- 1CIMMYT-JAAS Joint Center for Wheat Diseases, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
- 3Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
Fusarium head blight (FHB) of wheat is an important disease worldwide, affecting the yield, end-use quality and threatening food safety. Genetic resources or stable loci for FHB resistance are still limited in breeding programs. A panel of 265 bread wheat accessions from China, CIMMYT-Mexico and other countries was screened for FHB resistance under 5 field experiments in Mexico and China, and a genome-wide association analysis was performed to identify QTLs associated with FHB resistance. The major locus Fhb1 was significantly associated with FHB severity and Deoxynivalenol content in grains. FHB screening experiments in multiple environments showed that Fhb1-harbouring accessions Sumai3, Sumai5, Ningmai9, Yangmai18 and Tokai66 had low FHB index, disease severity and DON content in grains in response to different Fusarium species and ecological conditions in Mexico and China. Accessions Klein Don Enrique, Chuko and Yumai34 did not have Fhb1 but still showed good FHB resistance and low mycotoxin accumulation. Sixteen loci associated with FHB resistance or DON content in grains were identified on chromosomes 1A, 1B, 2B, 3A, 3D, 4B, 4D, 5A, 5B, 7A, and 7B in multiple environments, explaining phenotypic variation of 4.43–10.49%. The sources with good FHB resistance reported here could be used in breeding programs for resistance improvement in Mexico and China, and the significant loci could be further studied and introgressed for resistance improvement against FHB and mycotoxin accumulation in grains.
Introduction
Fusarium head blight (FHB) is one of the most devastating diseases of wheat and other small grain cereals, threatening global wheat production and food security and safety (Bai and Shaner, 2004; Ma et al., 2020). Many Fusarium spp. can infect wheat head, of which F. graminearum species complex (FGSC) is the predominant FHB pathogen in most wheat production areas (Van Der Lee et al., 2015). The distribution of various FGSC sub-species depends on geographies, climatic conditions and cropping systems, e.g., F. asiaticum is mainly in East Asia, while F. meridionale and F. boothii are mainly in South America and Africa (Zhang et al., 2012; Backhouse, 2014; Van Der Lee et al., 2015). FHB leads to severe yield loss, poor grain quality, and more importantly contamination of the infected grains with mycotoxins like deoxynivalenol (DON) or nivalenol, for which public concerns have prompted governments to set upper limits for DON in wheat grain and its products (Mesterházy et al., 1999; He et al., 2016; Ma et al., 2020).
Breeding for FHB resistance by using QTL/genes in genetic sources is one of the effective approaches to control this disease and prevent toxins contamination in grains after harvest. FHB resistance in wheat is a complex quantitative trait with strong genotype-by-environment interactions, resulting in different response to Fusarium pathogens across different environments (Mesterhazy, 2020). Host resistance to FHB involves five resistant types, i.e. Type I for initial infection, Type II for disease spread, Type III for toxin accumulation, Type IV for kernel infection, and Type V for yield reduction (Schroeder and Christensen, 1963; Miller and Amison, 1986; Mesterhazy, 1995). Type II and III resistance have been widely studied (Buerstmayr et al., 2020; Ma et al., 2020; Mesterhazy, 2020), and over 600 loci for Type II or III resistance have been mapped on all 21 wheat chromosomes (Venske et al., 2019; Zheng et al., 2021). Of the nominated Fhb genes, Fhb1 (Cuthbert et al., 2006; Liu et al., 2006), Fhb2 (Cuthbert et al., 2007), Fhb4 (Xue et al., 2010) and Fhb5 (syn. Qfhs.ifa-5A) (Xue et al., 2011; Buerstmayr et al., 2018; Steiner et al., 2019) are derived from common wheat, of which the former two mainly confer Type II resistance and the latter two mainly confer Type I resistance. The wild relatives of wheat have also contributed several resistance genes/loci, like Fhb3 (Qi et al., 2008) from Leymus racemosus, Fhb6 (Cainong et al., 2015) from Elymus tsukushiensis and Fhb7 (Guo et al., 2015) from Thinopyrum elongatum, all conferring Type II resistance. Type II and Type III resistance are usually highly correlated in field experiments as observed by most researchers, although QTL exclusively for Type III resistance have been reported (He et al., 2019).
Thousands of wheat accessions have been screened for FHB resistance throughout the world over the last decades, but resistant sources for FHB improvement in breeding programs remain limited. Fhb1 (Syn. Qfhs.ndsu-3BS) derived from Sumai3 or its derivative Ning7840 is recognized as a locus with major effect and stable resistance and has been widely used in wheat breeding programs worldwide (Zhu et al., 2019; Buerstmayr et al., 2020; Hao et al., 2020; Ma et al., 2020; Gaire et al., 2022b). Several research groups have cloned the candidate genes of Fhb1 (Rawat et al., 2016; Li et al., 2019; Su et al., 2019) and designed diagnostic markers for marker-assisted selection (MAS) (He et al., 2018; Su et al., 2018; Singh et al., 2019). Sumai3 that has Fhb1, Fhb2, and Fhb5 is a widely used donor for FHB resistance improvement, and more than 20 released spring wheat varieties have Sumai3 in their pedigrees in North America and Canada (Zhu et al., 2019; Ghimire et al., 2020). The donor parent of Fhb1 in the Chinese wheat breeding programs is, however, not Sumai3 but Ningmai9 (developed by Jiangsu Academy of Agriculture Sciences, JAAS) because of its better agronomic traits and high yield potential (Zhu et al., 2018). More recently, Fhb7 derived from Th. elongatum was reported to confer broad resistance to Fusarium species by detoxifying DON without yield penalty (Wang et al., 2020), and several locally adaptive lines with Fhb7 in their pedigrees have been tested in Regional Yield Trials in China (Prof. Hongwei Wang, Shandong Agricultural University, personal communication).
Despite the achievement, highly resistant sources and major loci for FHB resistance are scarce for wheat breeding, it is therefore worthwhile to identify additional FHB resistance sources and loci for gene pyramiding. The objectives of this study were to screen a collection of common wheat varieties and elite breeding lines of worldwide origin for FHB resistance in Mexico and China, and to conduct a genome-wide association study (GWAS) to identify QTL for FHB resistance in field experiments.
Materials and methods
Plant materials
In this study, 265 wheat accessions (CIMMYT-China panel) were screened for FHB resistance under field conditions in Mexico and China. The panel included commercial varieties and breeding lines of worldwide origin, i.e., 131 Chinese accessions mainly from the Yellow and Huai River Valley Region and Middle-lower Yangtze Valley Region, 71 from CIMMYT-Mexico, 41 from South America, 10 from North America, five each from Asia and Europe, and one each from Oceania and Africa (Supplementary Table S1). The accessions were mostly of spring type along with a few of winter type. In Mexico, Sumai3 and Heilo were used as resistant checks, while Gamenya and Ocoroni were included as susceptible checks. In China, Sumai3 and Yangmai158 served as resistant checks with Annong8455 and Jimai22 as susceptible checks.
Field trials and Fusarium head blight screening
The CIMMYT-China panel was evaluated for FHB resistance in CIMMYT’s El Batan research station (altitude of 2,240 masl, 19.5ºN/98.8ºW, with an average annual precipitation of 625 mm) in Mexico, and JAAS’s FHB nursery (altitude of 22 masl, 32.0ºN/118.8ºE, with an annual precipitation of 800–1034 mm) in Nanjing, China. In each location, the accessions were planted in 1-m double rows with two replications in randomized complete block design. The trials were conducted in the 2018 and 2019 cropping cycles (from May to September) in Mexico, and in the 2018–19, 2019–20, and 2020–21 cropping cycles (i.e. November to May) in China.
In Mexico, inoculum comprised a mixture of five F. graminearum isolates, CIMFU No. 85, 89, 108, 162, and 222, following the protocols described by (He et al., 2013). These isolates were obtained from naturally infected wheat heads in the El Batan station in 2017 and were selected based on their high DON productivity and high aggressiveness in greenhouse experiments. At anthesis, the field plots were sprayed with an inoculum of 50,000 spores/ml, and the procedure was repeated 2 days later to reinforce the infection. A misting system was set up in the nursery, operational from 9 am to 8 pm with 10 min of spraying each hour, to maintain a humid environment conducive for FHB infection. Field evaluation of FHB infection was carried out at 25 days post-inoculation (dpi) on the 10 spikes tagged at anthesis (Feekes 10.5.1). Numbers of total and infected spikelets of each spike were scored for the calculation of FHB index with the formula: FHB index = severity × incidence, where severity means the averaged percentage of diseased spikelets, and incidence the percentage of symptomatic spikes. About 20g grain sample was ground with a coffee mill and a 2g sub-sample was measured for DON quantification with the Ridascreen Fast DON ELISA kit (RBiopharm GmbH, Darmstadt, Germany) following the manufacturer’s instructions.
In China, F. asiaticum isolates Fa0609, Fa1312 and Fa0980 were used in this study. These isolates were isolated from different wheat plots in JAAS (Nanjing, China) in 2006, 2009 and 2013, respectively, and had shown strong virulence in greenhouse and field experiments. The fungal isolates were applied to wheat kernels to produce spawn inoculum, which was distributed on the soil surface with a density of 30 g/m2 on the 20th day before flowering and the inoculation was repeated 2 weeks later. All the plots were misted for 15 min per hour during the day to create a humid environment for fungal development and spread. The numbers of diseased spikelets of 10 random spikes were recorded to calculate the disease severity for each accession at the milky ripe stage (Feekes 11.2). The spawn inoculation experiments were performed only during 2018–19 and 2019–20 cropping cycles. Besides, point inoculation with a single isolate Fa0609 was also conducted to evaluate the Type II resistance of the panel during 2018–19, 2019–20, and 2020–21 cropping cycles. Ten spikes per accession were inoculated via injecting 10 μl suspension of F. asiaticum (100,000 spores/ml) into a central spikelet at anthesis (Feekes 10.5.1), and then the spikes were covered with a plastic bag for 3 days to meet the moisture requirement for fungal infection. The moisturizing measure was the same as that mentioned above. The numbers of diseased spikelets were recorded as FHB severity on the 21st day after inoculation. The inoculated spikes were harvested to detect DON content with DON ELISA test kit (Huaan Magnech Bio-tech, China) using an indirect competitive enzyme-labeled immunoassay.
The phenotypic dataset including the mean values of FHB index, FHB severity and DON content in grains in each environment for each accession were used in subsequent analysis. The datasets of Mexico and China were calculated separately. Pearson’s correlation coefficients among environments were computed by R to test the correlation of FHB responses across different environments (R Team, 2011). Turkey’s mean comparison tests were performed among subgroups using R (R Team, 2011).
Genotyping
The GWAS panel was genotyped with the DArTSeq technology at the Genetic Analysis Service for Agriculture (SAGA) at CIMMYT, Mexico. Markers with more than 10% missing data or minor allele frequency less than 1% were eliminated, resulting in 18,436 high quality markers, of which 14,195 SNPs with known physical positions in Chinese Spring reference genome v.1.0 were used for population structure and LD analysis. PCR-based marker JAASM395 was used to test whether an accession carried Fhb1 (Chinese patent, ZL201811515195.8), and the primer pair was as follows: JAASM395F: GTCTCCGTTCAATTCGGTGAGT; JAASM395R: GACAATGTGAAGGCGTTGTCTA. Analysis on population structure and linkage disequilibrium followed the method in (Wu et al., 2021). Briefly, a Bayesian model-based method was used to infer the number of subpopulation among all accessions with the software STRUCTURE v 2.3.4 (Pritchard et al., 2000). Five independent analyses for the assumed number of subpopulations (K value) from 1 to 8 were performed based on an admixture model with MCMC replications and burn-in time number set at 1×105 and 1×104, respectively (Pritchard et al., 2000). Optimal value of population size was inferred by CLUMPP based on the data of STRUCTURE (Jakobsson and Rosenberg, 2007). Pairwise LD (r2) for all pairwise comparisons in a distance of 10,000 kb was estimated by the software PLINK (Chang et al., 2015) and were plotted against the physical distances, and then a nonlinear regression was fitted in R (R Team, 2011). The critical r2 value was determined as the 95th quantile for all r2 values between unlinked SNPs. The intersection between the critical r2 value and the regression line was used to estimate the average size for LD blocks in this panel.
Genome-wide association analysis
Genome-wide association for FHB resistance across environments was performed using software TASSEL v5.0 pipeline command line interface (Bradbury et al., 2007). Mixed linear models (MLM) with K/Q and K/P matrices as covariates were chosen for all GWAS analyses. The Q matrix was calculated and estimated by STRUCTURE v2.3.4 (Pritchard et al., 2000). Physical positions of significant SNPs were determined by sequence alignments with “Chinese Spring” reference genome v1.0 on the website of EnsemblPlants (http://plants.ensembl.org/index.html) using BLAST program with default parameters. Physical positions of significant MTAs were compared to meta-QTL reported by (Zheng et al., 2021) to see their novelty.
Results
Fusarium head blight screening in Mexico and China
In spray inoculation experiments in Mexico, FHB index showed a skewed distribution toward the low disease direction and the panel exhibited a grand mean FHB index of 16.83%. The accession Ning894013 had the lowest FHB index of 0.97%, while Norseman had the maximum of FHB index of 62.87% (Figure 1A, Supplementary Table S1). DON content in grains showed a continuous distribution with a mean of 6.91 μg/g (Figure 1B, Supplementary Table S1). In point inoculation experiments in China, numbers of diseased spikelets evenly distributed ranging from 2 to 13 with a mean of 6.6 (Figure 1C, Supplementary Table S1). In spawn inoculation experiments, the mean numbers of diseased spikelets were higher than that in point inoculation experiments (Figure 1D). The DON content in grains in point inoculation experiments was skewed to the low content direction, though, the mean value 54.11 μg/g was much higher than that in spray inoculation experiments (Figure 1E, Supplementary Table S1).
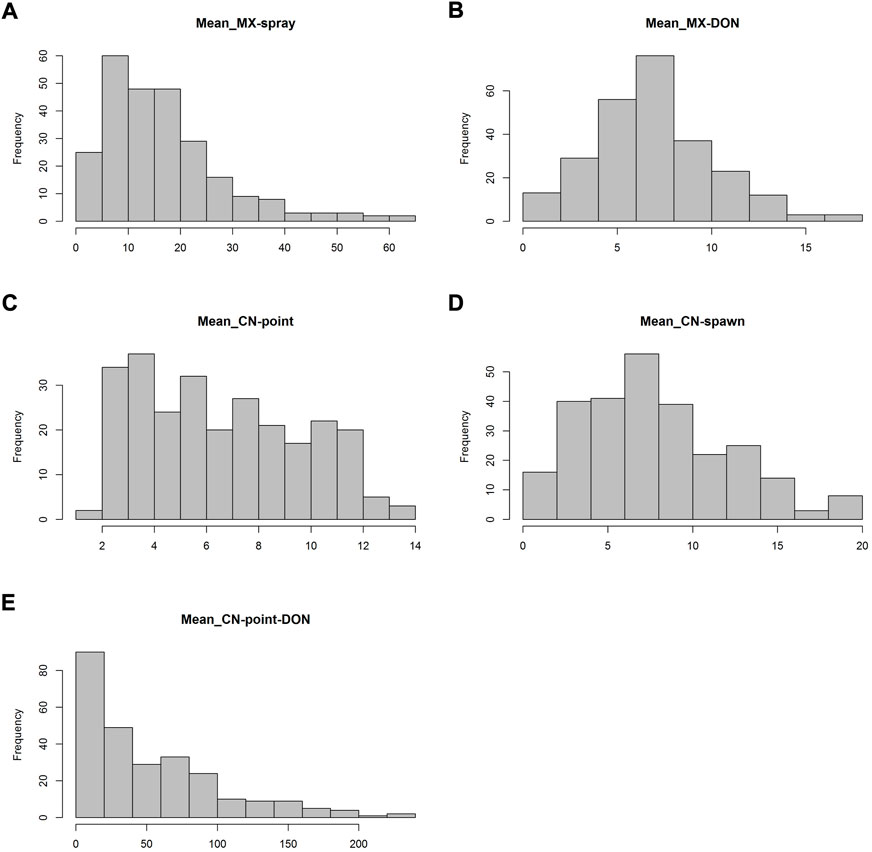
FIGURE 1. Frequency distribution of FHB index (A), number of diseased spikelets (C and D) and DON content in grains (B and E) based on the mean data in Mexico (MX, spray inoculation) and China (CN, point and spawn inoculation).
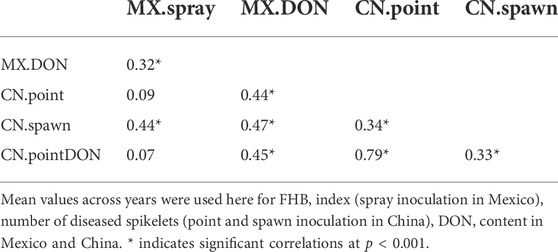
Phenotypic correlation varied greatly among different experiments, from low non-significant correlation (r = 0.07) to high significant correlation (r = 0.79). Generally, low to moderate correlations were observed among different inoculation methods, and the highest correlation was found between number of diseased spikelets and DON content in point inoculation experiments in China (Table 1).
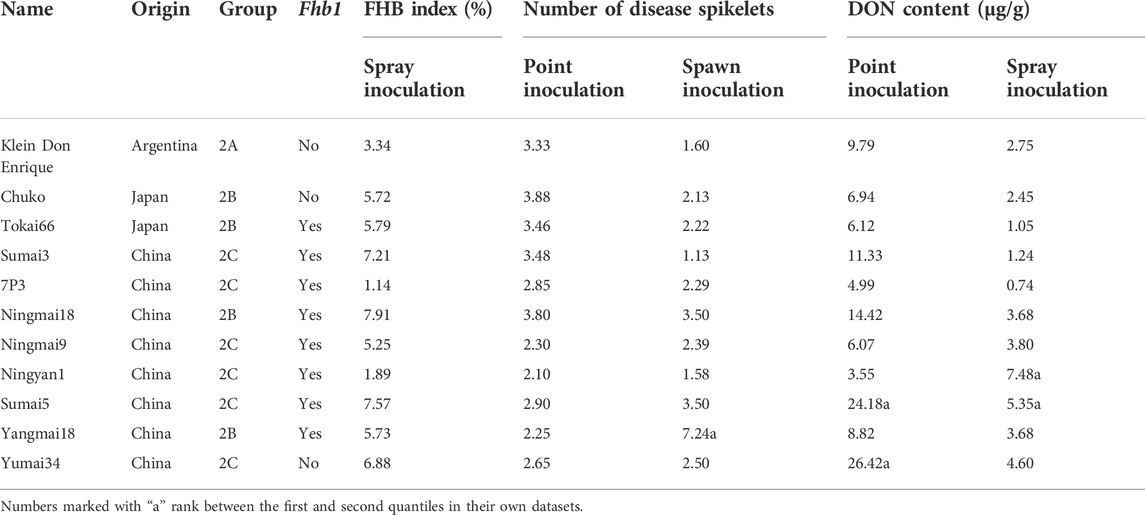
Genetic resources with moderate resistance to FHB and low DON content in grains were listed in Table 2. As expected, Fhb1-carriers Sumai3, Sumai5, Ningmai9, Yangmai18 and Tokai66 exhibited good resistance in all experiments in Mexico and China. Accessions Klein Don Enrique, Chuko and Yumai34 that do not have Fhb1 still showed good FHB resistance and low toxin accumulation.
Population structure analysis indicated that the panel could be divided into two subpopulations, which could be further divided into five groups 1A, 1B, 2A, 2B and 2C. The clustering of subpopulations mainly associated with geographical origins. Subpopulation 1 (130 accessions) involved accessions mainly from CIMMYT, South America, North America, and Europe, while most accessions in subpopulation 2 (135 accessions) were from Asian countries including China, Japan and India (Supplementary Table S1).
There were significant differences in FHB resistance or DON content in grains among the groups 1A, 1B, 2A, 2B and 2C (p < 0.01) (Figures 2A–E). In spray inoculation experiments in Mexico, group 2A had lower mean FHB index than 1B and 2C, and 2B had the lowest DON content in grains (Figures 2A,B). In the experiments in China, however, subpopulation 2 outperformed subpopulation 1 significantly in most cases, and significant differences were also found within subpopulations (Figures 2C–E).
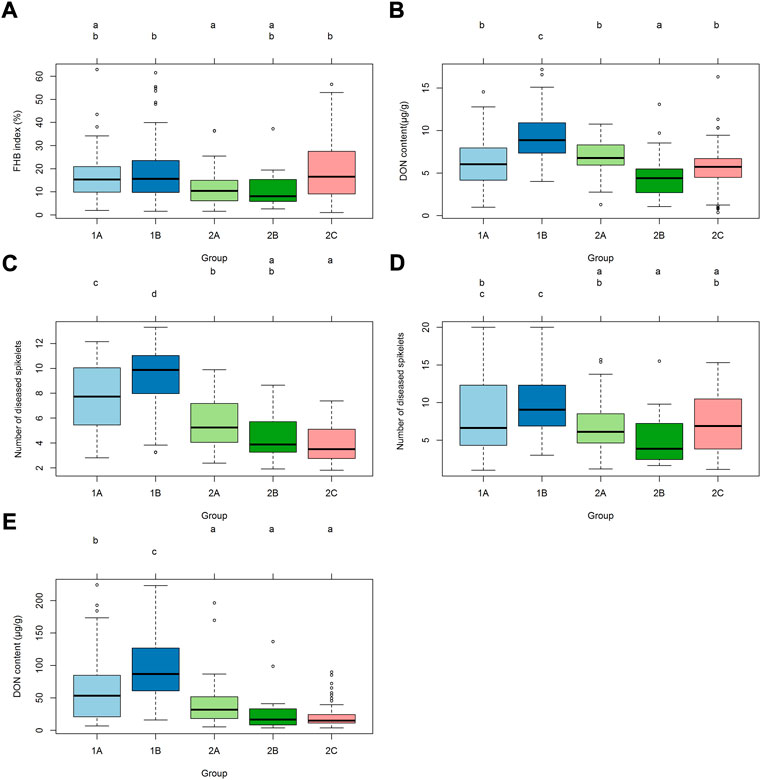
FIGURE 2. Distributions of FHB index, number of diseased spikelets and DON content among different groups. (A) Distribution of mean data of FHB index using spray inoculation among different groups in 2018 and 2019 in Mexico. (B) Distribution of mean data of DON content using spray inoculation among different groups in 2018 and 2019 in Mexico. (C) Distribution of mean data of FHB severity using point inoculation among different groups in 2018, 2019 and 2020 in China. (D) Distribution of mean data of FHB severity using spawn inoculation among different groups in 2018 and 2019 in China. (E) Distribution of mean data of DON content in grains using point inoculation among different groups in 2018, 2019 and 2020 in China.
Significant differences in FHB index or number of diseased spikelets were observed between Fhb1 and non-Fhb1 groups, where the former outperformed the latter; the same trend applied to DON content in both Mexico and China (Figure 3).
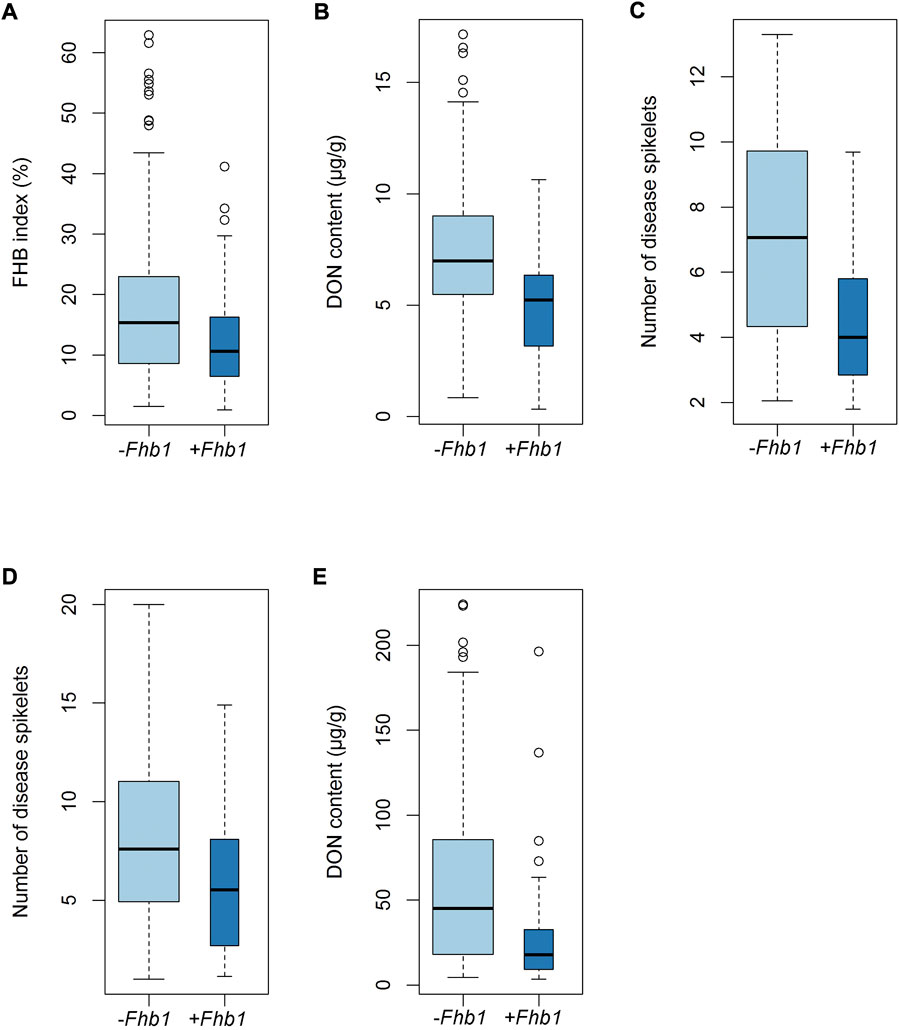
FIGURE 3. Differences in FHB index (A), number of diseased spikelets (C and D) and DON content (B and E) between Fhb1 and non-Fhb1 accessions.
Loci associated with Fusarium head blight resistance
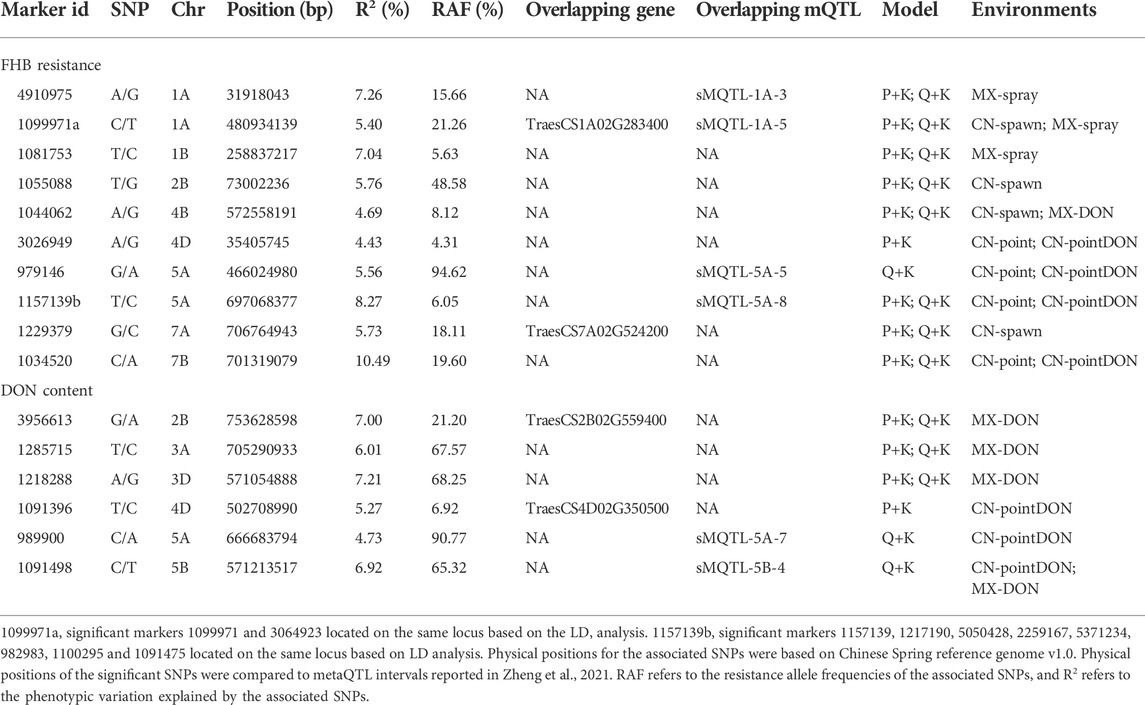
Marker-trait associations (MTAs) were tested separately for the dataset of mean value in Mexico and China, and a total of 16 MTAs (24 SNPs) were significantly associated with FHB resistance or DON content in grains (Figures 4, 5), explaining phenotypic variation between 4.43% and 10.49% (Table 3). Four of the SNPs located in annotated gene regions without known functions. Ten MTAs (18 SNPs) on chromosomes 1A, 1B, 2B, 7A, 4D, 4B, 5A and 7B were significantly associated with FHB resistance (p < 0.001), of which markers 3026949, 979146, 1157139 and 10334520 were significantly associated with both FHB resistance and DON content using point inoculation data in China. Marker 1044062 on 4B chromosome was identified in association with FHB resistance in China and DON content in Mexico. Only one marker 1099971 was significantly associated with FHB resistance in both China and Mexico, overlapping a reported mQTL sMQTL-1A-5. Six markers on chromosomes 2B, 3A, 3D, 4D, 5A and 5B were associated with only DON content across China and Mexico (Table 3). Significant markers for FHB resistance or DON content in individual environments were listed in Supplementary Table S2, explaining phenotypic variation of 4.34–11.33%.
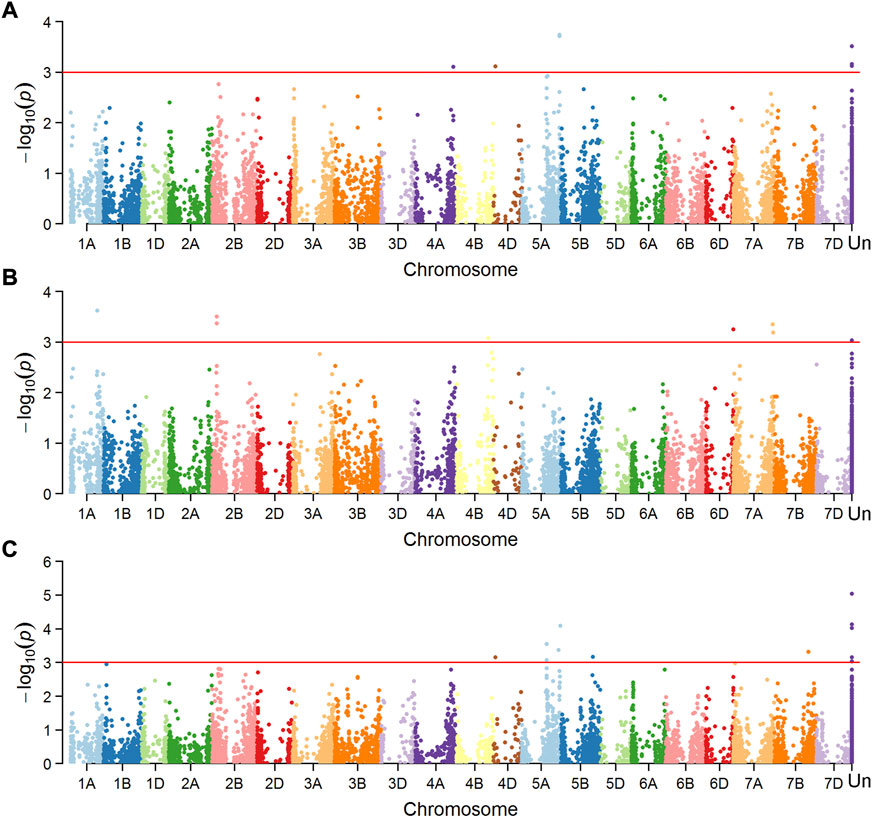
FIGURE 4. Manhattan plots showing SNPs associated with number of diseased spikelets using point inoculation (A) and spawn inoculation (B), and those for DON content using point inoculation (C) in China.
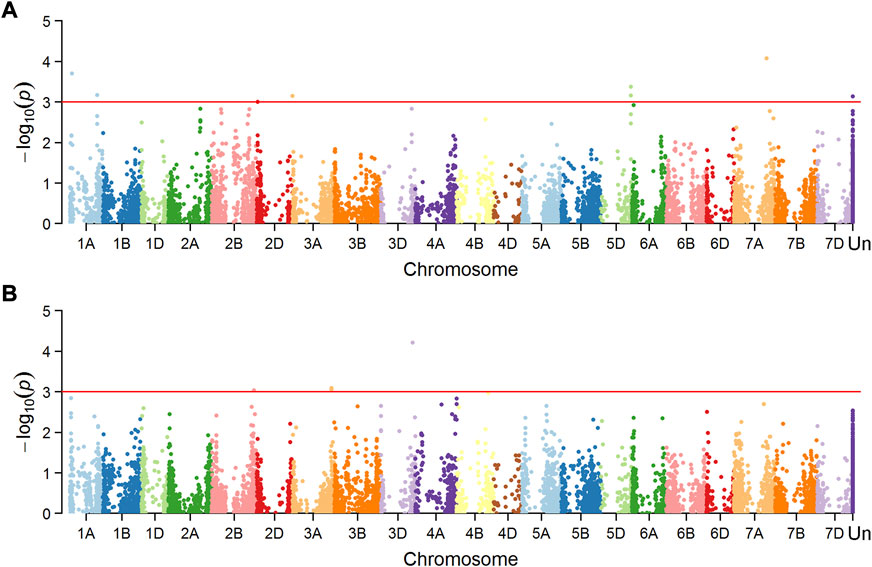
FIGURE 5. Manhattan plots showing SNPs associated with FHB index using spray inoculation (A) and those for DON content (B) using spray inoculation in Mexico.
Discussion
FHB of wheat is a serious disease in the temperate and humid regions around the world. Screening for resistant sources is a prerequisite for the improvement of FHB resistance, but many factors affect the development of locally adapted resistant varieties, including complex inheritance, multiple resistance types, difficulties on precise phenotyping, association of FHB resistance with undesirable traits (late and tall plant phenology etc.), and strong genotype-by-environment interaction. In the present study, 265 accessions from China, CIMMYT-Mexico and other countries were screened for FHB resistance in Mexico and China. FHB index and disease severity were used to evaluate FHB resistance of all the accessions in field trials, and then DON content in infected grains was detected. Spray inoculation simulates the process of disease development under natural conditions in Mexico, and the FHB index showed a combination of Types I and II resistance to FHB. Therefore, the data of FHB incidence does not refer to strictly Type I resistance due to the late scoring time. FHB severity is characterized only for Type II resistance by the point inoculation method in China, which is considered a stable and accurate screening method. There was no significant correlation between the data of FHB index in Mexico and FHB severity in China (Table 1), which was caused by accessions with contrasting resistance components in the two countries, e.g., several Chinese accessions showed good Type II resistance in China but poor Type I resistance in Mexico (Supplementary Table S1). The reason could be ascribed to the relatively high frequency of Fhb1 (34.1%) conferring good Type II resistance in Chinese accessions. Uncontrollable factors such as environmental change and inoculum content make accurate phenotyping of Type I resistance difficult. Type II resistance evaluation is mandatory before the release of Chinese varieties, while other resistance components including Type I resistance are optional in breeding program. However, accessions with good Type II resistance but poor Type I resistance could still suffer high yield loss and DON contamination under natural conditions with high Fusarium pressure. Pyramiding of Types I and II resistance to FHB will be a promising breeding strategy, and introgression of Fhb1, Fhb4 and Fhb5 into five modern Chinese wheat reduced FHB severity by 95% without penalty for agronomic traits and yield (Zhang et al., 2021). In Mexico, toxin content was moderately correlated with the FHB index, while in China, grain toxin levels were highly correlated with Type II resistance (Table 1). These data suggested that DON content in grains actually represented a combination of Type I/II and III resistance, which may explain why QTLs exclusively associated with Type III resistance are rarely mapped (He et al., 2019).
Most of the wheat varieties in the middle and lower reaches of the Yangtze river in China have Fhb1 that confers moderate resistance to FHB (data not shown), otherwise it would not be released in National Wheat Production Trials. Recently, the development of wheat germplasm combining Fhb1 and Sr2 in CIMMYT backgrounds would be used in breeding for both FHB and stem rust resistance (He et al., 2020). Large quantities of QTL mapping and omics data for FHB resistance have been released in the previous reports, it is widely accepted that Fhb1 on 3BS chromosome is a major and stable locus for Type II resistance to FHB (Cuthbert et al., 2006; Liu et al., 2006; Gunnaiah et al., 2012; Schweiger et al., 2013; Schweiger et al., 2016; Eldakak et al., 2018). Previous studies have shown that Fhb1 did not effectively increase resistance to FHB in certain genotypes (Pumphrey et al., 2007), and a similar case has been observed in our data, that is, several accessions harboring Fhb1 showed moderate susceptibility to FHB (Supplementary Table S1). The additive effect of minor loci might play an important role in FHB resistance, and non-Fhb1 accessions with moderate susceptibility to resistance could still be used in breeding or genetic analysis (Supplementary Table S1). Totally 11 accessions exhibited stable moderate resistance and low DON content in grains (Table 2). It was worth noting that three accessions Klein Don Enrique, Chuko and Yumai34 did not contain Fhb1, but were resistant to multiple Fusarium species from Mexico and China. These non-Fhb1 accessions with moderate resistance to FHB would facilitate their application in breeding and provide alternative options for FHB improvement.
The present study identified 16 genetic loci associated with FHB resistance and/or DON content in grains, 6 of which overlapped with reported metaQTL intervals (Zheng et al., 2021). Marker 4910975 is in the interval of previously reported smQTL-1A-3 (28–38 Mb) derived from “CJ9303” and two European winter wheats “History” and “Pirat” (Jiang et al., 2007; Holzapfel et al., 2008). Marker 1099971 is close to the flanking marker IWA7577 of smQTL-1A-5 (484–509 Mb) and overlapped with this QTL based on the LD data (Zheng et al., 2021). Marker 1099971 is found in the second exon of TraesCS1A02G283400, which is annotated as a copper ion-binding protein involved in lignin catalytic synthesis. Marker 979146 associated with Type II FHB resistance here overlaps with reported QTL interval of smQTL-5A-5 from “Wangshuibai” and CIMMYT wheat line “C615” (Ma et al., 2006; Yi et al., 2018). A cluster of SNPs tagged by marker 1157139 falls into the interval of smQTL-5A-8 (681–694 Mb), which is a minor QTL from Swiss winter wheat cultivar “Arina” (Paillard et al., 2004). A locus associated with DON content in grains tagged by marker 989900 is close to the flanking marker of smQTL-5A-7 (662-645 Mb) (Zheng et al., 2021), which has been associated with Types II and III resistance in different accessions by several independent research groups (Buerstmayr et al., 2011; Chu et al., 2011; Lu et al., 2013; Malihipour et al., 2017; Zhao et al., 2018). Similarly, marker 1091498 that was only associated with DON content in grains falls into a known QTL interval, too. Considering that these two loci were significant in point inoculation experiments, they are likely associated with Type II resistance, too (Zheng et al., 2021). The rest 10 loci are not tightly linked to the previously reported QTL through sequence alignments, implying that they may be novel for FHB resistance. Marker 1229379 associated with FHB resistance is located in the coding region of TraesCS7A02G524200 on 7A, which could play a role in defense responses to pathogens by maintaining cell membrane integrity.
Our results, together with the large number of loci reported in previous studies (Arruda et al., 2016a; He et al., 2016; Buerstmayr et al., 2020; Ma et al., 2020; Zheng et al., 2021), suggest that multiple genes/loci are involved in FHB resistance under different environments, although their effects are mostly small and could not be easily used in MAS. Four MTAs (3026949, 979146, 1157139 and 1034520) were associated with both Type II resistance and DON content, which was expected due to the high correlation between grain toxin levels and Type II resistance, as described in the present study and previous reports (He et al., 2019; Zheng et al., 2021). Markers 1099971, 1044062 and 1091498 were identified in experiments in both China and Mexico, which may suggest their roles in response to different environmental factors and Fusarium species.
Most significant MTAs reported here appeared as single SNP markers, possibly due to the low resolution of genotyping. Similar cases have been reported by (Arruda et al., 2016a) and (Miedaner et al., 2011). Based on MTAs, genomic selection has shown intermediate to high prediction accuracy for FHB resistance traits, including DON content in grains, Fusarium damaged kernels and FHB severity (Arruda et al., 2016b; Gaire et al., 2022a). The effects of the markers reported here could be re-estimated in larger breeding populations and used in genomic selection for FHB resistance improvement.
Conclusion
FHB screening experiments on 265 wheat accessions in Mexico and China showed that 11 accessions had stable FHB resistance or low DON content in grains, of which three accessions Klein Don Enrique, Chuko and Yumai34 did not contain Fhb1. Sixteen loci associated with FHB resistance or DON content in grains were identified on chromosomes 1A, 1B, 2B, 3A, 3D, 4B, 4D, 5A, 5B, 7A, and 7B in multiple environments, 6 of which overlapped with reported metaQTL. The genetic sources could be used in the breeding programs for FHB improvement, and the associated loci could be further mapped to identify better markers for MAS.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Raw genotypic data is accessible via https://hdl.handle.net/11529/10548748.
Author contributions
XZ and PS conceived and designed the experiments, XH and KX conducted the FHB screening in Mexico, LW conducted the FHB screening in China, YH and PJ assisted in genotypic analysis, LW and XH analyzed the data and wrote the first draft of the manuscript, and all authors reviewed and approved the final manuscript.
Funding
This research has been funded by National Key R&D Program of China (2017YFE0126700), CRP WHEAT, National Natural Sciences Foundation of China (31561143004), Jiangsu Agricultural Science and Technology Innovation Fund (CX (21)3099), Seed Industry Revitalization Project of Jiangsu Province (JBGS2021006), and Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry.
Acknowledgments
Technical support from Francisco Lopez in field trials and Nerida Lozano in inoculum preparation for trials in Mexico is highly acknowledged. Technical support from Chang Li and Lixuan Yu in field trials in China is also well appreciated.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.988264/full#supplementary-material
References
Arruda, M. P., Brown, P., Brown-Guedira, G., Krill, A. M., and Thurber, C. (2016a). Genome-wide association mapping of Fusarium head blight resistance in wheat using genotyping-by-sequencing. Plant Genome 9, 1–14. doi:10.3835/plantgenome2015.04.0028
Arruda, M. P., Lipka, A. E., Brown, P. J., Krill, A. M., Thurber, C., Brown-Guedira, G., et al. (2016b). Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 36, 84. doi:10.1007/s11032-016-0508-5
Backhouse, D. (2014). Global distribution of Fusarium graminearum, F. asiaticum and F. boothii from wheat in relation to climate. Eur. J. Plant Pathol. 139, 161–173. doi:10.1007/s10658-013-0374-5
Bai, G., and Shaner, G. (2004). Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi:10.1146/annurev.phyto.42.040803.140340
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi:10.1093/bioinformatics/btm308
Buerstmayr, M., Lemmens, M., Steiner, B., and Buerstmayr, H. (2011). Advanced backcross QTL mapping of resistance to Fusarium head blight and plant morphological traits in a Triticum macha ×T. aestivum population. Theor. Appl. Genet. 123, 293–306. doi:10.1007/s00122-011-1584-x
Buerstmayr, M., Steiner, B., and Buerstmayr, H. (2020). Breeding for Fusarium head blight resistance in wheat—progress and challenges. Plant Breed. 139, 12797. doi:10.1111/pbr.12797
Buerstmayr, M., Steiner, B., Wagner, C., Schwarz, P., Brugger, K., Barabaschi, D., et al. (2018). High-resolution mapping of the pericentromeric region on wheat chromosome arm 5AS harbouring the Fusarium head blight resistance QTL Qfhs.ifa-5A. Plant Biotechnol. J. 16, 1046–1056. doi:10.1111/pbi.12850
Cainong, J. C., Bockus, W. W., Feng, Y., Chen, P., Qi, L., Sehgal, S. K., et al. (2015). Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 128, 1019–1027. doi:10.1007/s00122-015-2485-1
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7. doi:10.1186/s13742-015-0047-8
Chu, C., Niu, Z., Zhong, S., Chao, S., Friesen, T. L., Halley, S., et al. (2011). Identification and molecular mapping of two QTLs with major effects for resistance to Fusarium head blight in wheat. Theor. Appl. Genet. 123, 1107–1119. doi:10.1007/s00122-011-1652-2
Cuthbert, P. A., Somers, D. J., and Brulé-Babel, A. (2007). Mapping of Fhb2 on chromosome 6BS: A gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 429–437. doi:10.1007/s00122-006-0439-3
Cuthbert, P. A., Somers, D. J., Thomas, J., Cloutier, S., and Brulé-Babel, A. (2006). Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 112, 1465–1472. doi:10.1007/s00122-006-0249-7
Eldakak, M., Das, A., Zhuang, Y., Rohila, J. S., Glover, K., and Yen, Y. (2018). A quantitative proteomics view on the function of Qfhb1, a major QTL for Fusarium head blight resistance in wheat. Pathogens 7, 7030058. doi:10.3390/pathogens7030058
Gaire, R., De Arruda, M. P., Mohammadi, M., Brown-Guedira, G., Kolb, F. L., and Rutkoski, J. (2022a). Multi-trait genomic selection can increase selection accuracy for deoxynivalenol accumulation resulting from fusarium head blight in wheat. Plant Genome 15, e20188. doi:10.1002/tpg2.20188
Gaire, R., Sneller, C., Brown-Guedira, G., Van Sanford, D. A., Mohammadi, M., Kolb, F. L., et al. (2022b). Genetic trends in Fusarium head blight resistance due to 20 years of winter wheat breeding and cooperative testing in the Northern US. Plant Dis. 106, 364–372. doi:10.1094/pdis-04-21-0891-sr
Ghimire, B., Sapkota, S., Bahri, B. A., Martinez-Espinoza, A. D., Buck, J. W., and Mergoum, M. (2020). Fusarium head blight and rust diseases in soft red winter wheat in the southeast United States: State of the art, challenges and future perspective for breeding. Front. Plant Sci. 11, 1080. doi:10.3389/fpls.2020.01080
Gunnaiah, R., Kushalappa, A. C., Duggavathi, R., Fox, S., and Somers, D. J. (2012). Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One 7, e40695. doi:10.1371/journal.pone.0040695
Guo, J., Zhang, X., Hou, Y., Cai, J., Shen, X., Zhou, T., et al. (2015). High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 128, 2301–2316. doi:10.1007/s00122-015-2586-x
Hao, Y., Rasheed, A., Zhu, Z., Wulff, B. B. H., and He, Z. (2020). Harnessing wheat Fhb1 for Fusarium resistance. Trends Plant Sci. 25, 1–3. doi:10.1016/j.tplants.2019.10.006
He, X., Brar, G. S., Bonnett, D., Dreisigacker, S., Hyles, J., Spielmeyer, W., et al. (2020). Disease resistance evaluation of elite CIMMYT wheat lines containing the coupled Fhb1 and Sr2 genes. Plant Dis. 104, 2369–2376. doi:10.1094/pdis-02-20-0369-re
He, X., Dreisigacker, S., Singh, R. P., and Singh, P. K. (2019). Genetics for low correlation between Fusarium head blight disease and deoxynivalenol (DON) content in a bread wheat mapping population. Theor. Appl. Genet. 132, 2401–2411. doi:10.1007/s00122-019-03362-9
He, X., Lillemo, M., Shi, J., Wu, J., Bjørnstad, Å., Belova, T., et al. (2016). QTL characterization of Fusarium head blight resistance in CIMMYT bread wheat line Soru#1. PLoS One 11, e0158052. doi:10.1371/journal.pone.0158052
He, X., Singh, P. K., Duveiller, E., Schlang, N., Dreisigacker, S., and Singh, R. P. (2013). Identification and characterization of international Fusarium head blight screening nurseries of wheat at CIMMYT, Mexico. Eur. J. Plant Pathol. 136, 123–134. doi:10.1007/s10658-012-0146-7
He, Y., Zhang, X., Zhang, Y., Ahmad, D., Wu, L., Jiang, P., et al. (2018). Molecular characterization and expression of PFT, an FHB resistance gene at the Fhb1 QTL in wheat. Phytopathology 108, 730–736. doi:10.1094/phyto-11-17-0383-r
Holzapfel, J., Voss, H. H., Miedaner, T., Korzun, V., Häberle, J., Schweizer, G., et al. (2008). Inheritance of resistance to Fusarium head blight in three European winter wheat populations. Theor. Appl. Genet. 117, 1119–1128. doi:10.1007/s00122-008-0850-z
Jakobsson, M., and Rosenberg, N. A. (2007). Clumpp: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi:10.1093/bioinformatics/btm233
Jiang, G. L., Shi, J., and Ward, R. W. (2007). QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor. Appl. Genet. 116, 3–13. doi:10.1007/s00122-007-0641-y
Li, G., Zhou, J., Jia, H., Gao, Z., Fan, M., Luo, Y., et al. (2019). Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 51, 1106–1112. doi:10.1038/s41588-019-0426-7
Liu, S., Zhang, X., Pumphrey, M. O., Stack, R. W., Gill, B. S., and Anderson, J. A. (2006). Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genomics 6, 83–89. doi:10.1007/s10142-005-0007-y
Lu, Q., Lillemo, M., Skinnes, H., He, X., Shi, J., Ji, F., et al. (2013). Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line 'Shanghai-3/Catbird. Theor. Appl. Genet. 126, 317–334. doi:10.1007/s00122-012-1981-9
Ma, H. X., Zhang, K. M., Gao, L., Bai, G. H., Chen, H. G., Cai, Z. X., et al. (2006). Quantitative trait loci for resistance to fusarium head blight and deoxynivalenol accumulation in Wangshuibai wheat under field conditions. Plant Pathol. 55, 739–745. doi:10.1111/j.1365-3059.2006.01447.x
Ma, Z., Xie, Q., Li, G., Jia, H., Zhou, J., Kong, Z., et al. (2020). Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 133, 1541–1568. doi:10.1007/s00122-019-03525-8
Malihipour, A., Gilbert, J., Fedak, G., Brûlé-Babel, A., and Cao, W. (2017). Mapping the A Genome for QTL conditioning resistance to Fusarium head blight in a wheat population with Triticum timopheevii background. Plant Dis. 101, 11–19. doi:10.1094/pdis-02-16-0144-re
Mesterhazy, A. (1995). Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 114, 377–386. doi:10.1111/j.1439-0523.1995.tb00816.x
Mesterhazy, A. (2020). Updating the breeding philosophy of wheat to Fusarium head blight (FHB): Resistance components, QTL identification, and phenotyping-A review. Plants (Basel) 9, 1702. doi:10.3390/plants9121702
Mesterházy, T., Mirocha, C. G., and Komoróczy, R. (1999). Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 118, 97–110. doi:10.1046/j.1439-0523.1999.118002097.x
Miedaner, T., Würschum, T., Maurer, H. P., Korzun, V., Ebmeyer, E., and Reif, J. C. (2011). Association mapping for Fusarium head blight resistance in European soft winter wheat. Mol. Breed. 28, 647–655. doi:10.1007/s11032-010-9516-z
Miller, J. D., and Amison, P. G. (1986). Degradation of deoxynivalenol by suspension cultures of the Fusarium head blight resistant wheat cultivar Frontana. Can. J. Plant Pathology 8, 147–150. doi:10.1080/07060668609501818
Paillard, S., Schnurbusch, T., Tiwari, R., Messmer, M., Winzeler, M., Keller, B., et al. (2004). QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 323–332. doi:10.1007/s00122-004-1628-6
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi:10.1093/genetics/155.2.945
Pumphrey, M. O., Bernardo, R., and Anderson, J. A. (2007). Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci. 47, 200–206. doi:10.2135/cropsci2006.03.0206
Qi, L. L., Pumphrey, M. O., Friebe, B., Chen, P. D., and Gill, B. S. (2008). Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor. Appl. Genet. 117, 1155–1166. doi:10.1007/s00122-008-0853-9
R Team (2011). R: A language and environment for statistical computing. Vienna, Austria: the R foundation for statistical computing. Available at: http://www.R-project.org/.
Rawat, N., Pumphrey, M. O., Liu, S., Zhang, X., Tiwari, V. K., Ando, K., et al. (2016). Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 48, 1576–1580. doi:10.1038/ng.3706
Schroeder, H. W., and Christensen, J. J. (1963). Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 53, 831–838.
Schweiger, W., Steiner, B., Ametz, C., Siegwart, G., Wiesenberger, G., Berthiller, F., et al. (2013). Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol. Plant Pathol. 14, 772–785. doi:10.1111/mpp.12048
Schweiger, W., Steiner, B., Vautrin, S., Nussbaumer, T., Siegwart, G., Zamini, M., et al. (2016). Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 129, 1607–1623. doi:10.1007/s00122-016-2727-x
Singh, L., Anderson, J. A., Chen, J., Gill, B. S., Tiwari, V. K., and Rawat, N. (2019). Development and validation of a perfect KASP marker for Fusarium head blight resistance gene Fhb1 in wheat. Plant Pathol. J. 35, 200–207. doi:10.5423/ppj.oa.01.2019.0018
Steiner, B., Buerstmayr, M., Wagner, C., Danler, A., Eshonkulov, B., Ehn, M., et al. (2019). Fine-mapping of the Fusarium head blight resistance QTL Qfhs.ifa-5A identifies two resistance QTL associated with anther extrusion. Theor. Appl. Genet. 132, 2039–2053. doi:10.1007/s00122-019-03336-x
Su, Z., Bernardo, A., Tian, B., Chen, H., Wang, S., Ma, H., et al. (2019). A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51, 1099–1105. doi:10.1038/s41588-019-0425-8
Su, Z., Jin, S., Zhang, D., and Bai, G. (2018). Development and validation of diagnostic markers for Fhb1 region, a major QTL for Fusarium head blight resistance in wheat. Theor. Appl. Genet. 131, 2371–2380. doi:10.1007/s00122-018-3159-6
Van Der Lee, T., Zhang, H., Van Diepeningen, A., and Waalwijk, C. (2015). Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 32, 453–460. doi:10.1080/19440049.2014.984244
Venske, E., Dos Santos, R. S., Farias, D. D. R., Rother, V., Da Maia, L. C., Pegoraro, C., et al. (2019). Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant Sci. 10, 727. doi:10.3389/fpls.2019.00727
Wang, H., Sun, S., Ge, W., Zhao, L., Hou, B., Wang, K., et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435. doi:10.1126/science.aba5435
Wu, L., He, X., Kabir, M. R., Roy, K. K., Anwar, M. B., Marza, F., et al. (2021). Genetic sources and loci for wheat head blast resistance identified by genome-wide association analysis. Crop J. 10, 793–801. doi:10.1016/j.cj.2021.07.007
Xue, S., Li, G., Jia, H., Xu, F., Lin, F., Tang, M., et al. (2010). Fine mapping Fhb4, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 121, 147–156. doi:10.1007/s00122-010-1298-5
Xue, S., Xu, F., Tang, M., Zhou, Y., Li, G., An, X., et al. (2011). Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 123, 1055–1063. doi:10.1007/s00122-011-1647-z
Yi, X., Cheng, J., Jiang, Z., Hu, W., Bie, T., Gao, D., et al. (2018). Genetic analysis of Fusarium head blight resistance in CIMMYT bread wheat line C615 using traditional and conditional QTL mapping. Front. Plant Sci. 9, 573. doi:10.3389/fpls.2018.00573
Zhang, H., Van Der Lee, T., Waalwijk, C., Chen, W., Xu, J., Xu, J., et al. (2012). Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS One 7, e31722. doi:10.1371/journal.pone.0031722
Zhang, Y., Yang, Z., Ma, H., Huang, L., Ding, F., Du, Y., et al. (2021). Pyramiding of Fusarium head blight resistance quantitative trait loci Fhb1,Fhb4, and Fhb5, in modern Chinese wheat cultivars. Front. Plant Sci. 12, 694023. doi:10.3389/fpls.2021.694023
Zhao, M., Leng, Y., Chao, S., Xu, S. S., and Zhong, S. (2018). Molecular mapping of QTL for Fusarium head blight resistance introgressed into durum wheat. Theor. Appl. Genet. 131, 1939–1951. doi:10.1007/s00122-018-3124-4
Zheng, T., Hua, C., Li, L., Sun, Z., Yuan, M., Bai, G., et al. (2021). Integration of meta-QTL discovery with omics: Towards a molcular breeding platform for improving wheat resistance to Fusarium head blight. Crop J. 9, 739–749. doi:10.1016/j.cj.2020.10.006
Zhu, Z., Hao, Y., Mergoum, M., Bai, G., Humphreys, G., Cloutier, S., et al. (2019). Breeding wheat for resistance to Fusarium head blight in the global North: China, USA, and Canada. Crop J. 7, 730–738. doi:10.1016/j.cj.2019.06.003
Keywords: Fusarium head blight, resistance, genetic sources, genome-wide association study, deoxynivalenol
Citation: Wu L, He X, He Y, Jiang P, Xu K, Zhang X and Singh PK (2022) Genetic sources and loci for Fusarium head blight resistance in bread wheat. Front. Genet. 13:988264. doi: 10.3389/fgene.2022.988264
Received: 07 July 2022; Accepted: 13 September 2022;
Published: 30 September 2022.
Edited by:
Sundeep Kumar, National Bureau of Plant Genetic Resources (ICAR), IndiaReviewed by:
Suraj Sapkota, United States Department of Agriculture, United StatesSpurthi N. Nayak, University of Agricultural Sciences, India
Copyright © 2022 Wu, He, He, Jiang, Xu, Zhang and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Zhang, eHV6aGFuZ0BqYWFzLmFjLmNu; Pawan K. Singh, cGsuc2luZ2hAY2dpYXIub3Jn
 Lei Wu
Lei Wu Xinyao He
Xinyao He Yi He
Yi He Peng Jiang
Peng Jiang Kaijie Xu
Kaijie Xu Xu Zhang
Xu Zhang Pawan K. Singh
Pawan K. Singh