
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 13 January 2023
Sec. Livestock Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1029200
This article is part of the Research TopicFisheries and Aquaculture GeneticsView all 18 articles
Transgenic technology and selective breeding have great potential for the genetic breeding in both edible fish and ornamental fish. The development of infertility control technologies in transgenic fish and farmed fish is the critical issue to prevent the gene flow with wild relatives. In this study, we report the genome editing of the dead end (dnd1) gene in the zebrafish model, using the CRISPR/Cas9 technology to achieve a loss-of-function mutation in both wild-type zebrafish and transgenic fluorescent zebrafish to develop complete infertility control technology of farmed fish and transgenic fish. We effectively performed targeted mutagenesis in the dnd1 gene of zebrafish with a single gRNA, which resulted in a small deletion (−7 bp) or insertion (+41 bp) in exon 2, leading to a null mutation. Heterozygotes and homozygotes of dnd1-knockout zebrafish were both selected by genotyping in the
Transgenic technology is a powerful and useful technique to establish transgenic organisms by insertion of a foreign gene into the genome of organisms to obtain or enhance the function of transgene in vivo under the control of ubiquitous promoter or tissue-specific promoter (Ozato et al., 1992; Gordon, 1997; Her et al., 2003). Since 1980, scholars have used transgenic technology to improve the performance of major farmed fish species (Du et al., 1992; Maclean, 1993). A transgenic Atlantic salmon, AquAdvantage Salmon, which expressing growth hormone of Chinook salmon was established under the control of a promoter from ocean pout and obtained the approval for consumption in the U.S. on November 2015 and 6-month later in Canada. AquAdvantage Salmon grows twice faster to market size than non-transgenic Atlantic salmon counterpart. Thus, transgenic technology has great potential for the development of aquaculture, as it significantly reduces farming costs and risks (Cook et al., 2000; Muir, 2004). With the subsequent simplification of the transgenic procedures and methods, the application of transgenic technology gradually became popular (Kawakami, 2007; Suster et al., 2009). Besides in the food fish aquaculture industry, transgenic technology has been applied in the ornamental industry (Gong et al., 2003). Fluorescent fish is a concept originated by scholars using the transgenic technique to visualize and calibrate tissues to study specific gene regulatory mechanisms and functions (Ju et al., 1999; Gong et al., 2001; Chudakov et al., 2010). According to the ornamental fish industry data, fluorescent fish are new varieties that meet consumers’ demands in terms of unique body colors, posing a huge potential business opportunity, which may enable the industry to become more competitive (Stewart, 2006). Transgenic fish applications are expected to increase exponentially in both the aquatic and ornamental fish industries globally. If these transgenic fish escape and interbreed with wild stock, this poses a potential threat to the ecosystem and environment (Hu et al., 2007). Recently, transgenic glowing zebrafish appear to be thriving after escape from fish farms into Brazilian streams and may threaten local biodiversity (Magalhaes et al., 2022; Moutinho, 2022). We believe that developing practical infertility control technology is the most effective way to avoid ecological risks, and it also promotes the development of an environmentally responsible aquaculture (Muir and Howard, 1999; Stigebrandt et al., 2004; Wong and Van Eenennaam, 2008).
The method of establishing infertile individuals has been continuously developed. The methods commonly used to produce infertile fish are triploidization and interspecies hybridization (Benfey et al., 1989; Arai, 2001; Cal et al., 2006); however, some treated individuals were found to maintain fertility and show limitations, depending on the species (Wagner et al., 2006; Piferrer et al., 2009). In our previous research, we tried to use the transgenic strategy, transferring the germ cells’ specific gene piwi promoter to be combined with the nitroreductase toxic protein (NTR). When the metronidazole (Mtz) bath immersion treatment succeeded, cell apoptosis began due to the conversion of Mtz into a cytotoxic compound, which may affect the germ cells and induce gonadal development disruptions. However, the effects of the NTR/Mtz system are not obvious. While the methods mentioned above failed, the disruption of the mechanisms of primordial germ cell formation and migration was successful in achieving complete infertility (Wong and Collodi, 2013; Wong and Zohar, 2015a; Wong and Zohar, 2015b).
Primordial germ cells (PGCs) are the progenitor cells of gametes. PGCs play an important role, as egg and sperm production relies on the formation, differentiation, and correct localization of PGCs (Yoon et al., 1997; Weidinger et al., 1999; Xu et al., 2010; Yamaha et al., 2010). In zebrafish, the development of PGCs has been studied extensively. PGC formation occurs during early embryogenesis maternally through a germplasm composed of maternal RNA-binding proteins and mRNAs (Braat et al., 1999; Extavour and Akam, 2003; Raz, 2003). In addition, the germ cell marker ddx4 also named as vasa, an evolutionarily conserved gene, was first discovered to label PGCs successfully, and it was shown to be a key factor in the migration of PGCs to the genital ridge (Yoon et al., 1997; Braat et al., 1999; Castrillon et al., 2000; Yoshizaki et al., 2000). However, by inhibiting the gene expression by gene silencing in different species, it did not induce a state of a complete loss of germ cells or infertility, resulting in a single sex (Raz, 2000; Li et al., 2009; Hartung et al., 2014). There is no evidence proving that no other cells can potentially contribute to the germline during normal development or when the number of ddx4-expressing cells is reduced. The dead end (dnd1) gene, encoding an RNA-binding protein DND1, was first identified in zebrafish (Weidinger et al., 2003) and is highly conserved in vertebrate species (Horvay et al., 2006; Aramaki et al., 2007; Liu et al., 2009; Nagasawa et al., 2013). The expression of some germline-specific mRNAs relied on the protection of DND1 from microRNA-mediated inhibition (Kedde et al., 2007a). The DND1 knockdown by translational inhibition of maternal dnd1 mRNA with Morpholino antisense oligonucleotides resulted in PGCs losing their ability to migrate actively and mis-migration, followed by apoptosis or transdifferentiation into somatic cells in zebrafish (Weidinger et al., 2003; Gross-Thebing et al., 2017). The dead end (dnd1) knockout fish by genome editing was first conducted in medaka with TALENs and showed germ cell-less gonads (Wang and Hong, 2014; Wang, 2015). The dnd1 knockout zebrafish was first established by genome editing with Zinc Finger Nucleases (ZFNs) as germ cells-less recipients of surrogate for germ cell transplantation (Li et al., 2017). However, compared to ZFN and TALEN, which rely on DNA binding domain to recognize DNA, the CRISPR/Cas9 system by using a single guide RNA (sgRNA) for DNA recognition is more efficient, convenient, and cost-effective (Doudna and Charpentier, 2014). Therefore, CRISPR/Cas9 technology had been applied in genome editing of several fish species such as Atlantic salmon, medaka, sterlet, and rainbow trout to target dnd1 to achieve infertility (Wargelius et al., 2016; Sawamura et al., 2017; Baloch et al., 2019; Baloch et al., 2021; Fujihara et al., 2022).
CRISPR/Cas9 technology has been widely used due to its ease of operation, which allows scholars to investigate gene function (Ran et al., 2013). Genome editing used the Cas9 protein and guide RNA (gRNA), and the 5’ of gRNA contained an 18–20 bp protospacer sequence that could identify the target sequence and guide Cas9 to cause a DNA double-strand break, thus inducing the mechanism of DNA repair, i.e., non-homologous end joining (NHEJ) and homology-directed repair (HDR) (Garneau et al., 2010; Horvath and Barrangou, 2010; Hwang et al., 2013). There is an opportunity to cause an insertion or deletion through the NHEJ repair mechanism, and an in-frame stop codon may emerge through the insertion or deletion performed by CRISPR/Cas9, causing the translation to stop early, which results in a loss of gene function due to early truncation of the protein (Chapman et al., 2012; Auer et al., 2014).
While transgenic technology has a huge potential to be applied in aquaculture, it poses unexpected risks to the ecosystem, as the organisms maintain the ability to crossbreed with wild stock. Developing effective and practical infertility control is the key to solving the problem. Although dnd1 knockout in zebrafish was achieved by ZFNs (Li et al., 2017), CRISPR/Cas9 technology was still not applied in dnd1 knockout of zebrafish, especially in new application of infertility control of transgenic fluorescent zebrafish as a model of fluorescent ornamental fish. The phenotypes of homozygous and heterozygous dnd1 knockout zebrafish by CRISPR/Cas like those by ZFNs are not novel but convince us of no off-targeting effect in dnd1 knockout zebrafish by both genome editing technologies. In this study, we demonstrate that the dnd1 knockout by CRISPR/Cas9 genome editing had been successfully applied to the infertility control of transgenic fluorescent zebrafish as a model of transgenic fluorescent ornamental fish to prevent potential impact on ecology when they escape into the wild field.
Wild-type (AB) strain zebrafish were used as experimental animals in this study. The zebrafish were maintained in a recirculating system, with the temperature maintained at 28°C and photoperiodism of 14 h of light and 10 h of dark. The animal use protocol of this research had been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Taiwan Ocean University. The IACUC Approval No. is 108042. The CRISPR/Cas9 system was used to establish the dnd1-knockout zebrafish line (Baloch et al., 2021). The sgRNA target site was designed using CHOPCHOP, using an off-target assay for analysis (Labun et al., 2016). The pCS2-nCas9n was a gift from Wenbiao Chen (Addgene plasmid # 47929; http://n2t.net/addgene:47929; RRID: Addgene_47929) for Cas9 mRNA synthesis by in vitro transcription (Jao et al., 2013). The zebrafish dnd1 target-specific DNA oligo (5′-TTCTAATACGACTCACTATAGTAACCCAAGTCAATGGGCAGGTTTTAGAGCTAGA-3′) was synthesized by “Genomics” company (Taiwan) for sgRNA synthesis. The sgRNA was synthesized based on the protocol of EnGen® sgRNA synthesis kit (New England Biolabs, United States). The zebrafish reproduction was performed under the conditions mentioned above. The embryos were collected to perform the microinjection. The microinjection was performed by co-injecting a mixture of reagents—300 ng/μL of Cas9 mRNA and 30 ng/μL of sgRNA—into one-cell-stage embryos. The injected embryos were cultivated in a Petri dish at 28°C for hatching. To predict the efficiency of the gRNA targeting, 10 individuals were randomly selected 24 h after fertilizing (hpf) the injected embryos for DNA extraction, and the T7 endonuclease I (T7E1) assay was used to evaluate the efficiency of the gRNA target site mutation (Urnov et al., 2005).
The
The transgenic fluorescent zebrafish line Tg(-2.4ckmb:TcCFP13), expressing the Taiwan coral (Acropora sp.) cyan fluorescent protein, TcCFP-13 cDNA (provided by Dr. Ming-Chyuan Chen, National Kaohsiung University of Science and Technology, Taiwan), driven by a novel zebrafish muscle-specific ckmb 2.4 kb promoter/enhancer (GenBank accession number HM347596), was previously established via the Tol2 transposon system (Gong et al., 2015). The fluorescent zebrafish strain was mated with the dnd1-knockout heterozygotes to establish the
The sense and antisense ddx4 riboprobes were designed in the open reading frame of zebrafish ddx4, and the primers of the probe were 5′-GCGTGTCCACCTGCTACCGGCTCTTCTGAA and 3′-TTCATCACGGGAGCCACTGCGAAAACC. The riboprobes were synthesized using the DIG RNA Labeling Kit (SP6/T7) (Roche) from linearized pGEM-T ddx4. The embryos were collected 24 h post-fertilization, followed by fixation with 4% paraformaldehyde. The signals were detected by NBT/BCIP staining (Brend and Holley, 2009). We selected and separated the embryos with a positive and negative signal under a Leica EZ4 microscope (Leica, Germany), after capturing the phenotypes. Then, the embryos were washed 5 times using 1× PBS, followed by DNA extraction with the MasterPure™ DNA Purification Kit (Epicentre, United States). PCR amplification was performed using the 5× PCR Dye Hot Start Master Mix (GeneMark, Taiwan). The genotype was analyzed by gel electrophoresis using a 2% agarose gel.
Three individuals from the wild types, heterozygotes, and homozygotes in the female and male groups were selected. The sample (whole fish) was fixed with Davidson’s fixation solution for 24 h; then, the sample was kept in 70% ethanol (Miki et al., 2018). The sample was dehydrated with ethanol and then embedded into paraffin blocks. Then, the sample was cut into 5 μm-thick sections, and the sections were mounted on glass slides in a 42°C thermostatic water bath. Staining was performed using hematoxylin and eosin (HE stains), and the sections were analyzed using an optical microscope (Blazer, 2002).
Three individuals from the wild types, heterozygotes in the female and male groups, and homozygotes were selected for sampling of the gonadal tissues. The total RNA of the gonads was extracted using Ambion™ TRIzol Reagent and purified with the PureLink™ RNA mini kit. Reverse transcription was performed to synthesize the corresponding cDNA. The primers used in this study were as follows: ddx4 (forward: 5′-ATGGATGACTGGGAGGAAGATCAGAGTCCCG-3’; reverse: 5′- TTCCATTTTCATCATTTTCATCACGGGA-3′); dazl (forward: 5′-ATGGTTCAGGGGG TTCAGTTACCCGTGT-3’; reverse 5′-TGATGGTGGGGCCAGGCCTGGAGGACAGCA-3′); nanos3 (forward: 5′-TTCTGGAATGACTATCTCGGCCTGTCCA-3’; reverse: 5′-ATTGTGTGCGCGTTGTCCCCGTTGGCACTGC-3′); tdrd7a: (forward: 5′-ATGGCGGACGAGGAACTGGTGAAGAA-3’; reverse: 5′-CCGCGACCTCCACCTCGCCCCAGACGTCCAC GG-3′); amh (forward: 5′-ATGCTTTTCCAGGCAAGATTTGGGCTGATG-3’; reverse 5′- ACTGTCTCCTTTAACAGGATTGACACTGAACAG-3′); and eef1α1l1 (forward: 5′-TGCCTTCG TCCCAATTTCAG-3’; reverse: 5′-CTCATGTCACGCACACAGCAAAG-3′). Semi-quantitative PCR was performed using the following procedure: 94°C for 2 min; 94°C for 30 s; 60°C for 30 s; 72°C for 1 min per kb (repeated for 25–35 cycles); 72°C for 5 min; the reaction was stopped at 4°C. The semi-quantitative PCR analysis was performed by gel electrophoresis using a 1% agarose gel.
The dnd1-knockout homozygotic individuals were selected and crossbred with wild-type female zebrafish randomly. The wild-type groups were selected randomly as a control. The photoperiodism was 14 h light and 10 h dark, and the observation was performed during breeding, collecting the eggs of every group and counting and recording photos every hour. The same process was used for the cyan fluorescent dnd1-knockout homozygotic groups.
According to previous studies on the dnd1 gene in zebrafish, the translation region that starts to form the key functional domain, the RNA-recognition motif (RRM), is located in exon 3. The target site of a single gRNA was designed to locate in the second exon of the dnd1 gene in zebrafish to disrupt the formation of the RNA-recognition motif (Figure 1A). The components of synthesized gRNA and Cas9 mRNA were injected into one cell of fertilized zebrafish embryos (Figure 1B). The injected embryos were cultivated in Petri dishes, and at 24 h post-fertilization (hpf), we randomly selected ten embryos to investigate the efficiency of the target site mutation generated by gRNA using the T7E1 assay. According to the T7E1 results, in nine out of 10 embryos, multiple banding patterns were observed, with some individuals showing significant second major band patterns, revealing DNA mismatches formed in a fraction of heteroduplexes and cleaved by T7E1 (Figure 1C). It was confirmed that the mutation was constituted through the gRNA, and dnd1 may be successfully disrupted by the CRISPR/Cas9 genome editing system in zebrafish. The
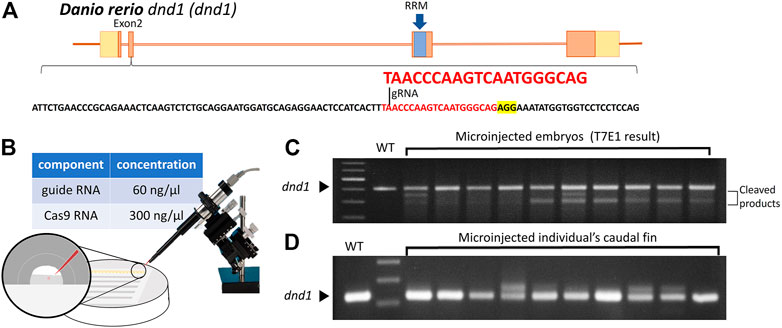
FIGURE 1. (A) Design of the CRISPR/Cas9 target sites of the guide RNA (gRNA) in dnd1. The gRNA was designed in exon 2, and the target site of the gRNA is indicated in red, while the sequence highlighted in yellow is PAM. The RNA-recognition motif (RRM) is the conserved functional domain encoded by exon 3 of the dnd1 gene. (B) Microinjection schematic with the component and concentration of the working solution. (C) Mutation identified in each
We further analyzed the mutation patterns of the
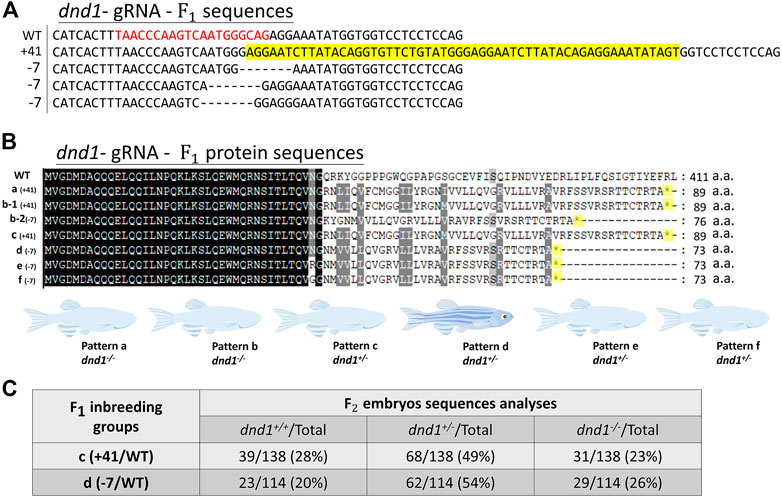
FIGURE 2. (A) Mutation sequences identified in the
In wild-type zebrafish, PGCs may be observed by whole-mount in situ hybridization in 24 hpf embryos localized on the genital ridge. We performed the inbreeding of pattern D heterozygotes to collect the fertilized embryos. To investigate whether the PGCs localized normally on the genital ridge in dnd1-knockout offspring, the germ cell marker ddx4 was used as an RNA probe label for PGC visualization. In the
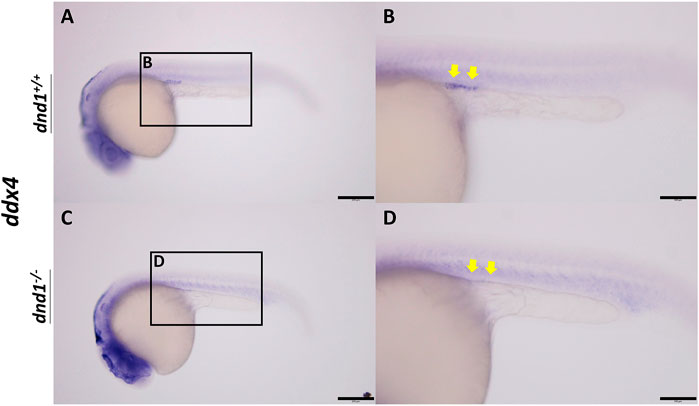
FIGURE 3. ddx4 expression visualized by whole-mount in situ hybridization in 24-h post-fertilized embryos of the
We separated the individuals according to the three groups of genotypes and randomly selected both male and female individuals from each genotype. No individual with the female phenotype was obtained from the
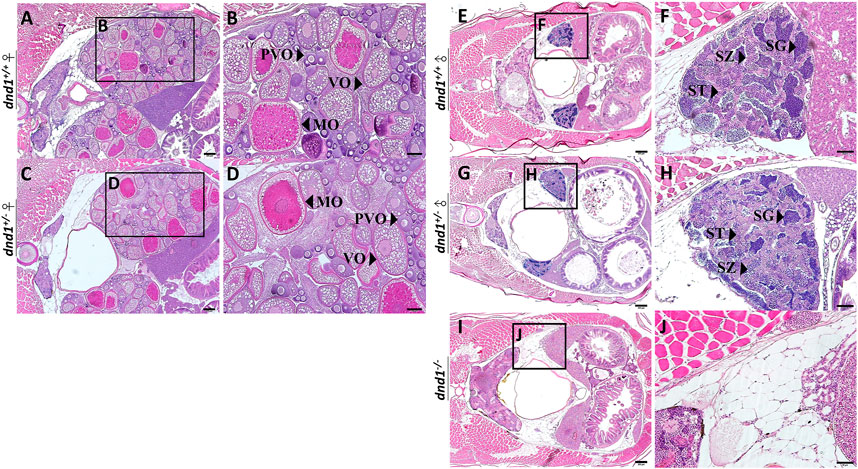
FIGURE 4. Histological analyses of zebrafish gonad. (A)-(B) Wild-type zebrafish ovary. (C)-(D) Heterozygote zebrafish ovary. (E)-(F) Wild-type zebrafish testis. (G)-(H) Heterozygote zebrafish testis. (I)-(J) Mutant zebrafish testis. PVO: previtellogenic oocytes; VO: vitellogenic oocytes; MO: mature oocyte; SC: spermatocytes; SG: spermatogonia; ST: spermatids. Bars = 200 μm (A, C, E, G, I), 100 μm (B, D) and 50 μm (F, H, J).
In the male individual groups, we observed that the
To determine whether the
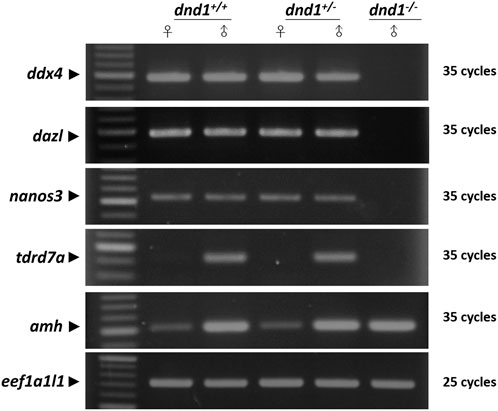
FIGURE 5. Analysis of the expression of germ-cell-related genes between the wild-type (
We analyzed the mating behavior and fertilization capabilities of the dnd1-knockout zebrafish. The results show that
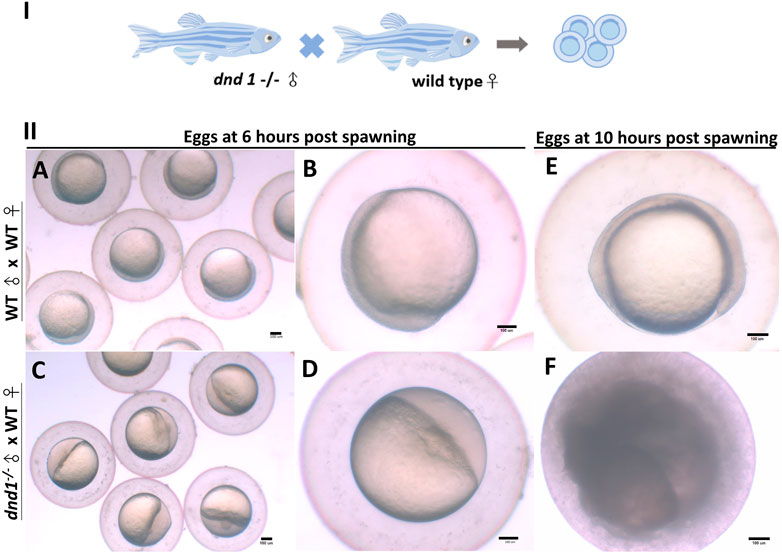
FIGURE 6. (I) dnd1−/− zebrafish were all male, with a normal courtship behavior. (II) Spawned eggs from WT female mating with dnd1−/− male and wild-type zebrafish. (A)-(B) Embryos spawned by wild-type zebrafish after 6 h and (E) after 10 h. (C)-(D) Eggs spawned by mutant male zebrafish and wild-type female zebrafish after 6 h and (F) after 10 h. Bars = 100 μm.
To confirm the results reported above, we sampled two
According to the results of the experiments above, we used wild-type zebrafish as a model to reveal the targeted mutagenesis in zebrafish dnd1, which may disrupt the germ cell formation, leading to a loss of germ cells and the
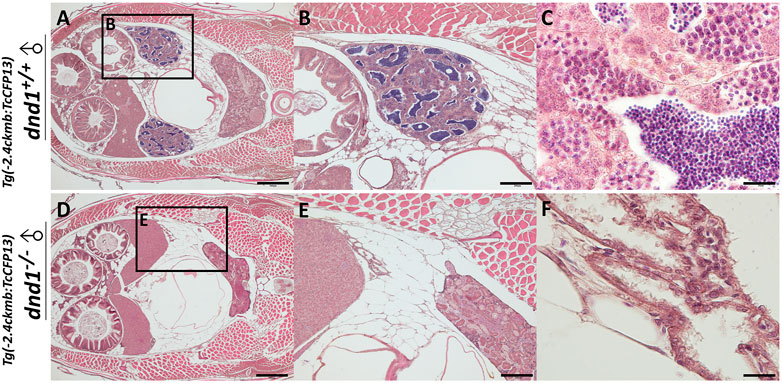
FIGURE 7. Histological analyses of fluorescent zebrafish testis structure. (A)-(C) dnd1+/+ fluorescent zebrafish. (D)-(F) dnd1-knockout homozygote fluorescent zebrafish. Bars = 500 μm (A, D), 200 μm (B, E), and 20 μm (C, F).
In case, the zebrafish which carry transgenes go into the river and mate with the wild type zebrafish, it causes widespread of the transgene as the transgenes are heritable. Therefore, we conduct the breeding experiment to simulate the escapement of the transgenic zebrafish, and to study the gene flow of the foreign gene. At the same time, we test whether the transgenes are completely inherited after performing dnd1 mutation treatment. For easy observation, we selected the strain with muscle florescent expression to perform. Both transgenic zebrafish, transgenic zebrafish with
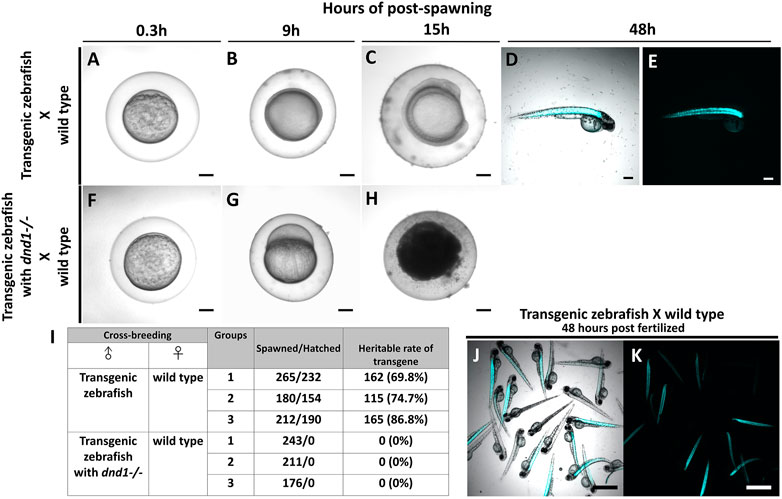
FIGURE 8. Transgenic zebrafish with dnd1 mutant can effectively avoid the transgene inherited. The eggs spawned by transgenic zebrafish and wild type zebrafish (A) .3 h (B) 9 h (C) 15 h (D) 48 h of post spawning under light view (E) 48 h of post spawning under dark view. The eggs spawned by transgenic zebrafish with dnd1 mutant and wild type zebrafish (F) .3 h (G) 9 h (H) 15 h. (I) The heritable rate counting of transgene in cross-breeding group. Top view of hatched offspring of cross-breeding by transgenic zebrafish and wild type zebrafish. (J) light view (K) dark view. Bars = 200 μm (A–H) and 500 μm (J, K).
Further, we calculated the total number of eggs that were spawned in each cross-breeding group, and also verified the heritable rate of transgenes. In the cross-breeding groups of transgenic zebrafish and wild type zebrafish, we collected 265, 180, and 212 spawned eggs, a total of 232, 154, and 190 eggs were hatched (Figure 8I). We observed the fluorescent signal under the microscope to calculate the heritable rate of the transgene, we found the transgenic zebrafish may have 69.8%, 74.7% and 86.8% of heritage (Figures 8I–K). In the cross-breeding groups of transgenic zebrafish with dnd1 mutant and wild type zebrafish, a total of 243, 211, and 176 eggs had collected, but all the eggs were found unfertilized (Figure 8I). From the result, the transgenic zebrafish are infertile after dnd1 mutant, and success to avoid the transgene hereditary.
Finally, we performed a larger scale of breeding experiment to confirm the infertility effect of transgenic fish with
As the demand for fishery resources has increased due to global population growth, marine fisheries are gradually starting to face a lack of resources. To reduce the threats relating to overfishing, to protect marine ecology, the supply of aquatic products has gradually shifted toward aquaculture (Tidwell and Allan, 2001). However, the demand for land resources and feed supplies has restrained the growth of the aquaculture industry (Cao et al., 2015). Therefore, nowadays, in the aquaculture industry, optimizing the growth, nutritional value, and disease resistance of farmed fish species, shortening their breeding times, and maximizing the production values are the main goals of development (Tidwell and Allan, 2001). To improve the quality of breeding livestock, selective breeding is commonly used in the industry. In addition, importing exotic species to adapt to cultural conditions is usually applied in the aquatic industry through global trade (Perez et al., 2003). This may increase cultivation and bring several advantages in terms of development (Gjedrem et al., 2012). However, if the artificially modified species escape from our cultural environment, the biodiversity and abundance of native species may be affected because the performance and adaptability of the selected individuals may be better than those of the native species (Lind et al., 2012).
According to previous studies, transgenic technology is a technique that could be applied in the aquatic industry for both ornamental fish and food fish, inserting exotic genes to provide the target species with the greatest performance, thus creating huge business opportunities (Gong et al., 2003; Clausen and Longo, 2012). Unfortunately, if the genetically modified species established using the transgenic technique that carry the exotic gene from the other species escape from our cultivation area, the potential risks posed to our ecosystem cannot be assessed. Thus, the application of transgenic technology has been regulated under GMO regulations. Based on the situation mentioned above, culturing sterile fish may allow the ecological risks to be avoided whenever an unexpected out-flow happens. We believe that applying effective and practical infertility control technologies is important.
DND1 is a vertebrate RNA-binding protein that was first discovered in zebrafish (Weidinger et al., 2003; Kedde et al., 2007b). Previous studies have shown that the amino acid sequence of DND1 contains six functional regions, including the N-terminal region (NR), the RNA-recognition motif (RRM), and four C-terminal regions (CR 1–4) (Slanchev et al., 2009). The RRM is the most important functional region in DND1 and helps to transfer DND1 from the nucleus to the germ cell granules. While CR has ATPase activity, required for the development of PGCs and involved in the mRNA protection of downstream genes, such as nanos3 and tdrd7a, mutations in this region will reduce the number of PGCs (Liu and Collodi, 2010). In previous research, the knockdown of dnd1 expression using a morpholino in zebrafish, Xenopus, medaka, Atlantic cod, and other species resulted in a loss of germ cells (Weidinger et al., 2003; Youngren et al., 2005; Horvay et al., 2006; Goto et al., 2012; Northrup et al., 2012; Skugor et al., 2014; Su et al., 2014; Linhartova et al., 2015; Su et al., 2015; Hong et al., 2016). In zebrafish, the knockdown of dnd1 resulted in a loss of germ cells due to the mis-migration and transdifferentiation of PGCs, which failed to protect the fate of the cells (Gross-Thebing et al., 2017). As dnd1 is a critical gene that has been proven to be important in the migration and cell fate stability of PGCs, several researchers have focused on dnd1 to perform gene disruption to explore the possibility of infertility control. In 2003, infertility control was achieved by the knockdown of the dnd1 gene’s expression using a morpholino, but performing a microinjection of one-cell fertilized eggs is an operation that is too technically complex to be applied in the industry (Weidinger et al., 2003). In 2015, researchers established the immersive bath method of Vivo-Morpholino-dnd1 and proposed the knockdown of dnd1. This technique is indeed simpler to operate than microinjection, but it may be too difficult to apply in industrial applications, as the Vivo-morpholino treatment is quite expensive and non-inheritable (Wong and Zohar, 2015a). The different conditions in terms of the various fish species and egg sizes may hinder the effectiveness of the technique, and it may therefore not be able to guarantee consistency in achieving complete infertility.
In this study, gene editing was performed using CRISPR/Cas9, and the target mutagenesis site for gRNA was located before the translation of functional regions. According to the results of this study, the in-frame stop codon generated by CRISPR/Cas9 showed an abnormal translation, resulting in the dnd1 gene showing a loss of function. This technology applied in model species, such as zebrafish, could help us to explore whether the lack of dnd1 generated by CRISPR/Cas9 could allow complete infertility to be achieved and a complete analytical procedure to be established in a shorter period of time. We found a loss of fertilization ability in both wild-type and fluorescent zebrafish, and the targeted mutagenesis of dnd1 using the CRISPR/Cas9 system resulted in a loss of germ cells in homozygotic individuals, which is consistent with the results presented above.
In previous studies, it was proved that nanos3 and tdrd7a were protected by DND1, which competed with the binding site of the 3′ untranslated region (UTR) of the gene’s mRNA to hinder its microRNA-induced degradation (Kedde et al., 2007a; Aguero et al., 2017). We analyzed dnd1-related genes, such as nanos3, tdrd7a, and ddx4, in dnd1-knockout individuals. In dnd1-knockout homozygotic individuals, germ-cell-related genes resulted in a lack of expression, but there was a gonadal somatic cell expression of the amh gene. We conjecture that the knockout of dnd1 caused a loss of germ cells, but with no significant effect on the gonadal somatic cells. According to research on dnd1 knockdown in medaka, injecting different concentrations of a morpholino leads to a direct effect on the number of PGCs (Hong et al., 2016). However, we did not observe germ-cell-related gene downregulation or histological evidence in heterozygotic individuals. We could only determine whether there were differences in the gene expression levels and the fertility capacity to determine whether there was a significant difference between the heterozygotes and wild types.
In this study, we found that the all-male phenotype in dnd1-knockout zebrafish by CRISPR/Cas9 was complete sterile, although the males still maintained normal courtship behavior. In the study of dnd1-knockdown zebrafish, we observed the same all-male phenomenon (Pradhan and Olsson, 2018). According to previous research, a lack of germ cells in zebrafish, medaka, and tilapia leads to their sex determination in the all-male phenotype, as the number of germ cells may affect the differentiation mechanism (Slanchev et al., 2005; Kurokawa et al., 2007; Li et al., 2014). In our study, CRISPR/Cas9 technology, which needs only prepare a single guide RNA and Cas9 mRNA to induce dsDNA break for genome editing, was used to knockout dnd1 gene for complete infertility control of transgenic fish. The CRISPR/Cas9 genome editing technology is a more easy, cheap and precise way than ZFNs and TALENs. As zebrafish as a model species, to establish an integrated infertility control procedure by CRISPR/Cas9 genome editing from the development to the analysis further is necessary. Also, this study mainly focused on the application of infertility control on transgenic fish, and we provided evidence to emphasize the practicality of this technique. We established dnd1-knockout zebrafish using CRISPR/Cas9 gene editing technology and achieved complete infertility in homozygotic individuals of transgenic cyan fluorescent zebrafish. While this application of preparatory work to perform gene editing is complicated, as long as heterozygous mutants are established, and the stable heritable knockout genome sequences are selected, followed by the interbreeding of heterozygotes, homozygotic individuals can be analyzed by fin cleaving, PCR amplification and genotyping (Figure 9). Besides the generation of completely infertile fish, infertility species may be used as germ cell transplantation carriers to produce the germ cells of different species (Saito et al., 2008; Yazawa et al., 2010; Yazawa et al., 2013; Pacchiarini et al., 2014; Morita et al., 2015; Lujic et al., 2018). However, the idea of infertility control by knockout strategy seems to be impractical when it is put into the application of in aquaculture scale. As the verification of genotype may require PCR amplification procedure and the inheritable homozygote ratio which is around 1/4 of the total offspring. As fertile dnd1 heterozygous mutants still can breed, they have to be kept in breeding company under strict regulation to prevent from escape. Only sterile transgenic fluorescent zebrafish with dnd1 homozygous mutations can be sold to local aquariums and consumers. There is no practical application if fertility cannot be restored in dnd1 homozygous mutants for mass production of sterile offspring. In Atlantic salmon, rescue of germ cells in dnd1 knockout embryos by CRISPR/Cas9 opened possibility to produce inherited sterility (Guralp et al., 2020). Development of practical mass production technology of sterile fish still need to be conquered by more studies and efforts. Nevertheless, practicality and stability are the most important elements in the infertility control technique. We propose that this technique may be suitable for the selective breeding of fish species to achieve a higher product value or to achieve transgenic individuals, which infertility control must be applied to ensure that the transgenic individuals are completely sterile.
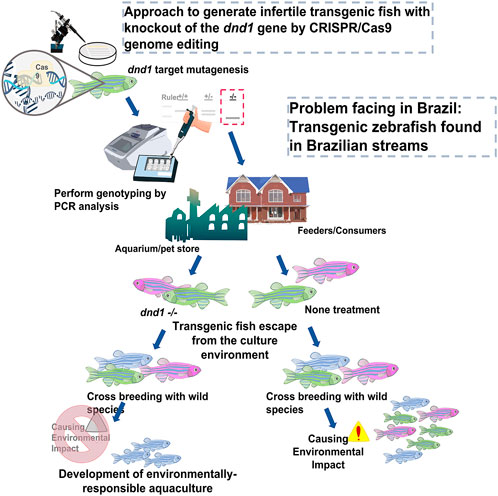
FIGURE 9. Approach to generate infertile transgenic fluorescent zebrafish with knockout out of the dnd1 gene by CRISPR/Cas9 genome editing. We demonstrate that the dnd1 knockout by CRISPR/Cas9 genome editing could be applied to the infertility control of transgenic fluorescent zebrafish to prevent them from cross-breeding with wild species when they escape into the wild field. It can promote the development of an environmentally responsible aquaculture.
Nowadays, the development of biotechnology is mature, and many important breeding fish species have developed breeding technologies with great application potential. In addition to being applied to the industry, these technologies must also pass numerous regulatory and analytical tests. Especially for the artificial selective breeding or genetic modified fish, the most concern of us is to protect our natural primitive ecology by avoiding the foreign gene or dominant species from invading or affecting the wild population, whereas the application of infertility control technique is one of the keys. In this study, we introduce a complete procedure from the development of dnd1 knockout candidates to the analyses of stable heritable rates of homozygotes zebrafish, with multiple breeding experiments to confirm the sterile fish which did not maintain their reproductive ability. It is important to develop and employ effective infertility control in industry practice, as this may allow us to protect our ecosystem and build up a responsible aquaculture environment. Although our study achieved infertility control by genome editing of the dead end gene, which has been reported in zebrafish by ZFNs, we still believed that using new gene editing technology ‘CRISPR/Cas9′ may help the progress of infertility control applicated in the industry, as the dead end gene is conserved between vertebrates and the accomplishment of the target mutagenesis is more simple, faster and precise. Zebrafish is the most important model species, and the completeness and systematic process description are vitally important for the reference of other species.
To avoid the ecological risks associated with escaping transgenic fish into the aquatic environment, the development of an effective sterilization strategy is essential. Dead end (dnd1) is a critical gene that plays an essential role in the migration, survival, and cell fate maintenance of primordial germ cells (PGCs). In this study, we report the targeted mutagenesis of the dnd1 gene in zebrafish by using CRISPR/Cas9 genome editing technology to achieve complete sterility in dnd1-gene-knockout non-transgenic zebrafish and transgenic fluorescent zebrafish, as the model for the infertility control in farmed ornamental fish and transgenic fish, especially in transgenic fluorescent ornamental fish.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Taiwan Ocean University under Approval No. 108042.
Conceptualization, H-YG and J-LW; Methodology, W-KC; Validation, W-KC; Formal analysis, W-KC; Investigation, W-KC and S-CH; Resources, H-YG and C-FC; Data curation, W-KC and S-CH; Writing—original draft preparation, W-KC; Writing—review and editing, H-YG and C-FC; Visualization, J-LW and C-FC; Supervision, H-YG and C-FC; Project administration, H-YG; Funding acquisition, H-YG. All authors have read and agreed to the published version of the manuscript.
The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
This study received funding (grant No.:MOST 109-2622-B-019- 002-CC1) from the Ministry of Science and Technology, Taiwan, and Jy Lin Trading Co., Ltd., in Taiwan, to H-YG.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1029200/full#supplementary-material
Aguero, T., Jin, Z., Chorghade, S., Kalsotra, A., King, M. L., and Yang, J. (2017). Maternal Dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development 144 (20), 3755–3765. doi:10.1242/dev.152611
Arai, K. (2001). Genetic improvement of aquaculture finfish species by chromosome manipulation techniques in Japan. Aquaculture 197 (1-4), 205–228. doi:10.1016/s0044-8486(01)00588-9
Aramaki, S., Sato, F., Kato, T., Soh, T., Kato, Y., and Hattori, M. A. (2007). Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 330 (1), 45–52. doi:10.1007/s00441-007-0435-1
Auer, T. O., Duroure, K., De Cian, A., Concordet, J. P., and Del Bene, F. (2014). Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24 (1), 142–153. doi:10.1101/gr.161638.113
Baloch, A. R., Franek, R., Saito, T., and Psenicka, M. (2021). Dead-end (dnd) protein in fish-a review. Fish. Physiol. Biochem. 47 (3), 777–784. doi:10.1007/s10695-018-0606-x
Baloch, A. R., Franěk, R., Tichopád, T., Fučíková, M., Rodina, M., and Pšenička, M. (2019). Dnd1 knockout in sturgeons by CRISPR/Cas9 generates germ cell free host for surrogate production. Anim. (Basel) 179 (4), 174. doi:10.3390/ani9040174
Benfey, T. J., Dye, H. M., Solar, , and Donaldson, E. M. (1989). The growth and reproductive endocrinology of adult triploid Pacific salmonids. Fish. Physiol. Biochem. 6 (2), 113–120. doi:10.1007/BF01875483
Bhattacharya, D., and Van Meir, E. G. (2019). A simple genotyping method to detect small CRISPR-Cas9 induced indels by agarose gel electrophoresis. Sci. Rep. 9 (1), 4437. doi:10.1038/s41598-019-39950-4
Blazer, V. S. (2002). Histopathological assessment of gonadal tissue in wild fishes. Fish Physiology Biochem. 26 (1), 85–101. doi:10.1023/a:1023332216713
Braat, A. K., Zandbergen, T., Van de Water, S., Goos, H. J. T., and Zivkovic, D. (1999). Characterization of zebrafish primordial germ cells: Morphology and early distribution of vasa RNA. Dev. Dynam 216 (2), 153–167. doi:10.1002/(SICI)1097-0177(199910)216:2<153::AID-DVDY6>3.0.CO;2-1
Brend, T., and Holley, S. A. (2009). Zebrafish whole mount high-resolution double fluorescent <em>in situ</em> hybridization. J. Vis. Exp. 25, 1229. doi:10.3791/1229
Cal, R. M., idal, S., Gomez, C., Alvarez-Blazquez, B., Martinez, P., and Piferrer, F. (2006). Growth and gonadal development in diploid and triploid turbot (Scophthalmus maximus). Aquaculture 251 (1), 99–108. doi:10.1016/j.aquaculture.2005.05.010
Cao, L., Naylor, R., Henriksson, P., Leadbitter, D., Metian, M., Troell, M., et al. (2015). Global food supply. China's aquaculture and the world's wild fisheries. Science 347 (6218), 133–135. doi:10.1126/science.1260149
Castrillon, D. H., Quade, B. J., Wang, T. Y., Quigley, C., and Crum, C. P. (2000). The human VASA gene is specifically expressed in the germ cell lineage. P Natl. Acad. Sci. U. S. A. 97 (17), 9585–9590. doi:10.1073/pnas.160274797
Chapman, J. R., Taylor, M. R. G., and Boulton, S. J. (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47 (4), 497–510. doi:10.1016/j.molcel.2012.07.029
Chudakov, D. M., Matz, M. V., Lukyanov, S., and Lukyanov, K. A. (2010). Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90 (3), 1103–1163. doi:10.1152/physrev.00038.2009
Clausen, R., and Longo, S. B. (2012). The tragedy of the commodity and the farce of AquAdvantage Salmon. Dev. Change 43 (1), 229–251. doi:10.1111/j.1467-7660.2011.01747.x
Cook, J. T., McNiven, M. A., Richardson, G. F., and Sutterlin, A. M. (2000). Growth rate, body composition and feed digestibility/conversion of growth-enhanced transgenic Atlantic salmon (Salmo salar). Aquaculture 188 (1-2), 15–32. doi:10.1016/s0044-8486(00)00331-8
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Du, S. J., Gong, Z. Y., Fletcher, G. L., Shears, M. A., King, M. J., Idler, D. R., et al. (1992). Growth enhancement in transgenic Atlantic salmon by the use of an "all fish" chimeric growth hormone gene construct. Biotechnol. (N Y) 10 (2), 176–181. doi:10.1038/nbt0292-176
Extavour, C. G., and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 130 (24), 5869–5884. doi:10.1242/dev.00804
Fujihara, R., Katayama, N., Sadaie, S., Miwa, M., Sanchez Matias, G. A., Ichida, K., et al. (2022). Production of germ cell-less rainbow trout by dead end gene knockout and their use as recipients for germ cell transplantation. Mar. Biotechnol. (NY) 24 (2), 417–429. doi:10.1007/s10126-022-10128-w
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 (7320), 67–71. doi:10.1038/nature09523
Gjedrem, T., Robinson, N., and Rye, M. (2012). The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture 350, 117–129. doi:10.1016/j.aquaculture.2012.04.008
Gong, H. Y., Hu, S. Y., and Wu, J. L. (2015). Progress of transgenic fluorescent ornamental fish and development of its infertility control technologies. Agric. Biotechnol. Ind. Q. 43, 34–48.
Gong, Z., Ju, B., and Wan, H. (2001). Green fluorescent protein (GFP) transgenic fish and their applications. Genetica 111 (1-3), 213–225. doi:10.1023/a:1013796810782
Gong, Z., Wan, H., Tay, T. L., Wang, H., Chen, M., and Yan, T. (2003). Development of transgenic fish for ornamental and bioreactor by strong expression of fluorescent proteins in the skeletal muscle. Biochem. Biophys. Res. Commun. 308 (1), 58–63. doi:10.1016/s0006-291x(03)01282-8
Gordon, J. W. (1997). Transgenic technology and laboratory animal science. ILAR J. 38 (1), 32–41. doi:10.1093/ilar.38.1.32
Goto, R., Saito, T., Takeda, T., Fujimoto, T., Takagi, M., Arai, K., et al. (2012). Germ cells are not the primary factor for sexual fate determination in goldfish. Dev. Biol. 370 (1), 98–109. doi:10.1016/j.ydbio.2012.07.010
Gross-Thebing, T., Yigit, S., Pfeiffer, J., Reichman-Fried, M., Bandemer, J., Ruckert, C., et al. (2017). The vertebrate protein dead end maintains primordial germ cell fate by inhibiting somatic differentiation. Dev. Cell 43 (6), 704–715. doi:10.1016/j.devcel.2017.11.019
Guralp, H., Skaftnesmo, K. O., Kjaerner-Semb, E., Straume, A. H., Kleppe, L., Schulz, R. W., et al. (2020). Rescue of germ cells in dnd crispant embryos opens the possibility to produce inherited sterility in Atlantic salmon. Sci. Rep. 10 (1), 18042. doi:10.1038/s41598-020-74876-2
Hartung, O., Forbes, M. M., and Marlow, F. L. (2014). Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reproduction Dev. 81 (10), 946–961. doi:10.1002/mrd.22414
Her, G. M., Chiang, C. C., Chen, W. Y., and Wu, J. L. (2003). In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 538 (1-3), 125–133. doi:10.1016/s0014-5793(03)00157-1
Hong, N., Li, M. Y., Yuan, Y. M., Wang, T. S., Yi, M. S., Xu, H. Y., et al. (2016). Dnd is a critical specifier of primordial germ cells in the medaka fish. Stem Cell Rep. 6 (3), 411–421. doi:10.1016/j.stemcr.2016.01.002
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327 (5962), 167–170. doi:10.1126/science.1179555
Horvay, K., Claussen, M., Katzer, M., Landgrebe, J., and Pieler, T. (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 291 (1), 1–11. doi:10.1016/j.ydbio.2005.06.013
Hu, W., Wang, Y., and Zhu, Z. (2007). Progress in the evaluation of transgenic fish for possible ecological risk and its containment strategies. Sci. China C Life Sci. 50 (5), 573–579. doi:10.1007/s11427-007-0089-y
Hwang, W. Y., Fu, Y. F., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31 (3), 227–229. doi:10.1038/nbt.2501
Jao, L. E., Wente, S. R., and Chen, W. B. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. P Natl. Acad. Sci. U. S. A. 110 (34), 13904–13909. doi:10.1073/pnas.1308335110
Ju, B. S., Xu, X. F., He, J. Y., Liao, J., Yan, T., Hew, C. L., et al. (1999). Faithful expression of green fluorescent protein (GFP) in transgenic zebrafish embryos under control of zebrafish gene promoters. Dev. Genet. 25 (2), 158–167. doi:10.1002/(SICI)1520-6408(1999)25:2<158::AID-DVG10>3.0.CO;2-6
Kawakami, K. (2007). Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. 8 (1), S7. doi:10.1186/gb-2007-8-s1-s7
Kedde, M., Strasser, M. J., Boldajipour, B., Oude Vrielink, J. A., Slanchev, K., le Sage, C., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131 (7), 1273–1286. doi:10.1016/j.cell.2007.11.034
Kedde, M., Strasser, M. J., Boldajipour, B., Vrielink, J. A. F. O., Le Sage, C., Nagel, R., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131 (7), 1273–1286. doi:10.1016/j.cell.2007.11.034
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Kurokawa, H., Saito, D., Nakamura, S., Katoh-Fukui, Y., Ohta, K., Baba, T., et al. (2007). Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl. Acad. Sci. U. S. A. 104 (43), 16958–16963. doi:10.1073/pnas.0609932104
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B., and Valen, E. (2016). CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44 (W1), W272–W276. doi:10.1093/nar/gkw398
Li, M., Yang, H., Zhao, J., Fang, L., Shi, H., Li, M., et al. (2014). Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 197 (2), 591–599. doi:10.1534/genetics.114.163667
Li, M. Y., Hong, N., Xu, H. Y., Yi, M. S., Li, C. M., Gui, J. F., et al. (2009). Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 126 (5-6), 366–381. doi:10.1016/j.mod.2009.02.004
Li, Q., Fujii, W., Naito, K., and Yoshizaki, G. (2017). Application of dead end-knockout zebrafish as recipients of germ cell transplantation. Mol. Reproduction Dev. 84 (10), 1100–1111. doi:10.1002/mrd.22870
Lind, C. E., Ponzoni, R. W., Nguyen, N. H., and Khaw, H. L. (2012). Selective breeding in fish and conservation of genetic resources for aquaculture. Reprod. Domest. Anim. 47 (4), 255–263. doi:10.1111/j.1439-0531.2012.02084.x
Linhartova, Z., Saito, T., Kaspar, V., Rodina, M., Praskova, E., Hagihara, S., et al. (2015). Sterilization of sterlet Acipenser ruthenus by using knockdown agent, antisense morpholino oligonucleotide, against dead end gene. Theriogenology 84 (7), 1246–1255. doi:10.1016/j.theriogenology.2015.07.003
Liu, L. X., Hong, N., Xu, H. Y., Li, M. Y., Yan, Y., Purwanti, Y., et al. (2009). Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr. Patterns 9 (7), 541–548. doi:10.1016/j.gep.2009.06.008
Liu, W., and Collodi, P. (2010). Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. FASEB J. 24 (8), 2641–2650. doi:10.1096/fj.09-148403
Lujic, J., Marinovic, Z., Bajec, S. S., Djurdjevic, I., Urbanyi, B., and Horvath, A. (2018). Interspecific germ cell transplantation: A new light in the conservation of valuable balkan trout genetic resources? Fish. Pysiol Biochem. 44 (6), 1487–1498. doi:10.1007/s10695-018-0510-4
Maclean, N. (1993). Transgenic induction in Salmonid and Tilapia fish. Methods Mol. Biol. 18, 95–107. doi:10.1385/0-89603-245-0:95
Magalhaes, A. L. B., Brito, M. F. G., and Silva, L. G. M. (2022). The fluorescent introduction has begun in the southern hemisphere: Presence and life-history strategies of the transgenic zebrafish Danio rerio (cypriniformes: Danionidae) in Brazil. Stud. Neotrop Fauna E. doi:10.1080/01650521.2021.2024054
Meeker, N. D., Hutchinson, S. A., Ho, L., and Trecle, N. S. (2007). Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 43 (5), 610, 612, 614–+. doi:10.2144/000112619
Miki, M., Ohishi, N., Nakamura, E., Furumi, A., and Mizuhashi, F. (2018). Improved fixation of the whole bodies of fish by a double-fixation method with formalin solution and Bouin's fluid or Davidson's fluid. J. Toxicol. Pathol. 31 (3), 201–206. doi:10.1293/tox.2018-0001
Morita, T., Morishima, K., Miwa, M., Kumakura, N., Kudo, S., Ichida, K., et al. (2015). Functional sperm of the yellowtail (Seriola quinqueradiata) were produced in the small-bodied surrogate, Jack Mackerel (Trachurus japonicus). Mar. Biotechnol. 17 (5), 644–654. doi:10.1007/s10126-015-9657-5
Moutinho, S. (2022). Transgenic fish invades Brazilian streams. Science 375 (6582), 704–705. doi:10.1126/science.ada1331
Muir, W. M., and Howard, R. D. (1999). Possible ecological risks of transgenic organism release when transgenes affect mating success: Sexual selection and the Trojan gene hypothesis. P Natl. Acad. Sci. U. S. A. 96 (24), 13853–13856. doi:10.1073/pnas.96.24.13853
Muir, W. M. (2004). The threats and benefits of GM fish. EMBO Rep. 5 (7), 654–659. doi:10.1038/sj.embor.7400197
Nagasawa, K., Fernandes, J. M. O., Yoshizaki, G., Miwa, M., and Babiak, I. (2013). Identification and migration of primordial germ cells in Atlantic salmon, Salmo salar: Characterization of Vasa, Dead End, and Lymphocyte antigen 75 genes. Mol. Reproduction Dev. 80 (2), 118–131. doi:10.1002/mrd.22142
Northrup, E., Zschemisch, N. H., Eisenblatter, R., Glage, S., Wedekind, D., Cuppen, E., et al. (2012). The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. Plos One 7 (5), e38001. doi:10.1371/journal.pone.0038001
Ozato, K., Wakamatsu, Y., and Inoue, K. (1992). Medaka as a model of transgenic fish. Mol. Mar. Biol. Biotechnol. 1 (4-5), 346–354.
Pacchiarini, T., Sarasquete, C., and Cabrita, E. (2014). Development of interspecies testicular germ-cell transplantation in flatfish. Reprod. Fertil. Dev. 26 (5), 690–702. doi:10.1071/RD13103
Perez, J. E., Alfonsi, C., Nirchio, M., Munoz, C., and Gomez, J. A. (2003). The introduction of exotic species in aquaculture: A solution or part of the problem? Interciencia 28 (4), 234–238.
Piferrer, F., Beaumont, A., Falguiere, J. C., Flajshans, M., Haffray, P., and Colombo, L. (2009). Polyploid fish and shellfish: Production, biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture 293 (3-4), 125–156. doi:10.1016/j.aquaculture.2009.04.036
Pradhan, A., and Olsson, P. E. (2018). Germ cell depletion in zebrafish leads to incomplete masculinization of the brain. Gen. Comp. Endocrinol. 265, 15–21. doi:10.1016/j.ygcen.2018.02.001
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 (11), 2281–2308. doi:10.1038/nprot.2013.143
Raz, E. (2003). Primordial germ-cell development: The zebrafish perspective. Nat. Rev. Genet. 4 (9), 690–700. doi:10.1038/nrg1154
Raz, E. (2000). The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 1 (3), REVIEWS1017. doi:10.1186/gb-2000-1-3-reviews1017
Saito, T., Goto-Kazeto, R., Arai, K., and Yamaha, E. (2008). Xenogenesis in teleost fish through generation of germ-line chimeras by single primordial germ cell transplantation. Biol. Reproduction 78 (1), 159–166. doi:10.1095/biolreprod.107.060038
Sawamura, R., Osafune, N., Murakami, T., Furukawa, F., and Kitano, T. (2017). Generation of biallelic F0 mutants in medaka using the CRISPR/Cas9 system. Genes cells. 22 (8), 756–763. doi:10.1111/gtc.12511
Skugor, A., Tveiten, H., Krasnov, A., and Andersen, O. (2014). Knockdown of the germ cell factor Dead end induces multiple transcriptional changes in Atlantic cod (Gadus morhua) hatchlings. Anim. Reprod. Sci. 144 (3-4), 129–137. doi:10.1016/j.anireprosci.2013.12.010
Slanchev, K., Stebler, J., de la Cueva-Mendez, G., and Raz, E. (2005). Development without germ cells: The role of the germ line in zebrafish sex differentiation. P Natl. Acad. Sci. U. S. A. 102 (11), 4074–4079. doi:10.1073/pnas.0407475102
Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E. (2009). Control of Dead end localization and activity--implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 126 (3-4), 270–277. doi:10.1016/j.mod.2008.10.006
Stewart, C. N. (2006). Go with the glow: Fluorescent proteins to light transgenic organisms. Trends Biotechnol. 24 (4), 155–162. doi:10.1016/j.tibtech.2006.02.002
Stigebrandt, A., Aure, J., Ervik, A., and Hansen, P. K. (2004). Regulating the local environmental impact of intensive marine fish farming. Aquaculture 234 (1-4), 239–261. doi:10.1016/j.aquaculture.2003.11.029
Su, B. F., Peatman, E., Shang, M., Thresher, R., Grewe, P., Patil, J., et al. (2014). Expression and knockdown of primordial germ cell genes, vasa, nanos and dead end in common carp (Cyprinus carpio) embryos for transgenic sterilization and reduced sexual maturity. Aquaculture 420, S72–S84. doi:10.1016/j.aquaculture.2013.07.008
Su, B. F., Shang, M., Grewe, P. M., Patil, J. G., Peatman, E., Perera, D. A., et al. (2015). Suppression and restoration of primordial germ cell marker gene expression in channel catfish, Ictalurus punctatus, using knockdown constructs regulated by copper transport protein gene promoters: Potential for reversible transgenic sterilization. Theriogenology 84 (9), 1499–1512. doi:10.1016/j.theriogenology.2015.07.037
Suster, M. L., Kikuta, H., Urasaki, A., Asakawa, K., and Kawakami, K. (2009). Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 561, 41–63. doi:10.1007/978-1-60327-019-9_3
Tidwell, J. H., and Allan, G. L. (2001). Fish as food: aquaculture's contribution - ecological and economic impacts and contributions of fish farming and capture fisheries. Embo Rep. 2 (11), 958–963. doi:10.1093/embo-reports/kve236
Urnov, F. D., Miller, J. C., Lee, Y. L., Beausejour, C. M., Rock, J. M., Augustus, S., et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435 (7042), 646–651. doi:10.1038/nature03556
Wagner, E. J., Arndt, R. E., Routledge, M. D., Latremouille, D., and Mellenthin, R. F. (2006). Comparison of hatchery performance, agonistic behavior, and poststocking survival between diploid and triploid rainbow trout of three different Utah strains. N. Am. J. Aquacult 68 (1), 63–73. doi:10.1577/a05-026.1
Wang, T. (2015). Gene editing of dnd by TALENs in medaka embryos and its role in PGC formation. Singapore: Ph.D. thesis. Department of Biological Sciencs, National University of Singapore.
Wang, T., and Hong, Y. (2014). Direct gene disruption by TALENs in medaka embryos. Gene 543 (1), 28–33. doi:10.1016/j.gene.2014.04.013
Wargelius, A., Leininger, S., Skaftnesmo, K. O., Kleppe, L., Andersson, E., Taranger, G. L., et al. (2016). Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 18, 21284. doi:10.1038/srep21284
Weidinger, G., Stebler, J., Slanchev, K., Dumstrei, K., Wise, C., Lovell-Badge, R., et al. (2003). Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 13 (16), 1429–1434. doi:10.1016/s0960-9822(03)00537-2
Weidinger, G., Wolke, U., Koprunner, M., Klinger, M., and Raz, E. (1999). Identification of tissues and patterning events required for distinct steps in early migration of zebrafish primordial germ cells. Development 126 (23), 5295–5307. doi:10.1242/dev.126.23.5295
Wong, A. C., and Van Eenennaam, A. L. (2008). Transgenic approaches for the reproductive containment of genetically engineered fish. Aquaculture 275 (1-4), 1–12. doi:10.1016/j.aquaculture.2007.12.026
Wong, T. T., and Collodi, P. (2013). Inducible sterilization of zebrafish by disruption of primordial germ cell migration. Plos One 8 (6), e68455. doi:10.1371/journal.pone.0068455
Wong, T. T., and Zohar, Y. (2015). Production of reproductively sterile fish by a non-transgenic gene silencing technology. Sci. Rep-Uk 5, 15822. doi:10.1038/srep15822
Wong, T. T., and Zohar, Y. (2015). Production of reproductively sterile fish: A mini-review of germ cell elimination technologies. Gen. Comp. Endocr. 221, 3–8. doi:10.1016/j.ygcen.2014.12.012
Xu, H. Y., Li, M. Y., Gui, J. F., and Hong, Y. H. (2010). Fish germ cells. Sci. China Life Sci. 53 (4), 435–446. doi:10.1007/s11427-010-0058-8
Yamaha, E., Goto-Kazeto, R., Saito, T., Kawakami, Y., Fujimoto, T., Adachi, S., et al. (2010). Primordial germ cell in teleost fish with special references to its specification and migration. J. Appl. Ichthyol. 26 (5), 816–822. doi:10.1111/j.1439-0426.2010.01548.x
Yazawa, R., Takeuchi, Y., Higuchi, K., Yatabe, T., Kabeya, N., and Yoshizaki, G. (2010). Chub mackerel gonads support colonization, survival, and proliferation of intraperitoneally transplanted xenogenic germ cells. Biol. Reprod. 82 (5), 896–904. doi:10.1095/biolreprod.109.081281
Yazawa, R., Takeuchi, Y., Morita, T., Ishida, M., and Yoshizaki, G. (2013). The Pacific bluefin tuna (Thunnus orientalis) dead end gene is suitable as a specific molecular marker of type A spermatogonia. Mol. Reprod. Dev. 80 (10), 871–880. doi:10.1002/mrd.22224
Yoon, C., Kawakami, K., and Hopkins, N. (1997). Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124 (16), 3157–3165. doi:10.1242/dev.124.16.3157
Yoshizaki, G., Sakatani, S., Tominaga, H., and Takeuchi, T. (2000). Cloning and characterization of a vasa-like gene in rainbow trout and its expression in the germ cell lineage. Mol. Reprod. Dev. 55 (4), 364–371. doi:10.1002/(SICI)1098-2795(200004)55:4<364::AID-MRD2>3.0.CO;2-8
Keywords: infertility control, fluorescent zebrafish, transgenic fish, genome editing, CRISPR/Cas9, Dead end, PGCs
Citation: Chu W-K, Huang S-C, Chang C-F, Wu J-L and Gong H-Y (2023) Infertility control of transgenic fluorescent zebrafish with targeted mutagenesis of the dnd1 gene by CRISPR/Cas9 genome editing. Front. Genet. 14:1029200. doi: 10.3389/fgene.2023.1029200
Received: 26 August 2022; Accepted: 02 January 2023;
Published: 13 January 2023.
Edited by:
Yuzine Esa, Putra Malaysia University, MalaysiaReviewed by:
Hong Wei Liang, Yangtze River Fisheries Research Institute (CAFS), ChinaCopyright © 2023 Chu, Huang, Chang, Wu and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Yi Gong, aHlnb25nQG1haWwubnRvdS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.