- 1Allergy Centre, Tampere University Hospital, Tampere, Finland
- 2Department of Pediatrics, Turku University Hospital, University of Turku, Turku, Finland
- 3Great Ormond Street Institute of Child Health, University College London, London, United Kingdom
- 4Human Genetics Department, Medical Research Institute, Alexandria University, Alexandria, Egypt
- 5PCD Diagnostic Team and Department of Paediatric Respiratory Medicine, Royal Brompton Hospita, London, United Kingdom
- 6School of Medicine, University of Dundee, Dundee, United Kingdom
- 7Department of Otorhinolaryngology, Head and Neck Surgery, Kuopio University Hospital and University of Eastern Finland, Kuopio, Finland
- 8Biochemistry Division, Chemistry Department, Faculty of Science, Zagazig University, Zagazig, Egypt
- 9Natural Resources Institute Finland (Luke), Helsinki, Finland
Primary ciliary dyskinesia (PCD) is a rare genetic condition characterized by chronic respiratory tract infections and in some cases laterality defects and infertility. The symptoms of PCD are caused by malfunction of motile cilia, hair-like organelles protruding out of the cell that are responsible for removal of mucus from the airways and organizing internal organ positioning during embryonic development. PCD is caused by mutations in genes coding for structural or assembly proteins in motile cilia. Thus far mutations in over 50 genes have been identified and these variants explain around 70% of all known cases. Population specific genetics underlying PCD has been reported, thus highlighting the importance of characterizing gene variants in different populations for development of gene-based diagnostics. In this study, we identified a recurrent loss-of-function mutation c.198_200delinsCC in CFAP300 causing lack of the protein product. PCD patients homozygous for the identified CFAP300 mutation have immotile airway epithelial cilia associated with missing dynein arms in their ciliary axonemes. Furthermore, using super resolution microscopy we demonstrate that CFAP300 is transported along cilia in normal human airway epithelial cells suggesting a role for CFAP300 in dynein complex transport in addition to preassembly in the cytoplasm. Our results highlight the importance of CFAP300 in dynein arm assembly and improve diagnostics of PCD in Finland.
Introduction
Primary ciliary dyskinesia (PCD) is an inherited, genetically and clinically heterogenous disorder caused by mutations in genes encoding proteins crucial for cilia motility. Motile cilia line the respiratory tract and are required for efficient mucus removal from the airways. Children with PCD suffer from early onset chronic airway infections and congestion leading to a distinctive, chronic wet cough (Lucas et al., 2020; Shoemark and Harman, 2021). If untreated, the condition may progress towards bronchiectasis in later life. During embryonic development, motile cilia determine the left-right positioning of body organs and nearly half of PCD patients have laterality problems. The brain ventricles are also lined with motile ependymal cilia, which are required for the flow of cerebrospinal fluid. However, hydrocephalus caused by defective ependymal cilia is rare in PCD patients. Motile cilia of the airways have a 9 + 2 axonemal core structure with a central pair of microtubules surrounded by nine peripheral doublet microtubules, which have outer and inner dynein arms (ODA and IDA, respectively) attached along their length. These ODA and IDA are motors that power cilia motility and are preassembled in the cytoplasm then transported via intraflagellar transport (IFT) to the cilium (Horani et al., 2018). Mutations in many preassembly factors have been identified as a cause for PCD in addition to mutations in structural genes of cilia components. Dynein arm preassembly factors DNAAF1 (Duquesnoy et al., 2009; Loges et al., 2009), DNAAF2 (Omran et al., 2008), DNAAF3 (Mitchison et al., 2012), DNAAF4 (Tarkar et al., 2013), DNAAF5 (Horani et al., 2012; Diggle et al., 2014), DNAAF11 (Kott et al., 2012; Horani et al., 2013; Inaba et al., 2016) and DNAAF6 (Olcese et al., 2017; Paff et al., 2017) are strictly localized in the cytoplasm, but interestingly ZMYND10 (Moore et al., 2013; Zariwala et al., 2013), CFAP298 (Austin-Tse et al., 2013; Jaffe et al., 2016) and CFAP300 (Fassad et al., 2018) are also found in the ciliary compartment, suggesting a combined role in assembly and transport. The core structure of the sperm flagellum is very similar to that of motile cilia, although some differences exist. Furthermore, male specific motile cilia are present in the efferent duct of the male reproductive tract and female specific motile cilia in the oviduct. Therefore, PCD is also associated with infertility, especially in men (Sironen et al., 2019), However, genotype/phenotype correlation between PCD and fertility is poorly understood and requires further studies.
PCD is inherited as an autosomal-recessive or rarely as X-linked disease. Thus far mutations in over 50 genes have been identified to cause PCD, explaining approximately 70% of cases. Over 300 genes encode essential components of the motile ciliary machinery and a large number of patients are still without a genetic diagnosis (Lucas et al., 2020). The aim of this study was to identify disease causing variants in the Finnish PCD population, where until now only two variants in DNAH11 have been previously reported (Schultz et al., 2020). It is important to increase the understanding of genetic causes for PCD, which can be most effectively traced in a genetically isolated population such as the Finns. This study contributes to the understanding of genetic causes of PCD by identification of a causative loss of function (LoF) mutation in CFAP300 in Finnish PCD patients. Population specific mutations play an important role in PCD and thus it is important to investigate the genetic variability in different populations (Fassad et al., 2020; Mitchison and Smedley, 2022). We also show that additional to a role in the cytoplasmic preassembly of dynein arms, human CFAP300 is involved in protein transport to motile cilia, which was previously only suggested in model organisms.
Methods
Subject
Blood samples were collected from thirteen PCD patients, recruited at the University Hospitals of Turku, Kuopio and Tampere after written informed consent was given. For variant segregation analysis saliva samples were collected from families (healthy parents and siblings when possible). Patients were interviewed and scored using the PICADAR questionnaire (Behan et al., 2016). The study was ethically approved by the University of Turku Ethics Committee (ETMK 69–2017), London Bloomsbury Research Ethics Committee approved by the Health Research Authority (08/H0713/82), and the referring hospitals.
Nasal nitric oxide analysis
For nasal nitric oxide (nNO) analysis a CLD 88sp analyser equipped with a Denox 88 module for flow control was used (Eco Physics, Dürnten, Switzerland). If cooperativity was established, three consecutive trials were taken, from which the highest value was recorded. Nasal nitric oxide analysis was repeated on two different occasions.
High-speed video microscopy analysis
Nasal epithelial cells were suspended in DMEM medium and evaluated under a differential-interference microscope (Zeiss, Oberkochen, Germany) at ×1,000 magnification and cilia beat was recorded with a digital high-speed video (DHSV) camera (Hamamatsu Orca Flash 4.0, Hamamatsu Japan) with a frame rate of 256 Hz. The detailed protocol for HSVM can be found in Schultz et al., 2022. DHSV video sequences were played back frame by frame and cilia beat frequency (CBF) was determined by calculating the mean of all recorded cilia beat cycles and the cilia beating pattern (CBP) was determined by two independent expert operators. HSVM was repeated on two different occasions.
Whole exome sequencing
The Nonacus Cell3 Exome panel was used for whole exome sequencing of patient samples (https://nonacus.com/cell3tm-target). Unmapped reads were aligned to the current Human reference genome (GRCh38 build) by Burrows-Wheeler Aligner tool (Bwa-mem2 version) (Li and Durbin, 2010), SAM files were produced and indexed and converted to BAM format previous to marking and removing duplicates using Picard (https://broadinstitute.github.io/picard/). Subsequent analysis was executed following best practices guidelines for GATK from the Broad Institute (https://gatk.broadinstitute.org/hc/en-us). Firstly, base quality score recalibration (BQSR) was done to numerically correct individual base calls. Variant discovery was carried out in a two-step pipeline: variant calling with HaplotypeCaller followed by joint genotyping with GenotypeGVCFs. Once obtained a multi-sample VCF file containing all definitive variant records, VariantRecalibrator was operated to fulfil Variant Quality Score Recalibration (VQSR) and refinement of the obtained variant callset.
Variants were individually selected for each sample. The called variants, including both single nucleotide variants (SNVs) and indels, were then annotated using ANNOVAR (http://www.openbioinformatics.org/annovar/). Such tool enables functional annotation, and thus obtaining final VCF files that contain detailed information for each variant site in the sample, such as their impact within a gene, predicted pathogenicity scores, minor allele frequency (MAF), zygosity status or reporting whether they have been recorded in large-scale databases like dbSNP.
Variant prioritization
Variants were filtered for MAF <1%, their predicted functional impact on the encoded protein (missense, splicing, frameshift or nonsense) and frequency (<0.001) in the Genome Aggregation Database (gnomAD, broadinstitute.org). A list of genes with high expression during motile cilia development (n = 652, reanalyzed gene list based on data in Marcet et al., 2011) was used to further filter variants in genes with potential roles in motile cilia. Finally, the pathogenicity of the identified variants was estimated with Combined Annotation Dependent Depletion tool (CADD, https://cadd.gs.washington.edu/snv) and a CADD score >20 was considered significant pathogenicity score. Due to the known inheritance pattern of PCD homozygous variants were prioritized, but output files were also analyzed for the presence of compound heterozygous variants.
Sanger sequencing
The identified NM_032930.3 CFAP300 variant was confirmed in the probands and segregation within family members by Sanger sequencing. Primers flanking the mutation were designed using the NCBI Primer-BLAST tool, forward GTATGTCAGTTGTTACGAAGGCAAT, reverse TGCTCTTATGTGTTAAGCCAGC. The genomic sequence was amplified using standard PCR conditions and predicted primer annealing temperature. The specificity of the PCR product was confirmed on agarose gel and purified using Exosap for Sanger sequencing.
Electron microscopy
Samples were fixed in 2.5% glutaraldehyde in cacodylate buffer and processed for electron microscopy. Defects were quantified using the method described by Shoemark et al. (2012). Briefly, cells were washed in sodium cacodylate buffer, post-fixed with 1% osmium tetroxide and centrifuged in agar or agarose to generate a pellet. Using a series of increasing concentrations of methanol followed by propylene oxide, cells were dehydrated before embedding in resin then 70–90 nm sections were cut using an ultramicrotome, mounted onto copper grids. Heavy metal staining was performed with uranyl acetate and lead citrate. Assessment of the respiratory epithelium and ciliary ultrastructure was made on a Jeol 1500 transmission electron microscope (TEM). Quantification of cells, microtubular arrangement in the axoneme and the presence of dynein arms was performed by a clinical electron microscopist blinded to the case information. Care was taken to assess cilia from a number of healthy cells from locations proximal and distal to the epithelial cell surface. Transverse sections of cilia were methodically quantified until either the entire section or >200 cilia had been counted.
Immunofluoresence
Nasal ciliated cells from nasal brushing of a PCD patient with CFAP300 mutations and a control sample were stained after blocking (10% BSA, in PBS) using DNAH5 (HPA037470, Cambridge Bioscience), DNAH7 (HPA037724, Atlas Antibodies), DNAI1 (HPA021649, Atlas Antibodies), RSPH4A (HPA031196, Sigma-Aldrich) and CFAP300 (c11orf70, HPA038585, Sigma-Aldrich) antibodies and colocalized with cilia marker alpha-tubulin (322588, Invitrogen). After washes with PBST the slides were incubated with secondary antibodies Alexa Fluor 488 anti-mouse and Alexa Fluor 594 anti-rabbit (1:500). Slides were imaged using Zeiss Axio Observer seven and deconvoluted using Huygens Deconvolution software (https://svi.nl/Huygens-Deconvolution). High resolution images were taken using a Zeiss LSM 880 upright confocal multiphoton microscope with AiryscanFast using ×63 objective and excitation lasers for 488 and 594. Images were processed using Zen software.
Statistical analysis
Unpaired t-test was used for statistical analysis between patient and control samples. p-value < 0.05 was considered statistically significant.
Results
Clinical identification of PCD patients
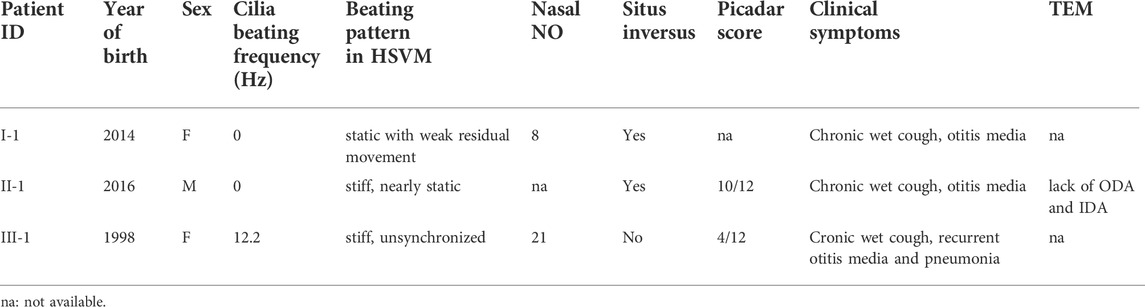
Patients were diagnosed based on low nasal NO, altered ciliary beating pattern in HSVM and clinical patient data (Table 1). Cystic fibrosis was excluded for all patients using a sweat test. All patients showed classic PCD symptoms early in life including chronic wet cough and otitis media, but to date no bronchiectasis has been observed (Table 1). Pneumonia was detected in one patient later in life, who had a low Picadar score (4/12, patient III-1). The low Picadar score was probably due to lack of situs defects in this patient. Defective beating of airway cilia was observed by HSVM showing stiff or static cilia in all patients. Some residual unsyncronized movement was detected in patient III-1. This variation in ciliary beating might be due to variable levels of dynein arms, however no samples for electron microscopy or immunofluorescence were available from patient III-1 preventing more detailed examination.
Whole exome sequencing identified patients carrying variants in CFAP300
We conducted whole exome sequencing of 13 Finnish PCD patients, which identified a homozygous LoF mutation in exon 3 of the CFAP300 gene. The variant was present in homozygous state in three unrelated Finnish patients with typical symptoms of PCD including chronic respiratory infections and static/stiff cilia in HSVM and two of the patients had situs inversus (Table 1). No other candidate homozygous or compound heterozygous variants were detected in these patients. The presence of the mutations was confirmed by Sanger sequencing and it also segregated consistently in two of the families where familial samples were available (Figure 1, samples unavailable for the third family). Sanger sequencing confirmed a 3bp deletion and insertion of CC at chr11:102058886-102058888, causing a frameshift c.198_200delinsCC predicted to cause a premature stop codon at position 76 in the protein sequence, p. Phe67fs (Figure 2A). The variant is not present in gnomAD and it seems to be specific to PCD patients as it has previously been reported in German, Israeli and Slavic populations (Höben et al., 2018; Zietkiewicz et al., 2019), providing strong evidence that the variant is disease causing based on American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015). Protein alignment showed the premature stop codon position after 75 aa (Figure 2B). The effect of the frameshift on protein structure was predicted by Phyre2 (PHYRE2 Protein Fold Recognition Server (ic.ac.uk)), which showed lack of helical structures in the patient protein prediction (Figure 2C). Based on the in silico analysis, it can be predicted that the identified frameshift mutation in CFAP300 is disease causing.
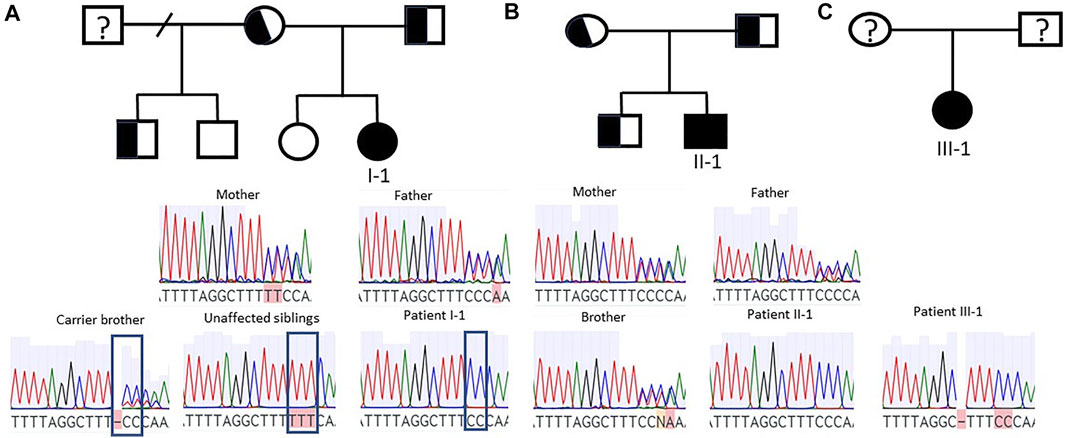
FIGURE 1. Segregation of CFAP300 mutations. Patients I-1 and II-1 were homozygous for the mutation while parents were heterozygous. (A). Patient I-1 family with carrier parents and one carrier brother and two unaffected siblings. The mutation site is indicated in carrier and unaffected siblings and in patient I-1. (B). Patient II-1 family with carrier parents and brother. (C). Sanger sequencing result for patient III-1.
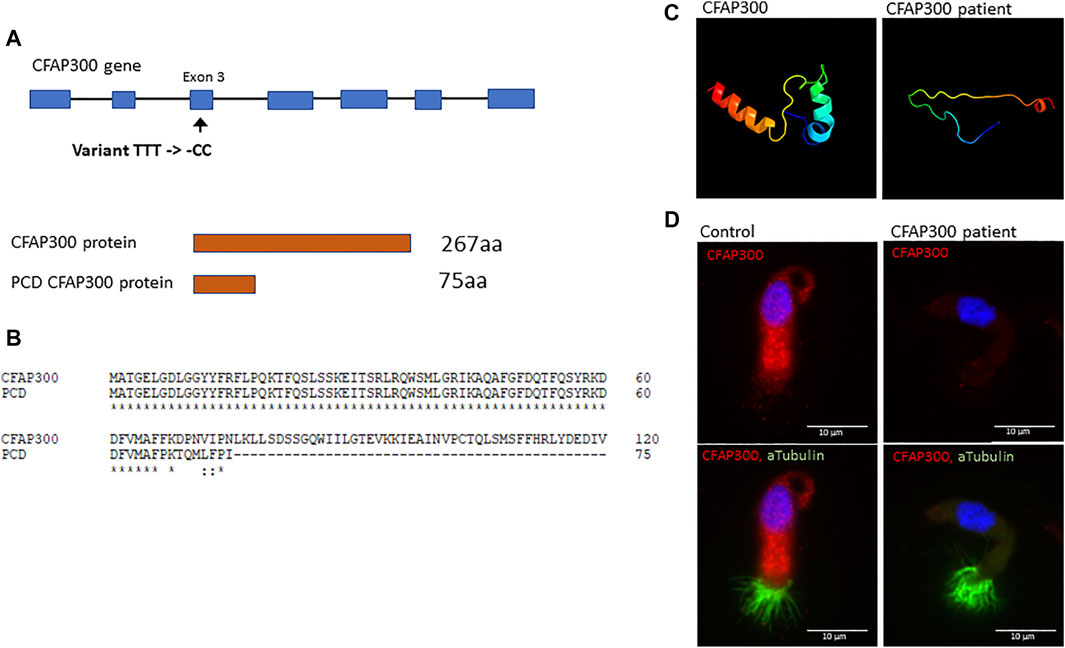
FIGURE 2. The effect of the CFAP300 variant on protein function. (A). The CFAP300 variant (C).198_200delinsCC results in premature stop codon at 76 aa and protein product. (B). Clustal Omega alignment of the control and PCD patient CFAP300 protein sequence. (C). Structural changes in the CFAP300 protein folding based on Phyre2. (D). Cytoplasmic localization of the CFAP300 protein product appears missing in the PCD patient (patient II-1). αTubulin was used as a marker for cilia and Dapi as a nuclear stain. Scale bar is 10 µm.
CFAP300 is missing in the PCD patient with a frameshift mutation resulting in a lack of dynein arms
To confirm the functional effect of the identified frameshift variant in CFAP300, we investigated the localization of the protein product in control and patient ciliated airway epithelial cells. In control ciliated airway epithelial cells CFAP300 localized in the cytoplasm, which is consistent with its predicted function as a dynein arm preassembly factor (Figure 2D). CFAP300 staining was not detected or extremely weak in cells from patients homozygous for the c.198_200delinsCC variant compared to the control, consistent with likely nonsense mediated decay of the defective mRNA transcripts (Figure 2D). The peptide sequence for CFAP300 antibody corresponds to 113-188 aa in the CFAP300 protein sequence (ENSP00000414390), which is expected to be affected by the predicted premature stop codon at position 75 aa. The weak staining detected in patient cells could potentially originate from a short protein isoform of CFAP300 containing 109-262 aa (ENST00000529204). TEM analysis showed a high number of ciliated cells and normal arrangement of microtubules in the PCD patient with CFAP300 LoF mutations. However, the orientations of the basal feet were disorganized (Figure 3A). For a synchronized beating pattern the basal feet at the motile cilia base all have to be organized to the same directions in all cilia (Nguyen et al., 2020). This defect may be secondary, due to the lack of cilia motility in the patient and additional studies are needed to confirm the order of events of basal foot orientation in the presence of the CFAP300 frameshift variant. The main structural change in cilia cross sections was the lack of dynein arms (Figure 3B). The cross-sectional morphology of cilia appeared abnormal with a partial to total absence of ODA and IDA. Both dynein arms were occasionally present as complete structures or truncated ‘horns’ in the case of ODA. Ciliary cross sections with majority of arms absent were counted as both arms absent. In the patient with CFAP300 mutation no cross sections with normal dynein arms were detected, which was statistically significant compared to control (>97% cross sections with normal dynein arms). No significant microtubular disorganization was detected in TEM. The lack of dynein arms was confirmed by immunofluorescence using antibodies against IDA protein DNAH7 and ODA proteins DNAI1 and DNAH5. These proteins appeared completely missing in patient cilia with strong staining retained in the cytoplasm (Figure 3C). In the control sample, DNAH7, DNAH5 and DNAI1 were specifically localized along the cilia. Furthermore, radial spoke protein RSPH4A was present in the patient cilia, although staining was also detected in the cytoplasm (Figure 3C). This may be due to inhibited transport of proteins to the cilium in patients with the CFAP300 mutation, but the mutational mechanism requires additional studies.
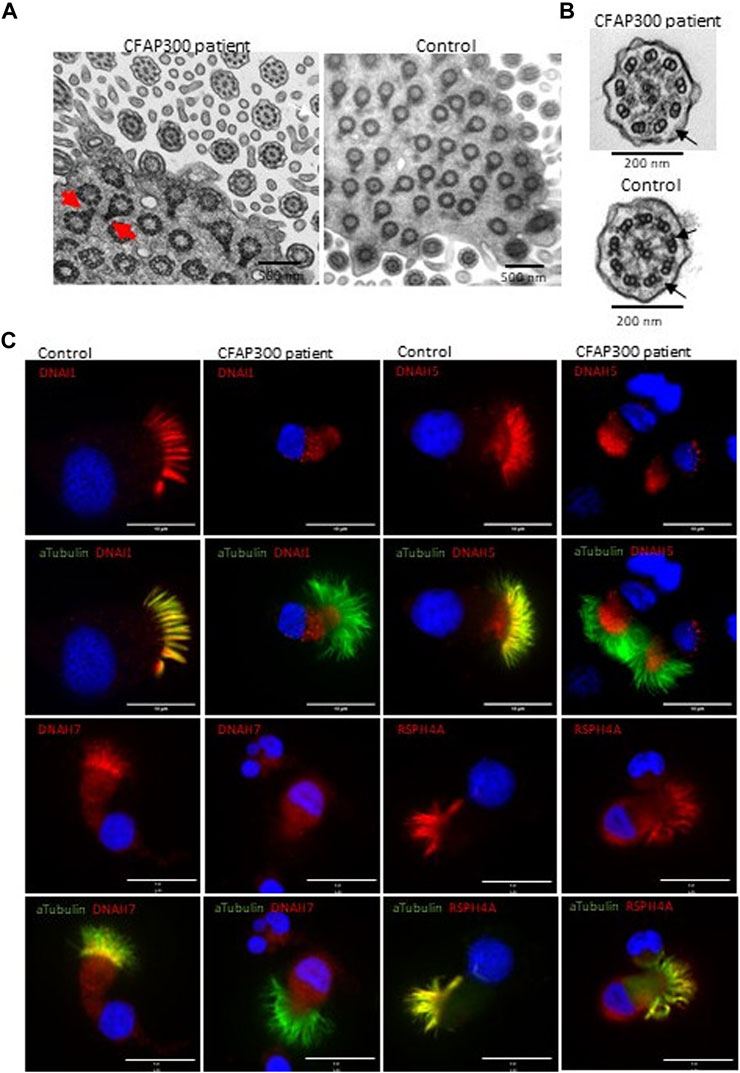
FIGURE 3. Localization of axonemal proteins in airway cilia of a control and PCD patient (patient II-1) with CFAP300 mutations. (A). High number of cilia cross sections were present in the nasal brushing sample of a PCD patient with CFAP300 c.198_200delinsCC mutations, but the orientation of the basal foot (red arrows) was disorganized compared to control. (B). The main finding in TEM of cilia cross sections in a patient with the CFAP300 variant was lack of dynein arms (black arrows). (C). ODA (DNAH5 and DNAI1) and IDA (DNAH7) proteins were missing from cilia in the patient airway epithelial cilia, while strong staining is present in the control sample. Radial spoke protein RSPH4A is present in the patient cilia. αTubulin was used as a marker for cilia and Dapi as a nuclear stain. Scale bar is 10 µm.
CFAP300 is transported to cilia in airway epithelial cells
Using Airyscan super resolution imaging we characterized the strong cytoplasmic staining of CFAP300 across the cytoplasm, which was present as granular spots in control ciliated airway epithelial cells. Previously, it has been shown in model species (Paramecium and Chlamydomonas) that a small amount of CFAP300 is also transported to the cilium (Fassad et al., 2018). To investigate the potential localization of CFAP300 in human cilia in addition to its strong staining in the cytoplasm (Figure 2D), we focused on imaging of CFAP300 along the cilium. We found that CFAP300 was present along the ciliary axoneme indicating that it is also transported to the cilium (Figure 4). Both cytoplasmic and cilia CFAP300 staining was depleted in the PCD patient sample with LoF mutations in CFAP300 (Figure 2D).
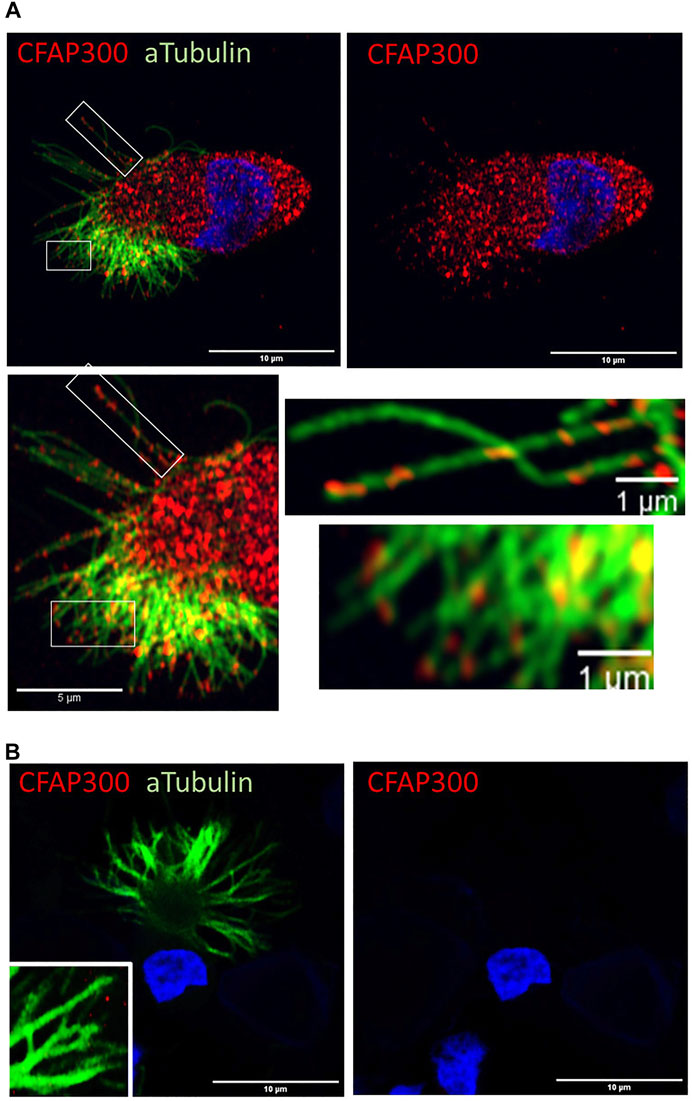
FIGURE 4. CFAP300 is transported along the motile airway epithelial cilium. (A). In normal airway epithelial cells most of the CFAP300 protein is localized in the cytoplasm, but some of the protein appears to be transported into the cilium. (B). No CFAP300 staining was detected in the patient airway epithelial cells.
Discussion
In this study we have identified a mutation in CFAP300 causative for PCD in Finland and analysed the effect of the mutation on motile cilia formation and function. The main defect in axonemal cross-sections was the lack of dynein arms as described in previous studies (Fassad et al., 2018; Höben et al., 2018; Zietkiewicz et al., 2019; Aprea et al., 2021; Bolkier et al., 2021; Yiallouros et al., 2021). Furthermore, we detected misorientation of the basal feet of the airway cilia, which we believe is most likely to be a secondary defect due to the lack of cilia motility and is also seen in PCD cases due to mutations in other genes. Previously it has been shown that cilia motility is required for correct polarization of basal bodies (Mitchell et al., 2007). However, it cannot be ruled out that CFAP300 has a more direct role in basal body polarization, as has been suggested for another dynein arm preassembly factor CFAP298 (Jaffe et al., 2016). We have demonstrated the main localization of CFAP300 to be in the cytoplasm of normal ciliated airway epithelial cells, but we find in addition the presence of small amount of CFAP300 along the ciliary length (Figure 4) suggesting that CFAP300 has a potential role in transport of dynein complexes into the cilium. These results are consistent with previous studies in model organisms Paramecium and Chlamydomonas, where CFAP300 orthologs have been shown to be mainly present in the cytoplasm with small amount along the cilia (Fassad et al., 2018). Gene silencing experiments indicated that localization of CFAP300 along the axoneme is IFT dependent (Fassad et al., 2018). These studies suggest that CFAP300 is involved in chaperone-mediated preassembly of dynein arms and transporting these complexes to motile cilia, which is supported by our results in human cells.
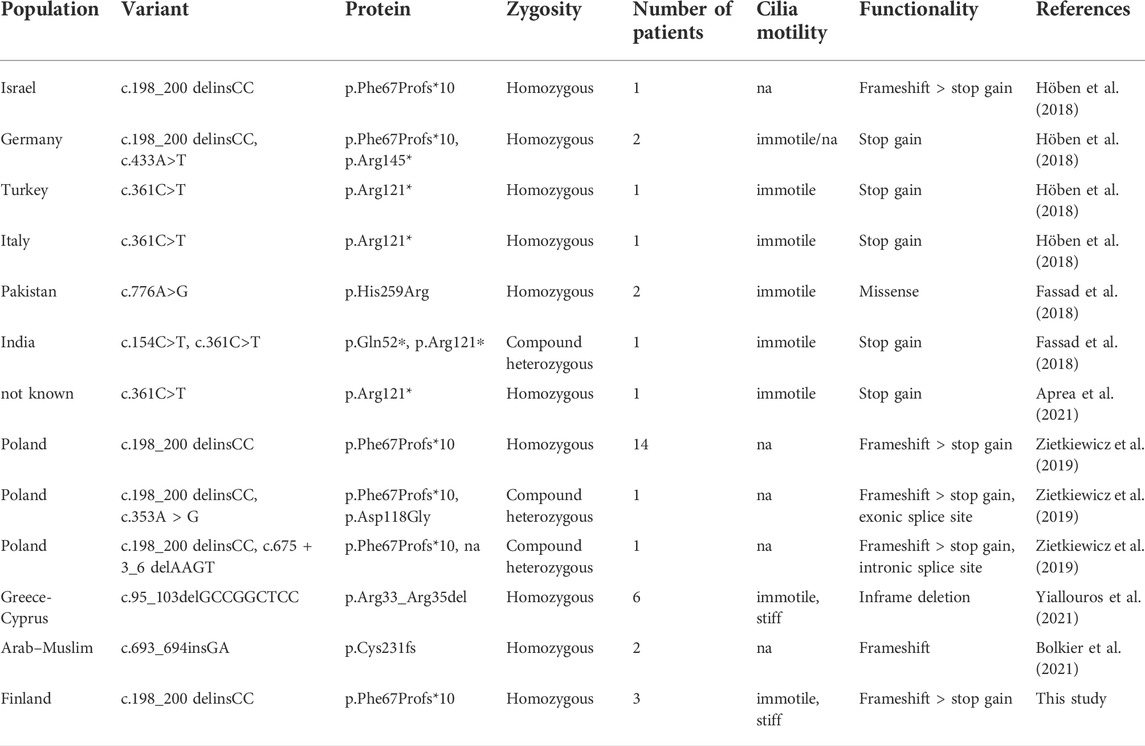
Previously children with CFAP300 mutations have been shown to share a consistent PCD phenotype from early life with laterality defects and immotile respiratory cilia displaying combined loss of IDA and ODA (Fassad et al., 2018). Finnish PCD patients with CFAP300 LoF mutations showed a similar phenotype with laterality defects in two of the patients and consistent chronic wet cough. The respiratory distress was already evident in the neonatal care unit and HSVM showed static or stiff cilia with loss of dynein arms as described previously. The variant identified in this study has also been previously reported in one German and Israeli case (Höben et al., 2018) and 14 PCD patients in the Slavic population (Zietkiewicz et al., 2019), which along with this study indicates that it is a reasonably common cause of CFAP300 PCD that could arise from an ancient European founder mutation. ODA loss was reported in 67–95% and IDA loss in 75–95% of axonemal cross sections in patients with homozygous missense mutations at c.776A>G (p.His259Arg) and compound heterozygous stop gain mutations at c.154C>T and c.361C>T (p.Gln52*, p. Arg121*), respectively (Fassad et al., 2018). DNAH5 and DNALI1 staining was missing in patient cilia, but RSPH4A and GAS8 staining was comparable to controls (Fassad et al., 2018). In a patient with a homozygous stop gain mutation c.361C>T (p.Arg121*) in CFAP300, DNAI1, DNAI2 and DNALI1 were reported to be missing in cilia and TEM reported only ODA defect (Aprea et al., 2021). In frame deletion c.98_106del (p.Arg33_Arg35del) in six patients of Greek-Cypriot population was associated with IDA and ODA loss in most cilia cross-sections based on TEM and there was an extremely stiff CBP (Yiallouros et al., 2021). The c.198_200delinsCC frameshift variant identified in this study caused lack or truncation of IDA or ODA in all axonemal cross sections based on TEM of patient II-1 and lack of DNAH5, DNAI1 and DNAH7 suggesting a severe defect in dynein arm assembly. Patient III-1 showed stiff cilia movement with a beat frequency of 12 Hz. Although we were unable to confirm the axonemal defects in this patient, the residual movement could be due to different levels of dynein arm formation as has been shown in previous studies (Fassad et al., 2018; Aprea et al., 2021; Yiallouros et al., 2021). Variable cilia beat frequency has also been reported for variant c.98_106del (0–9.1 Hz, Yiallouros et al., 2021). All the CFAP300 mutations reported to date are listed in Table 2.
In the Finnish population two novel mutations in DNAH11 have been previously reported, suggesting a distinct genetic pool for PCD in Finland. The CFAP300 mutation reported here in contrast appears to be common with Slavic, German and Israeli populations, suggesting a common genetic origin. It is important to establish the genetic causes of PCD in different populations including Finland to improve the genetic diagnosis of PCD. This can lead to development of a population specific gene panel for the diagnostic pipeline and the opportunity for targeted specific personalised medicines in future.
Our results and previously reported studies in model organisms and PCD patients underline the crucial role of CFAP300 in dynein arm preassembly and transport to motile cilia. The exact role of CFAP300 requires additional studies in mammalian species to establish the molecular mechanisms behind the preassembly and transport of dynein arm complexes. Interestingly, CFAP300 seems to be involved in transporting the complexes along the cilia possibly acting as a linker between the IFT machinery and dynein motor complexes.
Data availability statement
The data presented in the study are deposited in the UCL Research Data Repository DOI 10.5522/04/20793235.
Ethics statement
The studies involving human participants were reviewed and approved by University of Turku Ethics Committee (ETMK 69–2017) and London Bloomsbury Research Ethics Committee approved by the Health Research Authority (08/H0713/82). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
RS, VE, and TK diagnosed the patients and collected patient samples, MF analysed the exome data, GF and MM conducted the Sanger sequencing of patients and family members, AS and AR conducted cilia motility and ultrastructure assessment, JL did the immunofluorescence staining, HM supervised the study, AS analyzed the data, conducted microscopy imaging and wrote the manuscript.
Funding
Part of this research was funded by BBSRC grant BB/V011251/1. AS was funded by Marie Sklodowska-Curie individual fellowship No. 800556 from the European Union’s Horizon 2020 Research and Innovation Programme. MF was supported by a Wellcome Trust Collaborative Award in Science (210585/Z/18/Z).
Acknowledgments
We thank the patients and their families for participating in the study and Melissa Dixon (Royal Brompton Hospital) for her contributions to data used in this manuscript. Several study authors and data contributors participate in the BEAT-PCD clinical research collaboration, supported by the European Respiratory Society.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aprea, I., Raidt, J., Höben, I. M., Loges, N. T., Nöthe-Menchen, T., and Pennekamp, P., (2021). Defects in the cytoplasmic assembly of axonemal dynein arms cause morphological abnormalities and dysmotility in sperm cells leading to male infertility. PLoS Genet. 17, e1009306. doi:10.1371/journal.pgen.1009306
Austin-Tse, C., Halbritter, J., Zariwala, M. A., Gilberti, R. M., Gee, H. Y., and Hellman, N., (2013). Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 93, 672–686. doi:10.1016/j.ajhg.2013.08.015
Behan, L., Dimitrov, B. D., Kuehni, C. E., Hogg, C., Carroll, M., and Evans, H. J., (2016). Picadar: A diagnostic predictive tool for primary ciliary dyskinesia. Eur. Respir. J. 47, 1103–1112. doi:10.1183/13993003.01551-2015
Bolkier, Y., Barel, O., Marek-Yagel, D., Atias-Varon, D., Kagan, M., and Vardi, A., (2021). Whole-exome sequencing reveals a monogenic cause in 56% of individuals with laterality disorders and associated congenital heart defects. J. Med. Genet. 59, 691–696. doi:10.1136/jmedgenet-2021-107775
Diggle, C. P., Moore, D. J., Mali, G., zur Lage, P., Ait-Lounis, A., and Schmidts, M., (2014). HEATR2 plays a conserved role in assembly of the ciliary motile apparatus. PLoS Genet. 10, e1004577. doi:10.1371/journal.pgen.1004577
Duquesnoy, P., Escudier, E., Vincensini, L., Freshour, J., Bridoux, A. M., and Coste, A., (2009). Loss-of-function mutations in the human ortholog of chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 85, 890–89. doi:10.1016/j.ajhg.2009.11.008
Fassad, M. R., Patel, M. P., Shoemark, A., Cullup, T., Hayward, J., and Dixon, M., (2020). Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J. Med. Genet. 57 (5), 322–330. doi:10.1136/jmedgenet-2019-106501
Fassad, M. R., Shoemark, A., le Borgne, P., Koll, F., Patel, M., and Dixon, M., (2018). C11orf70 mutations disrupting the intraflagellar transport-dependent assembly of multiple axonemal dyneins cause primary ciliary dyskinesia. Am. J. Hum. Genet. 102, 956–972. doi:10.1016/j.ajhg.2018.03.024
Höben, I. M., Hjeij, R., Olbrich, H., Dougherty, G. W., Nöthe-Menchen, T., and Aprea, I., (2018). Mutations in C11orf70 cause primary ciliary dyskinesia with randomization of left/right body asymmetry due to defects of outer and inner dynein arms. Am. J. Hum. Genet. 102 (5), 973–984. doi:10.1016/j.ajhg.2018.03.025
Horani, A., Druley, T. E., Zariwala, M. A., Patel, A. C., Levinson, B. T., and Van Arendonk, L. G., (2012). Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 91, 685–693. doi:10.1016/j.ajhg.2012.08.022
Horani, A., Ferkol, T. W., Shoseyov, D., Wasserman, M. G., Oren, Y. S., and Kerem, B., (2013). LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS One 8, e59436. doi:10.1371/journal.pone.0059436
Horani, A., Ustione, A., Huang, T., Firth, A. L., Pan, J., and Gunsten, S. P., (2018). Establishment of the early cilia preassembly protein complex during motile ciliogenesis. Proc. Natl. Acad. Sci. U. S. A. 115, E1221–E1228. doi:10.1073/pnas.1715915115
Inaba, Y., Shinohara, K., Botilde, Y., Nabeshima, R., Takaoka, K., and Ajima, R., (2016). Transport of the outer dynein arm complex to cilia requires a cytoplasmic protein Lrrc6. Genes cells. 21, 728–739. doi:10.1111/gtc.12380
Jaffe, K. M., Grimes, D. T., Schottenfeld-Roames, J., Werner, M. E., Ku, T. S., and Kim, S. K., (2016). c21orf59/kurly controls both cilia motility and polarization. Cell Rep. 14, 1841–1849. doi:10.1016/j.celrep.2016.01.069
Kott, E., Duquesnoy, P., Copin, B., Legendre, M., Dastot-Le Moal, F., and Montantin, G., (2012). Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 91, 958–964. doi:10.1016/j.ajhg.2012.10.003
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi:10.1093/bioinformatics/btp698
Loges, N. T., Olbrich, H., Becker-Heck, A., Häffner, K., Heer, A., and Reinhard, C., (2009). Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 85, 883–889. doi:10.1016/j.ajhg.2009.10.018
Lucas, J. S., Davis, S. D., Omran, H., and Shoemark, A. (2020). Primary ciliary dyskinesia in the genomics age. Lancet. Respir. Med. 8, 202–216. doi:10.1016/S2213-2600(19)30374-1
Marcet, B., Chevalier, B., Luxardi, G., Coraux, C., Zaragosi, L. E., and Cibois, M., (2011). Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 13 (6), 693–699. doi:10.1038/ncb2241
Mitchell, B., Jacobs, R., Li, J., Chien, S., and Kintner, C. (2007). A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447, 97–101. doi:10.1038/nature05771
Mitchison, H. M., and Smedley, D. (2022). Primary ciliary dyskinesia: A big data genomics approach. Lancet. Respir. Med. 10 (5), 423–425. doi:10.1016/S2213-2600(22)00009-1
Mitchison, H. M., Schmidts, M., Loges, N. T., Freshour, J., Dritsoula, A., and Hirst, R. A., (2012). Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 44 (381-9), 381–389. doi:10.1038/ng.1106
Moore, D. J., Onoufriadis, A., Shoemark, A., Simpson, M. A., zur Lage, P. I., and de Castro, S. C., (2013). Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 93, 346–356. doi:10.1016/j.ajhg.2013.07.009
Nguyen, Q. P. H., Liu, Z., Albulescu, A., Ouyang, H., Zlock, L., and Coyaud, E., (2020). Comparative super-resolution mapping of basal feet reveals a modular but distinct architecture in primary and motile cilia. Dev. Cell 55, 209–223. doi:10.1016/j.devcel.2020.09.015
Olcese, C., Patel, M. P., Shoemark, A., Kiviluoto, S., Legendre, M., and Williams, H. J., (2017). X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 8, 14279. doi:10.1038/ncomms14279
Omran, H., Kobayashi, D., Olbrich, H., Tsukahara, T., Loges, N. T., and Hagiwara, H., (2008). Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 456, 611–616. doi:10.1038/nature07471
Paff, T., Loges, N. T., Aprea, I., Wu, K., Bakey, Z., and Haarman, E. G., (2017). Mutations in PIH1D3 cause X-linked primary ciliary dyskinesia with outer and inner dynein arm defects. Am. J. Hum. Genet. 100, 160–168. doi:10.1016/j.ajhg.2016.11.019
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., and Gastier-Foster, J., (2015). Laboratory quality assurance committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular PathologyTwo novel mutations in the DNAH11 gene in primary ciliary dyskinesia (CILD7) with considerable variety in the clinical and beating cilia phenotype. Genet. Med. MayBMC Med. Genet. 1721 (5), 405237–24020011712. doi:10.1038/gim.2015.3010.1186/s12881-020-01171-2
Schultz, R., Peromaa, T., Lukkarinen, H., and Elenius, V. (2022). High-speed video microscopy analysis for first-line diagnosis of primary ciliary dyskinesia. J. Vis. Exp. 19. doi:10.3791/63292
Shoemark, A., Dixon, M., Corrin, B., and Dewar, A. (2012). Twenty-year review of quantitative transmission electron microscopy for the diagnosis of primary ciliary dyskinesia. J. Clin. Pathol. 65, 267–271. doi:10.1136/jclinpath-2011-200415
Shoemark, A., and Harman, K. (2021). Primary ciliary dyskinesia. Semin. Respir. Crit. Care Med. 42, 537–548. doi:10.1055/s-0041-1730919
Sironen, A., Shoemark, A., Patel, M., Loebinger, M. R., and Mitchison, H. M. (2019). Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell. Mol. Life Sci. 77, 2029–2048. doi:10.1007/s00018-019-03389-7
Tarkar, A., Loges, N. T., Slagle, C. E., Francis, R., Dougherty, G. W., and Tamayo, J. V., (2013). DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 45, 995–1003. doi:10.1038/ng.2707
Yiallouros, P. K., Kouis, P., Kyriacou, K., Evriviadou, A., Anagnostopoulou, P., and Matthaiou, A., (2021). Implementation of multigene panel NGS diagnosis in the national primary ciliary dyskinesia cohort of cyprus: An island with a high disease prevalence. Hum. Mutat. 42, e62–e77. doi:10.1002/humu.24196
Zariwala, M. A., Gee, H. Y., Kurkowiak, M., Al-Mutairi, D. A., Leigh, M. W., and Hurd, T. W., (2013). ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 93, 336–345. doi:10.1016/j.ajhg.2013.06.007
Keywords: motile cilia, primary ciliary dyskinesia, CFAP300, dynein arm preassembly, diagnostics
Citation: Schultz R, Elenius V, Fassad MR, Freke G, Rogers A, Shoemark A, Koistinen T, Mohamed MA, Lim JSY, Mitchison HM and Sironen AI (2022) CFAP300 mutation causing primary ciliary dyskinesia in Finland. Front. Genet. 13:985227. doi: 10.3389/fgene.2022.985227
Received: 03 July 2022; Accepted: 24 August 2022;
Published: 30 September 2022.
Edited by:
Steven L. C. Pei, Yale University, United StatesReviewed by:
Shiaulou Yuan, Massachusetts General Hospital and Harvard Medical School, United StatesRaymond H. Kim, University of Toronto, Canada
Naoyuki Sone, Kyoto University, Japan
Copyright © 2022 Schultz, Elenius, Fassad, Freke, Rogers, Shoemark, Koistinen, Mohamed, Lim, Mitchison and Sironen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anu I. Sironen, YS5zaXJvbmVuQHVjbC5hYy51aw==
 Rüdiger Schultz
Rüdiger Schultz Varpu Elenius2
Varpu Elenius2 Mahmoud R. Fassad
Mahmoud R. Fassad Grace Freke
Grace Freke Amelia Shoemark
Amelia Shoemark Tiina Koistinen
Tiina Koistinen Hannah M. Mitchison
Hannah M. Mitchison Anu I. Sironen
Anu I. Sironen