Discussion
CUP was divided into two subgroups according to their clinical and pathological conditions in traditional assessment (Pavlidis et al., 2003). For the first subgroup, 15–20% of patients showed specific clinical and pathological characteristics, which strongly indicate the origin of tumor tissue (El Rassy et al., 2019; Rassy et al., 2019; Rassy et al., 2020a). This part of CUP has chemosensitive tumors with good prognoses when treated with native treatments (Fizazi et al., 2015). However, 80–85% of patients fall into the second subtype. This subtype cannot identify the primary tumor location and is managed using empirical chemotherapy, which has a poor prognosis (Alvi et al., 2017). It should be noted that there are obvious deficiencies in previous studies comparing the prognosis assessment of site-specific therapy with empirical chemotherapy. These deficiencies include patient accrual problems, study design limitations, heterogeneity among CUP classifiers, and incomparable treatments. Elie et al. recommended two comprehensive clinical trial designs, namely, visionary and pragmatic approaches that can implement the latest diagnostic and therapeutic advances to improve the prognosis of CUP patients.
It is gratifying to note that the subgroup with a poor prognosis can increase survival rates through histological diagnostic tools, site-specific treatment, and new therapeutic exploration (Mikhail et al., 2015). The primary tumor focus is helpful for physicians to design more targeted treatment plans, which are essential to identifying them accurately (Jardim et al., 2015; Schwaederle et al., 2015). With the increasing availability of high-throughput genomic and transcriptomic data, molecular biomarkers in The Cancer Genome Atlas are gradually enriched, including somatic mutation (Fernandez et al., 2012; Rassy et al., 2020b; Chebly et al., 2022), copy number variation (CNV) (Gatalica et al., 2018; Naing et al., 2020), gene expression (Rassy et al., 2021), microRNA expression, and DNA methylation (Bettegowda et al., 2014), which can be used to identify the primary cancer focus. Some studies have shown that the gene expression profile of the metastatic tumor is different from the tissue at the metastatic site and more similar to its primary site (Kim et al., 2018b; Gandara et al., 2018; Kim et al., 2022). It has been suggested that cancer always retains the gene expression characteristics of its tissue origin in tumor occurrence, development, and metastasis. Other studies have found that cancer cells are characterized by a massive overall loss of DNA methylation and that different tumor tissues have different epigenetic signatures depending on the tissue (Fernandez et al., 2012). DNA methylation is an effective biomarker for clinical diagnosis and can distinguish other tissues of origin (31). In addition, Küsters-Vandevelde et al. have found that specific CNVs may be associated with cancer metastasis (Naing et al., 2020). Liu et al. (2021) systematically compared the performance of three biomarkers (DNA methylation, gene expression profile, and somatic mutation) and their combinations in estimating the tissue origin of patients with CUP. The results showed that the accuracy of the gene expression profile was the highest, followed by DNA methylation, and the accuracy of somatic mutation was the worst of the three. However, Elie Rassy et al. (Rassy and Pavlidis, 2020) found that the prediction accuracy of methylation was 78%–87% and the gene expression profile was 54%–98%. More research is needed to explore which method is more accurate in identifying the origin of CUP. Positive hints have also appeared in the exploration of novel CUP therapies. A large study (32) demonstrated that the molecular patterns of BRAF/NRAS alterations were similar between patients with melanoma of unknown primary site and stage-matched melanoma of known prior site. Therapy with RAF/MEK inhibitors may improve survival for MUP. Chromosomal instability (CIN) has been considered a characteristic of some cancer cells but is not frequent in CUP (Rassy et al., 2020b; Chebly et al., 2022). It may provide therapeutic benefit from immune checkpoint inhibitors (ICIs) in patients with CUP. In addition, 28% of patients with CUP displayed one or more predictive biomarkers to ICI, such as PD-L1 expression on ≥5% cancer cells at 22.5% (≥1% at 34%) and lymphocytes at 58.7%, high microsatellite instability (MSI) at 1.8%, and tumor mutational burden (TMB) ≥17 mutations per megabase at 11.8% (Gatalica et al., 2018). A phase II trial, NCT02721732, showed that the clinical benefit rate of pembrolizumab for CUP patients was 54% (Naing et al., 2020).
In this case, the patient was confirmed to have a malignant nodule in the lower lobe of the left lung after a detailed case history, imaging, and cytopathological evaluation. However, its tissue origin could not be determined. Practitioners used lung puncture tissue samples to trace the source of tumor tissue by detecting 90 genes that are explicitly expressed in neoplastic tissue. Results from the gene expression profile showed that the malignant nodule in the left lower lobe of the lung originated from breast cancer but not from the lung where it occurred. At the same time, it also denied that it was a malignant tumor metastasis of the ascending colon. A review of postoperative pathological IHC outcomes in patients was performed 9 years ago, which suggested ER (+), PR (-), and HER2 (-). However, the histopathological indices of the left inferior pulmonary lobe were ER (-), PR (-), and HER2 (-). Through multidisciplinary discussion and literature research, we learned that ER, PR, and HER2 levels in primary lesions differed in metastatic lymph nodes and recurrent metastatic lesions. Changes in ER and PR levels may be related to the age of the initial onset and secondary endocrine therapy. In this case, exemestane was administered after primary lesion surgery until December 2021. It may be one of the underlying causes of triple-negative malignant nodules in the left lower lobe of the lung.
To find a personalized treatment schedule for this case, we conducted a high-depth sequencing analysis of circulating blood-derived tumor DNA (ctDNA). Unfortunately, next-generation sequencing has not detected effective drug targets. A previous study showed that CUP patients with TMB >10 mutations/Mb had a better prognosis trend when receiving ICI treatment. The TMB assessment, in this case, was 0.7 mutations/Mb (Rassy et al., 2021). It should be noted that this patient was bleeding during the second puncture, so further sampling was stopped. The evaluation of the patient’s chest structure revealed that it was not suitable for surgery. The blood sample was taken for testing. ctDNA in the blood was obtained from the release of tumor tissue. If tissue samples cannot be obtained for advanced-stage patients, ctDNA may be used for detection (Bettegowda et al., 2014; Kim et al., 2022). TMB can be accurately and repeatedly measured in plasma, which has been confirmed on the basis of POPLAR (NCT01903993) and OAK (NCT02008227) studies (Gandara et al., 2018). bTMB was positively correlated with the effectiveness of immune checkpoint inhibitors. B-F1RST research proved that patients with bTMB-H (bTMB ≥16) obtained clinical benefits in PFS, ORR, and OS (Kim et al., 2018b; Socinski et al., 2019). However, bTMB is still in the exploration stage. Currently, the tissue specimen is the preferred type for TMB detection. We developed a triple-negative breast cancer treatment plan for this case: paclitaxel albumin combined with capecitabine. It was gratifying that the malignant tumor in the left lower lobe of the lung was in remission under this treatment regimen. Next-generation ctDNA sequencing identified an unreported FAT1 p.G4570S mutation whose clinical significance was unknown. Some studies have suggested that mutations in the FAT1 gene accelerate tumorigenesis and malignant progression and promote the transformation of epithelial cells into mesenchymal cells (38, 39). FAT1 p.G4570S occurred in intrinsically disordered regions (IDRs) that require further research to explore the role in biological functions.
In summary, we present a patient with CUP whose location of tumor origin was successfully identified based on the results of a multiplex gene expression profiling analysis, and the patient benefited from specific treatment according to the tissue source. This case suggests that multiple gene expression profiling may be considered to assist in diagnosing tissue origin in CUP patients when conventional clinical testing methods fail to determine the source of tumor tissue. We believe that accurate diagnosis and specific CUP treatment are essential to improving patient outcomes.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Internal Review Board of Sir Run Run Shaw Hospital.
Author contributions
Conception and design: WDH and HH; Study development and methods: WDH, HH, and NY; Data collection and assembly: HH, QP, JYS, JLY, GXF, and FJT; Manuscript writing: WDH and HH.
Funding
This study was funded by the CSCO-BMS Cancer Immunotherapy Research Foundation (Grant No. Y-BMS2019-098) and the CSCO-Xinda Cancer Immunotherapy Research Foundation (Grant No. Y-XD2019-225).
Conflict of interest
NY was employed by the Company Dian Diagnostics Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1085549/full#supplementary-material
References
Alvi, M. A., Wilson, R. H., and Salto-Tellez, M. (2017). Rare cancers: The greatest inequality in cancer research and oncology treatment. Br. J. Cancer 117 (9), 1255–1257. eng. Epub 2017/09/22in: Pubmed; PMID 28934760. doi:10.1038/bjc.2017.321
Bettegowda, C., Sausen, M., Leary, R. J., Kinde, I., Wang, Y., Agrawal, N., et al. (2014). Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6 (224), 224ra24. 224ra24. eng. Epub 2014/02/21Cited in: Pubmed; PMID 24553385. doi:10.1126/scitranslmed.3007094
Chebly, A., Yammine, T., Boussios, S., Pavlidis, N., and Rassy, E. (2022). Chromosomal instability in cancers of unknown primary. Eur. J. Cancer 172, 323–325. eng. Epub 2022/07/13Cited in: Pubmed; PMID 35820241. doi:10.1016/j.ejca.2022.06.017
Chen, K., Zhang, F., Yu, X., Huang, Z., Gong, L., Xu, Y., et al. (2022). A molecular approach integrating genomic and DNA methylation profiling for tissue of origin identification in lung-specific cancer of unknown primary. J. Transl. Med. 20 (1), 158. eng. Epub 2022/04/07Cited in: Pubmed; PMID 35382836. doi:10.1186/s12967-022-03362-2
Daud, A. I. (2013). Removing the unknown from the carcinoma of unknown primary. J. Clin. Oncol. 31 (2), 174–175. eng. Epub 2012/12/13Cited in: Pubmed; PMID 23233703. doi:10.1200/jco.2012.45.7630
Dermawan, J. K., and Rubin, B. P. (2021). The role of molecular profiling in the diagnosis and management of metastatic undifferentiated cancer of unknown primary(✰): Molecular profiling of metastatic cancer of unknown primary. Seminars diagnostic pathology 38 (6), 193–198. eng. Epub 2020/12/15Cited in: Pubmed; PMID 33309276. doi:10.1053/j.semdp.2020.12.001
El Rassy, E., Kattan, J., and Pavlidis, N. (2019). A new entity of abdominal squamous cell carcinoma of unknown primary. Eur. J. Clin. investigation 49 (7), e13111. eng. Epub 2019/03/26Cited in: Pubmed; PMID 30908618. doi:10.1111/eci.13111
Fernandez, A. F., Assenov, Y., Martin-Subero, J. I., Balint, B., Siebert, R., Taniguchi, H., et al. (2012). A DNA methylation fingerprint of 1628 human samples. Genome Res. 22 (2), 407–419. eng. Epub 2011/05/27in: Pubmed; PMID 21613409. doi:10.1101/gr.119867.110
Fizazi, K., Greco, F. A., Pavlidis, N., Daugaard, G., Oien, K., Pentheroudakis, G., et al. (2015). Cancers of unknown primary site: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. official J. Eur. Soc. Med. Oncol. 26 (5), v133–v138. eng. Epub 2015/09/01Cited in: Pubmed; PMID 26314775. doi:10.1093/annonc/mdv305
Gandara, D. R., Paul, S. M., Kowanetz, M., Schleifman, E., Zou, W., Li, Y., et al. (2018). Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 24 (9), 1441–1448. eng. Epub 2018/08/08Cited in: Pubmed; PMID 30082870. doi:10.1038/s41591-018-0134-3
Gatalica, Z., Xiu, J., Swensen, J., and Vranic, S. (2018). Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur. J. Cancer 94, 179–186. eng. Epub 2018/03/24Cited in: Pubmed; PMID 29571084. doi:10.1016/j.ejca.2018.02.021
Jardim, D. L., Schwaederle, M., Wei, C., Lee, J. J., Hong, D. S., Eggermont, A. M., et al. (2015). Impact of a biomarker-based strategy on oncology drug development: A meta-analysis of clinical trials leading to fda approval. J. Natl. Cancer Inst. 107 (11), djv253. eng. Epub 2015/09/18Cited in: Pubmed; PMID 26378224. doi:10.1093/jnci/djv253
Kim, C. S., Hannouf, M. B., Sarma, S., Rodrigues, G. B., Rogan, P. K., Mahmud, S. M., et al. (2018). Survival outcome differences based on treatments used and knowledge of the primary tumour site for patients with cancer of unknown and known primary in Ontario. Curr. Oncol. Tor. Ont. 25 (5), 307–316. eng. Epub 2018/11/23Cited in: Pubmed; PMID 30464680. doi:10.3747/co.25.4003
Kim, E. S., Velcheti, V., Mekhail, T., Leal, T. A., Dowell, J. E., Tsai, M. L., et al. (2018). Primary efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC). Ann. Oncol. 29 (8), viii744. doi:10.1093/annonc/mdy424.067
Kim, E. S., Velcheti, V., Mekhail, T., Yun, C., Shagan, S. M., Hu, S., et al. (2022). Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: The phase 2 B-F1RST trial. Nat. Med. 28 (5), 939–945. eng. Epub 2022/04/16Cited in: Pubmed; PMID 35422531. doi:10.1038/s41591-022-01754-x
Li, M., Li, H., Hong, G., Tang, Z., Liu, G., Lin, X., et al. (2019). Identifying primary site of lung-limited Cancer of unknown primary based on relative gene expression orderings. BMC Cancer 19 (1), 67. eng. Epub 2019/01/16Cited in: Pubmed; PMID 30642283. doi:10.1186/s12885-019-5274-4
Liu, H., Qiu, C., Wang, B., Bing, P., Tian, G., Zhang, X., et al. (2021). Evaluating DNA methylation, gene expression, somatic mutation, and their combinations in inferring tumor tissue-of-origin. Front. Cell Dev. Biol. 9, 619330. eng. Epub 2021/05/21Cited in: Pubmed; PMID 34012960. doi:10.3389/fcell.2021.619330
Mikhail, S., Lustberg, M. B., Ruppert, A. S., Mortazavi, A., Monk, P., Kleiber, B., et al. (2015). Biomodulation of capecitabine by paclitaxel and carboplatin in advanced solid tumors and adenocarcinoma of unknown primary. Cancer Chemother. Pharmacol. 76 (5), 1005–1012. eng. Epub 2015/09/30Cited in: Pubmed; PMID 26416564. doi:10.1007/s00280-015-2877-6
Naing, A., Meric-Bernstam, F., Stephen, B., Karp, D. D., Habra, M. A., Rodon Ahnert, J., et al. (2020). Phase 2 study of pembrolizumab in patients with advanced rare cancers. J. Immunother. Cancer 8 (1), e000347. doi:10.1136/jitc-2019-000347
Pavlidis, N., Briasoulis, E., Hainsworth, J., and Greco, F. A. (2003). Diagnostic and therapeutic management of cancer of an unknown primary. Eur. J. Cancer 39 (14), 1990–2005. doi:10.1016/s0959-8049(03)00547-1
Pavlidis, N. (2007). Forty years experience of treating cancer of unknown primary. Acta Oncol. Stockh. Swed. 46 (5), 592–601. eng. Epub 2007/06/15Cited in: Pubmed; PMID 17562435. doi:10.1080/02841860701243095
Pu, X., Yang, S., Xu, Y., Chen, B., Wang, Q., Gong, Q., et al. (2021). Case report: Tissue origin identification for cancer of unknown primary: Gene expression profiling approach. Front. Oncol. 11, 702887. eng. Epub 2021/12/04Cited in: Pubmed; PMID 34858803. doi:10.3389/fonc.2021.702887
Rassy, E., Nicolai, P., and Pavlidis, N. (2019). Comprehensive management of HPV-related squamous cell carcinoma of the head and neck of unknown primary. Head. Neck 41 (10), 3700–3711. eng. Epub 2019/07/14Cited in: Pubmed; PMID 31301162. doi:10.1002/hed.25858
Rassy, E., Parent, P., Lefort, F., Boussios, S., Baciarello, G., and Pavlidis, N. (2020). New rising entities in cancer of unknown primary: Is there a real therapeutic benefit? Crit. Rev. oncology/hematology 147, 102882. eng. Epub 2020/02/28Cited in: Pubmed; PMID 32106012. doi:10.1016/j.critrevonc.2020.102882
Rassy, E., Assi, T., and Pavlidis, N. (2020). Exploring the biological hallmarks of cancer of unknown primary: Where do we stand today? Br. J. Cancer 122 (8), 1124–1132. eng. Epub 2020/02/12Cited in: Pubmed; PMID 32042068. doi:10.1038/s41416-019-0723-z
Rassy, E., Boussios, S., and Pavlidis, N. (2021). Genomic correlates of response and resistance to immune checkpoint inhibitors in carcinomas of unknown primary. Eur. J. Clin. investigation 51, e13583. doi:10.1111/eci.13583
Rassy, E., and Pavlidis, N. (20202020). Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat. Rev. Clin. Oncol. 17 (9), 541–554. doi:10.1038/s41571-020-0359-1
Richardson, A., Wagland, R., Foster, R., Symons, J., Davis, C., Boyland, L., et al. (2015). Uncertainty and anxiety in the cancer of unknown primary patient journey: A multiperspective qualitative study. BMJ Support. Palliat. care 5 (4), 366–372. eng. Epub 2014/03/20Cited in: Pubmed; PMID 24644189. doi:10.1136/bmjspcare-2013-000482
Schwaederle, M., Zhao, M., Lee, J. J., Eggermont, A. M., Schilsky, R. L., Mendelsohn, J., et al. (2015). Impact of precision medicine in diverse cancers: A meta-analysis of phase II clinical trials. J. Clin. Oncol. 33 (32), 3817–3825. eng. Epub 2015/08/26Cited in: Pubmed; PMID 26304871. doi:10.1200/jco.2015.61.5997
Socinski, M., Velcheti, V., Mekhail, T., Chae, Y. K., Leal, T. A., Dowell, J. E., et al. (2019). Final efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC). Ann. Oncol. 30, v919–v920. doi:10.1093/annonc/mdz394.081
Keywords: cancer of unknown primary, traceability of tumor tissue, gene expression profile, lung metastasis of breast cancer, case report
Citation: Hu H, Pan Q, Shen J, Yao J, Fu G, Tian F, Yan N and Han W (2023) The diagnosis and treatment for a patient with cancer of unknown primary: A case report. Front. Genet. 14:1085549. doi: 10.3389/fgene.2023.1085549
Received: 31 October 2022; Accepted: 05 January 2023;
Published: 19 January 2023.
Edited by:
Jitian Li, Henan Luoyang Orthopedic Hospital (Henan Provincial Orthopedic Hospital), ChinaReviewed by:
Nicholas Pavlidis, University of Ioannina, GreeceJidong Lang, Qitan Technology Co., Ltd., China
Copyright © 2023 Hu, Pan, Shen, Yao, Fu, Tian, Yan and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weidong Han, aGFud2RAIHpqdS5lZHUuY24=
 Hong Hu
Hong Hu Qin Pan
Qin Pan Jiaying Shen
Jiaying Shen Junlin Yao
Junlin Yao Guoxiang Fu3
Guoxiang Fu3 Na Yan
Na Yan Weidong Han
Weidong Han