- 1Department of Oral and Craniomaxillofacial Surgery, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2National Center for Stomatology, National Clinical Research Center for Oral Diseases, Shanghai Key Laboratory of Stomatology, Shanghai, China
- 3Shanghai Institute of Precision Medicine, Shanghai, China
Craniofacial development requires intricate cooperation between multiple transcription factors and signaling pathways. Six1 is a critical transcription factor regulating craniofacial development. However, the exact function of Six1 during craniofacial development remains elusive. In this study, we investigated the role of Six1 in mandible development using a Six1 knockout mouse model (Six1−/−) and a cranial neural crest-specific, Six1 conditional knockout mouse model (Six1f/f; Wnt1-Cre). The Six1−/− mice exhibited multiple craniofacial deformities, including severe microsomia, high-arched palate, and uvula deformity. Notably, the Six1f/f; Wnt1-Cre mice recapitulate the microsomia phenotype of Six1−/− mice, thus demonstrating that the expression of Six1 in ectomesenchyme is critical for mandible development. We further showed that the knockout of Six1 led to abnormal expression of osteogenic genes within the mandible. Moreover, the knockdown of Six1 in C3H10 T1/2 cells reduced their osteogenic capacity in vitro. Using RNA-seq, we showed that both the loss of Six1 in the E18.5 mandible and Six1 knockdown in C3H10 T1/2 led to the dysregulation of genes involved in embryonic skeletal development. In particular, we showed that Six1 binds to the promoter of Bmp4, Fat4, Fgf18, and Fgfr2, and promotes their transcription. Collectively, our results suggest that Six1 plays a critical role in regulating mandibular skeleton formation during mouse embryogenesis.
Introduction
The craniofacial development of vertebrates is precisely regulated by various genes and signaling pathways, including BMP, FGF, and WNT (Yin et al., 2015; Graf et al., 2016). Most craniofacial tissues are derived from cranial neural crest cells (CNCCs), which arise from the dorsal central nervous system and migrate into the developing craniofacial region (Liao et al., 2022). Within maxillary and mandibular prominences, CNCCs differentiate into ectomesenchymal cells, and the ectomesenchymal cells subsequently differentiate into various cell and tissue types, including the frontonasal skeleton, bone and cartilage of the jaw and middle ear (Liao et al., 2022). In contrast to other mesoderm-derived bones of the skeleton, the mandibular skeleton is generated during mandibular development via an intramembranous process in which ectodermal mesenchymal cells aggregate and then differentiate into bone (Parada and Chai, 2015; Liao et al., 2022).
The intricate regulation of craniofacial development and differentiation requires a number of transcription factors, such as the MSX family, DLX family, and the SIX family transcription factors, among others (Alappat et al., 2003; Takechi et al., 2013). The SIX family is a group of evolutionarily conserved transcription factors, which are expressed in multiple organs of humans, mice, drosophila, and other organisms, and play an essential role in the development of the craniofacial skeleton, kidney, ear, nose, brain, muscle, and gonads (Serikaku and O'Tousa, 1994). The mammalian SIX family consists of six members (SIX1-6). SIX family genetic mutations lead to various deformities, including craniofacial deformities, hearing disorders, visual disturbance, renal hypoplasia, and muscular dysplasia. (Kumar, 2009).
Six1 has been demonstrated to be a crucial member of the SIX family transcription factors in the embryonic development (Wu et al., 2014; Liu et al., 2019). Six1 knockout mice exhibited craniofacial deformity, hypoplastic kidneys (Xu et al., 2003), and severely dysplastic lungs (El-Hashash et al., 2011). Previous studies have shown that Six1 exerts versatile transcription regulatory effects by interacting with different molecular partners. SIX1 can form a complex with EYA1 and activate transcription (Li et al., 2003). Moreover, SIX1 can also form a transcription complex with members of the DACH family and repress the expression of downstream genes (Li et al., 2003). Six1 regulates Fgf10 and Bmp4 expression in the otic vesicle and interacts with Runx1 to regulate the cell fate of the Müllerian duct epithelium (Zheng et al., 2003; Terakawa et al., 2020).
It has been suggested that Six1 participates in the development of the craniofacial skeleton (Tavares et al., 2017). Six1 is widely expressed in craniofacial tissues of different origins, such as ectoderm, mesoderm, and endoderm (Liu et al., 2019). SIX1 mutation causes human branchio-oto-renal syndrome (BOR), characterized by hearing loss, auricular deformities, residual branchial arches, and renal abnormalities (Kumar et al., 2000; Ruf et al., 2004; Feng et al., 2021). However, the mechanisms by which SIX1 regulates craniofacial development, and skeletogenesis remain unclear.
In this study, we generated a Six1 knockout mouse model and conditional deletion of Six1 in cranial neural crest cells to investigate the role of Six1 in ectomesenchymal cells during murine embryonic mandibular development. We found that the mandibles of both Six1−/− and Six1f/f; Wnt1-Cre were significantly shortened, indicating that ectomesenchymal Six1 participates in mandibular skeletal development. Combining RNA-seq and immunofluorescence staining, we demonstrated that mandibular osteogenesis is impaired in E18.5 and E16.5 Six1−/− mice. In particular, mRNA expression levels of several key osteogenesis-related genes, such as Osteopontin (Opn), Osteocalcin (Ocn) and Osterix (Osx), were found to be downregulated. In vitro, the knockdown of Six1 in the mouse embryonic mesenchymal stem cell line C3H10 T1/2 resulted in decreased osteogenic differentiation capacity and dysregulation of ossification-related genes. By performing CUT&Tag, we further demonstrated that Six1 directly binds to the promoters of Bmp4, Fgfr2, Fgf18, and Fat4, all of which are critical genes involved in skeletal formation and regulates their expression (Hung et al., 2016; Crespo-Enriquez et al., 2019; Motch Perrine et al., 2019; Xu et al., 2021). Taken together, our data suggest that Six1 plays a critical role in the regulation of ossification during embryonic mandibular skeletal development and elucidates the potential Six1-dependent gene regulation networks involved in mandibular development.
Materials and methods
Animals
The Six1 knockout homozygous (Six1−/−) and Six1 conditional knockout (Six1f/f) mouse models were generated using the CRISPR/Cas9 system on a C57BL/6J mouse background by GemPharmatech Co., Ltd (Nanjing, China). The mouse strain creation strategy involved the knockout of exon1-2 of the Six1-201 (ENSMUST00000050029.7) transcript region. Wnt1-Cre mice were obtained from the Jackson Laboratory (Bar Harbor, ME, United States). Six1f/f mice were crossed with Six1f/+; Wnt1-cre mice to generate Six1f/f; Wnt1-Cre embryos. (Supplementary Figure 1; Supplementary Table S1).
Embryos were obtained for subsequent experiments at E18.5, E16.5, and E14.5 days. The day of the appearance of a vaginal plug was defined as E0.5, and the embryos were obtained at 12:30 on each day in question. All mice were maintained and used in experiments according to the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
Skeletal preparation
Skin and soft tissue were carefully removed from E18.5 embryos and the embryos were treated in 95% ethanol overnight, followed by staining with Alcian blue for 48 h at 37°C. Embryos were washed twice with 95% ethanol for 2 h each, treated with 1% KOH for 1 h, and stained with Alizarin red for 2 h. The embryonic bone tissue was soaked in a gradient mixture of 1% KOH in glycerol (75%, 50%, 25%) and photographed.
Histology and immunofluorescence, and TUNEL assay
The heads of the embryos were surgically isolated and fixed overnight with 4% PFA at 4°C, followed by gradient dehydration using an ethanol solution, embedded using paraffin. Hematoxylin–Eosin (HE) and Alcian blue staining were performed on 7 µm-thick paraffin sections. Immunofluorescence staining was performed with anti-Osteopontin Polyclonal antibody (22952-1-AP, Proteintech, Rosemont, IL, United States; 1:50), anti-Osterix antibody (ab209484, Abcam, Cambridge, United Kingdom; 1:200), or anti-Ki67 antibody (ab16667, Abcam; 1:100) followed by goat secondary antibody to rabbit IgG(A-11008, Thermo Fisher Scientific, Waltham, MA, United States; 1:500) following a previously described protocol (Ha et al., 2022). TUNEL staining was performed using In Situ Cell Death Detection Kit (11684795910, Roche, Mannheim, Germany) according to the manufacturer’s instructions. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured using an Olympus IX83 inverted microscope (Olympus, Tokyo, Japan).
Cell culture, osteogenic differentiation, alkaline phosphatase (ALP) staining and cell proliferation assay
C3H10 T1/2 cells were purchased from the Cell Bank of the Chinese Academy of Sciences, Shanghai. The cells were maintained at 37°C with 5% CO2, and were cultured in MEM-α containing 10% FBS (10099141C, Gibco™, Thermo Fisher Scientific), 1% penicillin/streptomycin, 1% Non-Essential Amino Acids, and 1% GlutaMAX™ Supplement, and the culture medium was replaced every 2 days. Osteogenic induction medium (MUXMT-90021, Cyagen Biosciences Inc., Guangzhou, China) was used in the process of cell osteogenic differentiation (Ma et al., 2021). C3H10 T1/2 cells were seeded into 6-well plates at 50,000 cells per well and incubated at 37°C, 5% CO2 for 24 h. Then the regular cell culture medium was replaced with osteogenic induction medium. The osteogenic induction medium was changed every 48 h. ALP staining and RNA extraction were performed 7 days after osteogenic induction. The cells were fixed for 30 min with 4% paraformaldehyde, and ALP staining was performed using the BCIP/NBT Alkaline Phosphatase color development kit following the manufacturer’s instructions (C3206, Beyotime Biotechnology, Beijing, China). Cell proliferation was analyzed using the Cell Counting Kit-8 according to the manufacturer’s instructions (C0037, Beyotime Biotechnology). Cells were seeded in a 96-well plate at a density of 1,000 cells per well and incubated at 37°C, 5% with complete MEM-α. At 24, 48, 72, 96, and 120 h, the medium was removed and added CCK-8 solution was added to the medium and incubated for 1 h. Then, the reaction solution was read with a multimode reader (BioTek, Winooski, VT, United States) to obtain the absorbance at 450 nm.
RNA extraction and quantitative RT-PCR
The RNA was extracted using a total RNA extraction kit (LS1040, Promega, Madison, WI, United States). Following the manufacturer’s instructions, 1 µg of total RNA was reverse transcribed into cDNA using Hifair first Strand cDNA Synthesis SuperMix (11141ES10, Yeasen Biotech, Shanghai, China) for RT-qPCR analysis. Quantitative PCR was performed on a Lightcycler 96 (Roche, Basel, Switzerland) with Hieff qPCR SYBR Green Master Mix (No Rox) (11201ES03; Yeasen Biotech). The relative expression was calculated for each gene by the 2−ΔΔCT method, normalized against GAPDH expression, and presented as fold changes relative to the control. The sequences of all the primers used in this study are shown in Supplementary Table S2.
Construction of knockdown short hairpin RNA vectors and cell infection
Short hairpin RNA (shRNA) targeting Six1 (NM_009189.3) was designed with the following sequence: GCTCATGTCCAGCTCAGAAGA. The shRNA was transfected into the pLV-shRNA-EGFP(2A) Puro vector. Six1-shRNA lentiviruses were packaged and amplified by co-transfecting recombinant vector together with pSPAX2 and pMD2G into 293T cells with lipo8000 and culturing for 48 h. Then the cell culture supernatants were collected and concentrated using a Universal Virus Concentration Kit (C2901M, Beyotime Institute of Biotechnology, Jiangsu, China). The virus concentrate was added to C3H10 T1/2 cells at an MOI of 50 with polybrene(12 μg/mL) and cultured for 6 h. Subsequently, the cell medium was changed and cells were cultured for a further 72 h. To obtain stably transfected cells, C3H10 T1/2 cells were cultured in MEM-α supplemented with puromycin dihydrochloride (10 μg/mL, Beyotime Institute of Biotechnology) for 7 days.
RNA sequencing
An RNA sequencing library was prepared using a NEBNext Ultra™ RNA Library Prep Kit for Illumina and was sequenced on an Illumina novaseq6000. Differentially-expressed genes (DEGs) were determined with log2 expression fold change(log2FC) > 1 and a p-value (padj) < 0.05. Gene Ontology (GO) enrichment analysis of DEGs was performed using the clusterProfiler package in R.
Cleavage under targets and tagmentation (CUT&Tag) library preparation
Cleavage under targets and tagmentation (CUT&Tag) libraries were prepared using an In-Situ ChIP Library Prep Kit (TD901, Vazyme Biotech, Nanjing, China). Adherent cultured cells were digested with 0.25% trypsin, and 50,000 C3H10 T1/2 cells per sample were used in two biological replicates. After centrifugation at 600 g for 5 min at room temperature, the cells were washed twice with Wash Buffer. Cells were bound to conA beads and incubated with anti-Six1 antibody (#12891, CST, Danvers, MA, United States; 1:50) overnight at 4°C. The primary antibody was removed, and then a secondary antibody (Goat Anti-Rabbit IgG, Vazyme) was diluted (1:100) in DIG Wash buffer and incubated with cells at room temperature in a shaker for 1 h. Next, cells were incubated with pA-Tn5 transposon complex (0.04 µM) at room temperature for 1 h. DNA was extracted and then purified using Hieff NGS® DNA Selection Beads (12601ES03, Yeasen Biotech). The libraries were sequenced on the Novaseq-150PE platform and 150-bp pair-end were generated. The sequencing depth was 10G base pair raw data.
Statistical analysis
GraphPad Prism v.9.0 for Windows (GraphPad Software, LaJolla, CA, United States) was used for statistical analysis. All numerical data are presented as means ± SD. Independent two-tailed Student’s t-tests were used for comparisons between two groups, and differences were considered statistically significant at a p-value < 0.05.
Results
The Six1 knockout mice exhibited craniofacial deformity
To explore the role of Six1 in craniofacial development, we generated Six1 knockout mice using the CRISPR/Cas9-based approach. To ensure the efficiency of Six1 knockout in Six1−/− mice, we examined the RNA-seq data at E18.5 and verified the Six1 expression level by RT-qPCR (Supplementary Figure S5). By comparing gross images and skeletal staining of Six1+/+ (n = 4) and Six1+/− (n = 4) embryos at E18.5, we found that the heterozygous embryos had no craniofacial deformities and no differences in mandibular length, and that heterozygous mice survived and reproduced normally (Figures 1A–D). Hence, we used Six1+/− to mate with each other to obtain Six1−/− pups, and the Six1−/− birth probability conformed to the Mendelian ratio (14/55). All the Six1−/− mice that died at birth exhibited a wide range of craniofacial deformities, including microsomia, high arched palate, and a small tongue. Morphologic observation and examination of skeletal preparations at E18.5 revealed that the mandible length of Six1−/− mice (n = 3) was significantly shorter than wild-type or heterozygous littermates. Analyses of HE staining of E16.5 embryos revealed that Six1−/− (n = 3) mice exhibited a high palate and small tongue with ankyloglossia (Figure 1B). The volume of tongue muscle of Six1−/− mice was significantly reduced. We also found that mice (1/3) exhibited bifurcated ribs, characterized by abnormal fusion between the upper and lower rib cartilage (Figure 1D). The mandibular length of Six1−/− (n = 3) embryos at E18.5 was significantly shorter than that of Six1+/+(n = 4) (p < 0.0001) (Figures 1C, E). Six1 knockout mice exhibited a stable phenotype of a short jaw with 100% penetrance (14/14). The craniofacial phenotype of Six1 knockout mice was similar to that reported in the literature (Liu et al., 2019), and the skeletal deformities were also as reported in the literature (Li et al., 2003). These results demonstrate that Six1 knockout mice were successfully constructed, and this model is suitable for studying the causes of the short mandible.
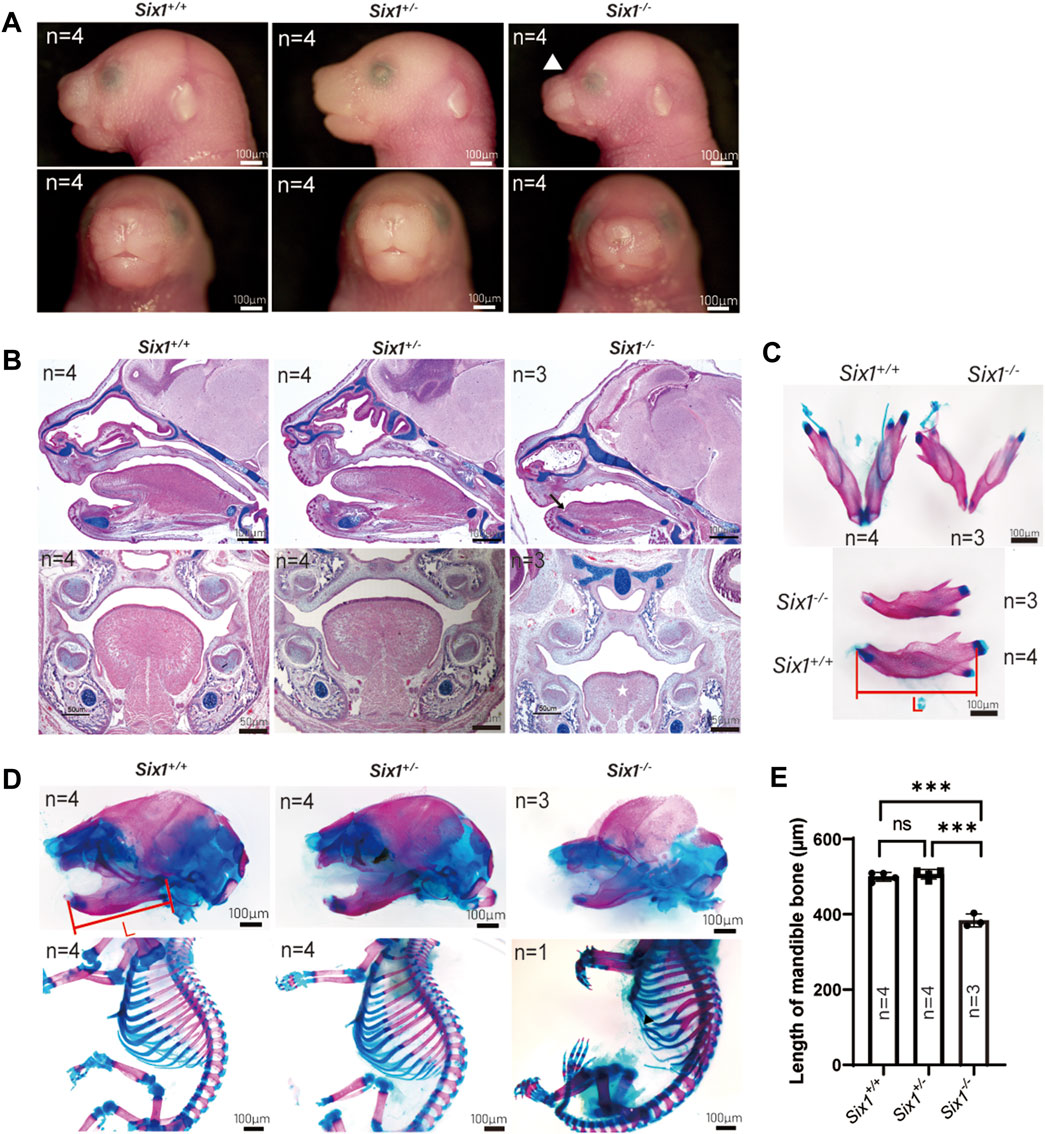
FIGURE 1. The Six1 deletion mice resulted in craniofacial deformity. (A) Lateral view (top) and frontal (bottom) gross morphology of E18.5 heads of the Six1−/−, Six1 ± and Six1+/+ embryos. Six1−/− embryos exhibit short mandible and classical abnormal curve between nose and forehead (white arrowhead). (B) Hematoxylin and eosin (HE) and Alcian blue staining of sagittal sections (top) and frontal (bottom) sections of Six1−/−, Six1+/− and Six1+/+ embryo at E16.5. Six1−/− embryos display a short mandible, ankyloglossia (black arrow), and uvula deformity (white star). (C) Skeletal staining of E18.5 mandible of the Six1−/− and Six1+/+ embryo. Six1−/− mice exhibit a shortened mandible. L: length of mandible. (D) Skeletal alizarin red and Alcian blue staining of E18.5 heads (top) and body (bottom) of the Six1−/−, Six1+/− and Six1+/+ embryo. The black arrowhead points to bifurcated ribs. (E) Quantification of the mandibular length from Six1−/−, Six1+/− and Six1+/+ embryos at E18.5.
Conditional knockout of Six1 in cranial neural crest cells caused microsomia and cleft palate
The mandible is derived from neural crest cell-derived tissues (Parada and Chai, 2015). To explicitly assess whether the craniofacial defects were caused by the loss of Six1 function in CNCC-derived ectomesenchyme, we generated Six1f/f mice and crossed them with Wnt1-Cre mice to conditionally knockout Six1 in CNCCs (Six1f/f; Wnt1-Cre). Wnt1-Cre pups were born in accordance with the Mendelian ratio. However, the majority of Six1f/f; Wnt1-Cre pups died at birth. By morphological analysis and examination of skeletal preparations at E18.5, we found that all Six1f/f; Wnt1-Cre (n = 3) pups exhibited microsomia compared with control littermates, and the phenotype was similar to that of Six1 knockout mice (Figure 2A).
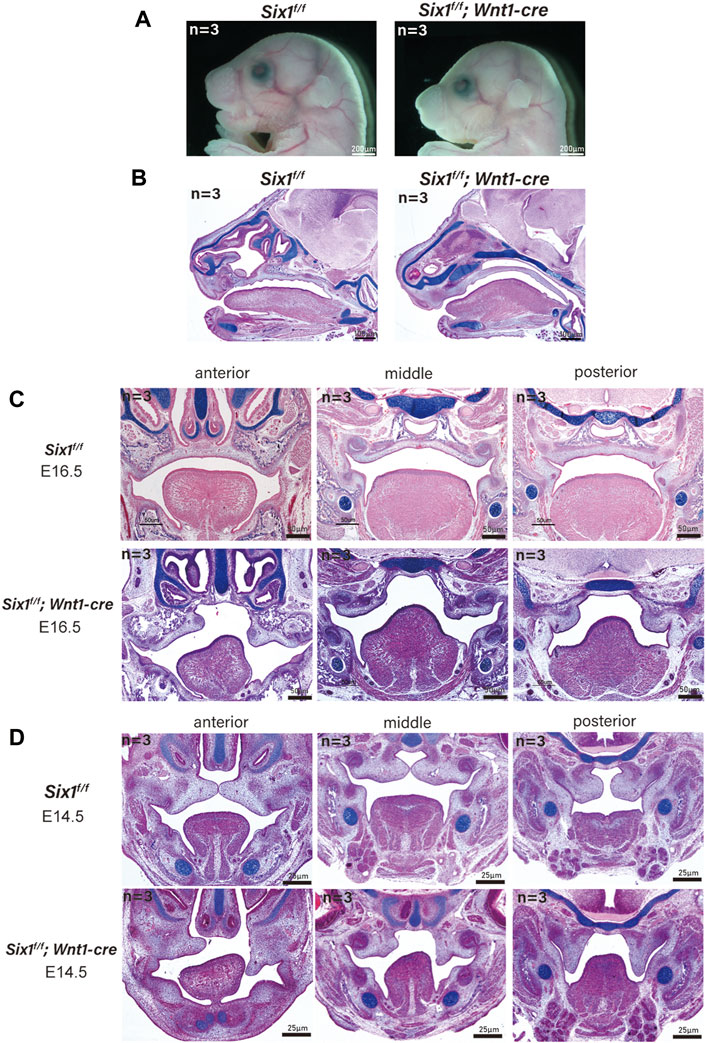
FIGURE 2. The conditional knockout of Six1 in cranial neural crest cells (CNCCs) results in microsomia. (A) Gross morphology of E18.5 heads of the Six1f/f; Wnt1-Cre and Six1f/f embryos. (B) HE and Alcian blue staining of sagittal sections showing the morphology of mandible of the Six1f/f; Wnt1-Cre and Six1f/f embryos at E18.5. Six1f/f; Wnt1-Cre embryo shows a short mandible. (C, D) HE and Alcian blue staining of frontal sections of the Six1f/f; Wnt1-Cre and Six1f/f embryos at E16.5 (C) and E14.5 (D). Six1f/f; Wnt1-Cre embryo shows cleft palate at E16.5 and E14.5.
Interestingly, a new phenotype, cleft palate (70%, 7/10) with ankyloglossia, was found in Six1f/f; Wnt1-Cre mice without atrophy of tongue muscle. We further studied the Six1f/f; Wnt1-Cre mice at E16.5 (n = 3) and E14.5 (n = 3), and found that the tongue muscle occupied the development space of the palate at E14.5, which prevented palatal lifting and led to the development of cleft palate (Figures 2C, D). Tongue connective tissue is derived from CNCCs, whereas the skeletal muscles originate from the myoblasts (Noden and Francis-West, 2006). The reduction in oral volume was due to the lack of mandibular development, while the unaffected tongue muscle volume resulted in an increased tongue muscle height. Therefore, the cleft palate phenotype of conditional knockout Six1 mice might be a secondary cleft palate. These data indicate that Six1 plays a crucial role in the growth and differentiation of CNCC-derived mesenchyme during craniofacial development.
Six1 knockout resulted in decreased mandibular bone formation and altered gene expression in mice
We reasoned that Six1 deletion might disrupt the complex gene expression pattern during craniofacial development. To reveal the key genes regulated by the transcription factor Six1 during mandibular development, we surgically isolated mandibular skeletal tissues and surrounding soft tissues from E18.5 Six1−/− or littermate control wild-type Six1+/+ mice and performed bulk RNA-seq on two independent biological replicates for each genotype. Analysis of the RNA-seq data revealed that the Six1 transcripts were completely absent in Six1−/−. Comparing the results of Six1−/− and Six1+/+ mice revealed that 196 genes exhibited significant expression changes (log2FC > 1, padj < 0.05). Among these, 172 genes were downregulated, and 24 genes were upregulated (Figure 3A; Supplementary Table S1). Correlation analysis of RNA-seq showed that Six1−/− and Six1+/+ were significantly different (Supplementary Figure S2). The uniquely mapped reads were all greater than 85%, indicating high-quality sequencing data (Supplementary Table S3). Notably, Six1−/− showed significantly downregulated expression of osteogenic and mineralization genes at E18.5, including Opn, Ocn, and Osx (Figure 3B). GO enrichment analysis of downregulated DEGs showed that multiple development-related biological processes were impacted, and the DEGs were significantly enriched in “ossification”, “biomineralization”, and “biomineral tissue development” (Figure 3C; Supplementary Table S2). Using immunofluorescence staining, we further verified that the level of Opn in the Six1−/− mandible was significantly lower than that in heterozygous littermates at E16.5 and E18.5 (Figure 3D). We also found a moderate downregulation in the mandibular region of Six1−/− mice by Osx immunofluorescence staining, which was consistent with the RT-qPCR results (Supplementary Figure S3). Six1−/− knockout mice had no significant effect on the proliferation and apoptosis of the mandible at E16.5 (Supplementary Figure S4). Collectively, these data suggest that the knockout of Six1 impaired mandibular bone formation by regulating the expression of critical genes involved in osteogenesis.
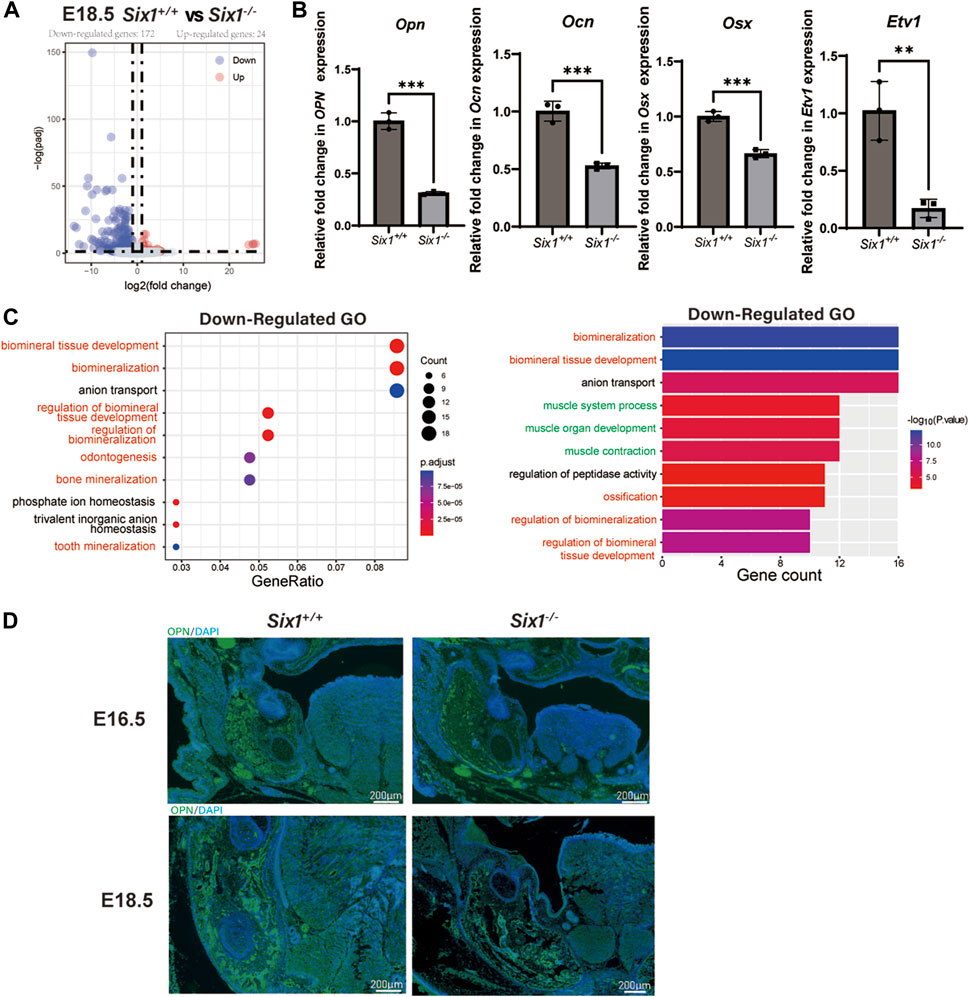
FIGURE 3. RNA-seq of the mandibular tissue from E18.5 Six1−/− and Six1+/+ embryos. (A) Volcano plots show differentially expressed genes between Six1−/− and Six1+/+ mandibular samples. (B) RT-qPCR analysis of Opn, Ocn, Osx and Etv1 in Six1−/− and Six1+/+ mandibular tissues at E18.5. (C) GO enrichment analysis of genes significantly downregulated in Six1−/− mandible. (D) Immunofluorescence staining of OPN in mandible of Six1−/− and Six1+/+ embryos at E16.5 and E18.5.
Interestingly, we also found that genes related to muscle development were significantly downregulated (Figure 3C). Observing the downregulated GO term “muscle organ development” revealed that their enriched genes include Etv1(Tenney et al., 2019), Tcap (Markert et al., 2010), Lbx1(Wang et al., 2022), Actn3 (Nicot et al., 2021), and Fos (Almada et al., 2021), which could explain the uvula deformity observed in Six1−/− mice. RT-qPCR showed that Etv1 expression was significantly reduced in the mandibular tissues of Six1−/− mice (Figure 3B). These results suggest that the craniofacial defects observed in Six1−/− mice result from profound dysregulation of genes related to skeletal and muscle development.
Six1 knockdown decreased the osteogenic differentiation capacity of C3H10 T1/2 cells
To further explore the role of Six1 during mandibular osteogenesis, we performed osteogenic induction assay on the mouse embryonic mesenchymal stem cell line (C3H10 T1/2) to investigate the potential mechanisms in vitro. By performing RT-qPCR, we showed that the expression of Six1 could be readily detected in C3H10 T1/2 cells (Figure 4A). We then performed Six1 knockdown by infecting C3H10 T1/2 cells with lentivirus expressing an shRNA specifically targeting Six1, and verified that the Six1 mRNA was markedly depleted in Six1 knockdown cells (p < 0.0001). RT-qPCR analysis showed that several critical osteogenic genes, including Osx, Runx2, Alp, and Dlk1, were downregulated in Six1 knockdown cells (Figure 4B). We further compared the osteogenic differentiation capacity of control and Six1 knockdown C3H10 T1/2 cells after osteogenic induction for 7 days by quantifying alkaline phosphatase (ALP) staining as well as measuring the mRNA levels of Alp, Osx, Opn, and Ocn by RT-qPCR (Figure 4C). Six1 knockdown C3H10 T1/2 cells exhibited lower ALP activity and downregulation of Osx, Opn, and Ocn expression. The proliferation activity of Six1 knockdown C3H10 T1/2 cells was inhibited (Supplementary Figure S4). These results indicate that Six1 knockdown leads to the decline of osteogenic marker genes expression and reduced osteogenic differentiation in osteogenesis.
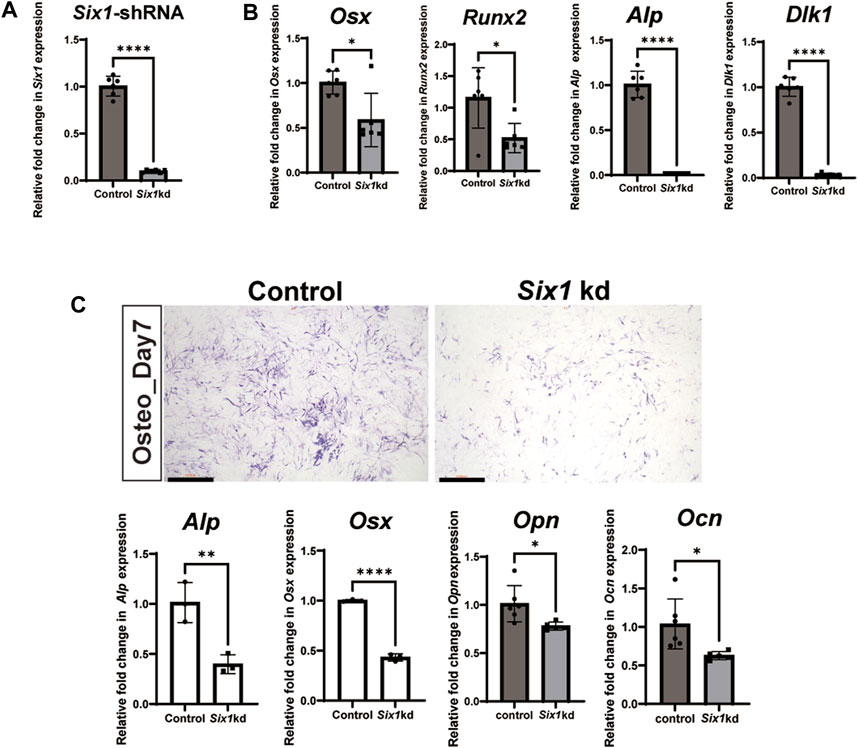
FIGURE 4. The effect of Six1 on the osteogenic differentiation of C3H10 T1/2 cells. (A) RT-qPCR analysis of C3H10 T1/2 cells transfected with negative control (Control) or Six1 knockdown cells (Six1kd) (n = 6). (B) RT-qPCR analysis of Osx, Runx2, Alp, and Dlk1 in control and Six1 knockdown cells (Six1kd). (C) Alkaline phosphatase (ALP) staining of negative control C3H10 T1/2 cells (control) and Six1kd cells after osteogenic induction for 7 days. RT-qPCR analysis of Alp, Osx, Opn, and Ocn after osteogenic induction for 7 days of control and Six1kd cells. Scale bar in C, 1000 µm.
Six1 promotes osteogenic function by regulating multiple osteogenesis-related genes
To investigate the underlying mechanism by which Six1 regulates osteogenic differentiation of C3H10 T1/2 cells, we analyzed the transcriptional effect of Six1 knockdown on C3H10 T1/2 cells by performing RNA sequencing on three biological replicates of control and Six1 knockdown cells. The knockdown and control groups showed a more significant correlation with each other, indicating good quality and repeatability of the RNA sequencing dataset (Figure 5B, Supplementary Figure S2). Analysis of the DEGs (log2FC > 1, padj<0.05) revealed that 662 genes were downregulated and 660 genes were upregulated in Six1 knockdown cells compared with that in control cells (Figure 5A; Supplementary Table S3). GO analysis of the downregulated DEGs showed that the knockdown of Six1 suppressed osteogenic differentiation through the regulation of biological processes associated with “ossification” and “muscle tissue development” (Figure 5C; Supplementary Table S4). We further validated the mRNA expression of several osteogenic differentiation related genes in Six1 knockdown C3H10 T1/2 cells. Consistent with the RNA-Seq results, the mRNA expression of Bmp4, Fat4, Fgf18, Fgfr2, and Runx1 significantly decreased (Figure 5D).
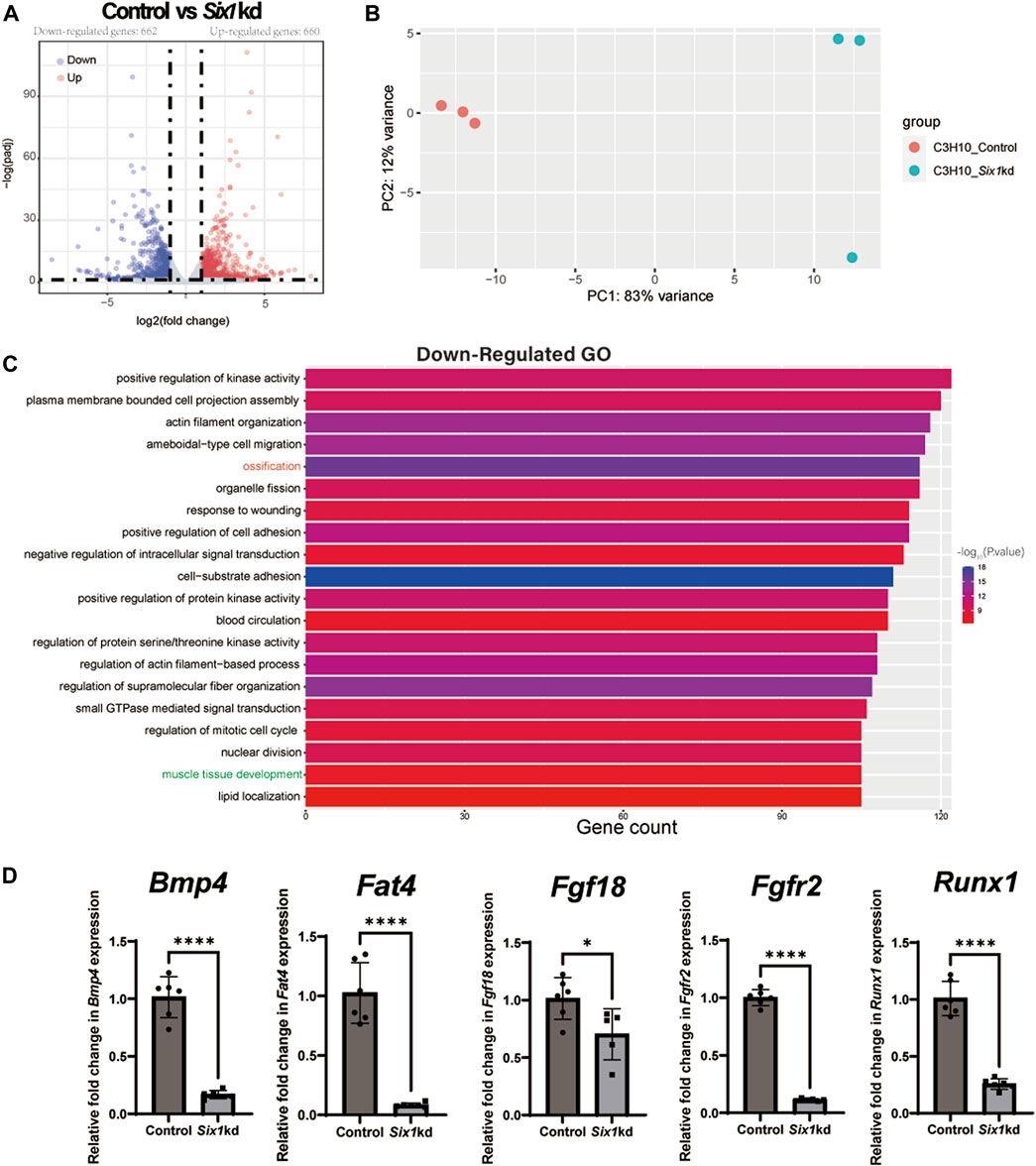
FIGURE 5. RNA-seq of C3H10 T1/2 cells transfected with negative control (Control) or Six1-shRNAs (Six1kd) lentivirus (n = 3). (A) Volcano plots for all the genes of the control and Six1kd groups. Dots on both sides indicate up and downregulated differentially expressed genes (DEGs; p-adj < 0.05). (B) PCA plot showing the correlation between RNA-seq replicates. (C) The terms associated with biological processes (p-adj < 0.05) involving the downregulated genes in the Six1kd group. (D) RT-qPCR analysis of Bmp4, Fat4, Fgf18, Fgfr2, and Runx1 in control and Six1kd cells.
SIX1 directly binds to the promoters of Bmp4, Fgfr2, Fgf18, and Fat4 and regulates their expression
To further explore the mechanism by which Six1 regulates osteogenesis, we examined the genome-wide occupancy of Six1 in C3H10 T1/2 cells by performing CUT&Tag. The IDR consistency test was performed on the two sets of CUT&Tag data, and a total of 19,728 peaks were obtained (Figure 6A; Supplementary Table S5). Among the Six1 peaks, 40.26% were located in the promoter region (≤ 1 kb from the TSS), while 24.87% were located in the distal intergenic region (Figure 6B). These CUT&Tag peaks were annotated to the 10,788 closest genes. In addition, CUT&Tag assay showed that Six1 directly regulated the promoters of Bmp4, Fgfr2, Fgf18, and Fat4, all of which have been reported to play important roles in osteogenesis and were downregulated in Six1 knockdown C3H10 T1/2 cells (Figure 6C). Importantly, nearly 3/4 (2,157/3,027) of the DEGs from RNA-seq were associated with the Six1 peaks (Figure 6D). GO enrichment analysis of these 2,157 Six1-bound DEGs again showed that the ossification function was significantly enriched (Figure 6E). Six1 was also showed to bind to the promoter of Etv1, a gene involved in muscle development (Tenney et al., 2019). Taken together, our data strongly suggest that Six1 regulates the expression of a group of genes involved in bone and muscle development by binding to their promoters or cis-regulatory regions, thereby influencing craniofacial development and morphogenesis.
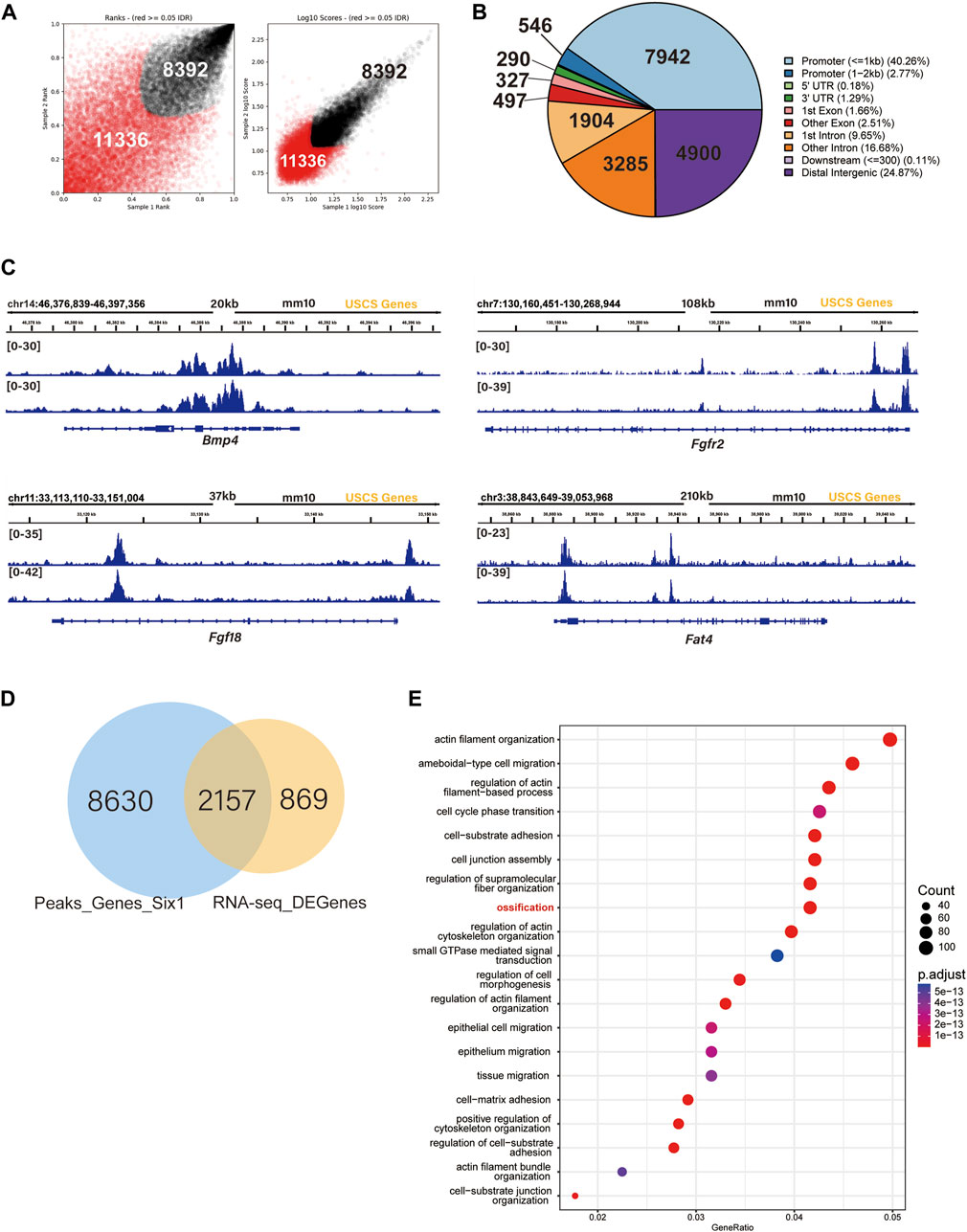
FIGURE 6. SIX1 directly regulates the promoter of osteogenic differentiation-related genes. (A) IDR tests the peaks of two biological replication. (B) Genomic distribution of Six1-enriched regions. (C) Six1 directly binds the promoter of Bmp4, Fgfr2, Fgf18, and Fat4. (D) A Venn diagram indicating overlap of Six1-binding genes and RNA-seq DEGs. (E) GO enrichment analysis of shared genes between Six1-binding genes and RNA-seq DEGs.
Discussion
Six1 plays an important role in embryonic development and is one of the pathogenic genes of human Branchio-oto-renal syndrome (BOR) (Shah et al., 2020). Children with BOR show hearing loss, renal abnormalities, and microsomia(Kochhar et al., 2007). Six1 is widely expressed in the mesenchymal and sensory epithelium of the craniofacial region (Liu et al., 2019; Li et al., 2020). Studies have revealed that Six1 regulates auditory sensory epithelial differentiation, and participates in ear development (Li et al., 2020). For craniofacial development, Six1-null mice exhibit abnormal craniofacial skeletal development, including microsomia and the formation of a novel bone in the zygomatic arch (Tavares et al., 2017; Liu et al., 2019). However, the mechanisms of Six1 during mandibular development remain unclear.
Tavares et al. found that Six1−/− mice upregulated Edn1 signaling in the first and second branchial epithelium, while Six1 was expressed in the adjacent mesenchymal region, suggesting that Six1 may participate in craniofacial development through epithelial-mesenchymal interaction (Tavares et al., 2017). We demonstrated that the conditional knockout of Six1 in mesenchyme largely phenocopied the underdevelopment of the mandible observed in Six1−/− mice, thus demonstrating that Six1 regulates development of the mandible in the ectodermal mesenchyme. Interestingly, Six1f/f; Wnt1-Cre mice showed normal tongue muscle but the cleft palate, a more severe craniofacial deformity. Tongue muscle originates from mesodermal myoblasts, and CNCC-derived mesenchyme in tongue development acts as a scaffold for the organization of migrating myoblasts into the myogenic core (Parada and Chai, 2015). Hence, Wnt1-cre does not knockout Six1 in the tongue muscle, but specifically knockout Six1 in the mandible. Six1f/f; Wnt1-Cre mice exhibited no tongue abnormalities, but showed a lack of Six1 expression in the mandible, resulting in reduced oral volume. We surmise that when the palate begins to fuse at E14.5, the insufficient oral volume in Six1f/f; Wnt1-Cre mice may cause the tongue to occupy the palatal space, thereby affecting the palatal lift and eventually leading to secondary cleft palate.
We found that the expression of osteogenesis-related genes, such as Opn, Ocn and Osx, was significantly downregulated in the mandible of Six1−/− mice at E18.5, suggesting that Six1 may regulate multiple osteogenesis-related genes. It was previously reported that Six1−/− mice showed increased Osx expression in the maxillary and hinge region, and zygomatic process hyperplasia which developed into a thicker rod-shaped bone (Tavares et al., 2017). However, in our study, Six1−/− mice showed reduced Osx expression in the mandible and defects in mandibular osteogenesis. Six1 does not affect the proliferation and apoptosis of mandibular development at the late stages of embryonic development. We propose that Six1 regulates different signaling pathways in the maxilla and mandible, thus producing different biological effects. More studies are needed further to explore the mechanism of Six1 during craniofacial skeletal development.
Our analyses of C3H10 T1/2 cells and mandibular tissue RNA-seq indicate that Six1 regulates the expression of multiple osteogenesis-related genes. The spatiotemporal expression of Bmp4 highly coincides with that of Six1, and it directly regulates the expression of Msx1 and other genes in the BMP family, and plays an important role in the process of mandibular osteogenesis(Xu et al., 2021). Bmp4f/f; Wnt1-Cre mutant pups exhibited short mandible (Xu et al., 2021). Similar phenotypes were observed in Fgf18−/− embryos (Hung et al., 2016). In addition, mice with deletion of Fgf18 in neural crest cells also exhibited a shortened mandible, suggesting that Six1 and Fgf18 in neural crest mesenchymal cells may be jointly involved in mandibular osteogenesis (Yue et al., 2021). Low expression of Fgfr2 is also closely related to cells’ decreased osteogenic ability (Jiang et al., 2019). The Dchs1-Fat4 signaling pathway is involved in the process of osteoblast differentiation in the mouse mandible and skull and plays a positive role in early Runx2 progenitors (Mao et al., 2016; Crespo-Enriquez et al., 2019). Our data suggest that Six1 regulating mandible development at least in part through regulating downstream genes Fgfr2, Fgf18, Bmp4, and Fat4. Future in vivo studies will shed more light on how Six1 coordinates the spatiotemporal expression of these genes to achieve proper craniofacial skeletal formation.
CUT&Tag assay showed that nearly half of the Six1 binding sites were located near the promoter of the downstream gene. Our results demonstrated that the changes in gene expression induced by Six1 knockdown were largely due to the direct regulation of Six1 on its downstream genes. For example, Six1 directly binds to the promoters of Fgfr2, Fgf18, Bmp4, and Fat4 and regulates their transcription. Interestingly, our results also showed that a significant fraction of Six1 peaks are located in the intergenic regions, which likely correspond to cis-regulatory elements such as enhancers. Increasing evidence suggests that the enhancers play critical roles in orchestrating the precise gene expression patterns during craniofacial development (Attanasio et al., 2013). Future investigation on these Six1-bound enhancers may open new avenues for studying the functions of Six1 in craniofacial development and abnormality.
Conclusion
In conclusion, our findings suggest that the transcription factor Six1 is critical for mandible development. Our Six1 knockout and conditional knockout mouse models provide valuable animal models for future studies of skeletal development during craniofacial development. By integrating RNA-Seq and CUT&Tag, we identified potential target genes of Six1 that are involved in osteogenic differentiation. Future studies building on these findings will further elucidate the mechanisms by which Six1 regulates mandibular osteogenesis during embryonic development.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), accession number GSE216761.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee approval (IACUC) of the Shanghai Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
Author contributions
SL and ZL conceived, designed, and performed the experiments, QB and XW designed the experiment and edited the manuscript. All authors agreed to be accountable for the content of this work.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82071096, No. 82001027, No. 31970585, and No. 32170544), the Rare Disease Registration Platform of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (JYHJB05), Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZLCX20211700); Opening Research fund from Shanghai Key Laboratory of Stomatology, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine (No.2022SKLS-KFKT007), Shanghai Clinical Research Center for Oral Diseases (19MC1910600), Shanghai Municipal Key Clinical Specialty (shslczdzk01601), Shanghai’s Top Priority Research Center (2022ZZ01017), and CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-037).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1082911/full#supplementary-material
References
Alappat, S., Zhang, Z. Y., and Chen, Y. P. (2003). Msx homeobox gene family and craniofacial development. Cell Res. 13 (6), 429–442. doi:10.1038/sj.cr.7290185
Almada, A. E., Horwitz, N., Price, F. D., Gonzalez, A. E., Ko, M., Bolukbasi, O. V., et al. (2021). FOS licenses early events in stem cell activation driving skeletal muscle regeneration. Cell Rep. 34 (4), 108656. doi:10.1016/j.celrep.2020.108656
Attanasio, C., Nord, A. S., Zhu, Y., Blow, M. J., Li, Z., Liberton, D. K., et al. (2013). Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342 (6157), 1241006. doi:10.1126/science.1241006
Crespo-Enriquez, I., Hodgson, T., Zakaria, S., Cadoni, E., Shah, M., Allen, S., et al. (2019). Dchs1-Fat4 regulation of osteogenic differentiation in mouse. Development 146 (14), dev176776. doi:10.1242/dev.176776
El-Hashash, A. H., Al Alam, D., Turcatel, G., Rogers, O., Li, X., Bellusci, S., et al. (2011). Six1 transcription factor is critical for coordination of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev. Biol. 353 (2), 242–258. doi:10.1016/j.ydbio.2011.02.031
Feng, H., Xu, H., Chen, B., Sun, S., Zhai, R., Zeng, B., et al. (2021). Genetic and phenotypic variability in Chinese patients with branchio-oto-renal or branchio-oto syndrome. Front. Genet. 12, 765433. doi:10.3389/fgene.2021.765433
Graf, D., Malik, Z., Hayano, S., and Mishina, Y. (2016). Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 27, 129–139. doi:10.1016/j.cytogfr.2015.11.004
Ha, N., Sun, J., Bian, Q., Wu, D., and Wang, X. (2022). Hdac4 regulates the proliferation of neural crest-derived osteoblasts during murine craniofacial development. Front. Physiol. 13, 819619. doi:10.3389/fphys.2022.819619
Hung, I. H., Schoenwolf, G. C., Lewandoski, M., and Ornitz, D. M. (2016). A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Dev. Biol. 411 (1), 72–84. doi:10.1016/j.ydbio.2016.01.008
Jiang, Q., Mei, L., Zou, Y., Ding, Q., Cannon, R. D., Chen, H., et al. (2019). Genetic polymorphisms in FGFR2 underlie skeletal malocclusion. J. Dent. Res. 98 (12), 1340–1347. doi:10.1177/0022034519872951
Kochhar, A., Fischer, S. M., Kimberling, W. J., and Smith, R. J. (2007). Branchio-oto-renal syndrome. Am. J. Med. Genet. A 143a (14), 1671–1678. doi:10.1002/ajmg.a.31561
Kumar, S., Deffenbacher, K., Marres, H. A., Cremers, C. W., and Kimberling, W. J. (2000). Genomewide search and genetic localization of a second gene associated with autosomal dominant branchio-oto-renal syndrome: clinical and genetic implications. Am. J. Hum. Genet. 66 (5), 1715–1720. doi:10.1086/302890
Kumar, J. P. (2009). The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol. Life Sci. 66 (4), 565–583. doi:10.1007/s00018-008-8335-4
Li, X., Oghi, K. A., Zhang, J., Krones, A., Bush, K. T., Glass, C. K., et al. (2003). Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426 (6964), 247–254. doi:10.1038/nature02083
Li, J., Zhang, T., Ramakrishnan, A., Fritzsch, B., Xu, J., Wong, E. Y. M., et al. (2020). Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 48 (6), 2880–2896. doi:10.1093/nar/gkaa012
Liao, J., Huang, Y., Wang, Q., Chen, S., Zhang, C., Wang, D., et al. (2022). Gene regulatory network from cranial neural crest cells to osteoblast differentiation and calvarial bone development. Cell Mol. Life Sci. 79 (3), 158. doi:10.1007/s00018-022-04208-2
Liu, Z., Li, C., Xu, J., Lan, Y., Liu, H., Li, X., et al. (2019). Crucial and overlapping roles of Six1 and Six2 in craniofacial development. J. Dent. Res. 98 (5), 572–579. doi:10.1177/0022034519835204
Ma, L., Wang, X., Zhou, Y., Ji, X., Cheng, S., Bian, D., et al. (2021). Biomimetic Ti-6Al-4V alloy/gelatin methacrylate hybrid scaffold with enhanced osteogenic and angiogenic capabilities for large bone defect restoration. Bioact. Mater 6 (10), 3437–3448. doi:10.1016/j.bioactmat.2021.03.010
Mao, Y., Kuta, A., Crespo-Enriquez, I., Whiting, D., Martin, T., Mulvaney, J., et al. (2016). Dchs1-Fat4 regulation of polarized cell behaviours during skeletal morphogenesis. Nat. Commun. 7, 11469. doi:10.1038/ncomms11469
Markert, C. D., Meaney, M. P., Voelker, K. A., Grange, R. W., Dalley, H. W., Cann, J. K., et al. (2010). Functional muscle analysis of the Tcap knockout mouse. Hum. Mol. Genet. 19 (11), 2268–2283. doi:10.1093/hmg/ddq105
Motch Perrine, S. M., Wu, M., Stephens, N. B., Kriti, D., van Bakel, H., Jabs, E. W., et al. (2019). Mandibular dysmorphology due to abnormal embryonic osteogenesis in FGFR2-related craniosynostosis mice. Dis. Model Mech. 12 (5), dmm038513. doi:10.1242/dmm.038513
Nicot, R., Raoul, G., Vieira, A. R., Ferri, J., and Sciote, J. J. (2021). ACTN3 genotype influences masseter muscle characteristics and self-reported bruxism. Oral Dis. 29, 232–244. doi:10.1111/odi.14075
Noden, D. M., and Francis-West, P. (2006). The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 235 (5), 1194–1218. doi:10.1002/dvdy.20697
Parada, C., and Chai, Y. (2015). Mandible and tongue development. Curr. Top. Dev. Biol. 115, 31–58. doi:10.1016/bs.ctdb.2015.07.023
Ruf, R. G., Xu, P. X., Silvius, D., Otto, E. A., Beekmann, F., Muerb, U. T., et al. (2004). SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. U. S. A. 101 (21), 8090–8095. doi:10.1073/pnas.0308475101
Serikaku, M. A., and O'Tousa, J. E. (1994). Sine oculis is a homeobox gene required for Drosophila visual system development. Genetics 138 (4), 1137–1150. doi:10.1093/genetics/138.4.1137
Shah, A. M., Krohn, P., Baxi, A. B., Tavares, A. L. P., Sullivan, C. H., Chillakuru, Y. R., et al. (2020). Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis. Model Mech. 13 (3), dmm043489. doi:10.1242/dmm.043489
Takechi, M., Adachi, N., Hirai, T., Kuratani, S., and Kuraku, S. (2013). The Dlx genes as clues to vertebrate genomics and craniofacial evolution. Semin. Cell Dev. Biol. 24 (2), 110–118. doi:10.1016/j.semcdb.2012.12.010
Tavares, A. L. P., Cox, T. C., Maxson, R. M., Ford, H. L., and Clouthier, D. E. (2017). Negative regulation of endothelin signaling by SIX1 is required for proper maxillary development. Development 144 (11), 2021–2031. doi:10.1242/dev.145144
Tenney, A. P., Livet, J., Belton, T., Prochazkova, M., Pearson, E. M., Whitman, M. C., et al. (2019). Etv1 controls the establishment of non-overlapping motor innervation of neighboring facial muscles during development. Cell Rep. 29 (2), 437–452. doi:10.1016/j.celrep.2019.08.078
Terakawa, J., Serna, V. A., Nair, D. M., Sato, S., Kawakami, K., Radovick, S., et al. (2020). SIX1 cooperates with RUNX1 and SMAD4 in cell fate commitment of Müllerian duct epithelium. Cell Death Differ. 27 (12), 3307–3320. doi:10.1038/s41418-020-0579-z
Wang, Y., Li, M., Chan, C. O., Yang, G., Lam, J. C., Law, B. C., et al. (2022). Biological effect of dysregulated LBX1 on adolescent idiopathic scoliosis through modulating muscle carbohydrate metabolism. Spine J. 22 (9), 1551–1565. doi:10.1016/j.spinee.2022.04.005
Wu, W., Huang, R., Wu, Q., Li, P., Chen, J., Li, B., et al. (2014). The role of Six1 in the Genesis of muscle cell and skeletal muscle development. Int. J. Biol. Sci. 10 (9), 983–989. doi:10.7150/ijbs.9442
Xu, P. X., Zheng, W., Huang, L., Maire, P., Laclef, C., and Silvius, D. (2003). Six1 is required for the early organogenesis of mammalian kidney. Development 130 (14), 3085–3094. doi:10.1242/dev.00536
Xu, J., Chen, M., Yan, Y., Zhao, Q., Shao, M., and Huang, Z. (2021). The effects of altered BMP4 signaling in first branchial-arch-derived murine embryonic orofacial tissues. Int. J. Oral Sci. 13 (1), 40. doi:10.1038/s41368-021-00142-4
Yin, X., Li, J., Salmon, B., Huang, L., Lim, W. H., Liu, B., et al. (2015). Wnt signaling and its contribution to craniofacial tissue homeostasis. J. Dent. Res. 94 (11), 1487–1494. doi:10.1177/0022034515599772
Yue, M., Lan, Y., Liu, H., Wu, Z., Imamura, T., and Jiang, R. (2021). Tissue-specific analysis of Fgf18 gene function in palate development. Dev. Dyn. 250 (4), 562–573. doi:10.1002/dvdy.259
Keywords: Six1, craniofacial development, mandibular skeletal development, cranial neural crest cells, osteogenic differentiation
Citation: Luo S, Liu Z, Bian Q and Wang X (2023) Ectomesenchymal Six1 controls mandibular skeleton formation. Front. Genet. 14:1082911. doi: 10.3389/fgene.2023.1082911
Received: 28 October 2022; Accepted: 24 January 2023;
Published: 08 February 2023.
Edited by:
Weimin Lin, Sichuan University, ChinaReviewed by:
Xiao-Jing Zhu, Hangzhou Normal University, ChinaZhonglin Jia, Sichuan University, China
Pranidhi Baddam, University of Alberta, Canada
Copyright © 2023 Luo, Liu, Bian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Bian, cWlhbmJpYW5Ac2hzbXUuZWR1LmNu; Xudong Wang, eHVkb25nd2FuZzcwQGhvdG1haWwuY29t
†These authors have contributed equally to this work and share first authorship
 Songyuan Luo
Songyuan Luo Zhixu Liu
Zhixu Liu Qian Bian
Qian Bian Xudong Wang
Xudong Wang