
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 17 April 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1049608
This article is part of the Research Topic Plant Growth-Promoting and Associated Microbes: Multi-Omics, Meta-Omics, and Applications View all 5 articles
 Seyyedeh Maryam Zamanzadeh-Nasrabadi1
Seyyedeh Maryam Zamanzadeh-Nasrabadi1 Fatemeh Mohammadiapanah1*
Fatemeh Mohammadiapanah1* Mehdi Hosseini-Mazinani2
Mehdi Hosseini-Mazinani2 Sajjad Sarikhan3
Sajjad Sarikhan3The application of plant growth-promoting bacteria (PGPB) is vital for sustainable agriculture with continuous world population growth and an increase in soil salinity. Salinity is one of the severe abiotic stresses which lessens the productivity of agricultural lands. Plant growth-promoting bacteria are key players in solving this problem and can mitigate salinity stress. The highest of reported halotolerant Plant growth-promoting bacteria belonged to Firmicutes (approximately 50%), Proteobacteria (40%), and Actinobacteria (10%), respectively. The most dominant genera of halotolerant plant growth-promoting bacteria are Bacillus and Pseudomonas. Currently, the identification of new plant growth-promoting bacteria with special beneficial properties is increasingly needed. Moreover, for the effective use of plant growth-promoting bacteria in agriculture, the unknown molecular aspects of their function and interaction with plants must be defined. Omics and meta-omics studies can unreveal these unknown genes and pathways. However, more accurate omics studies need a detailed understanding of so far known molecular mechanisms of plant stress protection by plant growth-promoting bacteria. In this review, the molecular basis of salinity stress mitigation by plant growth-promoting bacteria is presented, the identified genes in the genomes of 20 halotolerant plant growth-promoting bacteria are assessed, and the prevalence of their involved genes is highlighted. The genes related to the synthesis of indole acetic acid (IAA) (70%), siderophores (60%), osmoprotectants (80%), chaperons (40%), 1-aminocyclopropane-1-carboxylate (ACC) deaminase (50%), and antioxidants (50%), phosphate solubilization (60%), and ion homeostasis (80%) were the most common detected genes in the genomes of evaluated halotolerant plant growth-promoting and salinity stress-alleviating bacteria. The most prevalent genes can be applied as candidates for designing molecular markers for screening of new halotolerant plant growth-promoting bacteria.
Currently, climate change is the principal menace to sustainable agriculture. Biotic (30%) and abiotic (50%) stresses are the main restrictions for agriculture (Kumar and Verma, 2018; Sharma A. et al., 2021; Poria et al., 2022). Over 20% of the cultivable soil worldwide is influenced by salinity stress, and every year, about 1%–2% of arable lands are disqualified by the increased salinity (Arora et al., 2020; Noori et al., 2021). Salinity and drought as the most destructive abiotic stresses are causing secondary detrimental effects, including oxidative and osmotic stresses shared with both stresses; besides ionic stress in salinity (Chaudhry et al., 2022). Salinity affects plants at morphological, physiological, biochemical, and molecular levels. Salinity causes the decrease of the leaf area, and chlorophyll content of leaves, leaf thickening, decreased shoot and root weight, necrosis of plants, wilting, drying, the reduction in seed germination, seedling growth, flowering, and fruiting, less grain weight, oxidative damage, electrolyte leakage, reduced carbon fixation, membrane damage, loss of organelle function, closure of stomata, nutrient imbalance, reduction of photosynthesis, and phytohormones production (Saberi Riseh et al., 2021; Ullah et al., 2021). Different strategies are employed to ameliorate the crop resistance to stress, comprising breeding, genetic engineering, CRISPR/Cas9 technology, chemical priming, and biological priming (Godoy et al., 2021; Wang et al., 2023). The application of PGPB is cost-effective and the most capable strategy following the challenges related to the development of new tolerant plants due to the intricacy of abiotic stress tolerance mechanisms, the awareness of the toxicity of agrochemicals, and alternating green technologies (Mohammadipanah and Zamanzadeh, 2019; Alberton et al., 2020; Jiang et al., 2021; Saeed et al., 2021). Moreover, although beneficial effect of biochar amendments in agriculture have been demonstrated, biochar addition may not certainly play a positive role for all soils type, climate, and plants species, high rates application of biochar may have side effects on weed control, delay in flowering (Alberton et al., 2020), and biochar adsorbs essential nutrients such as nitrogen and Fe (Kavitha et al., 2018; Wang et al., 2020).
Among the assessed halotolerant PGPB in 40 articles, 54% belonged to Firmicutes, 39% to Proteobacteria, and 7% to Actinobacteria. The most dominant genera of halotolerant PGPB are Bacillus and Pseudomonas. PGPB are capable of offering cross-protection against several stresses and increasing plant growth via different direct and indirect mechanisms, including modification of root morphology, nutrient attainment, synthesis of exopolysaccharides, phytohormones, volatile compounds, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase, ion homeostasis, inducing aggregation of antioxidants and compatible solutes, induced systemic tolerance, and modulation of the stress-responsive genes (Mohammadipanah and Zamanzadeh, 2019; Leontidou et al., 2020). Despite extensive investigation on the mechanisms of action of PGPB, information on the molecular aspect of these mechanisms is slight (Balasubramanian et al., 2021). Applying next-generation sequencing (NGS), computational tools, and omics methods (meta (genomics) (transcriptomics) (proteomics) and metabolomics) can integrate data on molecular aspects of the plant-microbe reciprocal action or effect (Meena et al., 2017; Shelake et al., 2019). In recent years, the studies on molecular features of rhizobacteria have been increasing (Figure 1). However, there are many bacteria whose plant growth-promoting capabilities have been proven in the laboratory, pot, and field, but their genomes and molecular aspects of action have not been investigated. By assaying the genomes of these bacteria, new pathways and genes may be identified. Knowing the genes and pathways related to stress mitigation, which are identified so far, are essential for omics and meta-omics studies. In addition, the assignment of related gene sets to PGPB action assists in designing biomarkers of rapid screening of efficient plant growth-promoting strains. There are many whole genomes of the environmental bacteria, which can be screened by applying molecular markers to identify new plant growth-promoting taxa.
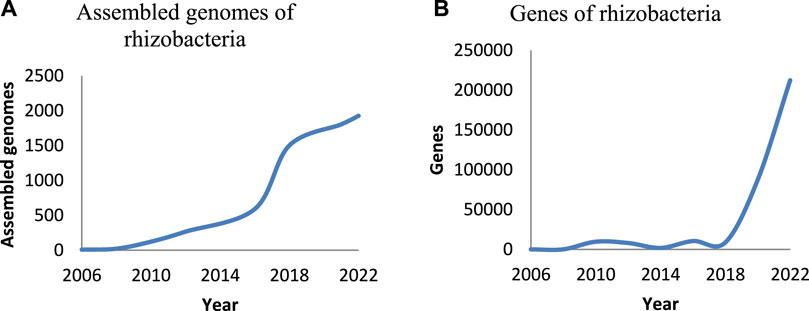
FIGURE 1. The studies on genomes and genes of rhizobacteria per year. The number of the genomes of rhizobacteria that were assembled (A), the number of represented genes of rhizobacteria (B) (information was.
While many studies have presented the mechanisms of action of PGPB, there is not any systematic review on genes conferring the plant growth-promoting trait to halotolerant symbiotic bacteria. In this survey, the molecular aspects of the action mechanisms of halotolerant PGPB are expressed. In addition, the identified pathways and genes related to plant growth promotion and salinity stress alleviation in the genomes of 20 halotolerant PGPB, including Pseudomonas fluorescens PCL1751 (Cho et al., 2015), Cronobacter muytjensii JZ38 (Eida et al., 2020), Klebsiella sp. KBG6.2 (Girma et al., 2020), Enterobacter roggenkampii ED5 (Guo et al., 2020), Jejubacter calystegiae (Jiang et al., 2021), Klebsiella sp. D5A (Liu et al., 2016), Bacillus megaterium STB1 (Nascimento et al., 2020), Pseudomonas thivervalensis SC5 (Nascimento et al., 2021), Pantoea agglomerans ANP8 (Noori et al., 2021), Hartmannibacter diazotrophicus E19T (Suarez et al., 2019), Bacillus flexus KLBMP 4941 (Wang et al., 2017), Enterobacter sp. SA187 (Andres-Barrao et al., 2017), Bacillus paralicheniformis ES-1 (Iqbal et al., 2022), Brevibacterium sediminis MG-1 (Lutfullin et al., 2022), Bacillus sp. BH32 (Belaouni et al., 2022), Pseudomonas chloritidismutans 6L11 (Zhou et al., 2022), Stenotrophomonas rhizophila IS26 (Dif et al., 2022), Pseudomonas sp. UW4 (Duan et al., 2013), Achromobacter xylosoxidans SQU-1 (Jana and Yaish, 2021), and Stenotrophomonas 169 (Ulrich et al., 2021) are assessed and these genes prevalence are characterized. Moreover, the studies conducted on the assay of transcriptome, proteome, and metabolome of PGPB in interaction with plants are presented.
For this review, an extensive search from the National Center for Biotechnology Information (bookshelf, PubMed, Assembly, BioProject, BioSample, and genome), Google Scholar, Scopus, ScienceDirect, and Springer was performed using relevant keywords such as halotolerant PGPB, salinity stress alleviation, PGPB genome mining, genome analysis, PGPR molecular action, PGPR pathways and genes, halotolerant rhizobacteria, genomes of rhizobacteria, genes of rhizobacteria, genome analysis tools, PGPB transcriptome, PGPB proteome, PGPB metabolome, and plant-bacteria interaction.
Omics and meta-omics methods (genomics, transcriptomics, proteomics and metabolomics) help decipher molecular mechanisms of PGPB action to reveal the intricate plant–bacteria interactions. It is essential to understand how bacterial metabolites affect plant–bacteria interactions. Genomic analysis of 20 halotolerant PGPB in this study present the related genes to plant-bacteria interaction in the genomes of these bacteria.
Metagenomic studies of bacterial communities provide the basic evidence of plants-bacteria interaction whichcan bepractical to be used in identifying novel species with special traits (Fadiji et al., 2022; Mukherjee, 2022). Multiple metagenomic analyses have been carried out for investigating the genes related to plant growth-promoting traits of bacterial community from soil samples of oil field at Wietze (Eze et al., 2021), from maize rhizosphere in the South Africa (Molefe et al., 2021), associated with the root of sugar beet (Tsurumaru et al., 2015), and the endophytes of Emilia sonchifolia (Linn.) DC (Urumbil and Anilkumar 2021). These metagenomic analyses showed structural and functional diversity of plant microbiomes and allowed identification of genes and taxa putatively related to plant growth promotion. Only one report is available on the metagenome of plant rhizospheric bacteria exposed to salinity stress. The metagenomic analysis of the rhizospheric microbial community of grapevine under salinity stress showed that the salinity stress tolerance of grapevines was associated to the composition and functions of the rhizospheric microbial community (Wang et al., 2023).
Studies on gene expression have mainly focused on the expression assay of genes in plants in interaction with PGPB (Sheibani-Tezerji et al., 2015). The expression of genes, proteome, and metabolome in different hosts while interacting with PGPB have been assessed in several studies (Wang et al., 2005; Kwon et al., 2016; Kazemi-Oskuei et al., 2018; Gamez et al., 2019; Yoo et al., 2019; Malviya et al., 2020; Gozalia et al., 2021; Kataoka et al., 2021; Li et al., 2021; Mellidou et al., 2021; Gao et al., 2022; Samaras et al., 2022; Wiggins et al., 2022; Xu et al., 2022; Yadav et al., 2022).
However, only a limited number of studies described below have investigated the gene expression and metabolites in PGPB during interaction with plants. Transcriptome analysis of Burkholderia phytofirmans PsJN colonizing potato under drought stress showed the upregulation of genes related to transcriptional regulation, homeostasis, and the detoxification of ROS (Sheibani-Tezerji et al., 2015).The transcriptome analysis of plant growth-promoting Paenibacillus polymyxa YC0136 showed 286 genes were up-regulated and 223 genes were down-regulated under interaction with tobacco (Liu et al., 2020). The expression of genes was assayed in three new plant growth-promoting bacterial strains (two Paenibacillus sp. strains and one Erwinia gerundensis strain) in interaction with barley (Li et al., 2021). The transcriptome of the plant growth-promoting bacterium Delftia acidovorans RAY209 was assayed during interaction with soybean and canola roots (Suchan et al., 2020). The gene expression of Azospirillum lipoferum 4B during interaction with rice roots was assayed (Drogue et al., 2014). A metatranscriptomic study was conducted to investigate the contributions of different nitrogen-fixing bacteria present in the maize inoculated liquid (Gomez-Godinez et al., 2018). Metabolomics was applied to explore the exo-metabolome of three PGPB (Pseudomonas putida IDE-01, Azospirillum brasilense IDE-06, and Bacilus megaterium IDE-14) in interaction with maize and rice (Garcia et al., 2022). No twodimentional study was found on gene expression and metabolome of salinity stress-alleviating PGPB in interaction with plants. Metaphenome is the product of expressed functions encoded in microbial genomes and the environmentwhich consistes of themeta-omics technologies, including metagenomics, metatranscriptomics, metaproteomics, and metabolomics. The rhizosphere metaphenomics remains a significant challenge that needs to be addressed (Jansson and Hofmockel, 2018; Azeem et al., 2022).
Exploring the complexity of plant-soil-bacteria interactions allows the application of bacteria in an efficient manner to increase productivity of crops. Interactomics is a comprehensive technology for determining the pathways associated with communication between host plant and PGPB under environmental stresses which need simultaneous analysis the interactions between different biomolecules including proteins, and enzymes from both plants and bacterial cells. (Arora et al., 2020).
Understanding molecular mechanisms related to stress tolerance in bacteria is vital for their application as plant stress protectors (Ahmed et al., 2018). Bacteria apply the cell surface extracytoplasmic function (ECF) sigma factors for sensing and responding to the surroundings. Mentioned signal transduction involves the outer membrane receptor, inner membrane-attached sigma factor regulator or anti-sigma factor, and ECF sigma factor. Anti-sigma factor firmly attaches to the ECF and maintains it inactive during lack of signal. In the presence of stress, the anti-sigma factor is decomposed. Consequently, the sigma factor is liberated and activates the expression of its related genes. Moreover, ECF sigma factors can be a factor in establishing the interactions between plants and bacteria (Sheibani-Tezerji et al., 2015).
The ability of osmotic stress mitigation in bacteria is evolved through horizontal gene transfer. The main adaptation mechanisms for tolerating salinity stress include ion homeostasis, accumulation of osmolytes, and production of universal proteins related to salt stress tolerance (Goyal et al., 2019). Na+/H+ antiporters are a group of transmembrane proteins that exist in the plasma membrane of nearly all cells and participate significantly in the conservation of intracellular pH, cellular sodium amount, homeostasis, and volume of the cell (Goyal et al., 2019). Five types of Na+/H+ antiporters are present in prokaryotes, including NhaA, NhaB, NhaC, NhaD, and NapA. Nhas perceive the ion concentration of the surroundings and moderate their action to preserve the homeostasis of cells (Kapoor and Kanwar, 2019). NhaA antiporter acts very selectively to exclude Na+ (Goyal et al., 2019). NhaA can be applied as an applicable marker to screen the salt tolerant strains (Kapoor and Kanwar, 2019). The genes of Nha proteins have been identified in the genomes of more than 80% of assessed halotolerant PGPB (Table 1).
Incitation of K+ uptake is the first quick reaction to an osmotic change by bacteria (Goyal et al., 2019). Three K+ uptake systems have different affinities: the Kup system, which is permanently active and keeps a little level of K+ absorption and is not regulated through osmolality, while Trk and Kdp systems are multipartite inducible systems, which activated afterward osmotic up shock (Goyal et al., 2019). Kdp was the most common gene related to K+ uptake systems, which has been identified in the genomes of 55% of assessed halotolerant PGPB (Table 1).
Osmolytes are accumulated either by intake from surroundings or by de novo synthesis (Mishra et al., 2018). The molecular aspect of the synthesis of osmolytes is discussed in Section 4.5.
Heat-shock proteins (HSPs) or chaperones, including DnaK, DnaJ, ClpX, ClpA, ClpB, GroES, GroEL, proteases, and sHSPs, are upregulated upon osmotic stress. The main role of chaperones is to control the folding and refolding process of stress-affected proteins (Bourque et al., 2016). Clp proteins are involved in regulating ATP-dependent proteolysis. ClpC gene expression is induced by several stresses in Bacillus subtilis (Goyal et al., 2019). Furthermore, a unique osmotolerance gene brpA encodes a carotenoid-modifying enzyme. The product of mazG is nucleoside triphosphate pyrophosphohydrolase which eliminates aberrant dNTPs from damaged DNA due to stress by hydrolyzing dNTPs to pyrophosphate and dNMPs (Goyal et al., 2019). Genes affiliated with the salinity tolerance in assayed halotolerant PGPB are presented in Table 1.
The transcriptome analysis of Chromohalobacter salexigens ANJ207 which is a halophilic PGPB showed the expression of genes related to HSPs synthesis increased (Srivastava et al., 2022). The gene expression assay of Azospirillum lipoferum 4B during interaction with rice roots showed several genes related to stress response (nhaA1, cspA2, and msrA) and genes related to HSPs (hspD2, groES1) were induced (Drogue et al., 2014).
Some PGPB show several plant-beneficial traits through the aggregation of the related genes which have been elected in these bacteria (Bruto et al., 2014). Additionally, PGPB or bacterial products offer cross-protection against other stresses due to the natural crosstalk between stress-response pathways (Rosier et al., 2018). PGPB act principally through the synthesis of numerous secondary metabolites, modulation of the transcription of several genes, and cellular communication via quorum sensing (Mokrani et al., 2020). PGPB colonize plants and confer salinity tolerance through alteration in root morphology, nutrient attainment, synthesis of exopolysaccharides, 1-aminocyclopropane-1-carboxylate (ACC) deaminase, phytohormones, and volatile composites, prompting accumulation of antioxidants and osmolytes, ion homeostasis, induced systemic tolerance and regulation of the stress response genes (Figure 2) (Mohammadipanah and Zamanzadeh, 2019; Etesami, 2020; Batista et al., 2021).
The mineral nutritional estate of plants influences their capability to adjust to stresses while salinity decreases the accumulation of plant nutrients (Carmen and Roberto, 2011; Shilev, 2020). Salinity-tolerant PGPB reduce the need for chemical fertilizers through the increasing accessibility of plant nutrients (Etesami, 2020). Furthermore, PGPB improve nutrient bioavailability indirectly by increasing the root surface area (Olenska et al., 2020). Genes related to nutrient attainment in salinity stress alleviating PGPB are exhibited in Table 2.
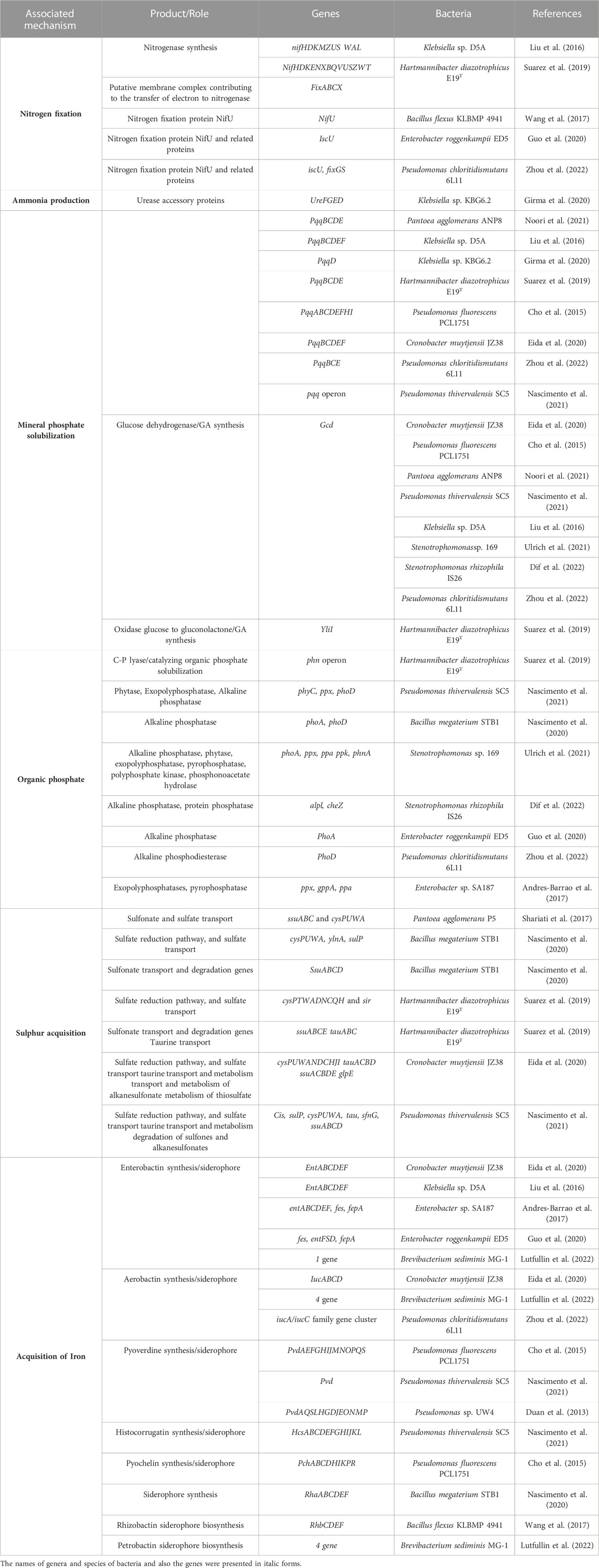
TABLE 2. Genes associated with nutrient acquisition in the 20 studied genomes of PGPB alleviating osmotic stress.
Nitrogen content of saline land is insufficient for normal plant growth (Kapoor and Kanwar, 2019). Bacteria can convert the non-available nitrogen to more available forms through mineralization, nitrification, and fixation. Mineralization entails a flow of microbial and enzymatic actions that transform soil organic nitrogen to inorganic (Olenska et al., 2020). PGPB can fix atmospheric nitrogen into ammonia form through nitrogen fixation (Vaishnav et al., 2016). Nitrogen can be converted naturally into ammonia by lightning or fires but is mainly biologically fixed by diazotrophs. Diazotrophs produce ammonia using nitrogenase encoded by nif genes located on chromosomes or plasmids like in most Rhizobium (Glick et al., 1999b; Olenska et al., 2020). The nif genes are organized in a single cluster along with seven discrete operons encode 20 diverse proteins (Glick, 2012). Nif genes contain regulatory genes (nifLA), structural genes (nifHDK), and supplementary genes (as nifBEMNQSUVW) (Glick et al., 1999b; Tomer et al., 2016). NifH gene can be applied as a valuable marker to illustrate the diazotrophs (Tomer et al., 2016). Among 20 assessed halotolerant PGPB, the nif gene cluster has been detected in the genomes of Klebsiella sp. D5A and Hartmannibacter diazotrophicus E19T (Table 2).
The transcriptome analysis of Paenibacillus sp. Strain S02 in interaction with barely showed the expression of the nif operon was upregulated (Li et al., 2021).
The metatranscriptome study of a group of different PGPB (Rhizobium phaseoli, Sinorhizobium americanum, Azospirillum brasilense, Bacillus subtillis, and Methylobacterium extorquens) that were inoculated to maize showed that the expression of nif genes of Azospirillum increased and it was the cause of nitrogen fixation in maize (Gomez-Godinez et al., 2018).
Numerous studies have shown PGPB-priming changes the expression of genes (Fadiji et al., 2022). A new non-coding RNA (ncRNA) was known in Pseudomonas stutzeri (NfiS), which controls the nif genes expression and presents on the core genome. NfiS acts through post-transcriptional modulation of dinitrogenase nifK mRNA and induces the RpoN/NtrC/NifA regulatory cascade, which is an activator of transcription of every nif operons by unknown procedures (Olenska et al., 2020).
In addition, some of the PGPB affect the root nutrient transport systems. Bacillus spp. trigger the expression of genes connected to nitrate and ammonium intake and transfer in Arabidopsis thaliana (Olenska et al., 2020).
Sulfur, as the secondary essential macronutrient, is only accessible for plants in the form of sulfate (5% of total soil S) (Vaishnav et al., 2016). Alkanesulfonates as the main ingredients of organosulfur mixtures in agricultural lands, are transmitted inside the cell using aliphatic sulfonate ABC transport (ssuABC) and transformed into sulfite by the alkanesulfonate monooxygenase (ssuD) and an NADPH-dependent FMN reductase (ssuE) (Suarez et al., 2019). There are multiple copies of the ssuABC genes on the Hartmannibacter diazotrophicus E19T genome. Remarkably, plasmid HDIAp1 encloses additional CDSs of ssuABC and ssuE. Furthermore, the genome of E19T carries the tauABC genes related to taurine transfer into the bacterial cell for higher decomposition of taurine to alanine and sulfoacetaldehyde using the taurinepyruvate aminotransferase (Tpa) (Suarez et al., 2019).
Potassium (K) is the third primary essential nutrient used by plants (Kumar et al., 2020). Nevertheless, most of the K is not accessible for plant absorption. Furthermore, salinity stress reduces K availability to plants. In this condition, K-solubilizing bacteria (KSB) are needed for plant survival (Vaishnav et al., 2016; Kumar et al., 2020). KSB solubilize K through chelation, acidolysis, synthesizing organic and inorganic acids, polysaccharides, and exchange reactions (Etesami et al., 2017).
Iron, as the fourth plentiful element utilized by the majority of living organisms is applied by almost 140 enzymes as a cofactor. In saline land, the solubility of ferric is lessened because of increased pH. Siderophores are iron-chelating means produced by most PGPB with a vast chemical diversity including peptidic, aminoalkane, and citric acid-based siderophores (Vaishnav et al., 2016). Siderophores are commonly made via non-ribosomal peptide synthetases (NRPSs) or polyketide synthase, which collaborates with NRPS modules (Olenska et al., 2020). The genes related to enterobactin (ent) and pyoverdine (pvd) synthesis have been identified with more prevalence in the genomes of assessed halotolerant PGPB (Table 2). The genes of enterobactin and pyoverdine synthesis were the most common identified genes related to siderophores synthesis in assessed bacteria belonging to Enterobacteriaceae (67%) and Pseudomonadaceae (75%) families, respectively.
The transcriptome analysis of Paenibacillus sp. in interaction with barely showed the expression of the siderophore cluster was upregulated (Li et al., 2021).
Phosphorus is a vital macronutrient for plants. Only around 4% of the phosphorus in soil is accessible to plants (Aslam et al., 2020; Olenska et al., 2020). Phosphate-solubilizing bacteria (PSB) hydrolyze unavailable forms of phosphorus into available forms by several mechanisms (Leontidou et al., 2020). Bacteria can do phosphate solubilization through releasing of H+ to the outer surface in interchange for cation intake, but phosphates are released mainly by soil acidification through organic acids. Bacterial organic acids are produced due to direct oxidation in the periplasmic space (Olenska et al., 2020). Organic acids chelate phosphate binding cations, causing a decrease in pH and providing phosphate anions (Olenska et al., 2020). The most prevalent organic acids consist of gluconic acid (GA) and 2-ketogluconic acid (Olenska et al., 2020). Glucose-1-dehydrogenase (gcd) synthesizes GA, and pyrrolo-quinolone quinine (PQQ) acts as its co-factor. Furthermore, GA dehydrogenase (gad) plays a role in GA production and its conversion to 2-ketogluconate (Shariati et al., 2017). Additionally, hydrogen sulfide (H2S) plays a role in the solubilization of phosphate. H2S, in reaction with ferric phosphate, produces ferrous sulfate and releases phosphate (Shariati et al., 2017). P-organic substrates enzymatically hydrolyze into inorganic kinds by PGPB (Khatoon et al., 2020). These enzymes are non-specific acid phosphatases (NSAPs), including phytases, acid and alkaline phosphomonoesterases (phosphatases), phosphonatases, and C-P lyases (Suarez et al., 2019; Olenska et al., 2020). The genes related to mineral phosphate solubilization (pqq and gcd) and organic phosphate solubilization have been identified in the genomes of 11 of assessed halotolerant PGPB (Table 2).
Phytohormones protect plants against abiotic stresses, and PGPB can modulate the level of endogenous phytohormones (Mohammadipanah and Zamanzadeh, 2019; Khan et al., 2020a). PGPB produce analogs of phytohormones, metabolize them, or influence the synthesis of plant hormone and signal transduction (Tsukanova et al., 2017; Olenska et al., 2020). PGPB hormones can trigger the division and growth of plant cells, alter root characteristics and play a significant role in organizing an array of genes, their regulators, and several signal transduction pathways when plants are exposed to abiotic stresses and make crops tolerant to the stresses (Mohammadipanah and Zamanzadeh, 2019; Etesami, 2020). The main phytohormones are abscisic acid, gibberellins, auxins, ethylene, salicylic acid, and cytokinins (Olenska et al., 2020). Genes of phytohormones synthesis in salinity stress mitigating PGPB are presented in Table 3.
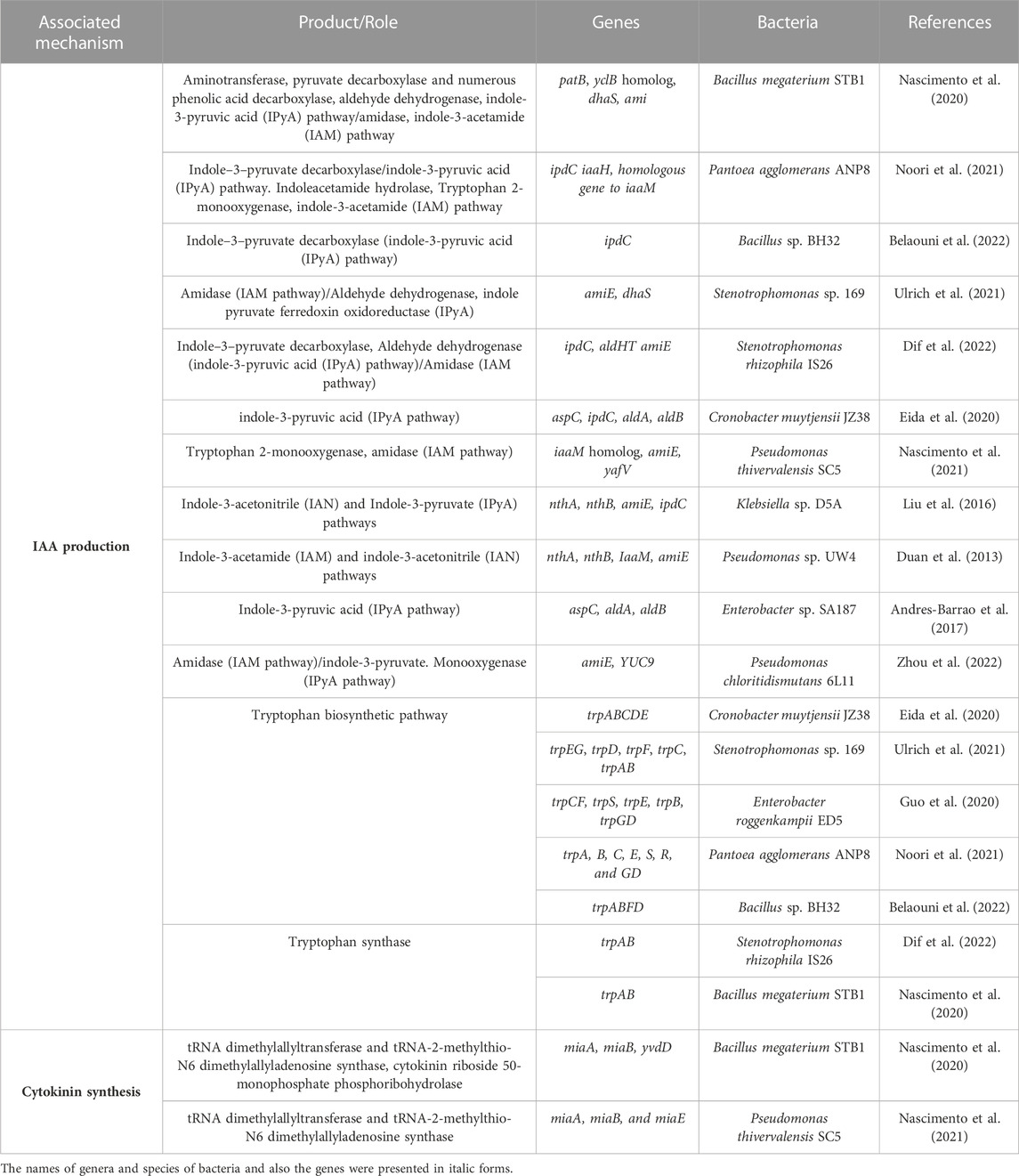
TABLE 3. Identified genes related to hormone synthesis in the 20 studied genomes of saline soil PGPB.
Abscisic acid (ABA) is recognized as a stress hormone and is upregulated in salinity stress (Kumar et al., 2020). ABA assists in the accumulation of osmolytes, Ca2+ and K+, modulates cell ion balance, prompts the roots elongation and the appearance of lateral roots, increases old leaf shedding, and controls leaf stomatal closure (Bhat et al., 2020; Kumar et al., 2020; Nian et al., 2021). ABA has two principal functions, regulates metabolism and transfer of itself through posttranslational modulation, and also it interacts with core transcription factors which are modulated through ABA and other phytohormones. The amount of ABA increases under osmotic stress through elevated expression of multiple genes of ABA production, including genes of aldehyde oxidase, zeaxanthin epoxidase, 9-cis-epoxycarotenoid dioxygenase, and molybdenum cofactor sulturase (Khan et al., 2020a). Several PGPB strains can synthesize and degrade the ABA (Tsukanova et al., 2017). Nevertheless, the production of ABA via bacteria or the excitation of ABA synthesis in plants has been scarcely studied (Gowtham et al., 2020).
Auxins are involved in nearly all aspects of plant physiology and the mitigation of abiotic stress (Olenska et al., 2020). Bacteria affect plant auxin homeostasis by synthesizing auxin, affecting the expression of the plant auxin production genes, and transport or signaling machinery (Tsukanova et al., 2017). Auxins are synthesized and excreted by above 80% of the rhizobacteria (Olenska et al., 2020). In bacteria, five of the six pathways for auxin production depend on tryptophan. These pathways have been categorized according to their intermediate including indole-3-pyruvate (IPyA), indole-3-acetamide (IAM), indole-3-acetonitrile, tryptophan side-chain oxidase, tryptamine, and tryptophan independent (Gamalero and Glick, 2011). IAM and IPyA are apparently two main microbial pathways (Spaepen, 2015; Gupta et al., 2016). At the IAM pathway, tryptophan is first transformed to IAM through tryptophan monooxygenase. Then, IAA is synthesized from IAM through an IAM hydrolase (Spaepen, 2015). Most PGPB apply the IPyA pathway (Gupta et al., 2016; Khatoon et al., 2020). In the IPyA pathway, tryptophan is transaminated to IPyA via a pyridoxal phosphate-dependent aromatic aminotransferase. Then IPyA decarboxylase (IPDC, ipdC gene) transforms IPyA to indole-3-acetaldehyde (IAAld). Lastly, IAAld is transformed into IAA (Glick et al., 1999a; Olenska et al., 2020). Multiple IAA biosynthetic pathways can be existent and active in one single organism as there are both the IAM and the IPyA pathway in the Pantoea agglomerans genome (Spaepen, 2015). The genes related to IAA biosynthesis have been identified in the genomes of more than 70% of assessed halotolerant PGPB and the genes related to IPyA and IAM pathways were more prevalent (80% and 60%, respectively). Over 63% of assessed IAA producing bacteria contain more than one pathway (Table 3).
The transcriptomic analysis of Paenibacillus polymyxa YC0136 in intraction with tobacco showed the expression of the ilvB gene in strain YC0136 was up-regulated. IlvB encodes aldehyde dehydrogenase, which transforms indole-3-acetaldehyde into IAA (Liu et al., 2020).
The expression of genes related to indole-3-acetic acid (IAA) synthesis including the genes of nitrile hydrolase, IAM hydrolase, and aldehyde dehydrogenase were found in transcriptome assay of Burkholderia phytofirmans PsJN colonizing potato under drought stress (Sheibani-Tezerji et al., 2015).
Gibberellins (GAs) are tetracyclic diterpenoid carboxylic acid derivatives that can alleviate the abiotic stress (Khatoon et al., 2020). PGPB affect the quantity of gibberellin in plants in a manner similar to auxin. Gibberellin-producing PGPB conserve plants from stress by modulating antioxidant levels, stimulating the absorption of calcium ions and other nutrients (Khatoon et al., 2020). Among 136 known GA structures, GA3 is most often synthesized by bacteria (Olenska et al., 2020). In recent years, the GA biosynthesis pathway in bacteria has been clarified (Salazar-Cerezoa et al., 2018). Bacterial synthesis is related to a cytochrome P450-rich operon which contains a cluster of core genes coding for eight enzymes (Figure 3). Numerous copies of the operon further than these core genes enclose an isopentenyl diphosphate isomerase (IDI) and CYP115. GA operon is typically located on plasmids, though; the basis of this genetic transfer has not been clarified (Nagel et al., 2018; Olenska et al., 2020). Some bacteria, such as Burkholderia phytofirmans PsJN don’t produce gibberellins but positively regulate the expression of the related genes in the synthesis of gibberellin in A. thaliana (Tsukanova et al., 2017).

FIGURE 3. Bacterial gibberellin synthetic core operon. Isoprenyl diphosphate synthase (IDS) synthesizes geranylgeranyl diphosphate (GGDP), the pair of diterpene cyclases (copalyl diphosphate (CDP) synthase (CPS) and ent-kaurene synthase (KS)) catalyze the synthesis of ent-kaurene from GGDP. Ent-kaurene is converted to GAs through several oxidation steps by a minimum of three cytochromes P450 (CYPs), CYP112, CYP114, and CYP117, short-chain alcohol dehydrogenase/reductase (SDR), and a ferredoxin (Fd) (created in BioRender.com).
Cytokinins are adenine derivatives involved in root differentiation, shoot formation, and stress alleviation (Numan et al., 2018). Many PGPB can synthesize cytokinin, which is started via isopentenyl transferase (ipt gene) that catalyzes the transportation of the isopentenyl moiety from dimethylallyl diphosphate to adenosine monophosphate. The influence of cytokinin in the case of pathogens is inhibitory on plants, and is stimulatory in the case of PGPB as the amounts of the synthesized cytokinin by PGPB are less than those from phytopathogens (Shilev, 2020).
Ethylene acts in several stages of plant ontogenesis and stress signaling pathways (Kumar et al., 2020). The ethylene synthesis is under tight control at transcriptional and post-transcriptional levels (Choudhary et al., 2015). The upper concentration of ethylene accumulation under stress condition can prevent plant growth and accelerate senescence (Olenska et al., 2020). Ethylene is produced through ACC oxidase (ACCO) from ACC (Olenska et al., 2020; Shilev, 2020). When ACCO synthesizes ethylene higher than its threshold amount can cause “stress ethylene” in the plant (Olanrewaju et al., 2017). The expression and activity of ACC synthase (ACCS) are increased by IAA increase in plants (Glick et al., 1999a; Olenska et al., 2020). ACC is exuded into the soil surrounding roots and is reabsorbed via the roots (Martinez-Viveros et al., 2010). PGPB mediate ethylene homeostasis by decreasing its amount within plants due to their rhizobitoxine and ACC deaminase enzyme (ACCD) generation (Olenska et al., 2020). Rhizobitoxine is an enol-ether amino acid and acts as a competitive inhibitor of ACCS and the production of ACCD. The activities of rhizobitoxine relate to the inhibition of β-cystathionase in the methionine (precursor of ethylene) synthesis and ACCS in the ethylene production pathway. PGPB decrease the deleterious effect of ethylene by eliminating ACC (Choudhary et al., 2015). PGPB may also affect plant ethylene concentration by influencing the expression of the genes connected to ethylene production; genes of ACCS and ACCO enzymes. For example, Burkholderia phytofirmans PsJN upregulates ACCS and ACCO genes in A. thaliana (Tsukanova et al., 2017).
Salicylic acid (SA) is a main phytohormone that modulates different aspects of plant growth, and biotic and abiotic stress responses. In bacteria, enzymes related to SA biosynthesis are encoded by the NRPS gene cluster. In first step which is common in both bacteria and plants, chorismate is converted to isochorismate by isochorismate synthase, and then SA is produced from isochorismate using isochorismate pyruvate lyase (Chen et al., 2019; Mishra and Baek, 2021).The metabolites related to the auxins and zeatins biosynthesis pathways were explored from three PGPB (Pseudomonas putida IDE-01, Azospirillum brasilense IDE-06, and Bacilus megaterium IDE-14) in interaction with maize and rice (Garcia et al., 2022).
Environmental stresses lead to increased ethylene production in the plant which hampers the growth of plants (Kumari et al., 2016). The ACCD-synthesizing bacteria can diminish the deleterious impact of the different stresses on plants by catabolizing ACC to
Many of the acdS genes have been principally modulated through the leucine-responsive regulatory protein (LrP) and AcdB protein. LrP is encoded by the ACCD regulatory (acdR) gene located upstream of the acdS gene (Figure 4) (Kumari et al., 2016). In addition, acdS gene expression is controlled through various regulatory proteins in diverse bacterial species for example, σ70 and LrP in Burkholderia sp. CCGE 1002 and Burkholderia phymatum STM 815, nifA1, nifA2, and σ54 in Mesorhizobium loti (Kumari et al., 2016). Moreover, some of the PGPB synthesize homologs of ACCD, d-cysteine desulfhydrase encoded via dcyD (Suarez et al., 2019). The potential genes for ACCD activity of Hartmannibacter diazotrophicus E19T are suggested as ygeX, and tdcB as their products have ammonia-lyase activity with pyridoxal phosphate dependencies like genes acdS and dcyD (Suarez et al., 2019) (Table 4).
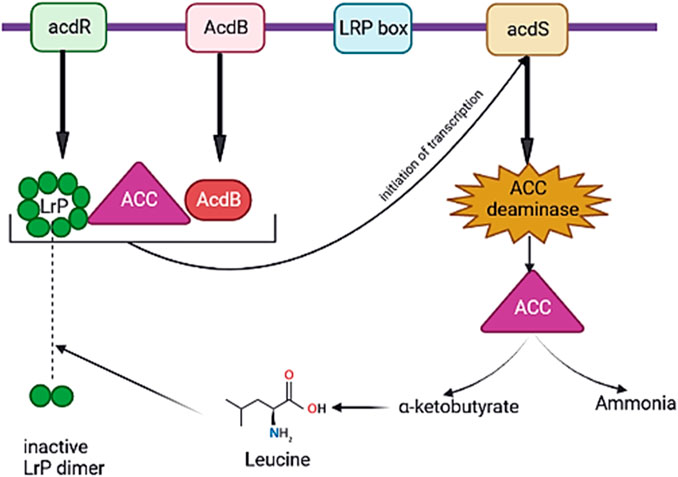
FIGURE 4. Regulation of expression of acdS (ACC deaminase synthase) gene through the leucine-responsive regulatory protein (LrP) and AcdB protein. When 1-aminocyclopropane-1-carboxylic acid (ACC) exists, active octamer of LrP is encoded through acdR gene. As a result of the interaction of active LrP with AcdB protein and ACC, a complex is generated, which starts the transcription of acdS gene, and ACCD is synthesized. The interaction of leucine as the amount of it rises in the bacteria with the active LrP octamer makes an inactive LrP dimer that causes stopping the transcription of the acdS gene (created in BioRender.com).
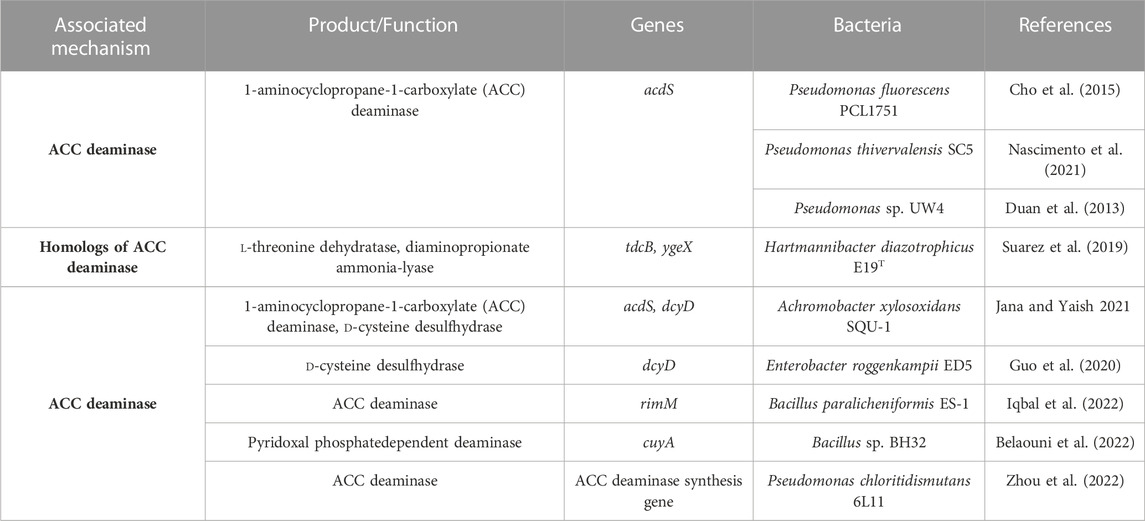
TABLE 4. Genes related to 1-aminocyclopropane-1-carboxylate (ACC) deaminase in the genomes of 20 assessed halotolerant PGPB.
Induced systemic tolerance is presented as physical and chemical changes triggered by PGPB and causes enhanced tolerance of plants to abiotic stress (Sharma et al., 2016). It has been proposed that adaptation to stresses is related to pre-existing or “memory” defenses. “Memory” defenses in plants are induced via priming with specified chemicals and microbes. The percept of plants to chemicals with external origin and microbe-associated molecular patterns (MAMPs) can prompt reaction versus abiotic stresses (Vaishnav et al., 2016). PGPB produce diverse signaling molecules comprising cyclodipeptides, cytokinins, volatile organic compounds (VOCs), quorum sensing molecules, antioxidants, and ACCD, which triggers systemic resistance in plants (Arora et al., 2020; Etesami, 2020).
The biosynthesis of osmolytes (compatible solutes) by PGPB and plants in reaction to stress operates in synergism to improve plant growth (Kaushal and Wani, 2016). Osmolytes include proline, quaternary ammonium compounds, sugars, betaines, polyamines, polyhydric alcohols, other amino acids, and water-stress proteins such as dehydrins (Vurukonda et al., 2016). Synthesis of osmolytes via PGPB is almost certainly faster than their related host plants, and plants prefer the uptake of osmolytes (Ilangumaran and Smith, 2017; Sekar et al., 2019).
Proline is a significant osmolyte having a multifunctional role, including maintenance of cytosolic pH, antioxidant activity, and function as molecular chaperone during osmotic stresses (Carmen and Roberto, 2011; Kaushal and Wani, 2016). Plants inoculated with bacteria illustrate a higher amount of proline but whether it is because of upregulation of the proline biosynthesis pathway or is absorbed from the rhizosphere has not been elucidated (Kaushal and Wani, 2016). In most of the bacteria, biosynthesis of proline implicates the united function of γ-glutamyl kinase, γ-glutamyl phosphate reductase, and 1-pyrroline-5-carboxylate reductase enzymes, which are encoded by proB, proA, and proC genes, respectively (Sunita et al., 2020). Nitrogen-fixing bacteria show elevated proline metabolism because of the enhanced acting of proline dehydrogenase (PDH) under the salinity stress condition (Sunita et al., 2020).
Glycine betaine (GB) (N,N,N-trimethyl glycine) is a quaternary ammonium synthesized by choline oxidase, aiding plants to tolerate stress through the stability of membranes, proteins, and the action of RuBisCO (Das et al., 2015; Kaushal and Wani, 2016). Bacillus subtilis can produce GB by oxidizing the choline to glycine betaine aldehyde through type III alcohol dehydrogenase, and finally, a glycine betaine aldehyde dehydrogenase synthesizes GB (Sunita et al., 2020). In Arthrobacter globiformis, the codA gene encodes choline oxidase (Kumar et al., 2020). More than 50% of assessed halotolerant PGPB contain the genes related to glycine betaine synthesis (Table 5).
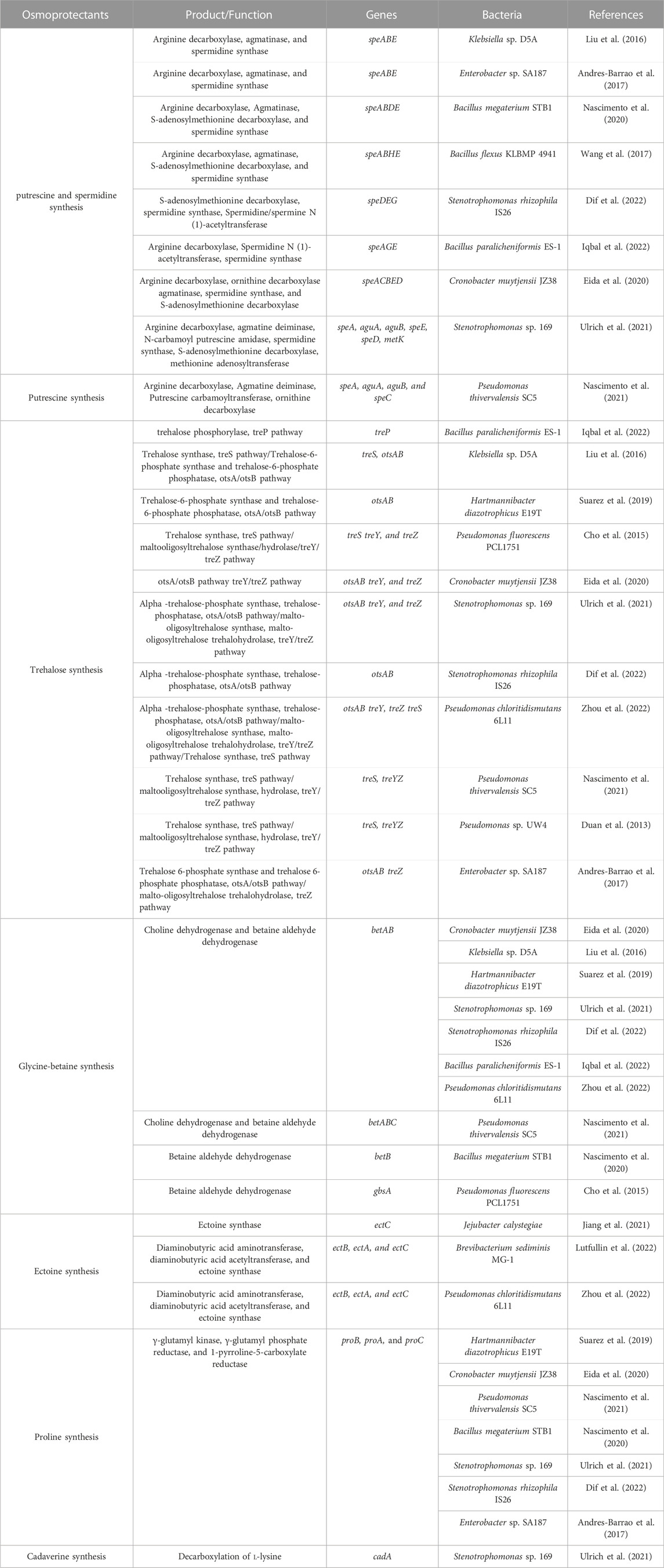
TABLE 5. Identified genes related to osmolyte production in the 20 studied genomes of halotolerant PGPB.
Polyamines are present in nearly all organisms and are low molecular weight composites with aliphatic nitrogen structure (Kaushal and Wani, 2016). Polyamines like spermine, putrescine, and spermidine are functional agents of PGPB (Nascimento et al., 2020). Furthermore, spermidine production has been revealed to reduce the action of the ACCO gene in the ethylene synthesis pathway in tobacco (Nascimento et al., 2020).
Ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid) accumulates in plants during salinity stress like other osmolytes and improves protein folding and protects biomolecules and even whole cells (Das et al., 2015; Sunita et al., 2020). Ectoine is synthesized by the products of ectABC. The ectA, ectB, and ectC genes code for diaminobutyric acid acetyltransferase, diaminobutyric acid aminotransferase, and ectoine synthase, respectively (Das et al., 2015; Sunita et al., 2020).
Trehalose is a non-reducing disaccharide, an
RNA-Seq analysis of Chromohalobacter salexigens ANJ207 revealed the expression of genes related to betaine and choline transport systems and synthesis of glycine betaine, coline, and proline increased (Srivastava et al., 2022).
Bacteria contribute to toxic ion homeostasis that ameliorates plant tolerance under salinity. These bacteria decrease the absorption of toxic ions by controlling the expression of plants ion transporter and the construction of rhizosheaths through the production of exopolysaccharides (EPS) (Vaishnav et al., 2016; Ilangumaran and Smith, 2017). EPS binds cations, including Na+, and limits the entry of Na+ into roots (Etesami, 2020). PGPB contribute to the nutrient status in plants through microbial activities like organic acid excretion, phosphate solubilization, and siderophore synthesis, as mentioned in section 3-1. These nutrients reduce toxic ion accumulation (Vaishnav et al., 2016; Ilangumaran and Smith, 2017). PGPB enhance the K+ absorption by upregulating the K+ transporter, reducing the accumulation of Na+ in leaves and enhancing Na+ exclusion at the roots leading to an increased K+/Na+ ratio (Bhat et al., 2020; Shilev, 2020). Limiting the Na+ uptake at the root surface induces the HKT1 expression in shoots which assists the recirculation of Na+ from shoot to roots and aids in keeping an elevated K+/Na+ ratio in plants. The inoculation of Helianthus annus by Bacillus subtilis leads to the downregulation of HKT1/K+ transporter expression (Bhat et al., 2020).
Bacterial VOCs are species-specific metabolites sensed by other bacteria, insects, plants, animals, and microorganisms participating in cell-to-cell signaling and growth promotion (Kaushal and Wani, 2016, Ryu, 2015). Furthermore, their role in regulating bacterial motility, controlling virulence factors, and production of osmolytes, phytohormones, and siderophores are reported (Vaishnav et al., 2016). VOCs synthesis also affects the modulation of the HKT1/K+ transporter and controls the Na+ homeostasis pathway in plants (Vaishnav et al., 2016; Sunita et al., 2020). VOCs work as signals for a systemic reaction inside the same or neighboring plants (Bhat et al., 2020).
Acetoin and 2,3-butanediol are synthesized when two pyruvate molecules are compressed into acetolactate and transformed to acetoin via acetolactate decarboxylase, and lastly, acetoin reductase catalyzes 2,3-butanediol from acetoin (Liu et al., 2016; Suarez et al., 2019). Hartmannibacter diazotrophicus E19T genome contains the encoding gene of acetolactate synthase (ilv) but not genes related to acetolactate decarboxylase and acetoin reductase. Acetoin can be synthesized in E19T through a spontaneous decarboxylation of acetolactate into diacetyl, when oxygen is present, and then reduction of diacetyl to acetoin through the diacetyl reductase (product of budC gene) (Table 6) (Suarez et al., 2019).
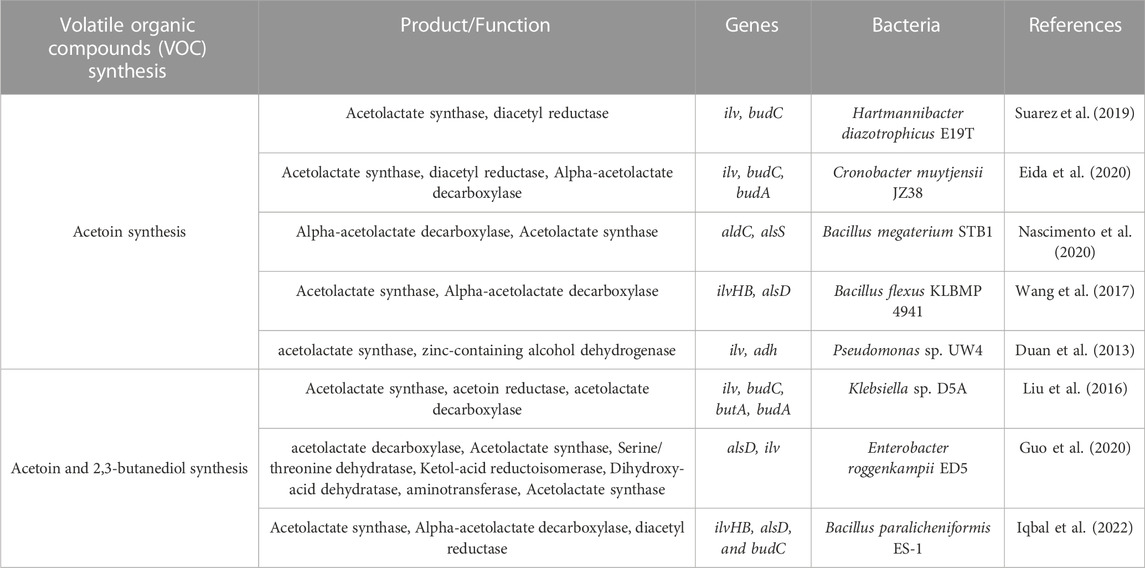
TABLE 6. Genes related to volatile organic compounds (VOCs) synthesis in the genomes of 20 studied halotolerant PGPB.
Plants are equipped with antioxidant defense systems, including enzymatic and non-enzymatic mechanisms against the harmful effects of ROS. Enzymatic elements comprise catalase (CAT), monodehydroascorbate reductase (MDHAR), superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione reductase (GR), and non-enzymatic compounds consist of ascorbate, cysteine, carotenoids, tocopherols, flavonoids, and glutathione (Arora et al., 2020; Kumar et al., 2020) (Table 7). PGPB can increase the functionality of antioxidant defense systems (Antar et al., 2021). Bacillus thuringiensis AZP2 induces SOD, CAT, and GR and enhances the tolerance of wheat to drought (Tiepo et al., 2020). Inoculated rice with Trichoderma asperellum and Pseudomonas fluorescens shows an increase in the action of APX, POD, CAT, and SOD (Bhat et al., 2020). Inoculation of Arabidopsis with Enterobacter sp. EJ01 caused higher APX activity (Mishra et al., 2018). Trichoderma, Pseudomonas, and their combination inoculation directly resulted in the upregulation of genes related to defense systems in plants, including phenylpropanoid (PAL), SODs, CAT, and APX (Singh et al., 2020). However, the mechanisms by which PGPB influence antioxidant enzymes are poorly understood (Figure 5).
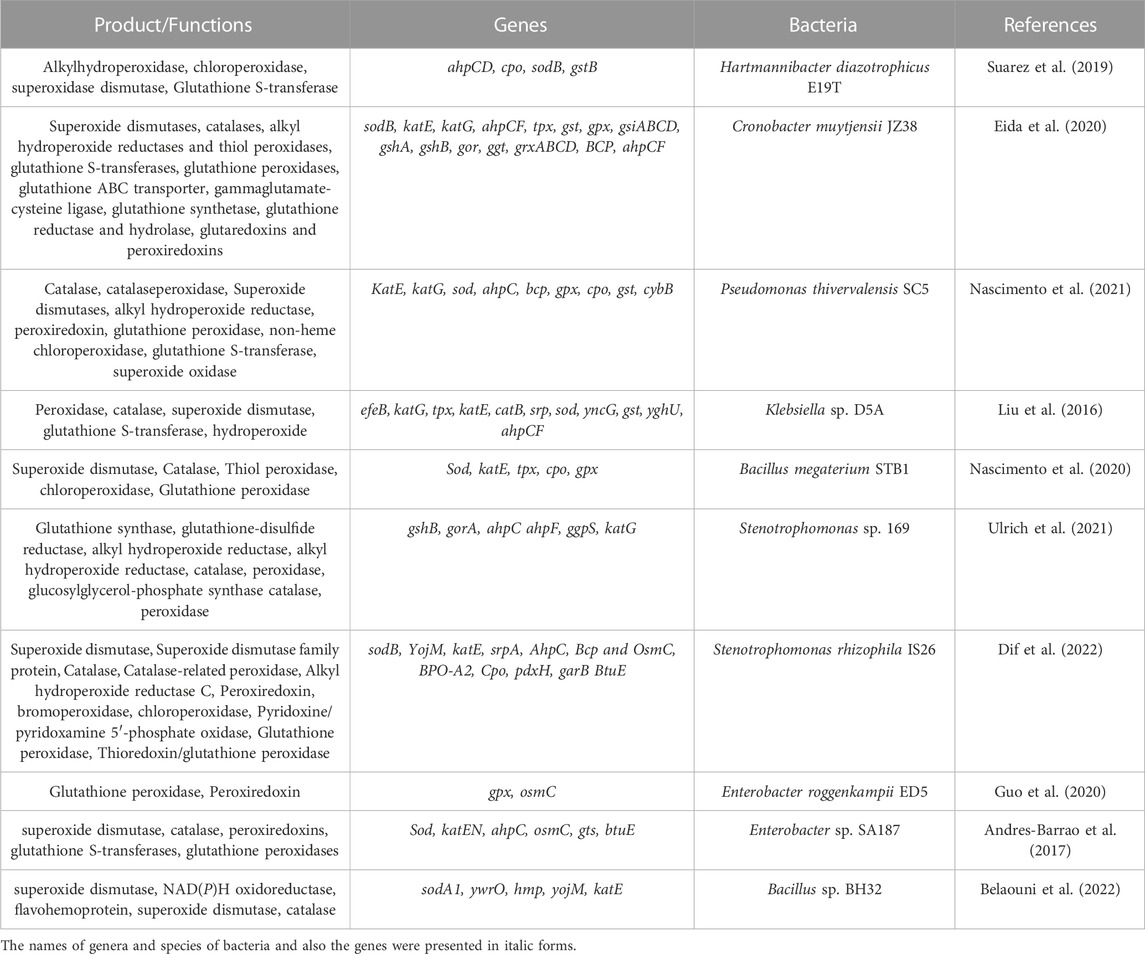
TABLE 7. Identified genes related to antioxidant defense in the genomes of studied halotolerant PGPB.
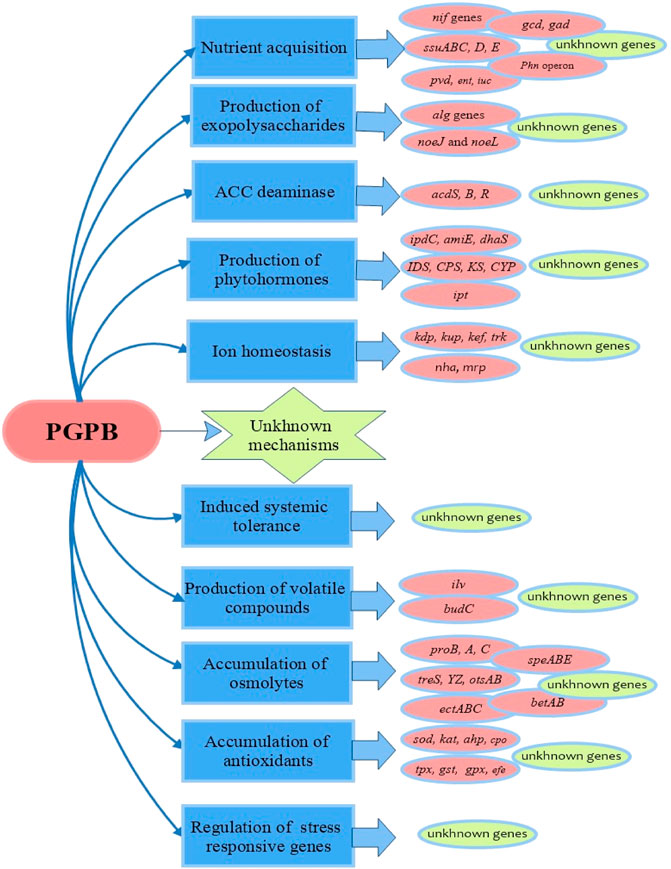
FIGURE 5. Mechanisms of salinity stress alleviation in plants by PGPB and so far assigned genes. There are some gaps and unknown mechanisms and genes that must be illustrated.
The comparisons of the expressed genes in Burkholderia phytofirmans PsJN from control plants and drought-stressed plants showed the genes related to glutaredoxin and alkyl hydroperoxide reductase were upregulated (Sheibani-Tezerji et al., 2015).
The transcriptome analysis of Chromohalobacter salexigens ANJ207 which is a halophilic PGPB showed the expression of genes related to catalase, thioredoxin reductase, oxidoreductase, methionine sulfoxide reductases, iron superoxide dismutase, peroxidase OsmC, peroxiredoxin, and an alkyl-hydroperoxide reductase, glutaredoxin, ferredoxin synthesis increased (Srivastava et al., 2022).
PGPB induce and up-regulate the expression of genes linked with tolerance to abiotic stresses in plants (Khan et al., 2020b; Sharma S. et al., 2021). Stress-responsive genes expression is mainly regulated via transcription factors and prompted through stress sensors that are exposed to complex control of phytohormones (Khan et al., 2020a). PGPB can augment tolerance of the plant to salt stress by regulating the expression of SOS genes, RAB18 (LEA), WRKY TFs, DRE (dehydration responsive element), RD29A, RD29B regulons of ABRE (ABA-responsive elements), besides transcription factor DRE binding proteins (DREB2b) (Chakdar et al., 2019; Etesami, 2020). PGPB also aids plants in reducing their cell water potential to continue absorbing water from saline soils by upregulating genes related to generating aquaporins. Enterobacter spp. upregulates salt stress-responsive genes including RAB18, DREB2b, RD29A, and RD29B in Arabidopsis in the salinity stress. TaMYB and TaWRKY genes expression are induced in inoculated wheat by Dietzia natronolimnaea (Kumar et al., 2020). Bacillus amyloliquefaciens SN13 upregulates SOS1, SERK1, EREBP, and NADP-Me2 genes and enhances the salt tolerance of rice (Chakdar et al., 2019).
Bacterial exopolysaccharides compose bound cell surface homo or hetero-polysaccharides in the form of capsule or slime, forming the skeleton of the biofilms. EPS production by PGPB under stress forms hydrophilic biofilms conferring desiccation protection, regulates nutrients and water flow across plant roots, binds to Na+ and decreases the bioavailability of the ion, aggregates root-adhering soils (RAS) and stabilizes soil aggregates (Mishra et al., 2018; Etesami, 2020; Jiang et al., 2021). The products of noeJ and noeL genes in Azospirillum brasilense Sp7 are mannose-6-phosphate isomerase, and GDP-mannose 4,6-dehydratase, which participate in EPS production. In P. aeruginosa, water scarcity condition induces alg genes of the alginate bioproduction gene cluster (Kaushal and Wani, 2016).
Nitric oxide (NO) plays role in plant–bacteria interactions. NO has a dual effect (harmful and beneficial effects). Produced NO by PGPB, promotes plant growth and health and plays important roles in the response to environmental stress. NO influences root growth and developmental processes as a mediator in auxin-regulated signaling cascades, induces antioxidant system and scavenges directly ROS. On the other hand, NO is implicated in the pathogenesis of bacterial phytopathogens (Santana et al., 2017). NO dualistic nature depends on the NO level. Phytopathogens synthesize a high NO level and stimulate disease, while a lower level of NO synthesized by PGPR promote plant growth and increase plant tolerance (Santana et al., 2017).
Nitric oxide (NO) is an intermediate of denitrification process in bacteria. A membrane-bound nitrate reductase (Nar) or a periplasmic nitrate reductase (Nap) reduces nitrate to nitrite. Nitrite is reduced to NO either by a heme (NirS) or copper (NirK) containing nitrite reductase (Santana et al., 2017). NO is also synthesized in bacteria through nitric oxide synthases (NOS) which produce NO through the oxidation of l-arginine to l-citrulline. Moreover, ammonia-oxidizing bacteria produce NO by couples ammonia oxidation and denitrification. At first ammonium is oxidized to hydroxylamine by ammonium monooxygenase. Hydroxylamine oxidoreductase oxidizes hydroxylamine to nitrite. Finally, nitrite is reduced to NO by nitrite reductase (Molina-Favero et al., 2007; Santana et al., 2017).
Molina-Favero et al., 2008 showed the synthesized NO by Azospirillum brasilense Sp245 induced lateral and adventitious root development in inoculated tomato (Molina-Favero et al., 2008). Application of Bacillus xiamenensis ASN-1 and sodium nitroprusside (SNP) as a NO donor synergistically alleviated salinity stress in sugarcane by preserving the relative water content, gas exchange parameters, osmolytes, electrolyte leakage, and Na+/K+ ratio, modulating the antioxidant enzyme activities and stress-related gene expression (Sharma A. et al., 2021).
Currently, due to the general requirement to feed a constantly growing world population, sustainable agriculture must be developed. This is despite the fact that the salinization of lands is one of the main obstacles to agricultural productivity. The application of salinity-tolerant PGPB and their metabolites improves the productivity of soils that are under salinity stress and has been a significant element in achieving the aim of food security and sustainability. Revealing the molecular mechanisms of action in PGPB and their interactions with plants is a crucial step toward targeting the use of PGPB as stress-alleviating biofertilizer. Accordingly, the number of genomes of rhizobacteria that were assembled, bioprojects, and articles on this subject in PubMed have increased 20, 7, and 6 times in 2020 compared to 2010, respectively. However, despite the development of new biotechnology tools and techniques, computational tools, and increasing studies that have been done on this issue, there are unknown gaps in molecular mechanisms of PGPB action because of the genetic and metabolic diversities of rhizobacteria and the intricacy of their interactions. Consequently, for evolving use of PGPB in sustainable agriculture, more studies must investigate the molecular aspect of PGPB action. To study genes, the PGPB genome analyzers need a better understanding of the genes and pathways related to plant growth promotion and stress alleviation that are known so far. In this study, the identified genes related to salinity stress mitigation and plant growth promotion in the genomes of 20 studied halotolerant PGPB are assessed. The genes related to the synthesis of IAA, siderophores, osmoprotectants, chaperons, ACC deaminase, and antioxidants, phosphate solubilization, and ion homeostasis are the most prevalent identified genes in the genomes of assayed halotolerant PGPB. The genes of dnaJK, groEL, groES, IPyA, and IAM pathways, and entABCDEF are the most commonly identified genes related to heat-shock proteins (HSPs), IAA, and enterobactin siderophore synthesis in the genomes of assessed halotolerant PGPB, respectively. Genes associated with the production of trehalose and glycine-betaine are the most prevalent identified osmoprotectants genes. In many of the assessed genomes of the halotolerant PGPB, there are two pathways of trehalose synthesis, and otsA/otsB, treY/treZ, and treS are the most common pathways. The presence of several trehalose biosynthesis pathways in the genomes of halotolerant PGPB can be due to the severe requirement to accumulate trehalose under stressful environmental conditions. There are several genes related to different antioxidant enzymes and non-enzymatic antioxidant mechanisms in the genomes of assessed halotolerant PGPB, which enabled them to tolerate stress. The identified genes related to gluconic acid synthesis (gcd and pqq) as the most prevalent agent for releasing phosphate and different phosphatases exist in the genomes of 11 of the assessed halotolerant PGPB. The existence of genes associated with phosphate solubilization in these halotolerant PGPB must be correlated to the decrease of the bioavailability of phosphorus in saline soil (Xie et al., 2022). Moreover, the genomes of assessed halotolerant PGPB contain the genes related to ion homeostasis, including kdp and nha, with high prevalence.
For the application of PGPB as stress-alleviating biofertilizers, determining the molecular aspects of action in PGPB and plant-bacteria interactions is a pivotal prerequisite. In this study, the identified genes and pathways related to plant growth promotion and stress alleviation in the genomes of 20 halotolerant PGPB (Pseudomonas and Bacillus genera, each with 20% prevalence as the most common genera) were assessed. The gene sets, pathways, and analog genes which have been identified in the genomes of assessed halotolerant PGPB can help the genome analyzers to mine the genomes of other plant-associated bacteria with higher accuracy. In addition, this data is useful for other omics and meata-omics studies. The genes related to synthesis of IAA (IPyA and IAM pathways), siderophores (based on the family of bacteria studied like ent and pvd), osmoprotectants (otsAB, treS, treY, treZ, and betAB), chaperons (dnaJK, groES, and groEL), ACC deaminase (acds and its homologs), and antioxidants (sod, kat, and ahp), phosphate solubilization (gcd and pqq), and ion homeostasis (kdp and nha) are the most prevalent attibuted genes in the genomes of these 20 halotolerant PGPB. Mentioned genes can be used as candidates for designing molecular markers to screen novel efficient PGPB for mitigating salinity stress. Genomics and metagenomics studies are only predictive of functional potential of bacteria. To determine the efficiency of PGPB, the expression of genes in them should be assayed in interaction with plants which reflect the inferiority of genomic interpretations versus interactomics with more realistic results. Moreover, due to the high intricacy of plant-bacteria interactions, and the genetic and metabolic diversities of plant-associated bacteria, further omics studies are needed to complete the gaps in molecular aspects of PGPB protective activities.
Further studies for illustrating the molecular detail of PGPB mechanisms of action will lead to the discovery of novel and diverse genes, pathways, and metabolites, and completion of gaps in the pathways. Furthermore, molecular analysis assists in identification of silent genes under lab conditions (because of lack of natural triggers or signals), introducing safety assessment markers (with analysis of presence or absence of pathogenic genes), easy and fast screening of more PGPB strains with the utilization of gene markers, finding strains with multiple capabilities such as bioremediation, neutralizing biotic/abiotic stress, and optimization of applying PGPB as biofertilizer. Furthermore, considering that about 1% of soil bacteria are culturable, metagenomic analysis of the rhizospheric bacteria will provide access to new and rich gene repositories for plant protection.) In combination with findings from applying other meta (omics) approaches in interaction studies between different bacteria, plants, and natural environment will unreveal the unknowns of pathways and mechanisms of actions. Molecular aspects of how PGPB cause induced systemic resistance (ISR) and regulate stress responsive genes are among the mechanisms that need to be studied more.
Accordingly, future advances in sequencing, mass spectrometry, and other metabolomics technologies, annotating and predicting platforms, and databases will facilitate the identification of molecular aspects with high throughput and coverage depth to open the current bottleneck in soil interactomics and metaphenomics.
SZ-N: Data collection and drafting of the manuscript FM: Conceptualization and manuscript edition MH-M: Manuscript edition SS: Manuscript edition.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
PGPB, plant growth-promoting bacteria; IAA, indole-3-acetic acid; NGS, next-generation sequencing; ECF, extracytoplasmic function; HSPs, Heat-shock proteins; KSB, K-solubilizing bacteria; NRPSs, non-ribosomal peptide synthetases; PSB, Phosphate-solubilizing bacteria; GA, gluconic acid; PQQ, pyrrolo-quinolone quinine; NSAPs, non-specific acid phosphatases; ABA, Abscisic acid; IPyA, indole-3-pyruvate; IAM, indole-3-acetamide; GAs, Gibberellins; SA, Salicylic acid; ACC, 1-aminocyclopropane-1-carboxylic acid; ACCO, ACC oxidase; ACCS, ACC synthase; ACCD, ACC deaminase enzyme; VOCs, volatile organic compounds; GB, Glycine betaine; EPS, exopolysaccharides; CAT, catalase; MDHAR, monodehydroascorbate reductase; SOD, superoxide dismutase; APX, ascorbate peroxidase; GR, glutathione reductase; RAS, root-adhering soils.
Ahmed, V., Verma, M. K., Gupta, S., Mandhan, V., and Chauhan, N. S. (2018). Metagenomic profiling of soil microbes to mine salt stress tolerance genes. Front. Microbiol. 9, 159. doi:10.3389/fmicb.2018.00159
Alberton, D., Valdameri, G., Moure, V. R., Monteiro, R. A., Pedrosa, F. O., Muller-Santos, M., et al. (2020). What did we learn from plant growth-promoting rhizobacteria (PGPR)-grass associations studies through proteomic and metabolomic approaches? Front. Sustain. Food Syst. 4, 607343. doi:10.3389/fsufs.2020.607343
Andres-Barrao, C., Lafi, F. F., Alam, I., de Zelicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8, 2023. doi:10.3389/fmicb.2017.02023
Antar, M., Gopal, P., Msimbira, L. A., Naamala, J., Nazari, M., Overbeek, W., et al. (2021). “Inter-organismal signaling in the rhizosphere,” in Rhizosphere biology: Interactions between microbes and plants. Editors V. V. S. R. Gupta, and A. K. Sharma (Singapore: Springer), 255–293. doi:10.1007/978-981-15-6125-2_13
Arora, N. K., Fatima, T., Mishra, J., Mishra, I., Verma, S., Verma, R., et al. (2020). Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 26, 69–82. doi:10.1016/j.jare.2020.07.003
Aslam, M. M., Akhtar, K., Karanja, J. K., Noor-ul-Ain, N., and Haider, F. U. (2020). “Understanding the adaptive mechanisms of plant in low phosphorous soil,” in Plant stress physiology. Editor A. Hossain (London: IntechOpen). Available at: https://www.intechopen.com./chapters/71489.10.5772/intechopen.91873.
Azeem, M., Soundari, P. G., Ali, A., Tahir, M. I., Imran, M., Bashir, S., et al. (2022). Soil metaphenomics: A step forward in metagenomics. Archives Agron. Soil Sci. 68 (12), 1645–1663. doi:10.1080/03650340.2021.1921743
Balasubramanian, V. K., Jansson, C., Baker, S. E., and Ahkami, A. H. (2021). “Molecular mechanisms of plant–microbe interactions in the rhizosphere as targets for improving plant productivity,” in Rhizosphere biology: Interactions between microbes and plants. Editors V. V. S. R. Gupta, and A. K. Sharma (Singapore: Springer), 295–338. doi:10.1007/978-981-15-6125-2_14
Batista, B. D., Bonatelli, M. L., and Quecine, M. C. (2021). “Fast screening of bacteria for plant growth promoting traits,” in The plant microbiome: Methods and protocols, methods in molecular biology. Editors L. C. Carvalhais, and P. G. Dennis (New York: Springer), 67–75. doi:10.1007/978-1-0716-1040-4_7
Belaouni, H. A., Compant, S., Antonielli, L., Nikolic, B., Zitouni, A., and Sessitsch, A. (2022). In-depth genome analysis of Bacillus sp. BH32, a salt stress-tolerant endophyte obtained from a halophyte in a semiarid region. Appl. Microbiol. Biotechnol. 106 (8), 3113–3137. doi:10.1007/s00253-022-11907-0
Bhat, M. A., Kumar, V., Bhat, M. A., Wani, I. A., Dar, F. L., Farooq, I., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 11, 1952. doi:10.3389/fmicb.2020.01952
Bourque, F. G., Bertrand, A., Claessens, A., Aliferis, K. A., and Jabaji, S. (2016). Alleviation of drought stress and metabolic changes in Timothy (Phleum pratense L.) colonized with Bacillus subtilis B26. Front. Plant Sci. 7, 584. doi:10.3389/fpls.2016.00584
Bruto, M., Prigent-Combaret, C., Muller, D., and Moenne-Loccoz, Y. (2014). Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 4, 6261. doi:10.1038/srep06261
Carmen, B., and Roberto, D. (2011). “Soil bacteria support and protect plants against abiotic stresses,” in Abiotic stress in plants-mechanisms and adaptations. Editor A. Shanker (InTech). ISBN: 978-953-307-394-1.
Chakdar, H., Borse, D. N., Verma, S., Choudhary, P., and Das, S. (2019). “Microbial management of crop salinity stress: Mechanisms, applications, and prospects,” in Salt stress, microbes, and plant interactions: Mechanisms and molecular approaches. Editor M. S. Akhtar (Singapore: Springer), 1–25. doi:10.1007/978-981-13-8805-7_1
Chaudhry, U. K., Gokce, Z. N. O., and Gokce, A. F. (2022). The influence of salinity stress on plants and their molecular mechanisms. Biol. Life Sci. Forum 31. doi:10.3390/IECPS2021-12017
Chen, H., Clinton, M., Guang, Q., Daowen, W., Liu, F., and Qing Fu, Z. (2019). Connecting the dots: A new and complete salicylic acid biosynthesis pathway. Mol. Plant 12, 1539–1541. doi:10.1016/j.molp.2019.11.007
Cho, S. T., Chang, H. H., Egamberdieva, D., Kamilova, F., Lugtenberg, B., and Kuo, C. (2015). Genome analysis of Pseudomonas fluorescens PCL1751: A rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS ONE 10 (10), e0140231. doi:10.1371/journal.pone.0140231
Choudhary, D. K., Kasotia, A., Jain, S., Vaishnav, A., Kumari, S., Sharma, K. P., et al. (2015). Bacterial-mediated tolerance and resistance to plants under abiotic and biotic stresses. J. Plant Growth Regul. 35, 276–300. doi:10.1007/s00344-015-9521-x
Das, P., Behera, B. K., Meena, D. K., Azmi, S. A., Chatterjee, S., Meena, K., et al. (2015). Salt stress tolerant genes in halophilic and halotolerant bacteria: Paradigm for salt stress adaptation and osmoprotection. Int. J. Curr. Microbiol. App. Sci. 4 (1), 642–658.
Dif, G., Belaouni, H. A., Yekkour, A., Goudjal, Y., Djemouai, N., Penazova, E., et al. (2022). Performance of halotolerant bacteria associated with sahara-inhabiting halophytes Atriplex halimus L. And lygeum spartum L. Ameliorate tomato plant growth and tolerance to saline stress: From selective isolation to genomic analysis of potential determinants. World J. Microbiol. Biotechnol. 38, 16. doi:10.1007/s11274-021-03203-2
Drogue, B., Sanguin, H., Borland, S., Prigent-Combaret, C., and Wisniewski-Dye, F. (2014). Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol. Ecol. 87 (2), 543–555. doi:10.1111/1574-6941.12244
Duan, J., Jiang, W., Cheng, Z., Heikkila, J. J., and Glick, B. R. (2013). The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS ONE 8 (3), e58640. doi:10.1371/journal.pone.0058640
Eida, A. A., Bougouffa, S., L’Haridon, F., Alam, I., Weisskopf, L., Bajic, V. B., et al. (2020). Genome insights of the plant-growth promoting bacterium Cronobacter muytjensii JZ38 with volatile-mediated antagonistic activity against Phytophthora infestans. Front. Microbiol. 11, 369. doi:10.3389/fmicb.2020.00369
Etesami, H. (2020). “Plant microbe interactions in plants and stress tolerance,” in Plant life under changing environment. Editor D. K. Tripathi (London: Academic Press, Elsevier), 355–396. doi:10.1016/B978-0-12-818204-8.00018-7
Etesami, H., Emami, S., and Alikhani, H. A. (2017). Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects - a review. J. Soil Sci. Plant Nutr. 17 (4), 897–911. doi:10.4067/s0718-95162017000400005
Eze, M. O., Thiel, V., Hose, G. C., George, S. C., and Daniel, R. (2021). Metagenomic insight into the plant growth-promoting potential of a diesel-degrading bacterial consortium for enhanced rhizoremediation application. bioRxiv 03 (26), 437261. doi:10.1101/2021.03.26.437261
Fadiji, A. E., Orozco-Mosqueda, M. d. C., Santos-Villalobos, S. d. l., Santoyo, G., and Babalola, O. O. (2022). Recent developments in the application of plant growth-promoting drought adaptive rhizobacteria for drought mitigation. Plants 11, 3090. doi:10.3390/plants11223090
Forni, C., Duca, D., and Glick, B. R. (2016). Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 410, 335–356. doi:10.1007/s11104-016-3007-x
Gamalero, E., and Glick, B. R. (2011). “Mechanisms used by plant growth-promoting bacteria,” in Bacteria in agrobiology: Plant nutrient management. Editor D. K. Maheshwari (Berlin Heidelberg: Springer-Verlag), 17–46. doi:10.1007/978-3-642-21061-7_2
Gamez, R. M., Rodriguez, F., Vidal, N. M., Ramirez, S., Alvarez, R. V., Landsman, D., et al. (2019). Banana (musa acuminata) transcriptome profiling in response to rhizobacteria: Bacillus amyloliquefaciens Bs006 and Pseudomonas fluorescens Ps006. BMC Genomics 20, 378. doi:10.1186/s12864-019-5763-5
Gao, Y., Feng, J., Wu, J., Wang, K., Wu, S., Liu, H., et al. (2022). Transcriptome analysis of the growth-promoting effect of volatile organic compounds produced by Microbacterium aurantiacum GX14001 on tobacco (Nicotiana benthamiana). BMC Plant Biol. 22, 208. doi:10.1186/s12870-022-03591-z
Garcia, C. J., Alacid, V., Tomas-Barberan, F. A., Garcia, C., and Palazon, P. (2022). Untargeted metabolomics to explore the bacteria exo-metabolome related to plant biostimulants. Agronomy 12, 1926. doi:10.3390/agronomy12081926
Girma, B., Panda, A. N., Mohanty, S., Ray, L., and Chowdhary, G. (2020). Draft genome sequence of the plant growth-promoting rhizobacterium Klebsiella sp. strain KBG6.2, imparting salt tolerance to rice. Microbiol. Resour. Announc 9, 004911–e520. doi:10.1128/MRA.00491-20
Glick, B. R., Patten, C. L., Holguin, G., and Penrose, D. M. (1999a). Auxin production. In Biochemical and genetic mechanisms used by plant growth promoting bacteria. London: Imperial College Press, 86–133. doi:10.1142/9781848160521_0004
Glick, B. R., Patten, C. L., Holguin, G., and Penrose, D. M. (1999b). Nitrogen fixation. Biochemical and genetic mechanisms used by plant growth promoting bacteria. London: Imperial College Press, 14–44. doi:10.1142/9781848160521_0002
Glick, B. R. (2012). Plant growth-promoting bacteria: Mechanisms and applications. Hindawi 2012, 963401. doi:10.6064/2012/963401
Godoy, F., Olivos-Hernandez, K., Stange, C., and Handford, M. (2021). Abiotic stress in crop species: Improving tolerance by applying plant metabolites. Plants (Basel) 20 (2), 186. doi:10.3390/plants10020186
Gomez-Godinez, L. J., Ormeno-Orrillo, E., and Martinez-Romero, E. (2018). Metatranscriptomics and nitrogen fixation from the rhizoplane of maize plantlets inoculated with a group of PGPRs. bioRxiv, 437087. doi:10.1101/437087
Gowtham, H. G., Duraivadivel, P., Ayusman, S., Sayani, D., Gholap, S. L., Niranjana, S. R., et al. (2020). ABA analogue produced by Bacillus marisflavi modulates the physiological response of host-plant under drought stress. bioRxiv 06 (12), 148049. doi:10.1101/2020.06.12.148049
Goyal, K., Kumar, T., Sharma, P., Rao, M., Ahmed, V., and Chauhan, N. S. (2019). “Crop improvement through microbial biotechnology: A cross talk,” in Salt stress, microbes, and plant interactions: Mechanisms and molecular approaches. Editor M. S. Akhtar (Singapore: Springer), 69–90. doi:10.1007/978-981-13-8805-7_4
Gozalia, E. N., Tjong, D. H., Renfiyeni, R., Jamsari, J., and Nova, B. (2021). RNA-sequencing for transcriptomic analysis of chili peppers induced by Serratia plymuthica strain UBCF_13. Ser. Earth Environ. Sci. 741, 012044. doi:10.1088/1755-1315/741/1/012044
Guo, D. J., Singh, R. K., Singh, P., Li, D. P., Sharma, A., Xing, Y. X., et al. (2020). Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 11, 580081. doi:10.3389/fmicb.2020.580081
Gupta, S., Seth, R., and Sharma, A. (2016). “Plant growth-promoting rhizobacteria play a role as phytostimulators for sustainable agriculture,” in Plant-microbe interaction: An approach to sustainable agriculture. Editor D. K. Choudhary (Singapore: Springer), 475–495. doi:10.1007/978-981-10-2854-0_22
Ilangumaran, G., and Smith, D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: A systems biology perspective. Front. Plant Sci. 8, 1768. doi:10.3389/fpls.2017.01768
Iqbal, S., Qasim, M., Rahman, H., Khan, N., Paracha, R. Z., Bhatti, M. F., et al. (2022). Genome mining, antimicrobial and plant growth-promoting potentials of halotolerant Bacillus paralicheniformis ES-1 isolated from salt mine. Mol. Genet. Genomics 298, 79–93. doi:10.1007/s00438-022-01964-5
Jana, G. A., and Yaish, M. W. (2021). Genome analysis of a salinity adapted Achromobacter xylosoxidans rhizobacteria from the date palm. Rhizosphere 19, 100401. doi:10.1016/j.rhisph.2021.100401
Jansson, J. K., and Hofmockel, K. S. (2018). The soil microbiome — From metagenomics to metaphenomics. Curr. Opin. Microbiol. 43, 162–168. doi:10.1016/j.mib.2018.01.013
Jaya, D. K., GiyantoNurhidayat, N., and Antonius, S. (2019). Isolation, identification, and detection of ACC deaminase gene-encoding rhizobacteria from rhizosphere of stressed pineapple. Indones. J. Biotechnol. 24 (1), 17–25. doi:10.22146/ijbiotech.39018
Jiang, L. M., Lee, Y. J., Han, H. L., Lee, M. H., Jeong, J. C., Kim, C. Y., et al. (2021). Genome insights into the novel species Jejubacter calystegiae, a plant growth-promoting bacterium in saline conditions. Diversity 13, 24. doi:10.3390/d13010024
Kapoor, R., and Kanwar, S. S. (2019). Genetic variations in salt tolerant and plant growth promoting rhizobacteria of the Western Himalayas. J. plant Biochem. Biotechnol. 28, 133–142. doi:10.1007/s13562-019-00489-0
Kataoka, R., Akashi, M., Taniguchi, T., Kinose, Y., Yaprak, A. E., and Turgay, O. C. (2021). Metabolomics analyses reveal metabolites affected by plant growth-promoting endophytic bacteria in roots of the halophyte Mesembryanthemum crystallinum. Int. J. Mol. Sci. 30 (21), 11813. doi:10.3390/ijms222111813
Kaushal, M., and Wani, S. P. (2016). Rhizobacterial-plant interactions: Strategies ensuring plant growth promotion under drought and salinity stress. Agric. Ecosyst. Environ. 231, 68–78. doi:10.1016/j.agee.2016.06.031
Kavitha, B., Reddy, P. V. L., Kim, B., Lee, S. S., Pandeye, S. K., and Kim, K. (2018). Benefits and limitations of biochar amendment in agricultural soils: A review. J. Environ. Manag. 227, 146–154. doi:10.1016/j.jenvman.2018.08.082
Kazemi-Oskuei, B., Bandehagh, A., Sarikhani, M. R., and Komatsu, S. (2018). Protein profiles underlying the effect of plant growth-promoting rhizobacteria on canola under osmotic stress. J. Plant Growth Regul. 37, 560–574. doi:10.1007/s00344-017-9754-y
Khan, N., Ali, S., Tariq, H., Latif, S., Yasmin, H., Mehmood, A., et al. (2020b). Water conservation and plant survival strategies of rhizobacteria under drought stress. Agronomy 10, 1683. doi:10.3390/agronomy10111683
Khan, N., Bano, A., Ali, S., and Babar, M. (2020a). Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 90, 189–203. doi:10.1007/s10725-020-00571-x
Khatoon, Z., Huang, S., Rafique, M., Fakhar, A., Kamran, M. A., and Santoyo, G. (2020). Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 273, 111118–118. doi:10.1016/j.jenvman.2020.111118
Kumar, A., Singh, S., Gaurav, A. K., Srivastava, S., and Verma, J. P. (2020). Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 11, 1216. doi:10.3389/fmicb.2020.01216
Kumar, A., and Verma, J. P. (2018). Does plant—microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 207, 41–52. doi:10.1016/j.micres.2017.11.004
Kumari, S., Varma, A., and Tuteja, N. (2016). “Bacterial ACC-deaminase: An eco-friendly strategy to cope abiotic stresses for sustainable agriculture,” in Plant-microbe interaction: An approach to sustainable agriculture. Editor D. K. Choudhary (Singapore: Springer), 165–185. doi:10.1007/978-981-10-2854-0_8
Kwon, Y. S., Lee, D. Y., Rakwal, R., Baek, S. B., Lee, J. H., Kwak, Y. S., et al. (2016). Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16 (1), 122–135. doi:10.1002/pmic.201500196
Leontidou, K., Genitsaris, S., Papadopoulou, A., Kamou, N., Bosmali, I., Matsi, T., et al. (2020). Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 10, 14857. doi:10.1038/s41598-020-71652-0
Li, T., Mann, R., Kaur, J., Spangenberg, G., and Sawbridge, T. (2021). Transcriptome analyses of barley roots inoculated with novel Paenibacillus sp. and Erwinia gerundensis strains reveal beneficial early-stage plant–bacteria interactions. Plants 10, 1802. doi:10.3390/plants10091802
Liu, H., Wang, J., Sun, H., Han, X., Peng, Y., Liu, J., et al. (2020). Transcriptome profiles reveal the growth-promoting mechanisms of Paenibacillus polymyxa YC0136 on tobacco (Nicotiana tabacum L.). Front. Microbiol. 11, 584174. doi:10.3389/fmicb.2020.584174
Liu, W., Wang, Q., Hou, J., Tu, C., Luo, Y., and Christie, P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 26710. doi:10.1038/srep26710
Lutfullin, M. T., Lutfullina, G. F., Pudova, D. S., Akosah, Y. A., Shagimardanova, E. I., Vologin, S. G., et al. (2022). Identification, characterization, and genome sequencing of Brevibacterium sediminis MG-1 isolate with growth-promoting properties. Biotech 12 (11), 326. doi:10.1007/s13205-022-03392-z
Malviya, M. K., Li, C. N., Solanki, M. K., Singh, R. K., Htun, R., Singh, P., et al. (2020). Comparative analysis of sugarcane root transcriptome in response to the plant growth-promoting Burkholderia anthina MYSP113. PLoS ONE 15 (4), e0231206. doi:10.1371/journal.pone.0231206
Martinez-Viveros, O., Jorquera, M. A., Crowley, D. E., Gajardo, G., and Mora, M. L. (2010). Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 10 (3), 293–319. doi:10.4067/s0718-95162010000100006
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 8, 172. doi:10.3389/fpls.2017.00172
Mellidou, I., Ainalidou, A., Papadopoulou, A., Leontidou, K., Genitsaris, S., Karagiannis, E., et al. (2021). Comparative transcriptomics and metabolomics reveal an intricate priming mechanism involved in PGPR-mediated salt tolerance in tomato. Front. Plant Sci. 12, 713984. doi:10.3389/fpls.2021.713984
Mishra, A. K., and Baek, K. H. (2021). Salicylic acid biosynthesis and metabolism: A divergent pathway for plants and bacteria. Biomolecules 11, 705. doi:10.3390/biom11050705
Mishra, J., Fatima, T., and Arora, N. K. (2018). “Role of secondary metabolites from plant growth-promoting rhizobacteria in combating salinity stress,” in Plant microbiome: Stress response, microorganisms for sustainability. Editors D. Egamberdieva, and P. Ahmad (Singapore: Springer), 127–163. doi:10.1007/978-981-10-5514-0_6
Mohammadipanah, F., and Zamanzadeh, M. (2019). “Bacterial mechanisms promoting the tolerance to drought stress in plants,” in Secondary metabolites of plant growth promoting rhizomicroorganisms. Editor H. B. Singh (Singapore: Springer), 185–224. doi:10.1007/978-981-13-5862-3_10
Mokrani, S., Nabti, E., and Cruz, C. (2020). Current advances in plant growth promoting bacteria alleviating salt Stress for sustainable agriculture. Appl. Sci. 10, 7025. doi:10.3390/app10207025
Molefe, R. R., Amoo, A. E., and Babalola, O. O. (2021). Metagenomic insights into the bacterial community structure and functional potentials in the rhizosphere soil of maize plants. J. Plant Interact. 16 (1), 258–269. doi:10.1080/17429145.2021.1936228
Molina-Favero, C., Monica, C. C., Simontacchi, M., Puntarulo, S., and Lamattina, L. (2008). Aerobic nitric oxide production by Azospirillum brasilense Sp245 and its influence on root architecture in tomato. MPMI 21, 1001–1009. doi:10.1094/MPMI-21-7-1001
Molina-Favero, C., Monica Creus, C., Lanteri, M. L., Correa-Aragunde, N., Lombardo, M. C., Barassi, C. A., et al. (2007). Nitric oxide and plant growth promoting rhizobacteria: Common features influencing root growth and development. Adv. Botanical Res. 46, 1–33. doi:10.1016/s0065-2296(07)46001-3
Mukherjee, A. (2022). What do we know from the transcriptomic studies investigating the interactions between plants and plant growthpromoting bacteria? Front. Plant Sci. 13, 997308. doi:10.3389/fpls.2022.997308
Nagel, R., Bieber, J. E., Schmidt-Dannert, M. G., Nett, R. S., and Peters, R. J. (2018). A third class: Functional gibberellin biosynthetic operon in beta-proteobacteria. Front. Microbiol. 9, 2916. doi:10.3389/fmicb.2018.02916
Nascimento, F. X., Hernandez, A., Glick, B., and Rossi, M. (2020). Plant growth-promoting activities and genomic analysis of the stress-resistant Bacillus megaterium STB1, a bacterium of agricultural and biotechnological interest. Biotechnol. Rep. 25, e00406. doi:10.1016/j.btre.2019.e00406
Nascimento, F. X., Uron, P., Glick, B. R., Giachini, A., and Rossi, M. J. (2021). Genomic analysis of the 1-aminocyclopropane-1-carboxylate deaminase-producing Pseudomonas thivervalensis SC5 reveals its multifaceted roles in soil and in beneficial interactions with plants. Front. Microbiol. 12, 752288. doi:10.3389/fmicb.2021.752288
Nian, L., Zhang, X., Yi, X., Liu, X., Ain, N. U., Yang, Y., et al. (2021). Genome-wide identification of ABA receptor PYL/RCAR gene family and their response to cold stress in Medicago sativa L. Physiol. Mol. Biol. Plants 27 (9), 1979–1995. doi:10.1007/s12298-021-01066-3
Nobre, A., Alarico, S., Fernandes, C., Empadinhas, N., and Costa, M. S. (2008). A unique combination of genetic systems for the synthesis of trehalose in rubrobacter xylanophilus: Properties of a rare actinobacterial TreT. J. Bacteriol. 190, 7939–7946. doi:10.1128/JB.01055-08
Noori, F., Etesami, H., Noori, S., Forouzan, E., Salehi, J. G., and Malboobi, M. A. (2021). Whole genome sequence of Pantoea agglomerans ANP8, a salinity and drought stress–resistant bacterium isolated from alfalfa (Medicago sativa L.) root nodules. Biotechnol. Rep. 29, e00600. doi:10.1016/j.btre.2021.e00600
Numan, M., Bashir, S., Khan, Y., Mumtaz, R., Shinwaric, Z. K., Khan, A. L., et al. (2018). Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 209, 21–32. doi:10.1016/j.micres.2018.02.003
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33, 197. doi:10.1007/s11274-017-2364-9
Olenska, E., Malek, W., Wojcik, M., Swiecicka, I., Thijs, S., and Vangronsveld, J. (2020). Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. total Environ. 743, 140682. doi:10.1016/j.scitotenv.2020.140682
Poria, V., Debiec-Andrzejewska, K., Fiodor, A., Lyzohub, M., Ajijah, N., Singh, S., et al. (2022). Plant GrowthPromoting bacteria (PGPB) integrated phytotechnology: A sustainable approach for remediation of marginal lands. Front. Plant Sci. 13, 999866. doi:10.3389/fpls.2022.999866
Rosier, A., Medeiros, F. H. V., and Bais, H. P. (2018). Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 428, 35–55. doi:10.1007/s11104-018-3679-5
Ryu, C. M. (2015). “Bacterial volatiles as airborne signals for plants and bacteria,” in Principles of plant-microbe interactions. Editor B. Lugtenberg (Berne: Springer), 53–64.
Saberi Riseh, R., Ebrahimi-Zarandi, M., Tamanadar, E., Moradi Pour, M., and Thakur, V. K. (2021). Salinity stress: Toward sustainable plant strategies and using plant growth-promoting rhizobacteria encapsulation for reducing it. Sustainability 13, 12758. doi:10.3390/su132212758
Saeed, Q., Xiukang, W., Haider, F. U., Kucerik, J., Mumtaz, M. Z., Holatko, J., et al. (2021). Rhizosphere bacteria in plant growth promotion, biocontrol, and bioremediation of contaminated sites: A comprehensive review of effects and mechanisms. Int. J. Mol. Sci. 22, 10529. doi:10.3390/ijms221910529
Salazar-Cerezoa, S., Martinez-Montiela, N., Garcia-Sancheza, J., Perez-y-Terronb, R., and Martinez-Contreras, R. D. (2018). Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 208, 85–98. doi:10.1016/j.micres.2018.01.010
Samaras, A., Kamou, N., Tzelepis, G., Karamanoli, K., Menkissoglu-Spiroudi, U., and Karaoglanidis, G. S. (2022). Root transcriptional and metabolic dynamics induced by the plant growth promoting rhizobacterium (PGPR) Bacillus subtilis Mbi600 on Cucumber Plants. Plants 11, 1218. doi:10.3390/plants11091218
Santana, M. M., Gonzalez, J. M., and Cruz, C. (2017). Nitric oxide accumulation: The evolutionary trigger for phytopathogenesis. Front. Microbiol. 8, 1947. doi:10.3389/fmicb.2017.01947
Sekar, J., Saharan, K., Raju, K., Singh, U., and Vaiyapuri, P. R. (2019). “Consequences of bioinoculants and intercropping approach to alleviate plant drought and salinity stress for sustainable agriculture,” in Salt stress, microbes, and plant interactions: Mechanisms and molecular approaches. Editor M. S. Akhtar (Singapore: Springer), 161–182. doi:10.1007/978-981-13-8805-7_8
Shariati, J. V., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owlia, P., and Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Available at: www.nature.com/scientificreports.
Sharma, A., Singh, R. K., Singh, P., Vaishnav, A., Guo, D. J., Verma, K. K., et al. (2021). Insights into the bacterial and nitric oxide-induced salt tolerance in sugarcane and their growth-promoting abilities. Microorganisms 22 (9), 2203. doi:10.3390/microorganisms9112203
Sharma, P., Khanna, V., and Kumari, S. (2016). “Abiotic stress mitigation through plant growth promoting rhizobacteria,” in Plant-microbe interaction: An approach to sustainable agriculture. Editor D. K. Choudhary (Singapore: Springer), 327–344. doi:10.1007/978-981-10-2854-0-15
Sharma, S., Chandra, D., and Sharma, A. K. (2021). “Rhizosphere plant–microbe interactions under abiotic stress,” in Rhizosphere biology: Interactions between microbes and plants, rhizosphere biology. Editors V. V. S. R. Gupta, and A. K. Sharma (Singapore: Springer), 195–216. doi:10.1007/978-981-15-6125-2_10
Sheibani-Tezerji, R., Rattei, T., Sessitsch, A., Trognitz, F., and Mitter, B. (2015). Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6 (5), 006211–e715. doi:10.1128/mBio.00621-15
Shelake, R. M., Pramanik, D., and Kim, J. Y. (2019). Exploration of plant-microbe interactions for sustainable agriculture in CRISPR Era. Microorganisms 7, 269. doi:10.3390/microorganisms7080269
Shilev, S. (2020). Plant-growth-promoting bacteria mitigating soil salinity stress in plants. Appl. Sci. 10, 7326. doi:10.3390/app10207326
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10, 4818. doi:10.1038/s41598-020-61140-w
Spaepen, S. (2015). “Plant hormones produced by microbes,” in Principles of plant-microbe interactions. Editor B. Lugtenberg (Switzerland: Springer), 247–256. doi:10.1007/978-3-319-08575-3_26
Srivastava, A. K., Srivastava, R., Sharma, A., Bharati, A. P., Yadav, J., Singh, A. K., et al. (2022). Transcriptome analysis to understand salt stress regulation mechanism of Chromohalobacter salexigens ANJ207. Front. Microbiol. 30 (13), 909276. doi:10.3389/fmicb.2022.909276
Suarez, C., Ratering, S., Hain, T., Fritzenwanker, M., Goesmann, A., Blom, J., et al. (2019). Complete genome sequence of the plant growth-promoting bacterium Hartmannibacter diazotrophicus strain E19T. Int. J. Genomics 2019, 7586430. doi:10.1155/2019/7586430
Suchan, D. M., Bergsveinson, J., Manzon, Lori., Pierce, A., Kryachko, Y., Korber, D., et al. (2020). Transcriptomics reveal core activities of the plant growth promoting bacterium Delftia acidovorans RAY209 during interaction with canola and soybean roots. Microb. Genomics 6, mgen000462. doi:10.1099/mgen.0.000462
Sunita, K., Mishra, I., Mishra, J., Prakash, J., and Arora, N. K. (2020). Secondary metabolites from halotolerant plant growth promoting rhizobacteria for ameliorating salinity stress in plants. Front. Microbiol. 11, 567768. doi:10.3389/fmicb.2020.567768
Tiepo, A. N., Constantino, L. V., Madeira, T. B., Goncalves, L. S. A., Pimenta, J. A., Bianchini, E., et al. (2020). Plant growth-promoting bacteria improve leaf antioxidant metabolism of drought-stressed Neotropical trees. Planta 251 (4), 83. doi:10.1007/s00425-020-03373-7
Tomer, S., Suyal, D. C., and Goel, R. (2016). “Biofertilizers: A timely approach for sustainable agriculture,” in Plant-microbe interaction: An approach to sustainable agriculture. Editor D. K. Choudhary (Singapore: Springer), 375–395. doi:10.1007/978-981-10-2854-0_17
Tsukanova, K. A., Сhеbоtаr, V. К., Meyer, J. J. M., and Bibikova, T. N. (2017). Effect of plant growth-promoting rhizobacteria on plant hormone homeostasis. South Afr. J. Bot. 113, 91–102. doi:10.1016/j.sajb.2017.07.007
Tsurumaru, H., Okubo, T., Okazaki, K., Hashimoto, M., Kakizaki, K., Hanzawa, E., et al. (2015). Metagenomic analysis of the bacterial community associated with the taproot of sugar beet. Microbes Environ. 30 (1), 63–69. doi:10.1264/jsme2.ME14109
Ullah, A., Bano, A., and Khan, N. (2021). Climate change and salinity effects on crops and chemical communication between plants and plant growth-promoting microorganisms under Stress. Front. Sustain. Food Syst. 5, 618092. doi:10.3389/fsufs.2021.618092
Ulrich, K., Kube, M., Becker, R., Schneck, V., and Ulrich, A. (2021). Genomic analysis of the endophytic Stenotrophomonas Strain 169 reveals features related to plant-growth promotion and stress tolerance. Front. Microbiol. 12, 687463. doi:10.3389/fmicb.2021.687463
Urumbil, S. K., and Anilkumar, M. (2021). Metagenomic insights into plant growth promoting genes inherent in bacterial endophytes of Emilia sonchifolia (Linn.) DC. Plant Sci. Today 8, 6–16. doi:10.14719/pst.1357
Vaishnav, A., Varma, A., Tutej, N., and Choudhary, D. K. (2016). “PGPR-mediated amelioration of crops under salt stress,” in Plant-microbe interaction: An approach to sustainable agriculture. Editor D. K. Choudhary (Singapore: Springer), 205–226. doi:10.1007/978-981-10-2854-0_10
Vurukonda, S. S. K. P., Vardharajula, S., Shrivastava, M., and SkZ, A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi:10.1016/j.micres.2015.12.003
Wang, B., Wang, X., Wang, Z., Zhu, K., and Wu, W. (2023). Comparative metagenomic analysis reveals rhizosphere microbial community composition and functions help protect grapevines against salt stress. Front. Microbiol. 14, 1102547. doi:10.3389/fmicb.2023.1102547
Wang, D., Jiang, P., Zhang, H., and Yuan, W. (2020). Biochar production and applications in agro and forestry systems: A review. Sci. Total Environ. 723, 137775. doi:10.1016/j.scitotenv.2020.137775
Wang, T. T., Ding, P., Chen, P., Xing, K., Bai, J. L., Wan, W., et al. (2017). Complete genome sequence of endophyte Bacillus flexus KLBMP 4941 reveals its plant growth promotion mechanism and genetic basis for salt tolerance. J. Biotechnol. 260, 38–41. doi:10.1016/j.jbiotec.2017.09.001
Wang, Y., Ohara, Y., Nakayashiki, H., Tosa, Y., and Mayama, S. (2005). Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. MPMI 18 (5), 385–396. doi:10.1094/MPMI-18-0385
Wiggins, G., Thomas, J., Rahmatallah, Y., Deen, C., Haynes, A., Degon, Z., et al. (2022). Common gene expression patterns are observed in rice roots during associations with plant growth-promoting bacteria, Herbaspirillum seropedicae and Azospirillum brasilense. Sci. Rep. 12, 8827. doi:10.1038/s41598-022-12285-3
Xie, W., Yang, J., Gao, S., Yao, R., and Wang, X. (2022). The Effect and influence mechanism of soil salinity on phosphorus availability in coastal salt-affected soils. Water 14, 2804. doi:10.3390/w14182804
Xu, H., Gao, J., Portieles, R., Du, L., Gao, X., and Borras-Hidalgo, O. (2022). Endophytic bacterium Bacillus aryabhattai induces novel transcriptomic changes to stimulate plant growth. PLoS ONE 17 (8), e0272500. doi:10.1371/journal.pone.0272500
Yadav, R., Chakraborty, S., and Ramakrishna, W. (2022). Wheat grain proteomic and protein–metabolite interactions analyses provide insights into plant growth promoting bacteria–arbuscular mycorrhizal fungi–wheat interactions. Plant Cell Rep. 41, 1417–1437. doi:10.1007/s00299-022-02866-x
Yoo, Y. H., Kim, M., Chandran, A. K. N., Hong, W. J., Ahn, H. R., Lee, G. T., et al. (2019). Genome-wide transcriptome analysis of rice seedlings after seed dressing with paenibacillus yonginensis dcy84t and silicon. Int. J. Mol. Sci. 20 (23), 5883. doi:10.3390/ijms20235883
Keywords: salinity stress alleviation, PGPB (plant growth-promoting bacteria), gene functions and pathways, molecular mechainsm, salinity stress, drought stress
Citation: Zamanzadeh-Nasrabadi SM, Mohammadiapanah F, Hosseini-Mazinani M and Sarikhan S (2023) Salinity stress endurance of the plants with the aid of bacterial genes. Front. Genet. 14:1049608. doi: 10.3389/fgene.2023.1049608
Received: 20 September 2022; Accepted: 23 March 2023;
Published: 17 April 2023.
Edited by:
Ntakadzeni Madala, University of Venda, South AfricaReviewed by:
Bhaskar Gupta, Government General Degree College, Singur, IndiaCopyright © 2023 Zamanzadeh-Nasrabadi, Mohammadiapanah, Hosseini-Mazinani and Sarikhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fatemeh Mohammadiapanah, Zm1vaGFtbWFkaXBhbmFoQHV0LmFjLmly
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.