- 1Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 2Department of Oncology, Anqing First People’s Hospital of Anhui Medical University/Anqing First People’s Hospital of Anhui Province, Anqing, China
- 3Institute of Bacterial Resistance, Anhui Medical University, Hefei, China
- 4Anhui Center for Surveillance of Bacterial Resistance, Hefei, China
Background: RAR-related orphan receptor C (RORC) plays an important role in autoimmune responses and inflammation. However, its function in cancer immunity is still unclear. Its potential value in cancer immunotherapy (CIT) needs to be further studied.
Methods: Expression and clinical data for 33 cancers were obtained from UCSC-Xena. The correlation between RORC expression and clinical parameters was analyzed using the limma software package to assess the prognostic value of RORC. Timer2.0 and DriverDBv3 were used to analyze the RORC mutation and methylation profiles. RORC-associated signaling pathways were identified by GSEA. The correlations of RORC expression with tumor microenvironment factors were further assessed, including immune cell infiltration (obtained by CIBERSORT) and immunomodulators (in pancancer datasets from the Tumor-Immune System Interactions and Drug Bank [TISIDB] database). In addition, the correlations of RORC with four CIT biomarkers (tumor mutational burden, microsatellite instability, programmed death ligand-1, and mismatch repair) were explored. Furthermore, three CIT cohorts (GSE67501, GSE168204, and IMvigor210) from the Gene Expression Omnibus database and a previously published study were used to determine the association between RORC expression and CIT response.
Results: RORC was differentially expressed in many tumor tissues relative to normal tissues (20/33). In a small number of cancers, RORC expression was correlated with age (7/33), sex (4/33), and tumor stage (9/33). Furthermore, RORC expression showed prognostic value in many cancers, especially in kidney renal clear cell carcinoma (KIRC), brain lower grade glioma (LGG), and mesothelioma (MESO). The mutation rate of RORC in most cancer types was low, while RORC was hypermethylated or hypomethylated in multiple cancers. RORC was associated with a variety of biological processes and signal transduction pathways in various cancers. Furthermore, RORC was strongly correlated with immune cell infiltration, immunomodulators, and CIT biomarkers. However, no significant association was found between RORC and CIT response in the three CIT cohorts.
Conclusion Our findings revealed the potential immunotherapeutic value of RORC for various cancers and provides preliminary evidence for the application of RORC in CIT.
Introduction
Every year, approximately 20 million new cases of cancer are diagnosed and 10 million cancer-related deaths occur around the world (Sung et al., 2021). The occurrence of cancer is closely related to autoimmunity and its progression reflects the inability of the immune system to control the growth of tumor cells. Over the past decades, cancer immunotherapy (CIT) has become an effective clinical strategy. The principle of CIT is to induce the immune system to eliminate tumors, but there are still many patients who are unresponsive or poorly responsive to CIT (Riley et al., 2019). Further investigation into the association of immunity-related factors with cancer will provide reference data for future studies.
To obtain a more suitable microenvironment for growth, tumor cells alter the normal immune microenvironment by secreting immunosuppressive factors and further regulating immune cells, among which CD4+ T cells play an important role (Lowery et al., 2022). CD4+ T cells can differentiate into various subsets such as T helper cells (Th1, Th2, Th9, Th17, and Th22), regulatory T cells (Tregs), and follicular helper T cells (Saravia et al., 2019). Recently, several studies have confirmed that Th17 and Treg play important roles in cancer and autoimmunity (Knochelmann et al., 2018).
RAR-related orphan receptor C (RORC) is a 24-kb protein-encoding gene located on chromosome 1 (1q21.3). RORγ T, a RORC-encoded protein, is the key transcription factor responsible for Th17 polarization and function, as well as thymocyte development (Yahia-Cherbal et al., 2019). Studies have shown that RORC plays an important role in autoimmunity and inflammation (IvanovMcKenzie et al., 2006; Michelini et al., 2021). Several studies have shown that RORC performs critical regulatory functions in cell proliferation, metastasis, and chemoresistance in various malignancies such as hematologic tumors (Subramanian et al., 2019), breast cancer (Oh et al., 2014), bladder cancer (Cao et al., 2019), and melanoma (Brożyna et al., 2016). The exact role that RORC plays in other cancers is still unknown.
In this study, RORC expression was evaluated in 33 cancer types and the effects of RORC on the tumor immune microenvironment were determined. Furthermore, the associations of RORC expression with many immunomodulators and dynamic immunological biomarkers were investigated. In brief, this study investigated the role of RORC in the pancancer immune microenvironment and explored the potential value of RORC in CIT, which may be helpful for future studies.
Materials and methods
Data collection
RNA-seq data on 33 human cancer types were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Abbreviations and full names of the 33 cancers are shown in Table 1. RORC expression data in normal tissues was downloaded from the Genotype-Tissue Expression (GTEx) database (https://commonfund.nih.gov/GTEx/). Corresponding clinical and prognostic data were acquired from the University of California Santa Cruz (UCSC) Xena Explorer. Additionally, RNA-seq and clinical data from the following three CIT cohorts were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and a previously published study (Mariathasan et al., 2018): 1) IMvigor 210 (34 patients with bladder cancer treated with the programmed death ligand 1 [PD-L1] inhibitor atezolizumab) (Mariathasan et al., 2018), 2) GSE67501 (patients with renal cell carcinoma treated with programmed cell death protein 1 [PD-1] inhibitor), and 3) GSE168204 (patients with metastatic melanoma treated with PD-1 inhibitor).
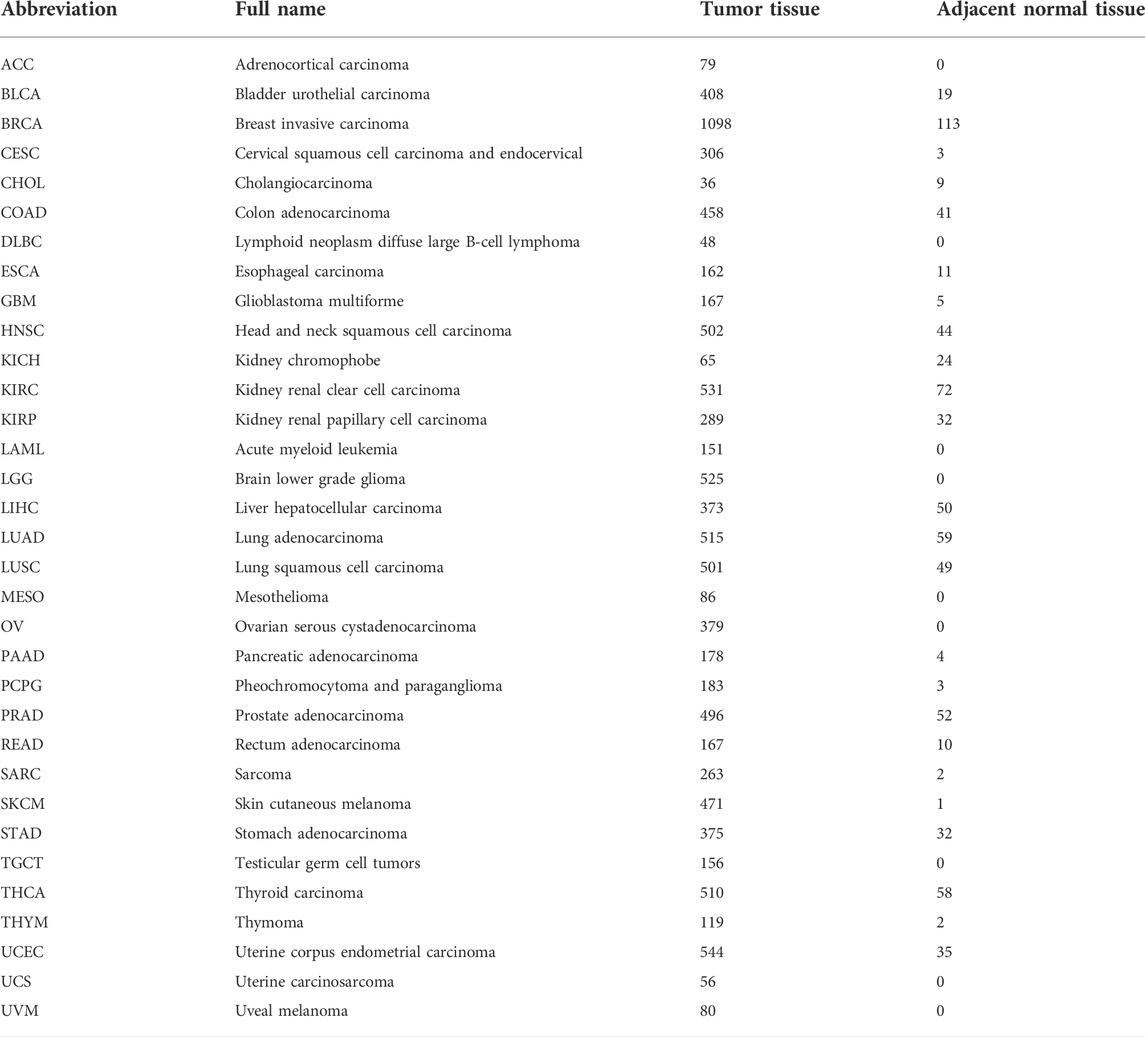
TABLE 1. Full names of 33 cancer types and the numbers of tumor and adjacent normal tissues from TCGA database.
RAR-related orphan receptor C expression in 33 cancer types
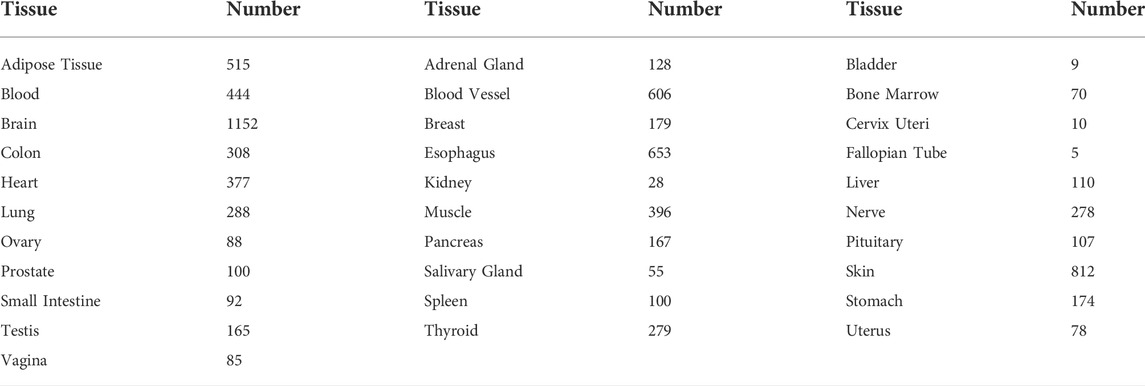
Differences in RORC expression between tumor and normal tissues for 33 cancer types were analyzed using the limma package in R Studio software. To further analyze the differences in RORC expression in tumor tissues and normal tissues, we combined the TCGA data with GTEx data. The numbers of “healthy tissue” samples from the GTEx database are presented in Table 2. The Wilcoxon test was also used to analyze differences in RORC expression stratified by various clinical characteristics (age, sex, and tumor stage).
Prognostic value of RAR-related orphan receptor C expression
Cox regression analysis was performed to investigate the prognostic value of RORC in 33 cancers, based on overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS). Hazard ratio (HR) > 1 represents a risk factor for death. p < 0.05 was considered significant. Kaplan–Meier survival curve analyses were also conducted for multiple cancer types. p < 0.05 was considered significant.
RAR-related orphan receptor C mutation and methylation profiles
TIMER2.0 (http://timer.comp-genomics.org/) was used to create a bar plot showing the RORC mutation rate for each TCGA cancer type (Li et al., 2020). DriverDBv3 (http://driverdb.tms.cmu.edu.tw/) is a cancer omics database that incorporates somatic mutation, methylation, copy number variation, RNA expression, miRNA expression and clinical data in addition to annotation bases (Liu et al., 2020). We explored the RORC mutation rates using both TIMER2.0 and DriverDBv3. Moreover, the RORC methylation types in multiple cancer types were obtained from DriverDBv3.
Gene set enrichment analysis (GSEA)
GSEA is commonly used to analyze differences in the levels of biological processes and pathways between two biological states in transcriptomics research (Powers et al., 2018). The gene set files c5. go.v7.4. symbols.gmt and c2. cp.kegg.v7.4. symbols were obtained from GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp). GSEA was then used to analyze the biological processes and signaling pathways associated with RORC in 33 cancer types.
Correlations of RAR-related orphan receptor C expression with immune-related factors
Estimation of STromal and Immune cells in MAlignant Tumors using Expression data (ESTIMATE) is an algorithm that uses gene expression signatures to infer the proportion of stromal and immune cells in tumor samples (Yoshihara et al., 2013). The immune score and stromal score were calculated by ESTIMATE. Spearman’s correlation coefficient was used to analyze the correlations of RORC expression with the immune and stromal scores.
Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) is a method for characterizing the cell composition of complex tissues based on gene expression profiles (Chen et al., 2018). We used the CIBERSORT algorithm to determine the infiltration of 22 lymphocyte subsets in the samples. Spearman’s correlation coefficient was used to analyze the correlations of RORC expression with the infiltration of the 22 lymphocyte subsets.
Tumor-Immune System Interactions and Drug Bank (TISIDB; http://cis.hku.hk/TISIDB/index.php) is a web portal dedicated to collecting data on the interactions between tumors and the immune system, and it integrates multiple heterogeneous datasets (Ru et al., 2019). The correlations of RORC expression with immunomodulators (immunoinhibitors, immunostimulators, and major histocompatibility complex (MHC) molecules) were investigated using TISIDB.
Additionally, PD-L1, tumor mutational burden (TMB), microsatellite instability (MSI), and mismatch repair (MMR) are crucial biomarkers that predict the CIT response (Rizzo et al., 2021). Therefore, the correlations of RORC expression with PD-L1, TMB, MSI, and MMR were further investigated using Spearman’s correlation coefficient. TMB was determined using the PERL programming language (version 5.32.1). MSI data were obtained from a previously published study (Bonneville et al., 2017).
Correlations of RAR-related orphan receptor C expression with CIT responses
Data on RORC expression and CIT responses [responsive, which was defined as complete response (CR) or partial response (PR), and non-responsive, which was defined as progressive disease (PD) or stable disease (SD)] were obtained from three relevant independent CIT cohorts (IMvigor210, GSE67501, and GSE168204). The Wilcoxon test was then used to analyze the differences in CIT response between the high and low RORC expression groups, and p < 0.05 was used to indicate a significant difference.
Statistical analysis
All statistical analyses were performed using R software (version 4.1.0). Correlation analyses were performed using Spearman’s correlation coefficient. Analyses of differential RORC gene expression were performed using Wilcoxon tests. p < 0.05 was considered to indicate statistical significance.
Results
RAR-related orphan receptor C expression in 33 cancers
We explored the differences in RORC expression between tumor and normal tissues in 33 human cancers. RORC was differentially expressed in 12 cancers compared to adjacent normal tissues (BRCA, CHOL, COAD, ESCA, HNSC, KIRP, LUAD, LUSC, PRAD, READ, STAD, and THCA), with RORC expression being significantly higher in BRCA, LUAD, and PRAD than in adjacent normal tissues (Figure 2A). Figures 2B,C show RORC expression in various cancers and various normal tissues (GTEx data), respectively. Figure 2D shows the results of the combined analysis of TCGA and GTEx data, indicating that RORC expression was higher in BRCA, COAD, LUAD, OV, UCEC, and UCS than in normal tissues, but lower in ACC, CHOL, HNSC, KIRP, LAML, LGG, LIHC, LUSC, PAAD, READ, SKCM, STAD, TGCT, and THCA.
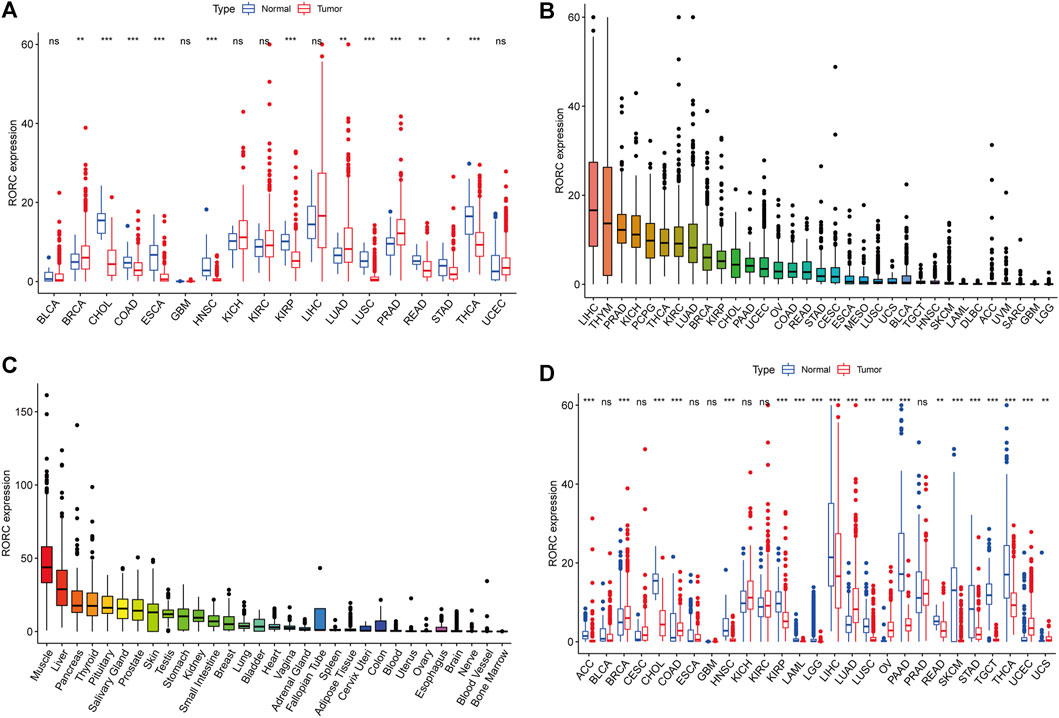
FIGURE 2. Differential RORC expression between cancer and normal tissues. (A) Differential RORC expression between cancer and adjacent normal tissues based on TCGA data (cancers with <5 adjacent normal tissue samples were discarded). (B) Mean RORC expression in 33 cancer tissues (from high to low). (C) Mean RORC expression in normal tissues based on GTEx data (from high to low). (D) Differential RORC expression between cancer and normal tissues in various cancers based on combined TCGA and GTEx data.
We also explored the associations between RORC expression and clinical features in 33 human cancers. RORC expression significantly differed by age (<65 vs. ≥ 65 years) in BLCA, BRCA, ESCA, LAML, LGG, PAAD, and THYM (Figure 3A), sex (higher in females) in KIRC, KIRP, LUSC, and READ (Figure 3B), and tumor stage in ACC, BLCA, COAD, ESCA, HNSC, KICH, KIRC, LIHC, and LUSC (Figure 3C).
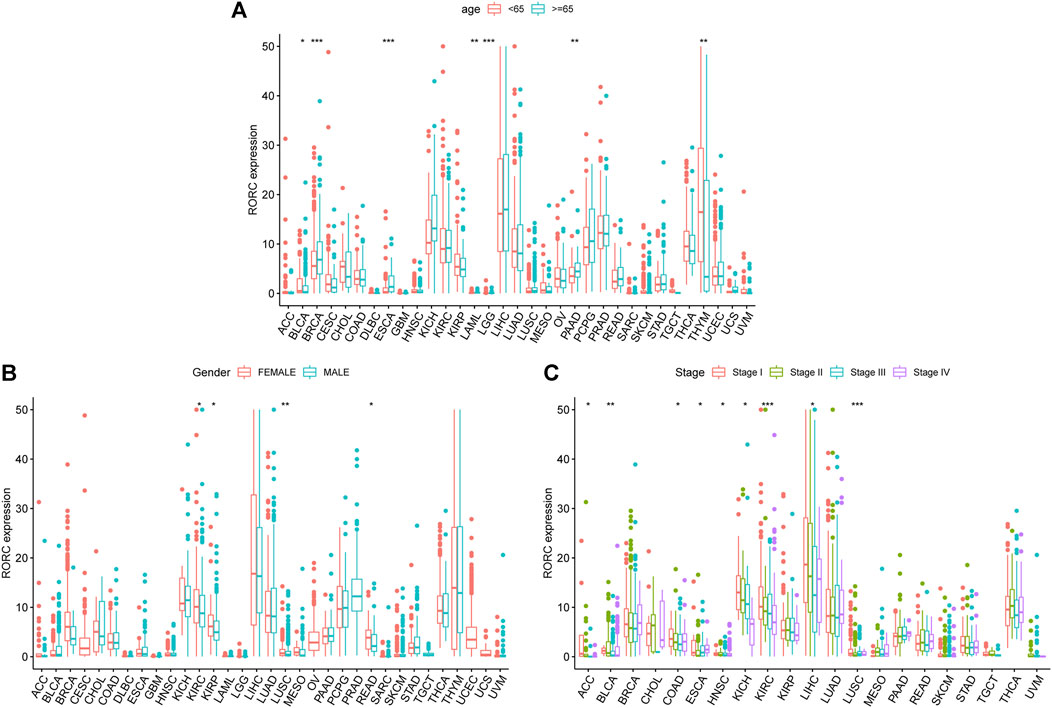
FIGURE 3. Associations of RORC with (A) age, (B) sex, and (C) tumor stage. *p < 0.05, **p < 0.01, and ***p < 0.001.
RAR-related orphan receptor C mutation and methylation profiles
To explore the mechanisms underlying differential RORC expression in various cancers, we explored the RORC mutation rate in multiple cancer types in the TCGA database using TIMER2.0 (Figure 4A) and DriverDBv3 (Figure 4B). TIMER2.0 showed that RORC had the highest mutation rate in SKCM (26/468) and the lowest in THCA (1/500). DriverDBv3 showed that the RORC mutation rate in most cancers was low. Figure 4C shows the methylation type of RORC in multiple cancer types. Interestingly, RORC was hypermethylated in CHOL, COAD, ESCA, KIRP, PAAD, and READ, but hypomethylated in LIHC, LUAD, LUSC, OV, PCPG, and UCEC.
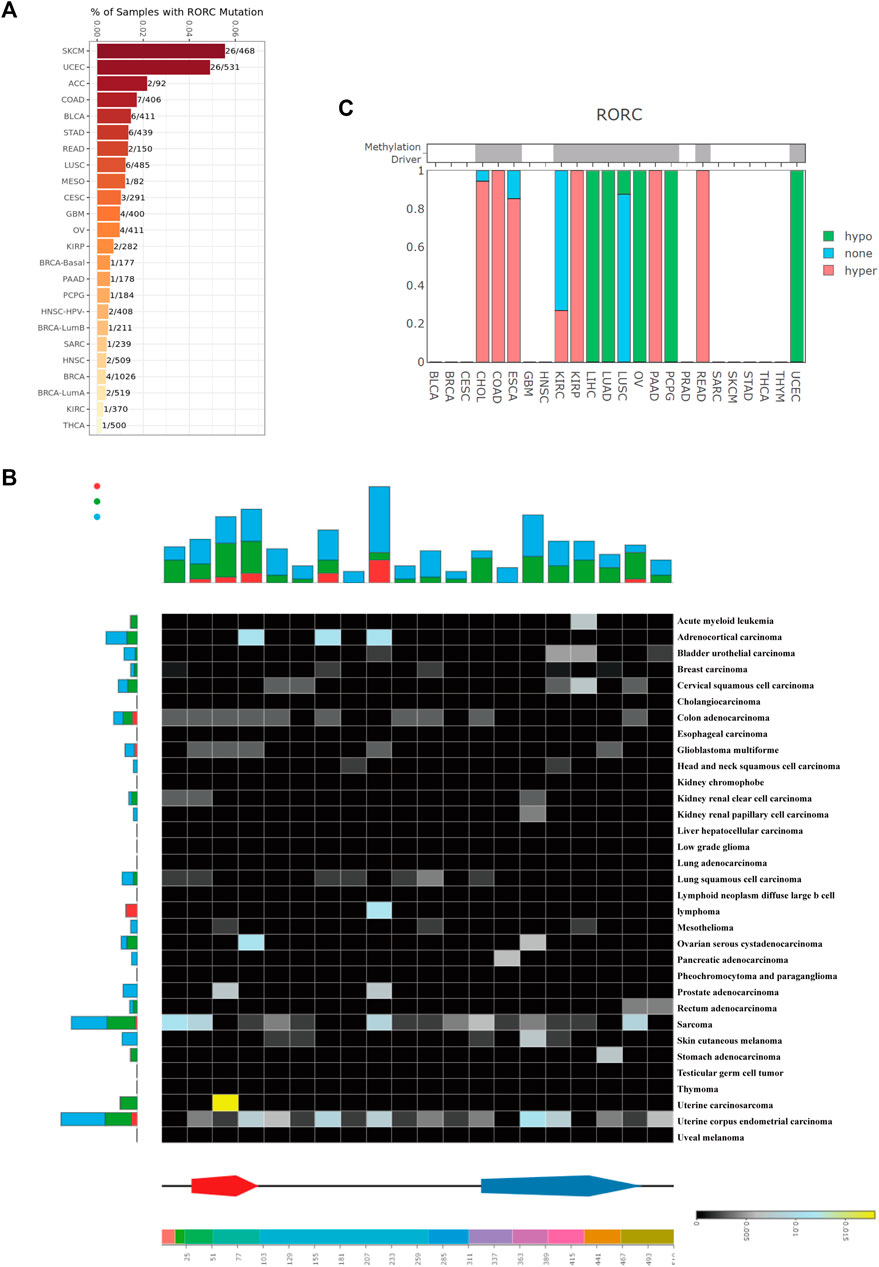
FIGURE 4. RORC mutation and methylation analysis in various cancers. RORC mutation analysis using (A) TIMER 2.0, and (B) DriverDBv3 data. (C) RORC methylation type in various cancers.
Prognostic value of RAR-related orphan receptor C expression
To determine the correlation of RORC expression with prognosis (OS, DFS, DSS, and PFS) in 33 tumors, we used the survival and survminer R packages. RORC expression was a risk factor for OS in LGG patients, but a protective factor in KIRC, LIHC, MESO, SKCM, and THYM (Figure 5A). It was a risk factor for DFS in UCEC but a protective factor in PCPG, PRAD, and THCA (Figure 5B). It was a risk factor for DSS in LGG but a protective factor in BLCA, KIRC, LUAD and MESO (Figure 5C). It was a risk factor for PFS in LGG and CESC but a protective factor in BLCA, KIRC, LUAD, MESO, PCPG, PRAD, SKCM, and UVM (Figure 5D).
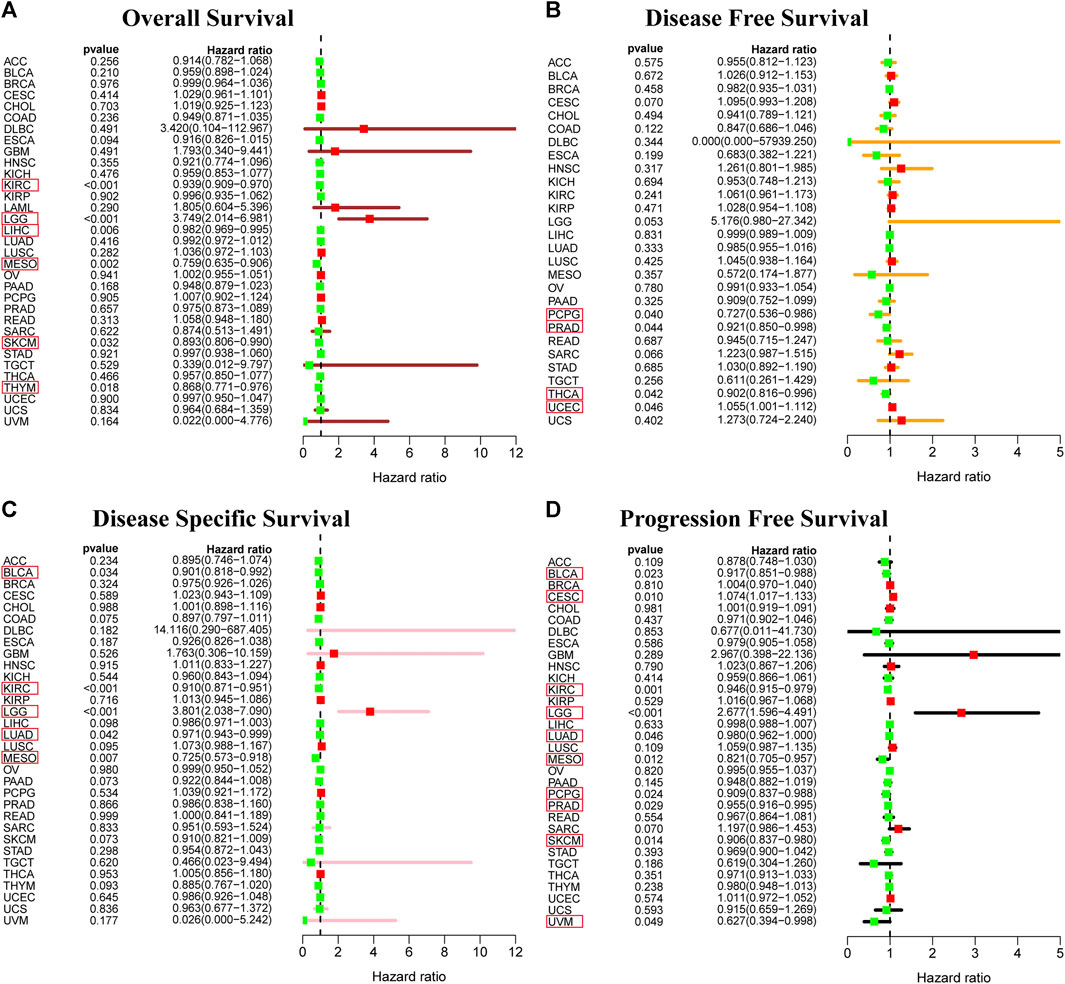
FIGURE 5. Prognostic value of RORC. Correlations of RORC expression with (A) OS, (B) DFS, (C) DSS, and (D) PFS. RORC expression was significantly correlated with the prognosis of the cancers in red box (p < 0.05). Hazard ratio >1 indicates that RORC expression was a risk factor for death.
To analyze the relationship between RORC expression and cancer prognosis more comprehensively, Kaplan-Meier survival curve analyses for 33 cancer types were conducted. RORC was significantly associated with OS in ACC, BLCA, GBM, KIRC, LIHC, MESO, THCA, THYM, and UVM (Supplementary Figure 1A), DFS in CESC, HNSC, PAAD, and THCA (Supplementary Figure 1B), DSS in ACC, BLCA, GBM, KIRC, LIHC, LUSC, MESO, PAAD, and THYM (Supplementary Figure 1C), and PFS in ACC, BLCA, CESC, KIRC, LGG, MESO, PAAD, THCA, and UVM (Supplementary Figure 1D).
Correlations of RAR-related orphan receptor C expression with immune cell infiltration
Tumor-infiltrating immune cells, as an important part of the tumor microenvironment, are closely related to the occurrence, progression, and metastasis of cancer. We explored the correlation of RORC with the immune and stromal scores, and on immune cell infiltration (thresholds: correlation coefficient >0.4, p < 0.001). RORC expression was positively correlated with the immune score in TGCT and ACC and the stromal score in TGCT (Figure 6). RORC expression was positively correlated with Treg infiltration in ESCA and TGCT and mast cell infiltration in MESO. In contrast, it was negatively correlated with mast cell, B lymphocyte, M0 macrophage, and M1 macrophage infiltration in THYM, but positively correlated with plasma cell, Treg, and naive CD4+ T cell infiltration.
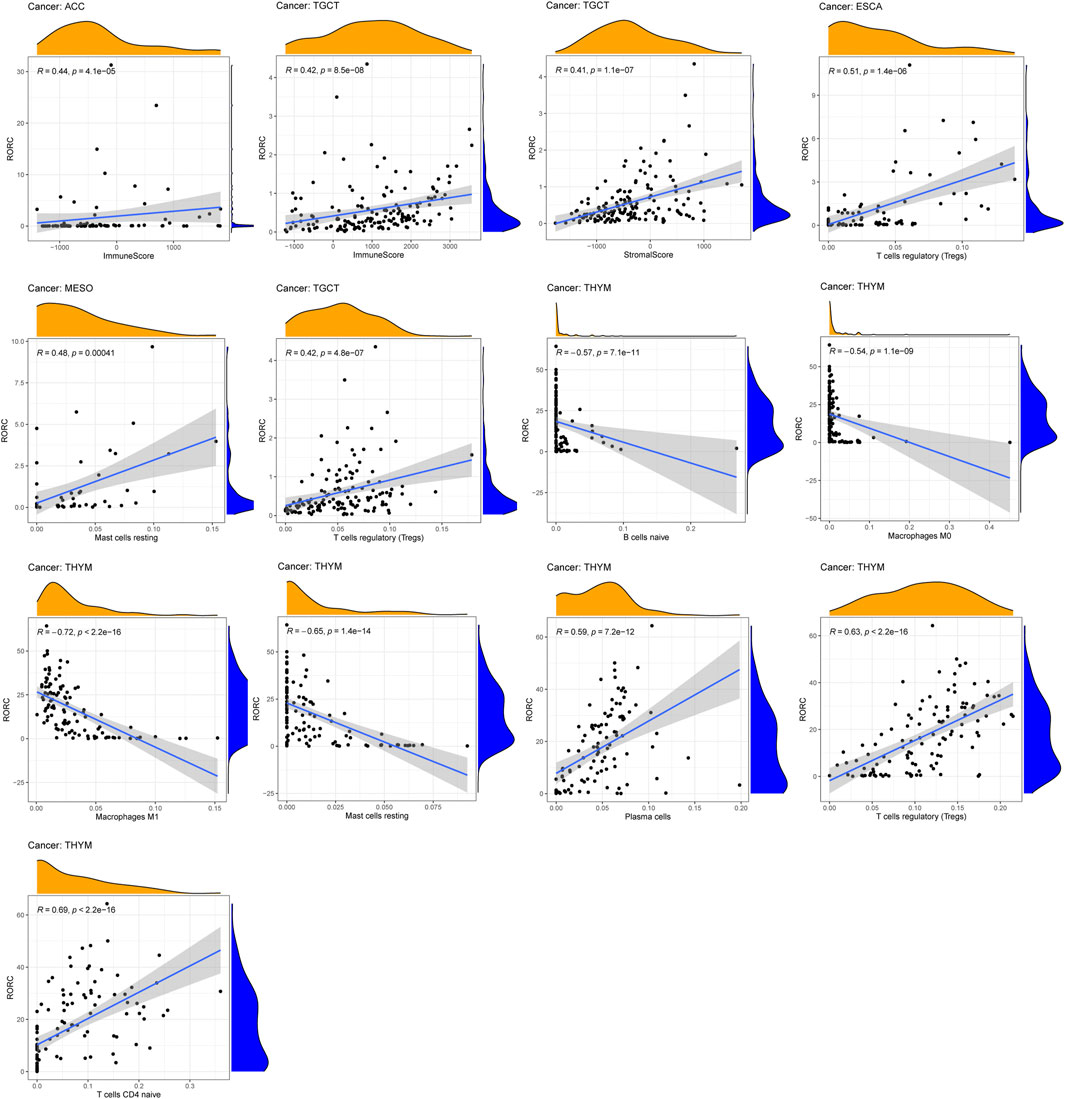
FIGURE 6. Correlations of RORC expression with ESTIMATE scores and immune cell infiltration. Correlations are shown if R > 0.5 and p < 0.05.
Correlations of RAR-related orphan receptor C expression with immunomodulators
To further investigate the potential value of RORC in CIT, we explored the correlations of RORC expression with immunomodulators. Figure 7A demonstrates the correlations between RORC expression and 24 immunoinhibitors in a pancancer analysis. RORC expression exhibited the strongest positive correlation with IL10RB in TGCT, while it exhibited the strongest negative correlation with TGFB1 in ESCA. Figure 7B shows the correlations between RORC and 45 immunostimulators in a pancancer analysis. RORC expression exhibited the strongest positive correlation with TNFSF13 in ESCA, while it exhibited the strongest negative correlation with NT5E in MESO. Figure 7C shows the correlations between RORC expression and 21 MHC molecules in a pancancer analysis. RORC expression exhibited the strongest positive correlation with HLA-F in ACC, while it exhibited the strongest negative correlation with B2M in UVM.
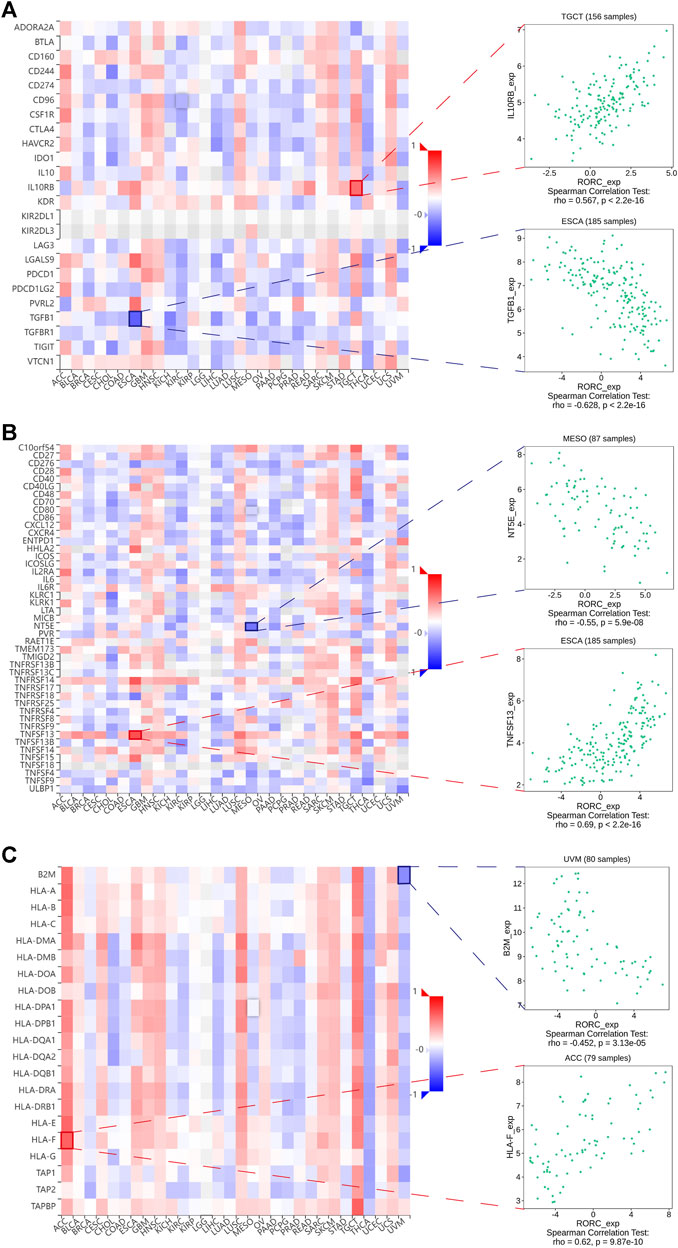
FIGURE 7. Correlations of RORC expression with (A) immunoinhibitors, (B) immunostimulators, and (C) MHC molecules. Red indicates positive correlation; blue indicates negative correlation. The strongest negative and positive correlations are presented on the right.
Taken together, our results revealed particularly strong correlations between RORC and immunomodulators in ACC, ESCA, MESO, TGCT, and UVM. Interestingly, the biological processes involving RORC (according to GSEA) differed among these five cancers (Figure 8A). In particular, RORC was associated with T cell activation in ACC, and activation of the immune response and the adaptive immune response in TGCT. Additionally, the signaling pathways involving RORC (according to GSEA) were strikingly different (Figure 8B). Remarkably, RORC was associated with the Toll-like receptor (TLR) and Rig-I-like receptor (RLR) signaling pathways in MESO, but was also associated with the intestinal immune network.
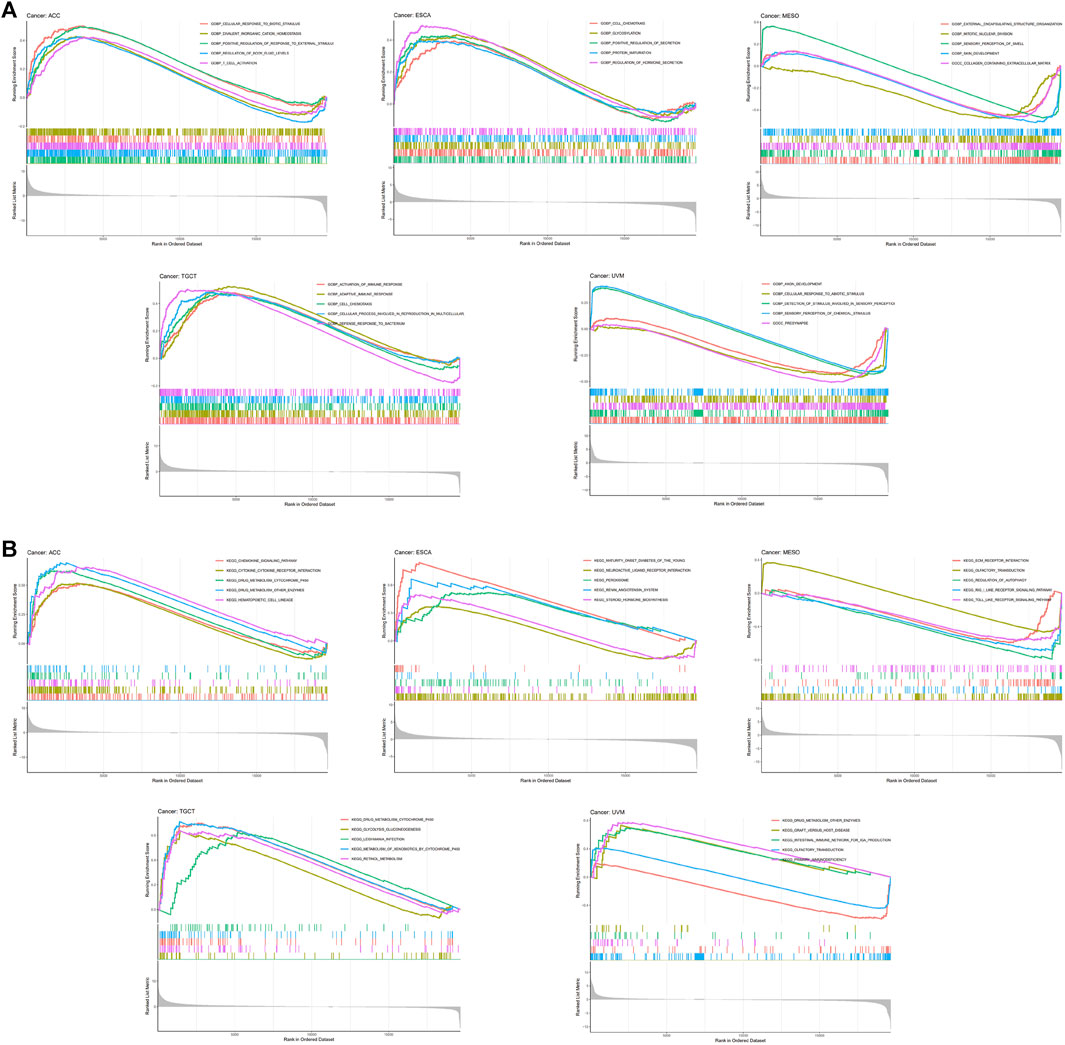
FIGURE 8. GSEA showing the associations of RORC expression with (A) biological processes and (B) signaling pathways in five cancers.
Potential value of RAR-related orphan receptor C in CIT
TMB, PD-L1, and MSI are effective CIT biomarkers. The correlations between RORC expression and these three CIT biomarkers are shown in Figure 9A. RORC was positively correlated with TMB in LIHC, LGG, and ESCA, but negatively correlated in ACC, THYM, THCA, TGCT, SKCM, OV, LUSC, HNSC, GBM, DLBC, and BRCA. RORC expression was positively correlated with PD-L1 expression in ACC, UCS, TGCT, SKCM, SARC, LGG, LAML, and HNSC, but negatively correlated in THYM, THCA, STAD, READ, PRAD, PAAD, LUAD, ESCA, COAD, CESC, and BRCA. Interestingly, RORC expression was not significantly correlated with MSI in most cancers, but was negatively associated with MSI in TGCT, LUSC, and HNSC. The correlations of RORC expression with mismatch repair (MMR) were also investigated. RORC expression was highly correlated with MMR-related genes (including MLH1, MSH2, MSH6, PMS2, and EPCAM) in BRCA, ESCA, LUSC, and THYM (Figure 9B).
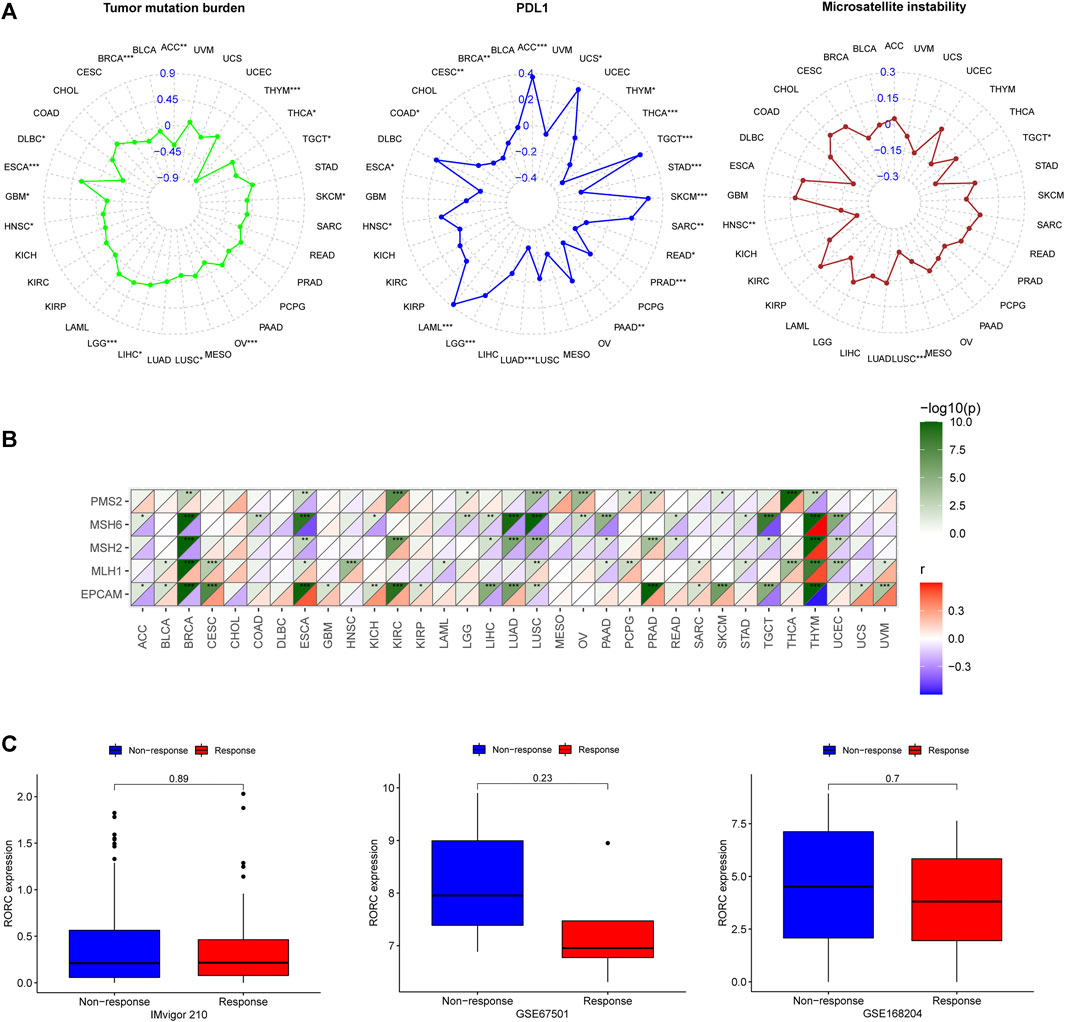
FIGURE 9. Correlations of RORC expression with (A) cancer immunotherapy (CIT) biomarkers (TMB, PD-L1, and MSI) and (B) MMR-related genes (PMS2, MSH6, MSH2, MLH1, and EPCAM) and (C) differences in RORC expression between responder and non-responder groups in three CIT cohorts. *p < 0.05, **p < 0.01, ***p < 0.001.
To verify the role of RORC in CIT, we investigated the association of RORC expression with CIT responses in three independent CIT cohorts (IMvigor 210, GSE67501, and GSE168204). In all three cohorts, RORC expression did not differ significantly between the responder and nonresponder groups (Figure 9C). However, there was a non-significant trend indicating that low RORC expression rendered tumors more responsive to CIT.
Discussion
Cancer has always been a major public health problem worldwide and is the second leading cause of death after cardiovascular diseases (Global Burden of Disease Cancer et al., 2022). Currently, the main cancer treatment methods include surgery, radiotherapy, chemotherapy, and targeted therapy. It is difficult to completely eradicate tumors with conventional surgical treatment, which is always accompanied by a high risk of postoperative recurrence. Radiotherapy and chemotherapy kill normal cells as well as tumor cells and cause many side effects (Islam et al., 2019; Williams et al., 2020). Although targeted drugs may reduce adverse drug reactions, many tumors are prone to develop resistance against these drugs, leading to tumor recurrence (Holohan et al., 2013). With the development of molecular biology and oncobiology, CIT has become a new type of cancer treatment that effectively overcomes certain shortcomings of traditional treatment methods (McNutt, 2013). However, there are various ways in which cancer cells can evade the immune system and become unresponsive or poorly responsive to CIT.
RORγ T, a specific transcription factor encoded by RORC, plays a major regulatory role in Th17 cell polarization and function and is associated with autoimmunity and inflammation (IvanovMcKenzie et al., 2006). Recently, several studies have found that RORC is involved in the regulation of hematological tumors, breast cancer, bladder cancer, and melanoma (Oh et al., 2014; Brożyna et al., 2016; Cao et al., 2019; Subramanian et al., 2019). However, the exact role of RORC in other cancers remains unknown, although comprehensive studies of differences in RORC expression between tumor and normal tissues have revealed its potential value in CIT for a variety of cancer types.
Our study showed that there were significant differences in RORC expression between tumor and normal tissues (adjacent normal tissues [TCGA data] and healthy tissues [GTEx data]) in some cancers. Specifically, RORC expression was significantly higher in BRCA, COAD, LUAD, OV, UCEC, and UCS. Similarly, Lin et al. (Lin et al., 2015) found that RORC expression increases in BRCA, while Dong et al. (Dong et al., 2021) found that RORC expression in LUAD was higher than in normal tissues.
It is well known that gene mutation and methylation can alter gene expression (Yang et al., 2014; Jia and Zhao, 2017). Our results showed that the RORC mutation rate in most cancer types was low, whereas RORC methylation significantly differed between cancer and normal tissues in several cancers. Interestingly, abnormal RORC methylation occurred in the majority of cancers with differential RORC expression, which indicates that differential RORC expression may be caused by abnormal methylation in some cancers.
We found that RORC expression was higher in older patients (≥65 years) than younger patients (<65 years) in LAML. Notably, Subramanian et al. (Subramanian et al., 2019) found that RORC expression decreased with age in human T lymphocytic virus (HTLV)-1-infected patients who did not develop adult T-cell leukemia (which can occur decades after HTLV-1 infection (Nakahata et al., 2011)), demonstrating that an age-dependent decline in RORC expression indicates a possible early event in HTLV-1-driven leukemogenesis. We also found that RORC expression was not sex-specific in most cancers, and RORC expression varied with tumor stage in some cancers. Similarly, a previous study reported that RORC expression in BLCA decreases with increasing tumor stage (Cao et al., 2019). Notably, RORC expression had prognostic value in some cancers, especially KIRC, LGG, and MESO. Consistently, Ait Ssi et al. (Ait Ssi et al., 2021) found that RORC expression is upregulated in LGG patients with a poor prognosis. In conclusion, RORC expression can be an important regulator of cancers and RORC has prognostic value in some cancers.
The immune microenvironment is a vital feature of tumors; it can be used to predict the clinical prognosis of patients and it also plays an important role in predicting the response to CIT (Hanahan and Weinberg, 2011). Several studies (Paijens et al., 2021; Zheng et al., 2021) have confirmed that tumors are locally infiltrated with various immune cell subsets, including macrophages, dendritic cells, mast cells, natural killer cells, CD4+ T cells, and CD8+ T cells. Different types of tumors have different levels of immune cell infiltration, and even different patients with the same type of tumor have different levels of immune cell infiltration, which is one reason why patients respond differently to CIT (Wu and Dai, 2017).
The results of our study showed that RORC expression was positively correlated with immune and stromal scores as well as Tregs. Tregs play a crucial role in maintaining immune tolerance and repressing antitumor immunity (Knochelmann et al., 2018). Siska et al. (Siska et al., 2017) showed there was higher Treg infiltration in advanced TGCT. Moreover, the authors suggested that an increase in Treg infiltration may lead to a poor CIT response in nonspermatogonial germ cell tumors. Interestingly, RORC expression was positively correlated with Tregs, naive CD4+ T cells, and plasma cells in THYM, but was negatively correlated with naive B cells, M0 macrophages, M1 macrophages, and resting mast cells in THYM. In cases of THYM, the imbalance between Th17/Tregs in THYM may be the pathological mechanism underlying the development of myasthenia gravis (MG) (Chen et al., 2021a). Furthermore, the increase in plasma cell levels in MG patients has previously been confirmed (Kohler et al., 2013). Moreover, impaired macrophage polarization affects T cell apoptosis and T cell-mediated autoimmunity (Ghosh et al., 2015; Veremeyko et al., 2018). Thus, RORC may contribute to the development of MG by altering the infiltration of immune cells in THYM. Thus, RORC may be a potential target for the treatment of MG in patients with THYM.
We also investigated the biological processes and signaling pathways involving RORC in cancers. TLRs and RLRs play a vital role in cancer immunity (Bourquin et al., 2020). Notably, TLR and RLR signaling pathways were enriched in the high RORC expression subgroup of MESO patients. Similarly, a previous study demonstrated that TLR and Rig-I contribute to the progression of malignant MESO (Wörnle et al., 2009), which means that RORC may be a potential CIT target for use in MESO patients. In short, RORC is an important gene in the immune microenvironment in cancer patients.
Regarding the negative correlations between RORC expression and immunostimulators, RORC expression exhibited the strongest negative correlation with NT5E in MESO. NT5E encodes CD73, which is an ectonucleotidase present on cancer cells that helps cancer cells convert ATP into adenosine (Kordaß et al., 2018). Adenosine is an effective immunomodulator that inhibits antitumor immune responses and promotes metastasis (Allard et al., 2017). Al-Taei et al. (Al-Taei et al., 2017) reported the regulatory function of CD73 in the immune microenvironment in MESO, demonstrating the promising role of RORC in CIT for the treatment of MESO. Regarding the positive correlations between RORC expression and immunostimulators, RORC expression exhibited the strongest positive correlation with TNFSF13 in ESCA. TNFSF13 is a proliferation-inducing ligand of the tumor necrosis factor (TNF) superfamily. It plays an important role in autoimmunity and has also been reported as a potential molecular target to overcome immunosuppression (Chen et al., 2021b). Studies have shown that high TNFSF13 expression in tumor cells and fibroblasts is associated with a poor prognosis of non-small cell lung cancer, and TNFSF13 also promotes the development of LAML (Qian et al., 2014; Chapellier et al., 2019). Furthermore, TNFSF13 plays a crucial role in triple-negative breast cancer, laryngeal squamous cell carcinoma, gastric cancer, and glioma (Wang et al., 2016; Lin et al., 2020; Chen et al., 2021b; Zhang et al., 2021).
Regarding the negative correlations between RORC expression and immunoinhibitors, RORC expression exhibited the strongest negative correlation with TGFB1 in ESCA. Recently, Talukdar et al. demonstrated that TGFB1 expression influences the prognosis of ESCA (Talukdar et al., 2021). Furthermore, several studies have reported that high TGFB1 expression is associated with poor prognosis in many cancers (Liang et al., 2020; de Streel and Lucas, 2021). These findings further confirmed that RORC may be a potential CIT target for treating ESCA. Regarding the positive correlations between RORC expression and immunoinhibitors, RORC expression exhibited the strongest positive correlation with IL10RB in TGCT. As Hanna et al. (Hanna et al., 2021) reported, IL10RB can promote antitumor immunity by maintaining the PD-1intTCF-1+CD8+ T cell population.
Regarding the correlations between RORC expression and MHC molecules (which are closely associated with cancer occurrence and development (Cornel et al., 2020)), RORC expression exhibited the strongest positive correlation with HLA-F in ACC, and the strongest negative correlation with B2M in UVM. Although an association between HLA-F and ACC has rarely been reported, the differential HLA-F expression in STAD, BRCA, and COAD (relative to normal tissue) has been confirmed (Ishigami et al., 2015; Huang et al., 2020; Wuerfel et al., 2020). B2M is a critical molecule in tumorigenesis and immune monitoring and is considered a latent CIT target (Wang et al., 2021). On the basis of the correlations between RORC expression and the immunomodulators mentioned above, we propose that RORC may be a potential CIT target for treating certain cancers.
PD-L1 expression is considered an effective biomarker that can predict the clinical efficacy of PD-1/PD-L1 inhibitors (Davis and Patel, 2019), and MSI and TMB are also effective CIT biomarkers (Le et al., 2015; Romero, 2019). Therefore, we further studied the correlations of RORC expression with PD-L1, TMB, and MSI. RORC was negatively associated with PD-L1 and TMB in BRCA, THCA, and THYM, and negatively correlated with TMB and MSI in LUSC. Notably, RORC was correlated (mostly negatively) with PD-L1 in most cancers. Lytle et al. (Lytle et al., 2019) found that inhibiting RORC expression hinders the progression of PAAD, which they believe may represent a new treatment strategy for PAAD. Interestingly, our results showed that in PAAD, RORC expression was negatively correlated with PD-L1, suggesting that high RORC expression may be a reason for the insensitivity to anti-PD-L1 treatment in PAAD. These findings suggest that low RORC expression may improve CIT responses in certain cancers. There was a non-significant tendency for the low RORC expression subgroups to exhibit a better CIT response than the high RORC expression subgroups in the three independent CIT cohorts. Because only three relevant cohorts were explored in this study, it is difficult to comprehensively elucidate the CIT response in relation to RORC expression levels. More CIT cohorts should be studied in the future.
In addition, we must acknowledge that our study has several further limitations. In particular, the data on RORC gene expression in tumor and normal tissues, as well as the data showing the association between RORC expression and cancer prognosis, were obtained retrospectively from public databases. Although our study has obtained useful findings, validation involving animal experiments and prospective multicenter, large-sample studies is required.
Conclusions
This study comprehensively demonstrates the important role that RORC plays in immunity in many cancers. These findings provide preliminary evidence for the application of RORC in CIT.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
JL and YL were responsible for the design of the study. SH, JY, and YS drafted and completed the manuscript. SH, JY, WS, MT, and BM were responsible for data download and analysis. All authors contributed to the article and agreed to the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (no. 81973983), Collaborative Tackling and Public Health Collaborative Innovation Project in Anhui Province (no. GXXT-2020-018), Joint Construction Project of Clinical Medicine University and Hospital (no. 2021lcxk006), and Natural Science Research Project of Universities in Anhui Province (no. KJ 2020A0176).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.969476/full#supplementary-material
References
Ait Ssi, S., Chraa, D., El Azhary, K., Sahraoui, S., Olive, D., and Badou, A. (2021). Prognostic gene expression signature in patients with distinct glioma grades. Front. Immunol. 12, 685213. doi:10.3389/fimmu.2021.685213
Al-Taei, S., Salimu, J., Spary, L. K., Clayton, A., Lester, J. F., and Tabi, Z. (2017). Prostaglandin E(2)-mediated adenosinergic effects on CD14(+) cells: Self-amplifying immunosuppression in cancer. Oncoimmunology 6 (2), e1268308. doi:10.1080/2162402X.2016.1268308
Allard, B., Longhi, M. S., Robson, S. C., and Stagg, J. (2017). The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 276 (1), 121–144. doi:10.1111/imr.12528
Bonneville, R., Krook, M. A., Kautto, E. A., Miya, J., Wing, M. R., Chen, H. Z., et al. (2017). Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017, 73. doi:10.1200/PO.17.00073
Bourquin, C., Pommier, A., and Hotz, C. (2020). Harnessing the immune system to fight cancer with Toll-like receptor and RIG-I-like receptor agonists. Pharmacol. Res. 154, 104192. doi:10.1016/j.phrs.2019.03.001
Brożyna, A. A., Jóźwicki, W., Skobowiat, C., Jetten, A., and Slominski, A. T. (2016). RORα and RORγ expression inversely correlates with human melanoma progression. Oncotarget 7 (39), 63261–63282. doi:10.18632/oncotarget.11211
Cao, D., Qi, Z., Pang, Y., Li, H., Xie, H., Wu, J., et al. (2019). Retinoic acid-related orphan receptor C regulates proliferation, glycolysis, and chemoresistance via the PD-L1/ITGB6/STAT3 signaling Axis in bladder cancer. Cancer Res. 79 (10), 2604–2618. doi:10.1158/0008-5472.CAN-18-3842
Chapellier, M., Peña-Martínez, P., Ramakrishnan, R., Eriksson, M., Talkhoncheh, M. S., Orsmark-Pietras, C., et al. (2019). Arrayed molecular barcoding identifies TNFSF13 as a positive regulator of acute myeloid leukemia-initiating cells. Haematologica 104 (10), 2006–2016. doi:10.3324/haematol.2018.192062
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M., and Alizadeh, A. A. (2018). Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 1711, 243–259. doi:10.1007/978-1-4939-7493-1_12
Chen, R., Wang, X., Dai, Z., Wang, Z., Wu, W., Hu, Z., et al. (2021). TNFSF13 is a novel onco-inflammatory marker and correlates with immune infiltration in gliomas. Front. Immunol. 12, 713757. doi:10.3389/fimmu.2021.713757
Chen, Y., Zhang, X. S., Wang, Y. G., Lu, C., Li, J., and Zhang, P. (2021). Imbalance of Th17 and Tregs in thymoma may be a pathological mechanism of myasthenia gravis. Mol. Immunol. 133, 67–76. doi:10.1016/j.molimm.2021.02.011
Cornel, A. M., Mimpen, I. L., and Nierkens, S. (2020). MHC class I downregulation in cancer: Underlying mechanisms and potential targets for cancer immunotherapy. Cancers 12 (7), E1760. doi:10.3390/cancers12071760
Davis, A. A., and Patel, V. G. (2019). The role of PD-L1 expression as a predictive biomarker: An analysis of all US food and drug administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 7 (1), 278. doi:10.1186/s40425-019-0768-9
de Streel, G., and Lucas, S. (2021). Targeting immunosuppression by TGF-β1 for cancer immunotherapy. Biochem. Pharmacol. 192, 114697. doi:10.1016/j.bcp.2021.114697
Dong, Y., Chen, C., Chen, C., Zhang, C., Zhang, L., Zhang, Y., et al. (2021). Stigmasterol inhibits the progression of lung cancer by regulating retinoic acid-related orphan receptor C. Histol. Histopathol. 36 (12), 1285–1299. doi:10.14670/HH-18-388
Ghosh, S., Mukherjee, S., Choudhury, S., Gupta, P., Adhikary, A., Baral, R., et al. (2015). Reactive oxygen species in the tumor niche triggers altered activation of macrophages and immunosuppression: Role of fluoxetine. Cell. Signal. 27 (7), 1398–1412. doi:10.1016/j.cellsig.2015.03.013
Global Burden of Disease Cancer, C., Kocarnik, J. M., Compton, K., Dean, F. E., Fu, W., Gaw, B. L., et al. (2022). Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. JAMA Oncol. 8, e216987.
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Hanna, B. S., Llaó-Cid, L., Iskar, M., Roessner, P. M., Klett, L. C., Wong, J. K. L., et al. (2021). Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity 54 (12), 2825–2841. doi:10.1016/j.immuni.2021.11.004.2825
Holohan, C., Van Schaeybroeck, S., Longley, D. B., and Johnston, P. G. (2013). Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 13 (10), 714–726. doi:10.1038/nrc3599
Huang, Y., Sun, H., Ma, X., Zeng, Y., Pan, Y., Yu, D., et al. (2020). HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 254, 117180. doi:10.1016/j.lfs.2019.117180
Ishigami, S., Arigami, T., Okumura, H., Uchikado, Y., Kita, Y., Kurahara, H., et al. (2015). Human leukocyte antigen (HLA)-E and HLA-F expression in gastric cancer. Anticancer Res. 35 (4), 2279–2285.
Islam, K. M., Anggondowati, T., Deviany, P. E., Ryan, J. E., Fetrick, A., Bagenda, D., et al. (2019). Patient preferences of chemotherapy treatment options and tolerance of chemotherapy side effects in advanced stage lung cancer. BMC cancer 19 (1), 835. doi:10.1186/s12885-019-6054-x
Ivanov, , McKenzie, B. S., Zhou, L., Tadokoro, C. E., Lepelley, A., Lafaille, J. J., et al. (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126 (6), 1121–1133. doi:10.1016/j.cell.2006.07.035
Jia, P., and Zhao, Z. (2017). Impacts of somatic mutations on gene expression: An association perspective. Brief. Bioinform. 18 (3), 413–425. doi:10.1093/bib/bbw037
Knochelmann, H. M., Dwyer, C. J., Bailey, S. R., Amaya, S. M., Elston, D. M., Mazza-McCrann, J. M., et al. (2018). When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell. Mol. Immunol. 15 (5), 458–469. doi:10.1038/s41423-018-0004-4
Kohler, S., Keil, T. O., Swierzy, M., Hoffmann, S., Schaffert, H., Ismail, M., et al. (2013). Disturbed B cell subpopulations and increased plasma cells in myasthenia gravis patients. J. Neuroimmunol. 264 (1-2), 114–119. doi:10.1016/j.jneuroim.2013.09.006
Kordaß, T., Osen, W., and Eichmüller, S. B. (2018). Controlling the immune suppressor: Transcription factors and MicroRNAs regulating CD73/nt5e. Front. Immunol. 9, 813. doi:10.3389/fimmu.2018.00813
Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372 (26), 2509–2520. doi:10.1056/NEJMoa1500596
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48 (W1), W509–w514. doi:10.1093/nar/gkaa407
Liang, C., Xu, J., Meng, Q., Zhang, B., Liu, J., Hua, J., et al. (2020). TGFB1-induced autophagy affects the pattern of pancreatic cancer progression in distinct ways depending on SMAD4 status. Autophagy 16 (3), 486–500. doi:10.1080/15548627.2019.1628540
Lin, H. Y., Kuei, C. H., Lee, H. H., Lin, C. H., Chen, Y. L., Chen, C. L., et al. (2020). TNFSF13 upregulation confers chemotherapeutic resistance via triggering autophagy initiation in triple-negative breast cancer. J. Mol. Med. 98 (9), 1255–1267. doi:10.1007/s00109-020-01952-5
Lin, M. L., Patel, H., Remenyi, J., Banerji, C. R., Lai, C. F., Periyasamy, M., et al. (2015). Expression profiling of nuclear receptors in breast cancer identifies TLX as a mediator of growth and invasion in triple-negative breast cancer. Oncotarget 6 (25), 21685–21703. doi:10.18632/oncotarget.3942
Liu, S. H., Shen, P. C., Chen, C. Y., Hsu, A. N., Cho, Y. C., Lai, Y. L., et al. (2020). DriverDBv3: A multi-omics database for cancer driver gene research. Nucleic Acids Res. 48 (D1), D863–D870. doi:10.1093/nar/gkz964
Lowery, F. J., Krishna, S., Yossef, R., Parikh, N. B., Chatani, P. D., Zacharakis, N., et al. (2022). Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Sci. (New York, NY) 375 (6583), 877–884. doi:10.1126/science.abl5447
Lytle, N. K., Ferguson, L. P., Rajbhandari, N., Gilroy, K., Fox, R. G., Deshpande, A., et al. (2019). A multiscale map of the stem cell state in pancreatic adenocarcinoma. Cell 177 (3), 572–586. doi:10.1016/j.cell.2019.03.010.e522
Mariathasan, S., Turley, S. J., Nickles, D., Castiglioni, A., Yuen, K., Wang, Y., et al. (2018). TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554 (7693), 544–548. doi:10.1038/nature25501
McNutt, M. (2013). Cancer immunotherapy. Sci. (New York, NY) 342 (6165), 1417. doi:10.1126/science.1249481
Michelini, S., Ricci, M., Serrani, R., Stuppia, L., Beccari, T., Veselenyiova, D., et al. (2021). Possible role of the RORC gene in primary and secondary lymphedema: Review of the literature and genetic study of two rare causative variants. Lymphat. Res. Biol. 19 (2), 129–133. doi:10.1089/lrb.2020.0030
Nakahata, S., Saito, Y., Marutsuka, K., Hidaka, T., Maeda, K., Utsunomiya, A., et al. (2011). Clinical significance of CADM1/TSLC1/IgSF4 expression in adult-T cell leukemia/lymphoma (ATLL): Identification of various types of ATLL cells. Retrovirology (Auckl). 8, A49. doi:10.1186/1742-4690-8-s1-a49
Oh, T. G., Bailey, P., Dray, E., Smith, A. G., Goode, J., Eriksson, N., et al. (2014). PRMT2 and RORγ expression are associated with breast cancer survival outcomes. Mol. Endocrinol. 28 (7), 1166–1185. doi:10.1210/me.2013-1403
Paijens, S. T., Vledder, A., de Bruyn, M., and Nijman, H. W. (2021). Tumor-infiltrating lymphocytes in the immunotherapy era. Cell. Mol. Immunol. 18 (4), 842–859. doi:10.1038/s41423-020-00565-9
Powers, R. K., Goodspeed, A., Pielke-Lombardo, H., Tan, A. C., and Costello, J. C. (2018). GSEA-InContext: Identifying novel and common patterns in expression experiments. Bioinforma. Oxf. Engl. 34 (13), i555–i564. doi:10.1093/bioinformatics/bty271
Qian, Z., Qingshan, C., Chun, J., Huijun, Z., Feng, L., Qiang, W., et al. (2014). High expression of TNFSF13 in tumor cells and fibroblasts is associated with poor prognosis in non-small cell lung cancer. Am. J. Clin. Pathol. 141 (2), 226–233. doi:10.1309/AJCP4JP8BZOMHEAW
Riley, R. S., June, C. H., Langer, R., and Mitchell, M. J. (2019). Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 18 (3), 175–196. doi:10.1038/s41573-018-0006-z
Rizzo, A., Ricci, A. D., and Brandi, G. (2021). PD-L1, TMB, MSI, and other predictors of response to immune checkpoint inhibitors in biliary tract cancer. Cancers 13 (3), 558. doi:10.3390/cancers13030558
Romero, D. (2019). TMB is linked with prognosis. Nat. Rev. Clin. Oncol. 16 (6), 336. doi:10.1038/s41571-019-0206-4
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). Tisidb: An integrated repository portal for tumor-immune system interactions. Bioinforma. Oxf. Engl. 35 (20), 4200–4202. doi:10.1093/bioinformatics/btz210
Saravia, J., Chapman, N. M., and Chi, H. (2019). Helper T cell differentiation. Cell. Mol. Immunol. 16 (7), 634–643. doi:10.1038/s41423-019-0220-6
Siska, P. J., Johnpulle, R. A. N., Zhou, A., Bordeaux, J., Kim, J. Y., Dabbas, B., et al. (2017). Deep exploration of the immune infiltrate and outcome prediction in testicular cancer by quantitative multiplexed immunohistochemistry and gene expression profiling. Oncoimmunology 6 (4), e1305535. doi:10.1080/2162402X.2017.1305535
Subramanian, K., Dierckx, T., Khouri, R., Menezes, S. M., Kagdi, H., Taylor, G. P., et al. (2019). Decreased RORC expression and downstream signaling in HTLV-1-associated adult T-cell lymphoma/leukemia uncovers an antiproliferative IL17 link: A potential target for immunotherapy? Int. J. Cancer 144 (7), 1664–1675. doi:10.1002/ijc.31922
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Talukdar, J., Kataki, K., Ali, E., Choudhury, B. N., Baruah, M. N., Bhattacharyya, M., et al. (2021). Altered expression of TGF-β1 and TGF-βR2 in tissue samples compared to blood is associated with food habits and survival in esophageal squamous cell carcinoma. Curr. Probl. Cancer 45 (1), 100617. doi:10.1016/j.currproblcancer.2020.100617
Veremeyko, T., Yung, A. W. Y., Dukhinova, M., Kuznetsova, I. S., Pomytkin, I., Lyundup, A., et al. (2018). Cyclic AMP pathway suppress autoimmune neuroinflammation by inhibiting functions of encephalitogenic CD4 T cells and enhancing M2 macrophage polarization at the site of inflammation. Front. Immunol. 9, 50. doi:10.3389/fimmu.2018.00050
Wang, H., Liu, B., and Wei, J. (2021). Beta2-microglobulin(B2M) in cancer immunotherapies: Biological function, resistance and remedy. Cancer Lett. 517, 96–104. doi:10.1016/j.canlet.2021.06.008
Wang, R., Guo, Y., Ma, H., Feng, L., Wang, Q., Chen, X., et al. (2016). Tumor necrosis factor superfamily member 13 is a novel biomarker for diagnosis and prognosis and promotes cancer cell proliferation in laryngeal squamous cell carcinoma. Tumour Biol. 37 (2), 2635–2645. doi:10.1007/s13277-015-4016-8
Williams, P. A., Cao, S., Yang, D., and Jennelle, R. L. (2020). Patient-reported outcomes of the relative severity of side effects from cancer radiotherapy. Support. Care Cancer 28 (1), 309–316. doi:10.1007/s00520-019-04820-2
Wörnle, M., Sauter, M., Kastenmüller, K., Ribeiro, A., Roeder, M., Mussack, T., et al. (2009). Role of viral induced vascular endothelial growth factor (VEGF) production in pleural effusion and malignant mesothelioma. Cell Biol. Int. 33 (2), 180–186. doi:10.1016/j.cellbi.2008.10.011
Wu, T., and Dai, Y. (2017). Tumor microenvironment and therapeutic response. Cancer Lett. 387, 61–68. doi:10.1016/j.canlet.2016.01.043
Wuerfel, F. M., Huebner, H., Häberle, L., Gass, P., Hein, A., Jud, S. M., et al. (2020). HLA-G and HLA-F protein isoform expression in breast cancer patients receiving neoadjuvant treatment. Sci. Rep. 10 (1), 15750. doi:10.1038/s41598-020-72837-3
Yahia-Cherbal, H., Rybczynska, M., Lovecchio, D., Stephen, T., Lescale, C., Placek, K., et al. (2019). NFAT primes the human RORC locus for RORγt expression in CD4(+) T cells. Nat. Commun. 10 (1), 4698. doi:10.1038/s41467-019-12680-x
Yang, X., Han, H., De Carvalho, D. D., Lay, F. D., Jones, P. A., and Liang, G. (2014). Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 26 (4), 577–590. doi:10.1016/j.ccr.2014.07.028
Yoshihara, K., Shahmoradgoli, M., Martínez, E., Vegesna, R., Kim, H., Torres-Garcia, W., et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. doi:10.1038/ncomms3612
Zhang, Q., Ni, Y., Zhi, X., Wang, J., Li, Z., Tang, J., et al. (2021). Involvement of APRIL in Helicobacter pylori-related gastric cancer. J. Cancer Res. Clin. Oncol. 147 (6), 1685–1697. doi:10.1007/s00432-021-03574-x
Keywords: RAR-related orphan receptor C, pan-cancer, prognosis, immune microenvironment, immunotherapy
Citation: He S, Yu J, Sun W, Sun Y, Tang M, Meng B, Liu Y and Li J (2022) A comprehensive pancancer analysis reveals the potential value of RAR-related orphan receptor C (RORC) for cancer immunotherapy. Front. Genet. 13:969476. doi: 10.3389/fgene.2022.969476
Received: 15 June 2022; Accepted: 29 August 2022;
Published: 15 September 2022.
Edited by:
Jinhui Liu, First Affiliated Hospital, Nanjing Medical University, ChinaReviewed by:
Katia Aviña-Padilla, University of Illinois at Urbana-Champaign, United StatesXing Jiahua, Chinese PLA General Hospital, China
Copyright © 2022 He, Yu, Sun, Sun, Tang, Meng, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Liu, bGl1eWFueWFuNzI1QDE2My5jb20=; Jiabin Li, bGlqaWFiaW5AYWhtdS5lZHUuY24=
†These authors have contributed equally to this work
 Shengfu He
Shengfu He Jiawen Yu
Jiawen Yu Weijie Sun
Weijie Sun Yating Sun
Yating Sun Mingyang Tang
Mingyang Tang Bao Meng
Bao Meng Yanyan Liu
Yanyan Liu Jiabin Li
Jiabin Li