
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 22 August 2022
Sec. Genomic Assay Technology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.967288
This article is part of the Research Topic Multi-Omics Analysis During the Animal Development View all 4 articles
 Liwei Sun1,2†
Liwei Sun1,2† Keya Tong1,2†
Keya Tong1,2† Weiwei Liu1,2
Weiwei Liu1,2 Yin Tian1,2
Yin Tian1,2 Sheng Yang1,2
Sheng Yang1,2 Danni Zhou1,2
Danni Zhou1,2 Dongyun Liu1,2
Dongyun Liu1,2 Guoning Huang1,2*
Guoning Huang1,2* Jingyu Li1,2*
Jingyu Li1,2*Background: This study aims to describe clinical and diagnostic phenotype and identify pathogenic variants of a female with unknown causes of infertility.
Methods: Clinical assessment was performed for the phenotype diagnosis. Whole-exome sequencing (WES) and the followed cDNA-PCR sequencing were applied to identify the pathogenic variant and investigate the potentially aberrant mRNA splicing event. The pathogenicity of the variant was analysed using multiple in silico prediction tools, including the 3D protein remodelling. Quantitative RT-PCR (qRT-PCR) was performed to measure PATL2 mRNA expression in the peripheral blood leukocytes of the proband and controls.
Results: The proband was diagnosed with the female infertility due to oocyte germinal vesicle (GV) arrest. A novel homozygous splice site variant of PATL2 (NM_001145112.2, c.871-1G>A), inherited from her asymptomatic heterozygous parents, was detected by WES. Sequencing of cDNA amplification products demonstrated that this variant resulted in the exon 10 skipping and in-frame loss of 54 nucleotides in the PATL2 transcript. Quantitative RT-PCR suggested that the mutant transcript escape the mRNA degradation.
Conclusion: We identified a novel pathogenic homozygous splice site of PATL2 (c.871-1G>A) underlying the oocyte GV arrest phenotype and elucidated its molecular mechanism. This study expands the variant spectrum of PATL2 and benefits our understanding of its genotype-phenotype correlations.
Infertility is a reproductive disorder that affects approximately 10–15% of couples worldwide (Maddirevula et al., 2020). A fundamental prerequisite for reproduction is oocyte maturation, which involves the stages from germinal vesicle (GV) to metaphase I (MI) and ultimately to metaphase II (MII) oocytes. Accordingly, oocyte maturation arrest (OMA) can occur at these different stages, including GV arrest, MI arrest, MII arrest, or mixed arrest, causing female infertility. OMA causes recurrent in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) failure; however, no therapeutic approaches are available due to our limited understanding of OMA causes (Beall et al., 2010). The phenotype of OMA was first described in 1990 (Rudak et al., 1990). The genetic causes underlying OMA were largely unknown until 2016, when variants in TUBB8 (MIM: 616768) were reported to cause oocyte MI arrest (Feng et al., 2016a; Feng et al., 2016b; Chen et al., 2017a). Subsequently, TRIP13 (MIM: 604507) was also identified as the causative gene for OMA (Zhang et al., 2020). It was also reported that variants in PADI6 (MIM: 610363) cause female infertility characterized by early embryonic arrest (Xu et al., 2016). Variants in WEE2 (MIM: 614084) are responsible for fertilization failure (Sang et al., 2018). Variants in BTG4 (MIM: 605673) can cause zygotic cleavage failure and female infertility (Zheng et al., 2020).
Moreover, variants in PATL2 (MIM: 614661; NM_001145112.2) can also cause female infertility. PATL2 is an orthologue of S. cerevisiae Pat1. PATL2 is located on 15q21.1 and encodes the protein PAT1 homologue 2, which is a mRNA-binding protein specifically expressed in immature oocytes that inhibits the post-transcriptional process in cells. As the oocyte matures, the expression of PATL2 gradually disappears (Chen et al., 2017b). To date, 26 PATL2 (NM_001145112.2) variants, including 21 nonsense variants and missense variants, 2 splicing variants, and 3 small deletions have been identified. However, pathogenic variants in PATL2 were identified to result in GV arrest initially (Chen et al., 2017b; Maddirevula et al., 2017; Christou-Kent et al., 2018; Huang et al., 2018; Liu et al., 2020), and variable phenotypes were subsequently reported, including GV arrest, MI arrest, fertilization failure, and early embryonic arrest, indicating its high phenotypic heterogeneity (Wu et al., 2019). Although cases involving PATL2 variants are accumulating, their molecular and pathophysiological mechanisms remains largely unknown. Therefore, more cases need to be investigated to illustrate phenotype-genotype relations and understand the molecular characteristics of PATL2 variants.
Here, a novel homozygous splice site variant in PATL2 that causes GV arrest was identified, and we revealed that this variant can result in exon skipping via aberrant splicing. Our study expands the variant spectrum of PATL2 and benefits our understanding of its genotype-phenotype correlations and molecular aetiology.
In our study, the patients of primary female infertility with unknown reasons and control individuals (women with normal fertility) were recruited. All cases and control individuals with normal fertility were obtained from the Women and Children’s Hospital of Chongqing Medical University. Clinical information and peripheral blood samples were collected from the family after obtaining individual written informed consent. Genetic testing was performed in accordance with the Helsinki Declaration and approved by the ethics committee of Women and Children’s Hospital of Chongqing Medical University.
Genomic DNA was extracted from peripheral blood samples using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the standard protocol. To detect the genetic causes of female infertility with oocyte germinal vesicle arrest, genetic variants were screened by whole-exome sequencing (WES) and confirmed by Sanger sequencing. Briefly, genomic DNA was captured using the Agilent SureSelectXT Human All Exon kit (Agilent Technologies, CA, United States) and sequenced on the MGISEQ-2000 platform with a 100 X read depth. Paired-end reads were aligned to the GRCh37/hg19 reference sequence using Burrows Wheeler Aligner software (BMA: version 0.7.8-r455). Variant calling was performed using SAMtools software (version 1.0). After variant detection, Annotate Variation (ANNOVAR) was used for variant annotation. Pathogenicity analysis of variants was performed according to the American College of Medical Genetics and Genomics (ACMG) practice guidelines. Variants were filtered based on the following criteria: 1) occurring in coding regions and/or splice sites; 2) nonsynonymous; 3) a frequency of less than 0.1% [Single Nucleotide Polymorphism database (dbSNP), Exome Variant Server, Genome Aggregation Database (gnomAD)]; 4) segregation with the phenotype. Sanger sequencing was used to verify the variants we screened and analyse the segregation data in the family. Pathogenicity prediction results of computational software (Human Splicing Finder 3.1 software (HSF), PROVEAN, MutationTaster, and CADD) were used to evaluate the effects of the variants on splicing.
Total RNA was isolated from the peripheral blood leukocytes of the proband and control individuals by TRIzol LS reagent (Invitrogen, CA, United States). RNA was reverse-transcribed using a TaKaRa PrimeScript reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. To investigate the abnormal splicing of the variant of PATL2, we amplified PATL2 cDNA using primers spanning exons 8 to 15 in the proband and a normal control individual (NC). Then, the obtained PCR products were analysed by gel electrophoresis on a 1% agarose gel, and the further Sanger sequencing was performed on the cDNA amplified product. The PCR products were sequenced using ABI BigDye3.1 (Applied Biosystems, Foster City, CA, USA) and analysed using an ABI 3730XL sequencer.
Quantitative RT–PCR (qRT–PCR) was conducted in a CFX96 real-time PCR detection system (Bio–Rad, Hercules, CA) by a final volume of 20 μl using SYBR Premix Ex Taq (Takara, Dalian, China) according to the manufacturer’s protocol. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was served as an endogenous control. The relative mRNA expression levels were calculated using the 2−ΔΔCt method. All reactions were run in triplicate at a minimum, and data are presented as the means ± SD. Information on amplification for PATL2 genomic splicing variant validation, cDNA sequencing and qPCR primers are shown in Supplementary Table S1.
The 3D modelled structures of the PATL2 protein for the wild-type and mutant types were prepared using homology modelling in SWISS-MODEL (https://swissmodel.expasy.org/). Structural analysis and attribution of the residue interaction networks to the protein function were analysed and visualized using the PyMOL software (https://pymol.org/2/).
An individual diagnosed with primary infertility was recruited. The proband (II-2) was 34 years old and suffered from primary infertility for 9 years. Infertility-related examination did not reveal any physical abnormalities. Then, 225 IU of recombinant human follitropin was administered, and gonadotropin (Gn) was administered for 9 days. In addition, nine follicles with a diameter of at least 14 mm and one follicle with a diameter of 13 mm were achieved after human chorionic gonadotrophin (HCG) trigger administration, and eight oocytes were obtained. However, seven of them were arrested at the GV stage, and one oocyte was degenerated (Table 1).
A novel homozygous splice site variant in the acceptor splice site of intron nine of PATL2 (c.877-1G>A, NM_001145112.2) was detected in the proband, the WES analysis procedure was shown (Figure 1). Her father and mother were heterozygous carriers of the identified splice site variant (Figure 2A,B). The oocytes characteristics of IVF attempt of the proband was shown (Figure 2C). Locations of the splice site variant was denoted in the genomic structure and protein domains of PATL2. The PATL2 splicing variant has a minor allele frequency of 6.76 × 10–6 within the global population and a minor allele frequency of 9.78 × 10–5 within the East Asian population in the gnomAD. The variant was considered deleterious by pathogenicity analysis using several in silico prediction tools, including HSF, MutationTaster, and CADD, which also predicted that the variant destroyed the acceptor site and most likely affected splicing. This variant detected in affected individual is also evolutionarily conserved across different species (Figure 2D).
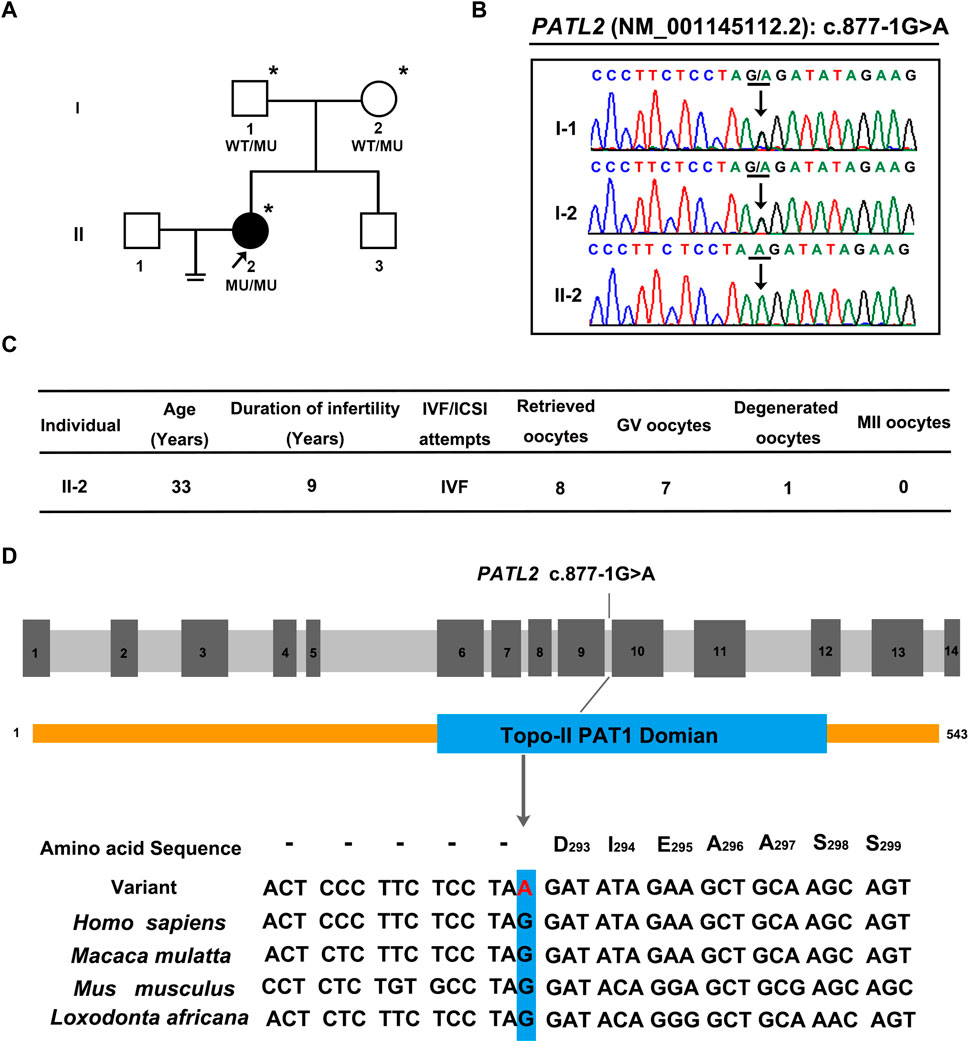
FIGURE 2. Pedigrees, clinical characteristics, and detection of variants in the female infertility family with oocyte germinal vesicle arrest. (A) Pedigrees of the affected family. The squares, circles, blackened, and open symbols indicate males, females, and affected and unaffected individuals, respectively. The arrows indicate the proband (II-2), and the asterisks denote the individuals who underwent genotyping. (B) Sequencing chromatograms of affected individuals who were homozygous for PATL2 variant. Her father and mother were heterozygous carriers of the identified splicing variant. The black arrows indicate the sites of the variants. (C) clinical characteristic of affected individual and the retrieved oocytes in the IVF cycle. (D) The detected variant (C)871-1G>A in PATL2 (MIM: 614661; GenBank: NM_001145112.2) in the affected family is evolutionarily conserved across different species. The site of PATL2 variant is indicated in blue rectangle.
To investigate the abnormal splicing result of PATL2 mRNA, Sanger sequencing of the PATL2 cDNA was performed, and the exon 10 skipping was observed (Figure 3A). The aberrant splicing of this novel splice site variant was also denoted (Figure 3B). Furthermore, three pairs of PATL2 primers were designed for qPCR, including one pair of primer downstream at the exon 10 region, and two pairs of primers spanning the N-termination and C-termination which do not include the variant region, respectively. As a result, no detectable mRNA expression level was showed in the proband at the exon 10 region, and the splicing variant would not introduce premature translation stop codons or result in PATL2 mRNA decay, which further confirmed the abnormal exon 10 skipping results (Figure 3C).
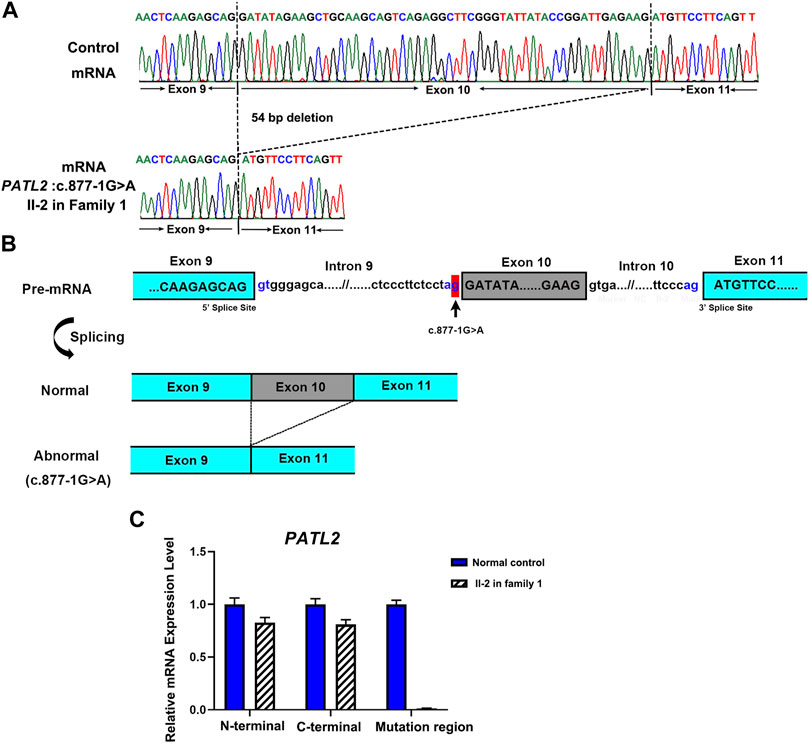
FIGURE 3. Functional analysis of the identified novel PATL2 splice site variant. (A) Sanger sequencing traces of the RT–PCR products showed the wild-type (normal control, top) and PATL2 exon 10 skipped transcripts (II-2 in the family, bottom). (B) Schematic representation of exon 9 to exon 11 of PATL2 showing normal or abnormal splicing. The boxed regions denote exons, whereas the connecting lines indicate introns. (C) Quantitative RT–PCR (qRT–PCR) of the relative PATL2 mRNA expression in peripheral blood lymphocytes from the affected individual (II-2) and the normal female controls (NC). For all qRT–PCR assays, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. NC was set to 1.0, and data are presented as the mean ± SD (n ≥ 3).
To explore the phenotype-genotype relationship, we located the reported PATL2 variants in the PATL2 protein and summarized genotype-phenotype correlations based on previous reports (Figure 4A). In addition to the result that the homozygous splicing variant c.871-1G>A causes the exon 10 skipping at the mRNA level, protein structure modelling also revealed that the alteration in the three-dimensional positioning of the splicing variant causes steric hindrance to the formation of α-helix structures in the PATL2 protein, which are highlighted in pink (Figure 4B). The PATL2 variants reported and the phenotype including the ratio of GV arrest were characterized (Table 2). Effect of reported PATL2 variants on the protein structure were also modelled (Figure 4C).
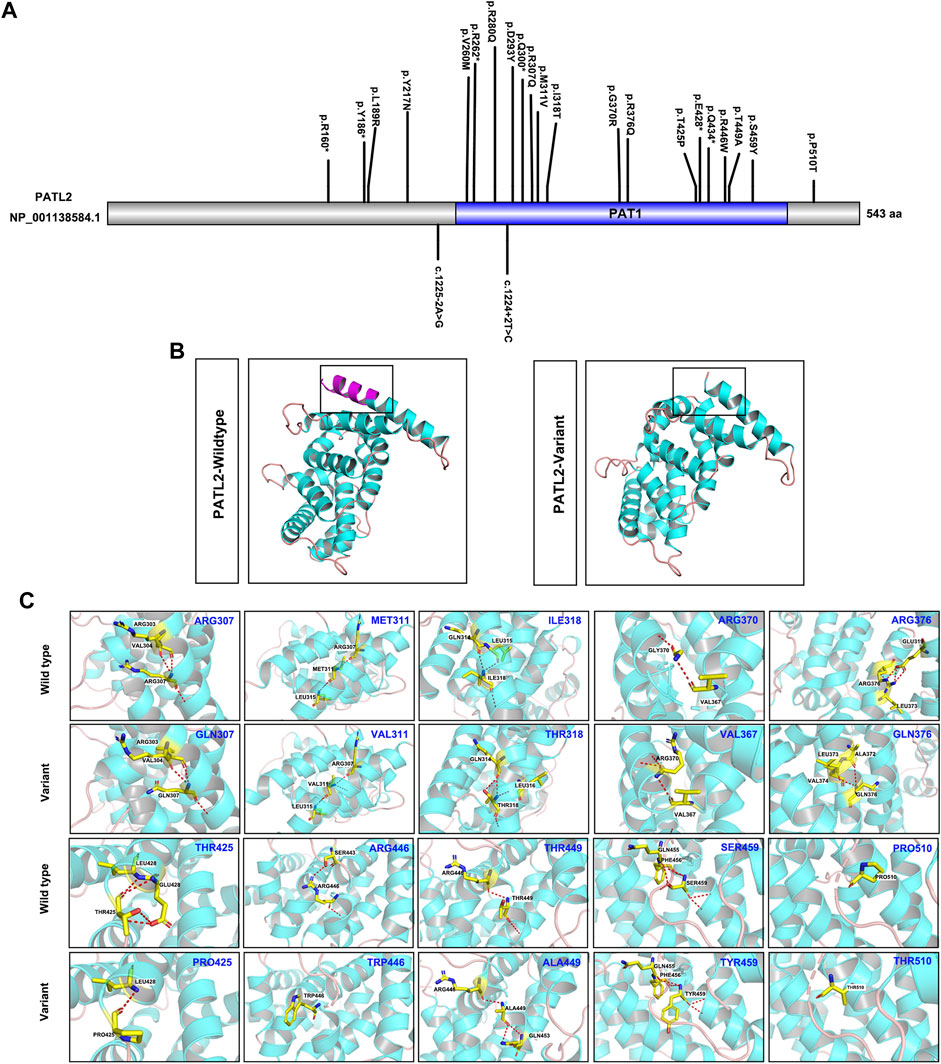
FIGURE 4. Schematic diagrams of the locations of reported PATL2 variants and molecular modelling of wild-type and mutant PATL2 proteins. (A) The variants marked are the PATL2 variants reported to date, and the novel variant identified in the present study is marked and indicated in red. (B) Three-dimensional schematic of the structure of normal PATL2 protein (on the left) and the mutant PATL2 (on the right). A 18-amino-acid in-frame deletion p. (Asp293_Lys310) in the PAT1 domain of the PATL2 protein is the putative result of the homozygous splicing variant (C)871-1G>A. The alteration in the three-dimensional positioning of the splicing variant causes steric hindrance to the formation of α-helix structures in the PATL2 protein, which are highlighted in pink. (C) Three-dimensional schematic of the structure of the previous reported missense variants of PATL2.
Several clinical reports on pathogenic variants of PATL2 in OMA patients have been published. In our study, the majority of the oocytes retrieved from the proband were arrested at the GV stage, which was consistent with the typical phenotype from previously reported PATL2 variants (Chen et al., 2017b; Maddirevula et al., 2017; Christou-Kent et al., 2018; Huang et al., 2018; Wu et al., 2019; Liu et al., 2020; Cao et al., 2021). To date, only two splicing variants have been reported, including c.1225-2A>G and c.1224 + 2T>C. The splicing variant c.1225-2A>G was predicted to abolish the canonical splice acceptor site of exon 13, and c.1224 + 2T>C was predicted to contribute to PATL2 protein truncation (Chen et al., 2017b; Huang et al., 2018). The two reported splicing variants were all compound heterozygous variants. To the best of our knowledge, we report a novel homozygous splicing variant for the first time. To understand the molecular pathogenesis of this novel PATL2 splicing variant of c.877-1G>A, we demonstrated that this variant may cause exon 10 of PATL2 to be skipped at the mRNA level and lead to a 18-amino-acid in-frame deletion p. (Asp293_Lys310) in the PAT1 domain of the PATL2 protein, which may result in loss of the RNA-binding ability. Additionally, we also confirmed that this variant did not introduce a premature termination codon (PTC) or trigger nonsense-mediated decay (NMD) of the truncated mRNA. Immunostaining analysis using oocytes from the affected individual may help to evaluate the possible effects of the novel variant on the PATL2 protein level. However, it is unfortunate that no oocyte was available for such analyses in the present study.
PATL2 was initially regarded as a mRNA-binding protein (mRNP) associated with other mRNPs, such as Xp54, xRAP55, and CPEB (Radford et al., 2008; Nakamura et al., 2010). Previous studies have reported that variants in PATL2 lead to female infertility with oocyte maturation arrest; however, the mechanisms by which PATL2 variants affect meiotic maturation remain unclear and controversial. It was confirmed that affected individuals with PATL2 variants harbour loss-of-function variants, and the oocytes of some individuals showed decreased on PATL2 protein levels. These findings imply that the variants cause accelerated protein degradation instead of activation (Chen et al., 2017b). Another study recently revealed that some PATL2 variants lead to abnormally increased PATL2-bound mRNAs in Mos, an upstream activator of mitogen-activated protein kinase (MAPK), to regulate its translational activity and subsequently impair the MAPK signalling pathway and oocyte meiosis (Cao et al., 2021). These results highlighted the role of PATL2 in translational regulation of Mos and its association with the MAPK signalling pathway during oocyte meiotic maturation. Subsequently, we analysed the clinical characteristics of the cases in the present study and published cases associated with PATL2 variants. Therefore, combined with our finding that the novel homozygous variants c.877-1G>A in PATL2 reduce PATL2 mRNA expression levels, we inferred that the diverse pathogenic mechanisms of PATL2 over expression, deletion or bidirectional effects of PATL2 may be involved in the oocyte maturation process. These hypotheses are worthy of further investigation to elucidate the genetic mechanisms involved.
In conclusion, we have identified a novel homozygous splice site variant of PATL2 in a female individual affected by oocyte maturation arrest with GV arrest. We also provided evidence for the effects of the splice site variant on PATL2 mRNA expression levels to molecularly characterize its role in the oocyte GV arrest phenotype. Our study expands the variant spectrum of oocyte maturation arrest patients with PATL2 variants and benefits our understanding of genotype-phenotype correlations of PATL2.
The data presented in the study are deposited in the Genome Sequence Archive (GSA) repository, accession number subHRA003831.
The studies involving human participants were reviewed and approved by the Chongqing Health Center for Women and Children, Women and Children’s Hospital of Chongqing Medical University. The patients/participants provided their written informed consent to participate in this study.
LS and KT designed the study and performed the experiments. WL, YT, and SY collected the clinical samples. KT, DZ, and DL analysed the clinical data of patients. JL, LS, and KT conducted the manuscript writing with the help from all authors. JL and GH conceived the study and supervised the study progress. All authors read and approved the final manuscript.
This study was supported by the Science Foundation for Post Doctorate of Women and Children’s Hospital of Chongqing Medical University and the General Project of Chongqing Health Center for Women and Children, Women and Children’s Hospital of Chongqing Medical University (2021YJMS05).
We would like to thank all the individuals for their collaboration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.967288/full#supplementary-material
Beall, S., Brenner, C., and Segars, J. (2010). Oocyte maturation failure: A syndrome of bad eggs. Fertil. Steril. 94 (7), 2507–2513. doi:10.1016/j.fertnstert.2010.02.037
Cao, Q., Zhao, C., Wang, C., Cai, L., Xia, M., Zhang, X., et al. (2021). The recurrent mutation in PATL2 inhibits its degradation thus causing female infertility characterized by oocyte maturation defect through regulation of the mos-MAPK pathway. Front. Cell Dev. Biol. 9, 628649. doi:10.3389/fcell.2021.628649
Chen, B., Li, B., Li, D., Yan, Z., Mao, X., Xu, Y., et al. (2017a). Novel mutations and structural deletions in TUBB8: Expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum. Reprod. 32 (2), 457–464. doi:10.1093/humrep/dew322
Chen, B., Zhang, Z., Sun, X., Kuang, Y., Mao, X., Wang, X., et al. (2017b). Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am. J. Hum. Genet. 101 (4), 609–615. doi:10.1016/j.ajhg.2017.08.018
Christou-Kent, M., Kherraf, Z. E., Amiri-Yekta, A., Le Blevec, E., Karaouzene, T., Conne, B., et al. (2018). PATL2 is a key actor of oocyte maturation whose invalidation causes infertility in women and mice. EMBO Mol. Med. 10 (5), e8515. doi:10.15252/emmm.201708515
Feng, R., Sang, Q., Kuang, Y., Sun, X., Yan, Z., Zhang, S., et al. (2016a). Mutations in TUBB8 and human oocyte meiotic arrest. N. Engl. J. Med. 374 (3), 223–232. doi:10.1056/NEJMoa1510791
Feng, R., Yan, Z., Li, B., Yu, M., Sang, Q., Tian, G., et al. (2016b). Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J. Med. Genet. 53 (10), 662–671. doi:10.1136/jmedgenet-2016-103891
Huang, L., Tong, X., Wang, F., Luo, L., Jin, R., Fu, Y., et al. (2018). Novel mutations in PATL2 cause female infertility with oocyte germinal vesicle arrest. Hum. Reprod. 33 (6), 1183–1190. doi:10.1093/humrep/dey100
Liu, Z., Zhu, L., Wang, J., Luo, G., Xi, Q., Zhou, X., et al. (2020). Novel homozygous mutations in PATL2 lead to female infertility with oocyte maturation arrest. J. Assist. Reprod. Genet. 37 (4), 841–847. doi:10.1007/s10815-020-01698-6
Maddirevula, S., Awartani, K., Coskun, S., AlNaim, L. F., Ibrahim, N., Abdulwahab, F., et al. (2020). A genomics approach to females with infertility and recurrent pregnancy loss. Hum. Genet. 139 (5), 605–613. doi:10.1007/s00439-020-02143-5
Maddirevula, S., Coskun, S., Alhassan, S., Elnour, A., Alsaif, H. S., Ibrahim, N., et al. (2017). Female infertility caused by mutations in the oocyte-specific translational repressor PATL2. Am. J. Hum. Genet. 101 (4), 603–608. doi:10.1016/j.ajhg.2017.08.009
Nakamura, Y., Tanaka, K. J., Miyauchi, M., Huang, L., Tsujimoto, M., and Matsumoto, K. (2010). Translational repression by the oocyte-specific protein P100 in Xenopus. Dev. Biol. 344 (1), 272–283. doi:10.1016/j.ydbio.2010.05.006
Radford, H. E., Meijer, H. A., and de Moor, C. H. (2008). Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 1779 (4), 217–229. doi:10.1016/j.bbagrm.2008.02.002
Rudak, E., Dor, J., KiMchiM., , Goldman, B., Levran, D., and MaShiach, S. (1990). Anomalies of human oocytes from infertile women undergoing treatment by in vitro fertilization. Fertil. Steril. 54 (2), 292–296. doi:10.1016/s0015-0282(16)53706-6
Sang, Q., Li, B., Kuang, Y., Wang, X., Zhang, Z., Chen, B., et al. (2018). Homozygous mutations in WEE2 cause fertilization failure and female infertility. Am. J. Hum. Genet. 102 (4), 649–657. doi:10.1016/j.ajhg.2018.02.015
Wu, L., Chen, H., Li, D., Song, D., Chen, B., Yan, Z., et al. (2019). Novel mutations in PATL2: Expanding the mutational spectrum and corresponding phenotypic variability associated with female infertility. J. Hum. Genet. 64 (5), 379–385. doi:10.1038/s10038-019-0568-6
Xu, Y., Shi, Y., Fu, J., Yu, M., Feng, R., Sang, Q., et al. (2016). Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am. J. Hum. Genet. 99 (3), 744–752. doi:10.1016/j.ajhg.2016.06.024
Zhang, Z., Li, B., Fu, J., Li, R., Diao, F., Li, C., et al. (2020). Bi-Allelic missense pathogenic variants in TRIP13 cause female infertility characterized by oocyte maturation arrest. Am. J. Hum. Genet. 107 (1), 15–23. doi:10.1016/j.ajhg.2020.05.001
Keywords: female infertility, germinal vesicle arrest, PATL2, novel variant, aberrant splicing
Citation: Sun L, Tong K, Liu W, Tian Y, Yang S, Zhou D, Liu D, Huang G and Li J (2022) Identification and characterization of a novel homozygous splice site variant of PATL2 causing female infertility due to oocyte germinal vesicle arrest. Front. Genet. 13:967288. doi: 10.3389/fgene.2022.967288
Received: 12 June 2022; Accepted: 13 July 2022;
Published: 22 August 2022.
Edited by:
Duancheng Wen, Cornell University, United StatesReviewed by:
Yiren Qin, Icahn School of Medicine at Mount Sinai, United StatesCopyright © 2022 Sun, Tong, Liu, Tian, Yang, Zhou, Liu, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoning Huang, Z25odWFuZzIxN0BzaW5hLmNvbQ==; Jingyu Li, Y3F0bmxqeUBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.