- 1Center for Advanced Studies and Technology (CAST), G.d’Annunzio University of Chieti-Pescara, Chieti, Italy
- 2Eusoma Breast Centre, “G. Bernabeo” Hospital, Ortona, Italy
- 3UOSD Genetica Oncoematologica, Dipartimento di Oncologico-Ematologico, Pescara, Italy
Introduction: A considerable number of families with pedigrees suggestive of a Mendelian form of Breast Cancer (BC), Ovarian Cancer (OC), or Pancreatic Cancer (PC) do not show detectable BRCA1/2 mutations after genetic testing. The use of multi-gene hereditary cancer panels increases the possibility to identify individuals with cancer predisposing gene variants. Our study was aimed to evaluate the increase in the detection rate of pathogenic mutations in BC, OC, and PC patients when using a multi-gene panel.
Methods: 546 patients affected by BC (423), PC (64), or OC (59) entered the study from January 2020 to December 2021. For BC patients, inclusion criteria were i) positive cancer family background, ii) early onset, and iii) triple negative BC. PC patients were enrolled when affected by metastatic cancer, while OC patients were all submitted to genetic testing without selection. The patients were tested using a Next-Generation Sequencing (NGS) panel containing 25 genes in addition to BRCA1/2.
Results: Forty-four out of 546 patients (8%) carried germline pathogenic/likely pathogenic variants (PV/LPV) on BRCA1/2 genes, and 46 (8%) presented PV or LPV in other susceptibility genes.
Discussion: Our findings demonstrate the utility of expanded panel testing in patients with suspected hereditary cancer syndromes, since this approach increased the mutation detection rate of 15% in PC, 8% in BC and 5% in OC cases. In absence of multi-gene panel analysis, a considerable percentage of mutations would have been lost.
1 Introduction
In these years of personalized medicine, the study of individual’s genotype is an important part of the determination of his specific susceptibility to several diseases, including cancer. The National Comprehensive Cancer Network Breast Cancer Risk Panel (NCCN) has for years been updating with publishing the indications for genetic testing of cancer patients and their family members (Sorscher 2019). The majority of Breast Cancer (BC), Ovarian Cancer (OC) and Pancreatic Cancer (PC) cases are sporadic (75%–80%), ∼15%–20% are considered familial types and 5%–10% are hereditary (Russo et al., 2009; Antonucci et al., 2017b; Incorvaia et al., 2020). Over the past 20–30 years, molecular diagnosis of hereditary BC, OC or PC has focused primarily on two high-penetrance genes, BRCA1 and BRCA2 (Antonucci et al., 2017a). The identification of germline deleterious variants in BRCA1/BRCA2 has a significant impact on clinical management of both affected individuals and their family members (Babore et al., 2019; Lombardi et al., 2019; 2022). Nevertheless, an increasing number of families with pedigrees suggestive of a Mendelian form of BC, OC or PC have not detectable mutations in BRCA1/BRCA2. The problem of “missing heritability” can be explained with the presence of pathogenic gene variants in other susceptibility genes involved with low frequency or with reduced penetrance, usually not included in the diagnostic flowchart of patients with hereditary cancer, mainly due to the costs and the time required for the analysis in the Sanger sequencing era. Therefore, it has become mandatory to study many genes in a brief time and in an economic way. In this scenario, advances in genetic technology and implementation of NGS in clinical oncology have accelerated the discovery of new cancer-related genes revolutionizing cancer research, diagnosis and therapies (Rossi et al., 2022). The advent of NGS allows the simultaneous sequencing of multiple samples and genes (Fountzilas et al., 2018). Because of the advantage from cost-benefit reduction, this approach provides a powerful enforcement for patients with LPVs and PVs in other genes, beyond BRCA1/2. Several germline PVs in susceptibility genes as CDH1, PALB2, PTEN, STK11, TP53, ATM, CHEK2, BARD1, BRLP1, RAD51C, and RAD51D (Shah et al., 2016; Fanale et al., 2020) can be associated with hereditary tumors. Most of these genes are involved in cell cycle checkpoint and DNA damage repair mechanism, and function together in these physiological pathways (Nielsen et al., 2016; Piombino et al., 2020; Neiger et al., 2021); therefore, a fundamental comprehension of the disease drivers in the cascades would facilitate the accurate evaluation of the genetic risk of cancer development (Yoshimura et al., 2022). In our study we used a multi-gene panel including 27 genes in the diagnostic iter of 546 patients with BC, OC or PC (Table 1). The aims of this work were: 1) to investigate the prevalence of PVs or LPVs in susceptibility genes implicated in hereditary cancer predisposition, and 2) to assess the utility of carrying out a multi-gene panel testing in BC, OC or PC individuals who fulfill specific criteria on their familiar and personal history of tumor.
2 Materials and methods
2.1 Study population
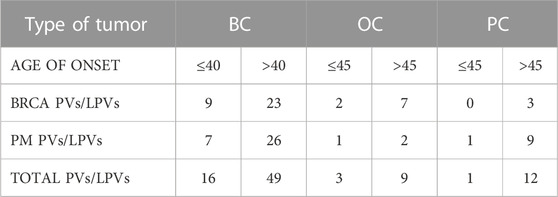
Our study includes a cohort of individuals who referred to our Center between January 2020 and December 2021. We collected and analyzed DNA samples from 546 patients with BC (423), PC (64) or OC (59), averaging 54 years (range 25–70). For BC patients, inclusion criteria were 1) positive cancer family background, 2) early onset and 3) triple negative BC. PC patients were enrolled when affected by metastatic cancer, while OC patients were all submitted to genetic testing without selection. PC and OC patients were classified into 2 groups related to the age of disease onset: 1) early onset cancer (age at diagnosis ≤45 years) and 2) late onset cancer (age at diagnosis >45 years), while for BC patients the considered age of onset was 40 years. Among BC patients, 64 had early onset cancer and 359 had late onset cancer; among OC patients 9 had early onset cancer and 50 had late onset cancer; among PC patients 2 had early onset cancer and 62 had late onset cancer. Starting from 423 BC patients, 27 (6.4%) had triple-negative breast cancer (TNBC), including 25 patients with late onset BC and only 2 with early onset BC. Genetic counseling was performed in the presence of a geneticist and a psychologist to acquire the clinical personal and familiar history of patients. In addition, data about histological cancer type, any surgical operations and current therapies were acquired. All subjects signed an informed consent about the significance of the molecular genetic test.
2.2 Next-generation sequencing (NGS)
Genomic DNA of BC, OC and PC patients were collected using buccal swabs and extracted through MagPurix instrument and Forensic DNA Extraction Kit (Zinexts Life Science Corp.- CodZP01001) according to the manufacturer’s protocol. NGS was executed by the Ion Torrent S5 system (Thermo Fisher Scientific, Waltham, MA, United States) after automatic library preparation using Ion Chef (Thermo Fisher Scientific, Waltham, MA, United States). Ion Chef consists of fragmentation and adapter ligation onto the PCR products, clonal amplification. The DNA libraries were quantified with Real-Time Step One PCR System (Thermo Fisher Scientific, Waltham, MA, United States) and the prepared samples were loaded onto an Ion 530™ chip by Ion Chef (Thermo Fisher Scientific, Waltham, MA, United States). Ion S5™ Plus (Thermo Fisher Scientific, Waltham, MA, United States) instrument was used for the sequencing. Specific plugins as “SampleId” and “Coverageanalysis” were used for NGS data analysis on the Torrent Suite 5.14.0 platform. The uniformity of base coverage was over 98% in all batches, and base coverage was over ×20 at all target regions. This NGS method cannot detect variations outside the +/−10 nucleotide coding sequence.
2.3 Sanger sequencing
Sanger Sequencing was performed using SeqStudio Genetic Analyzer System (Thermo Fisher Scientific) and BigDye Terminator 3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) to confirm all the PV/LPVs identified with NGS multi-gene panel.
2.4 Classification of the genetic variants
The genetic variants found in patients were classified into five classes: benign (C1), likely benign (C2), variant of uncertain significance (VUS, C3), likely pathogenic (C4), and pathogenic (C5), according to the guidelines of Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) (https://enigmaconsortium.org/). We focused on the LPVs and PVs that can be used for clinical purposes. Variants were referred in according to the nomenclature recommendations of the Human Genome Variation Society (https://www.hgvs.org). The clinical significance of the genetic variants found in this study was evaluated according to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), Varsome (https://varsome.com), Franklin Genoox (https://franklin.genoox.com) and, for some susceptibility genes (APC, MLH1, MSH2, MSH6, PMS2, EPCAM, MUTYH, CDH1), according to LOVD-InSIGHT (https://www.insight-group.org/variants/databases/).
3 Results
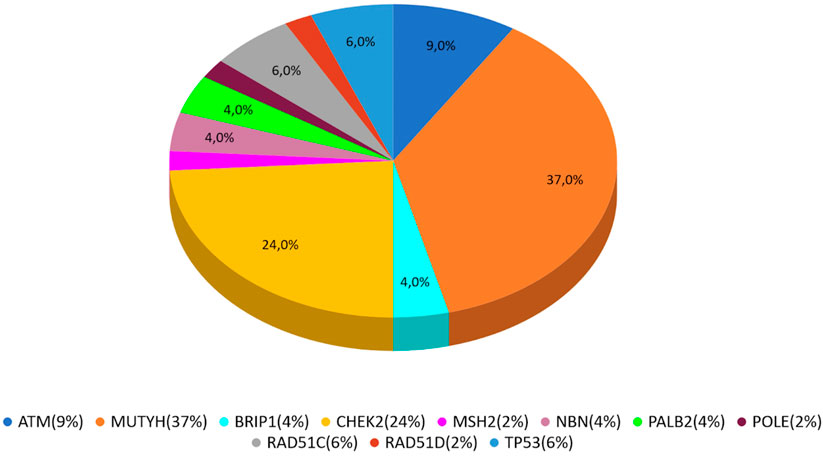
In our study, 546 cases with BC, OC, or PC were enrolled from January 2020 to December 2021. PVs or LPVs on BRCA1/2 genes were detected in 44 patients (8%), specifically 32/423 (7%) with BC, 9/59 (15%) with OC and 3/64 (5%) with PC. On the other hand, 46 patients (8%), namely 33/423 (8%) with BC, 3/59 (5%) with OC and 10/64 (16%) with PC harbored germline PVs/LPVs in other cancer susceptibility genes, as follows: 17 (37%) in MUTYH, 11 (24%) in CHEK2, 4 (9%) in ATM, 3 (6%) in RAD51C and TP53, 2 (4%) PALB2, BRIP1, and NBN. In addition, a single PV in POLE, MSH2, and RAD51D was detected in two patients (Table 2).
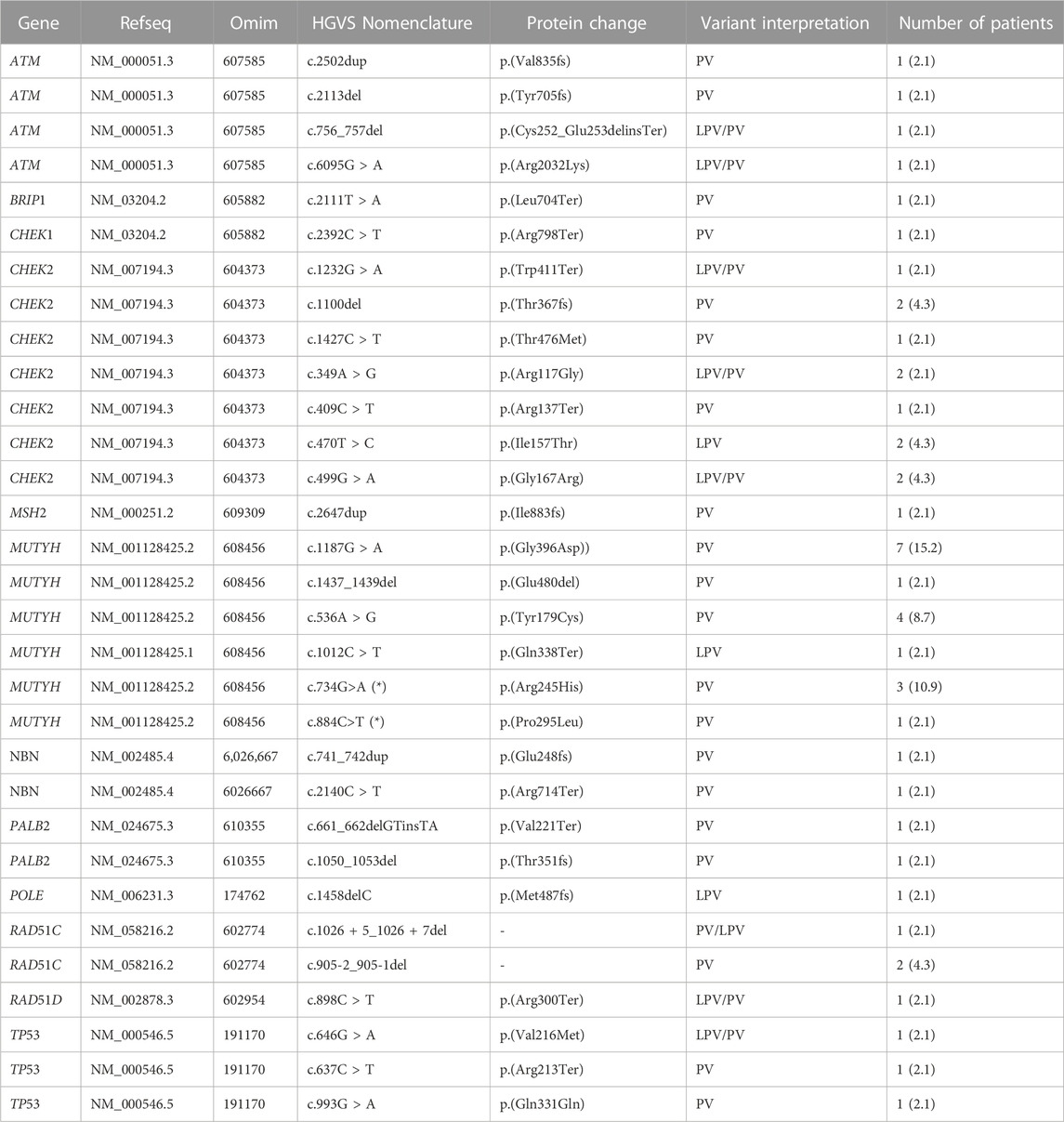
TABLE 2. All single PVs/LPVs recurrent in patients analyzed by multi-gene panel. All variants reported in the Table 2 are in heterozygous, except only one subject that had two PVs/LPVs on MUTYH gene (*).
Seven subjects enrolled showed two pathogenic variants in the genes analyzed.
According to age of onset, we found PVs/LPVs in 20 early onset patients (≤45 for PC and OC, ≤40 for BC) and in 70 late onset patients (>45 for PC and OC, >40 for BC). Eleven early onset patients with BC (14%) had PVs or LPVs mutations in BRCA1 or BRCA2 genes, whereas 17 patients (11%) reported mutations in one of the other genes included in the multi-gene panel. On the other hand, 27 late onset patients with BC (36%) had PVs or LPVs mutations in BRCAor BRCA2 genes, whereas 30 patients (40%) reported mutations in one of the several genes included in the multi-gene panel. On the OC and PC patients groups 2 early onset subjects (18%) had a PV or LPV in BRCA1/2, while 2 patients (18%) had PV or LPV in other gene. In the late onset group 10 patients (9%) had a PV or LPV in BRCA1/2 and 11(10%) with pathogenic variant in other gene. The distribution of PVs/LPVs in BRCA1/2 or in other genes in the different groups of patients is reported in Table 3.
MUTYH resulted as the gene with the higher percentage of mutation within the group analyzed by the multigene panel (16 out of 46 detected mutations), with the second most recurrent involved genes represented by CHEK2 with 11 cases (Table 2; Figure 1; Figures 2A, B). All MUTYH variants reported in this study are in heterozygous, except only one subject that had two PVs/LPVs on MUTYH gene, respectively c.734G>A and c.884C>T.
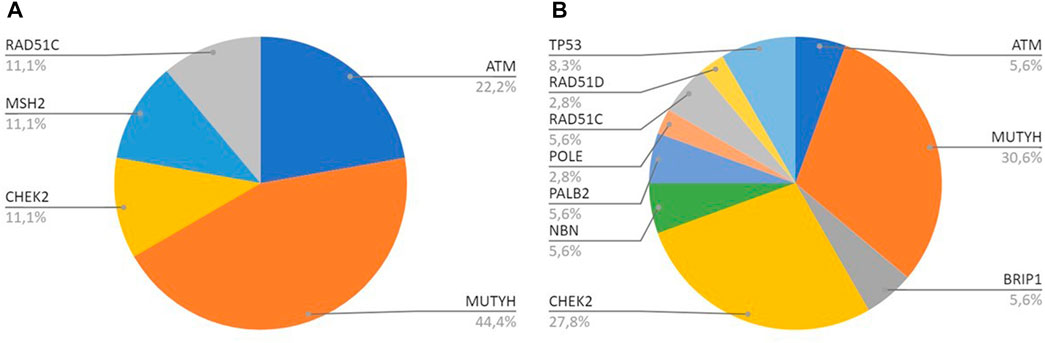
FIGURE 2. (A) All genes with PV/LPV in Breast Cancer cases; (B) All genes with PV/LPV in Pancreatic Cancer cases.
As to PVs/LPVs, the most frequent PV was c.1187G>A p.(Gly396Asp) of MUTYH gene, located in coding exon 13 and causing the substitution of a Glycine with Aspartate in codon position 396. This alteration, found in seven patients (15.2%) with BC, PC and OC, is frequently reported as founder mutation in multiple populations. M. Nielsen et al. have shown that this missense variant change the function of MUTYH protein (M. Nielsen et al., 2009).
The second recurrent PV found on the MUTYH gene was c.734G>A p. (Arg245His), in coding exon 9, results from the substitution of a Guanine to Adenine, and consequently the replacement of the arginine with a histidine at codon 245. Literature’s data supports that this missense variant has a deleterious effect on protein structure/function (Viel et al., 2017). We found this mutation in five patients (10.9%).
Analyzing the second most gene mutated, CHEK2, the other most recurrent PVs were: c.1100delC p. (Thr367fs), c.470T>C p. (Ile157Thr) and c.499G>A p. (Gly167Arg). All subjects with CHEK2 variant, are carriers of only one PV/LPV.
Specifically, CHEK2 c.1100delC caused deletes of one Cytosine from exon 11 in position 1,100 causing a frameshift at codon 367, and a premature translational stop signal p. (Thr367fs). This is expected to result in an absent or disrupted protein product. (Weischer et al., 2008). This variant is linked to increased risk of BC and OC.
4 Discussion
The identification of PV or LPV in genes responsible for hereditary cancers plays a key role in the prognosis, prevention and therapy of these conditions. In fact, cancer patients carriers of such gene variants must undergo specific protocols for the prevention of additional cancers but they can also benefit from specific drug therapies, such as those based on PARP inhibitors (PARPi), which represent a successful example of precision medicine (Slade, 2020). On the other hand, unaffected family members of a cancer patient carrier of a PV/LPV should be tested for the presence of the same variant and, when positive, specific prevention protocols, different from the common cancer screening programs used for the general populations, should be offered. In this view, a critical issue is represented by the number of genes to analyze in each condition, mainly in order to maintain a balanced cost/benefit ratio. While in a first moment it was suggested that each different type of cancer was related to one or a few specific genes (e.g., BRCA1/2 for BC and OC, APC for familial adenomatous polyposis, etc.), our study revealed that often there is not correspondence between tumor type and the associated mutated gene, raising the question about the need for more genes to be analyzed in hereditary cancers. Interestingly, our study showed that 94% of MUTYH carriers had a heterozygous variant. PVs/LPVs in MUTYH are associated with colorectal adenomatous polyposis autosomal recessive, while recent literature data revealed the association between monoallelic MUTYH variants and several type of cancer (Dell’Elice et al., 2021). BC, PC and OC, together with colon and prostate cancer, are the major tumors linked to clinical familiar history, as well as the major BRCA-associated cancers (Daly et al., 2021). Nevertheless, many of these patients result negative to the genetic testing for BRCA1/2 genes PVs and LPVs, even in presence of an evident familiar and/or personal cancer’s background. This has been confirmed by data obtained in the present study, showing that no more that 8% of BC, OC or PC cancer show BRCA1/2 mutations even in the group of early onset cases.; that the use of multi-gene hereditary cancer panels increases the possibility to identify individuals with cancer predisposing gene variants (Shin et al., 2020; Hu et al., 2021). In an association analysis involving 113,000 women, the Breast Cancer Association Consortium, Dorling L, Carvalho S, et al. define the susceptibility genes that are most clinically useful for inclusion on panels for the prediction of breast cancer risk (Breast Cancer Association Consortium et al., 2021). By extending the test using a multi-gene panel, we found an additional 8% mutations in different susceptibility genes, such as MUTYH, CHEK2, ATM, NBN, BRIP1, and TP53 involved in several hereditary cancer syndromes (Desmond et al., 2015; Tsaousis et al., 2019; N; Tung et al., 2015). These results confirmed the studies already performed in 2021 by Bono et al., where a considerable percentage of PVs/LPVs have been lost without the use of multi-gene panel (Bono et al., 2021). Thus, our results evidenced that both in early and late onset cancer patients, using the classical approach of BRCA1/2 testing, we would have lost a large number of cases resulted BRCA1/2 negative, but actually carriers of a PV/LPV in other genes. In addition to the increased detection rate, the use of multigenic panel test allow the identification of specific prevention strategies based on the gene involved, in a precision medicine approach. For example, we diagnosed three patients with Li-Fraumeni syndrome (LFS) associated with PV/LPV in TP53 on chromosome 17p13.1. This syndrome represents a severe condition inherited in an autosomal dominant manner with very high penetrance. Prevention strategies of this condition are different from the one used for BRCA1/2 mutation carriers, since LFS component tumors also include soft tissue sarcomas, osteosarcoma, brain tumors, and adrenocortical carcinomas. Interestingly, in these patients no strong familiar history was found, but they all showed early onset cancer (≤35). In one case, a “de novo” origin of the mutation was demonstrated, allowing to suggest that the age of onset of the disease could be considered as a more reliable indicator of the presence of a genetic condition than the familiarity itself. Oncology therapy putting forth the concept of selective targeting of cancer cells thanks to precision medicine. According to our goal, one of the most interesting future perspectives is the therapy with poly-adenosine diphosphate-ribose polymerase (PARP). PARP inhibitors (PARPi) were a significant example of precision medicine (Slade, 2020). The identification of specific mutations in genes different from BRCA1/2 is relevant also for the therapeutical strategies. In fact, while the benefits of PARP inhibition have been well characterized for BRCA1/2 (Risdon et al., 2021), the efficiency of this therapy in carriers of other mutations is so far a question of debate. For the therapy of metastatic breast cancer (MBC), is in progress a phase II study that are showing the efficacy of PARPi’s Olaparib, in patients with germline/somatic (g/s) mutations in related genes (PALB2, ATM and CHEK2) other than BRCA1/2 (N. M. Tung et al., 2020). Responses were seen only with gPALB2 mutations, while there are not evidences for ATM or CHEK2 mutations respectively. For this reason, Olaparib could be used in patients with gPALB2 mutation beyond in gBRCA1/2 mutation carriers, significantly expanding the number of patients with MBC who would benefit from PARPi (Pommier et al., 2016; Lord and Ashworth., 2017; Cortesi et al., 2021). In conclusion, the multi-gene panel approach could be useful for targeting therapy in oncology patients that are carriers of mutations in susceptibility genes, beyond BRCA1/2.
Data availability statement
The datasets presented in this study can be found in online repositories. The link to the data can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA927294. Accession to cite for these SRA data: PRJNA927294.
Author contributions
Writing—original draft preparation, FA and LP, writing—review and editing FA, LP, AD, and RF; performed the genetic analysis, FA, LP, AD, and RF; data curation RF; conceptualization and supervision, LS and IA; clinical investigation SGr, LMF, CM, SGi, GC, LS, and IA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antonucci, I., Provenzano, M., Sorino, L., Balsamo, M., Battista, P., Euhus, D., et al. (2017a). Comparison between CaGene 5.1 and 6.0 for BRCA1/2 mutation prediction: A retrospective study of 150 BRCA1/2 genetic tests in 517 families with breast/ovarian cancer. J. Hum. Genet. 62 (3), 379–387. doi:10.1038/jhg.2016.138
Antonucci, I., Provenzano, M., Sorino, L., Rodrigues, M., Palka, G., and Stuppia, L. (2017b). A new case of “de Novo” BRCA1 mutation in a patient with early-onset breast cancer. Clin. Case Rep. 5 (3), 238–240. doi:10.1002/ccr3.718
Babore, A., Bramanti, S. M., Lombardi, L., Stuppia, L., Trumello, C., Antonucci, I., et al. (2019). The role of depression and emotion regulation on parenting stress in a sample of mothers with cancer. Support Care Cancer 27 (4), 1271–1277. doi:10.1007/s00520-018-4611-5
Bono, M., Fanale, D., Incorvaia, L., Cancelliere, D., Fiorino, A., Calò, V., et al. (2021). Impact of deleterious variants in other genes beyond BRCA1/2 detected in breast/ovarian and pancreatic cancer patients by NGS-based multi-gene panel testing: Looking over the hedge. ESMO Open 6 (4), 100235. doi:10.1016/j.esmoop.2021.100235
Breast Cancer Association Consortium, Dorling, L., Carvalho, S., Allen, J., González-Neira, A., Craig, L., Wahlström, C., et al. (2021). Breast cancer risk genes - association analysis in more than 113,000 women. N. Engl. J. Med. 384 (5), 428–439. doi:10.1056/NEJMoa1913948
Cortesi, L., Piombino, C., and Toss, A. (2021). Germline mutations in other homologous recombination repair-related genes than BRCA1/2: Predictive or prognostic factors? J. Personalized Med. 11 (4), 245. doi:10.3390/jpm11040245
Daly, M. B., Tuya Pal, M. P., Buys, S. S., Dickson, P., Domchek, S. M., Ahmed, E., et al. (2021). Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. JNCCN 19 (1), 77–102. doi:10.6004/jnccn.2021.0001
Dell’Elice, A., Cini, G., Fornasarig, M., Franco, A., Barana, D., Bianchi, F., et al. (2021). Filling the gap: A thorough investigation for the genetic diagnosis of unsolved polyposis patients with monoallelic MUTYH pathogenic variants. Mol. Genet. Genomic Med. 9 (12), e1831. doi:10.1002/mgg3.1831
Desmond, A., Kurian, A. W., Gabree, M., Mills, M. A., Anderson, M. J., Kobayashi, Y., et al. (2015). Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 1 (7), 943–951. doi:10.1001/jamaoncol.2015.2690
Fanale, D., Incorvaia, L., Filorizzo, C., Bono, M., Fiorino, A., Calò, V., et al. (2020). Detection of germline mutations in a cohort of 139 patients with bilateral breast cancer by multi-gene panel testing: Impact of pathogenic variants in other genes beyond BRCA1/2. Cancers 12 (9), E2415. doi:10.3390/cancers12092415
Fountzilas, C., and Kaklamani, V. G. (2018). Multi-gene panel testing in breast cancer management. Cancer Treat. Res. 173, 121–140. doi:10.1007/978-3-319-70197-4_8
Hu, C., Hart, S. N., Gnanaolivu, R., Huang, H., Lee, K. Y., Na, J., et al. (2021). A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 384 (5), 440–451. doi:10.1056/NEJMoa2005936
Incorvaia, L., Fanale, D., Bono, M., Calò, V., Cancelliere, D., Castiglia, M., et al. (2020). Hereditary breast and ovarian cancer in families from southern Italy (Sicily)-Prevalence and geographic distribution of pathogenic variants in BRCA1/2 genes. Cancers 12 (5), E1158. doi:10.3390/cancers12051158
Lombardi, L., Trumello, C., Stuppia, L., Antonucci, I., Brandão, T., and Babore, A. (2022). BRCA1/2 pathogenetic variant carriers and reproductive decisions: Gender differences and factors associated with the choice of preimplantation genetic diagnosis (PGD) and prenatal diagnosis (PND). J. Assisted Reproduction Genet. 39 (7), 1433–1443. doi:10.1007/s10815-022-02523-y
Lombardi, L., Bramanti, S. M., Babore, A., Stuppia, L., Trumello, C., Antonucci, I., et al. (2019). Psychological aspects, risk and protective factors related to BRCA genetic testing: A review of the literature. Support Care Cancer 27 (10), 3647–3656. doi:10.1007/s00520-019-04918-7
Lord, C. J., and Ashworth., A. (2017). PARP inhibitors: Synthetic lethality in the clinic. Sci. (New York, N.Y.) 355 (6330), 1152–1158. doi:10.1126/science.aam7344
Neiger, H. E., Siegler, E. L., and Shi, Y. (2021). Breast cancer predisposition genes and synthetic lethality. Int. J. Mol. Sci. 22 (11), 5614. doi:10.3390/ijms22115614
Nielsen, F. C., Hansen, T. O., and Sørensen, C. S. (2016). Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer 16 (9), 599–612. doi:10.1038/nrc.2016.72
Nielsen, M., Jones, N., Vogt, S., Carli, M., Vasen, H. F. A., Sampson, J. R., et al. (2009). Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-associated polyposis. Gastroenterology 136 (2), 471–476. doi:10.1053/j.gastro.2008.10.056
Piombino, C., Cortesi, L., Lambertini, M., Kevin, P., Grandi, G., and Toss, A. (2020). Secondary prevention in hereditary breast and/or ovarian cancer syndromes other than BRCA. J. Oncol. 2020, 6384190. doi:10.1155/2020/6384190
Pommier, Y., O’Connor, M. J., and de Bono, J. (2016). Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8 (362), 362ps17. doi:10.1126/scitranslmed.aaf9246
Risdon, E. N., Chau, C. H., Price, D. K., Sartor, O., and Figg, W. D. (2021). PARP inhibitors and prostate cancer: To infinity and beyond BRCA. Oncol. 26 (1), e115–e129. doi:10.1634/theoncologist.2020-0697
Rossi, C., Cicalini, I., Cufaro, M. C., Ada, C., Upadhyaya, P., Sala, G., et al. (2022). Breast cancer in the era of integrating “omics” approaches. Oncogenesis 11 (1), 17. doi:10.1038/s41389-022-00393-8
Russo, A., Calò, V., Bruno, L., Schirò, V., Agnese, V., Cascio, S., et al. (2009). Is BRCA1-5083del19, Identified in Breast Cancer Patients of Sicilian Origin, a Calabrian Founder Mutation? Breast Cancer Res. Treat. 113 (1), 67–70. doi:10.1007/s10549-008-9906-7
Shah, P. D., Patil, S., Dickler, M. N., Offit, K., Hudis, C. A., and Robson, M. E. (2016). Twenty-one-gene recurrence score assay in BRCA-associated versus sporadic breast cancers: Differences based on germline mutation status. Cancer 122 (8), 1178–1184. doi:10.1002/cncr.29903
Shin, H-C., Lee, H-B., Yoo, T-K., Lee, E-S., Kim, R. N., Park, B., et al. (2020). Detection of germline mutations in breast cancer patients with clinical features of hereditary cancer syndrome using a multi-gene panel test. Cancer Res. Treat. 52 (3), 697–713. doi:10.4143/crt.2019.559
Slade, D. (2020). PARP and PARG inhibitors in cancer treatment. Genes & Dev. 34 (5–6), 360–394. doi:10.1101/gad.334516.119
Sorscher, S. (2019). Universal multigene panel testing in all breast cancer patients. Am. J. Med. 132 (11), e765–e766. doi:10.1016/j.amjmed.2019.03.012
Tsaousis, G.N., Papadopoulou, E., Apessos, A., Agiannitopoulos, K., Pepe, G., Kampouri, S., et al. (2019). Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 19 (1), 535. doi:10.1186/s12885-019-5756-4
Tung, N., Battelli, C., Allen, B., Kaldate, R., Bhatnagar, S., Bowles, K., et al. (2015). Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 121 (1), 25–33. doi:10.1002/cncr.29010
Tung, N. M., Robson, M. E., Ventz, S., Santa-Maria, C. A., Nanda, R., Marcom, P. K., et al. (2020). Tbcrc 048: Phase II study of Olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J. Clin. Oncol. 38 (36), 4274–4282. doi:10.1200/JCO.20.02151
Viel, A., Bruselles, A., Meccia, E., Fornasarig, M., Quaia, M., Canzonieri, V., et al. (2017). A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine 20, 39–49. doi:10.1016/j.ebiom.2017.04.022
Weischer, M., Bojesen, S. E., Ellervik, C., Tybjaerg-Hansen, A., and Nordestgaard, B. G. (2008). CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J. Clin. Oncol. 26 (4), 542–548. doi:10.1200/JCO.2007.12.5922
Keywords: NGS, hereditary cancer, BRCA, cancer predisposition gene, multi-gene panel testing, breast cancer, ovarian cancer, pancreatic cancer
Citation: Anaclerio F, Pilenzi L, Dell’Elice A, Ferrante R, Grossi S, Ferlito LM, Marinelli C, Gildetti S, Calabrese G, Stuppia L and Antonucci I (2023) Clinical usefulness of NGS multi-gene panel testing in hereditary cancer analysis. Front. Genet. 14:1060504. doi: 10.3389/fgene.2023.1060504
Received: 03 October 2022; Accepted: 04 January 2023;
Published: 01 February 2023.
Edited by:
Pasquale Pisapia, University of Naples Federico II, ItalyReviewed by:
Laura Cortesi, University Hospital of Modena, ItalyRicharda Maria De Voer, Radboud University Medical Centre, Netherlands
Copyright © 2023 Anaclerio, Pilenzi, Dell’Elice, Ferrante, Grossi, Ferlito, Marinelli, Gildetti, Calabrese, Stuppia and Antonucci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rossella Ferrante, cm9zc2VsbGFmZXJyYW50ZUB5YWhvby5pdA==
†These authors have contributed equally to this work and share first authorship
 Federico Anaclerio
Federico Anaclerio Lucrezia Pilenzi
Lucrezia Pilenzi Anastasia Dell’Elice
Anastasia Dell’Elice Rossella Ferrante
Rossella Ferrante Simona Grossi2
Simona Grossi2 Liborio Stuppia
Liborio Stuppia