- 1Fujian Key Laboratory on Conservation and Sustainable Utilization of Marine Biodiversity, College of Geography and Oceanography, Minjiang University, Fuzhou, China
- 2Longyan Fisheries Technology Extension Center, Longyan, China
- 3Liancheng Jiming Fish Farming Company, Longyan, China
Xenocyprinae is a cyprinid subfamily that not only has a discrete geographic distribution but also has a long history dating to the Early Miocene. However, it is controversial whether systematic classification and some species validity of Xenocyprinae exist, as well as its phylogenetic relationships and evolutionary history. In the present study, we first reviewed the description and taxonomic history of Xenocyprinae, and then the complete mitochondrial genome of Distoechodon compressus, an endemic and locally distributed species belonging to Xenocyprinae, was sequenced and annotated. Finally, all the mitogenomes of Xenocyprinae were compared to reconstruct the phylogenetic relationship and estimate the divergence time. The results showed that the mitogenomes are similar in organization and structure with 16618–16630 bp length from 12 mitogenomes of eight species. Phylogenetic analysis confirmed the monology of Xenocyprinae and illustrated three clades within the Xenocyprinae to consist of ambiguous generic classification. Plagiognathops is a valid genus located at the base of the phylogenetic tree. The genus Xenocypris was originally monophyletic, but X. fangi was excluded. Divergence time estimation revealed that the earliest divergence within Xenocyprinae occurred approximately 12.1 Mya when Plagiognathops separated from the primitive Xenocypris. The main two clades (Xenocypris and (Distoechodon + Pseudobrama + X. fangi)) diverged 10.0 Mya. The major divergence of Xenocyprinae species possibly occurred in the Middle to Late Miocene and Late Pliocene, suggesting that speciation and diversifications could be attributed to the Asian monsoon climate. This study clarifies some controversial issues of systematics and provides essential information on the taxonomy and phylogeny of the subfamily Xenocyprinae.
Introduction
Xenocyprinae is a cyprinid subfamily that not only has a discrete geographic distribution (restricted to East Asia, especially in China) but also has a long history (its extant genera and species became dominant and continued to develop in the beginning of the Early Pliocene) (Chang et al., 1996). The subfamily Xenocyprinae made it possible to distinguish populations and species in response to both profound and more recent historical environmental changes in East Asia (Xiao et al., 2012). Moreover, many of the species have been domesticated into excellent aquacultural varieties with high economic value (Lin et al., 2021), and they could also be used for ecological restoration of rivers to improve water quality and control the algae bloom in freshwater because most of them graze on benthic algae and debris (Zhang et al., 2015). Hence, the taxonomic status, historical evolution, and phylogeny of Xenocyprinae have become subjects of increasing concern.
The subfamily Xenocyprinae was established by Günther (1868) and subsequently discussed in terms of systematics and phylogeny. With further studies on the systematic status, different views on its classification have emerged. It appeared to be similar to Chondrostoma because of the sharp horny beak of the lower jaw and six main teeth of the lower pharyngeal teeth (Chu, 1935; Yang, 1964). However, Howes (1981) gave a contrary report because the superior pharyngeal process handle of the basal occipital was different, with many different signs and no overlap in distribution. Chen et al. (1984) incorporated Hypophthalmichthyinae into Xenocyprinae based on skeletal characteristics. The latest version of fishes of the world also supported that Xenocyprinae included Xenocypris, Hypophthalmichthys, and Aristichthys. In the current classification, Fauna Sinica is proposed to be a Xenocyprinae, including 10 species in four genera, as a sister group to Hypophthalmichthyinae (Liu, 1998). Zhao et al. (2009) reviewed them and proposed 12 species (previously 13, but Distoechodon multispinnis later seemed to be a subspecies of D. tumirostris) in four or five genera, which are the current systematics of Xenocyprinae. Molecular evidence of the subfamily Xenocyprinae is consistent with the results of previous studies (He et al., 2004; Tang et al., 2013), and the two main clusters and the genus Xenocyprioides are at the base of the phylogenetic trees. Furthermore, Xenocyprioides was established by Chen (1982) in Guangxi Province, China, but it is similar to Danioninae in morphological characteristics (Liu, 1998) and not closely related to other genera of Xenocyprinae based on phylogenetic analyses (Fang et al., 2009; Xiao et al., 2012) and differences in habitats and habits. Therefore, Xenocyprioides were not included in the present study.
Based on the results of the aforementioned molecular studies, there is still ambiguity regarding the phylogenetic positions and evolutionary history of some species, and the systematic questions have not been adequately answered after decades of discussion. For example, the genus status of D. hupeinensis remains controversial. Based on its morphology, early development, and substitution of pharyngeal teeth, Shan (1998) proposed the movement of D. hupeinensis from Distoechodon to Xenocypris and renamed it as X. hupeinensis. Phylogenetic analysis from mitochondrial evidence supports this point of view (Zhang et al., 2022). However, the current taxonomic characteristics between genera of Xenocyprinae were in accordance with the number of rows of lower pharyngeal teeth and folds of the pelvic fin base still supported it as the Distoechodon genus (Liu, 1998). Another controversy is whether Plagiognathops is a valid genus. Based on the three rows of pharyngeal teeth, X. microlepis was classified into the genus Xenocypris (Liu, 1998). However, Zhao et al. (2009) reclassified it as Plagiognathops because the lateral line scales were more than 70. Both views are supported by different morphological evidence and phylogenetic studies based on different molecular markers that show different topologies (He et al., 2004; Xiao et al., 2012; Zhang et al., 2022). Moreover, it is still debated whether D. compressus is a valid species in both morphological and molecular studies (support as a valid species: Yang, 1964; Cao and Meng, 1992; Liu, 2002a; Liu et al., 2021; Zhao et al., 2009; seem as just a subspecies: Bănărescu, 1970; He et al., 1989; Liu, 1998; Xiao et al., 2012). Although there is growing evidence to support the validity of D. compressus (Lin et al., 2020; Chen et al., 2021), there are no systematic characteristics or methods for classification, and even common molecular markers cannot distinguish it from its related species (D. tumirostris) (Zhang et al., 2022). Therefore, the direct aim of this study was to understand the validity of D. compressus and to further identify the variation in mitogenomes within Xenocyprinae.
Mitochondrial DNA (mtDNA) is a closed-circular molecule found in most animals and is independent of the nuclear genome. Animal mtDNA is generally small (15–20 kb) and contains 37 genes, 13 protein-coding genes (PCGs), 22 tRNAs, and 2 rRNAs (Macey et al., 1997; Satoh et al., 2016; Kobayashi et al., 2021). Mitochondria play an essential role in oxidative phosphorylation, which is essential for the production of ATP and a variety of other biochemical functions (Chaban et al., 2014). Mitochondrial DNA has the advantages of matrilineal inheritance and evolutionary conservation and has been used as a genetic marker to detect genetic differentiation and molecular evolution in population genetics (Avise, 2000; Miya and Nishida, 2015). Notably, the complete mtDNA genome has been proven to be more informative at deep phylogenetic levels (Kong et al., 2020; Wang et al., 2021) and is useful in recovering internal nodes with high statistical support compared to partial mtDNA genes (Yu et al., 2022). With rapid advances in sequencing technologies, complete mitogenome sequences are becoming available, providing abundant data on comparative analyses of mitogenomes and phylogenetic reconstruction (Shi et al., 2018). From single conservative protein-coding genes, such as Cox1 and Cyt b, to multi-gene combination, phylogenetic studies have entered the omics era based on complete mitochondrial genomes or even the whole genomes (Ellegren, 2014; Koepfli et al., 2015). Phylogenetic analyses based on mitogenome sequences often result in better resolution than analyses that use few gene sequences (Bleidorn et al., 2009; Kobayashi et al., 2021). At present, except D. compressus and D. hupeinensis, the complete mitochondrial genomes of other species of the subfamily Xenocyprinae have been reported (Saitoh et al., 2006; He et al., 2008; Liu et al., 2014; Hu et al., 2015a, 2015b; Xu et al., 2019; Xue et al., 2019; Liu et al., 2021). The same phylogenetic results were analyzed by complete mitogenomes, and X. fangi clustered with the genus Distoechodon (Liu et al., 2021). Hence, the phylogenetic relationships and speciation within the subfamily Xenocyprinae need further discussion.
In the present study, we systematically reviewed the description and taxonomic history of the subfamily Xenocyprinae (in Abstract). We then sequenced the complete mitogenome of D. compressus and compared the mitogenomes of Xenocyprinae to reconstruct the phylogenetic relationship and estimate divergence time among the subfamily Xenocyprinae. The aim of this study was to understand the phylogenetic relationships and evolutionary history of the subfamily Xenocyprinae, confirm the genus classification and the validity of D. compressus, and provide references for the systematics of cyprinid fish.
Materials and methods
Sampling, DNA extraction, polymerase chain reaction, and sequencing
Specimens were collected using gill nets from a tributary of the Ting River (116.62°E, 25.35°N) in Liancheng County, Fujian Province, China, from April to July 2021. Thirty specimens were identified following Zhao et al. (2009). Muscle tissues were preserved in 95% ethanol and stored at the Institute of Oceanology, Minjiang University. Total genomic DNA was extracted using the salt extraction method (Aljanabi and Martinez, 1997). The complete mitochondrial genome of D. compressus was amplified based on the two-step process (six universal primers and six specific primers described in Supplementary Table S1). Polymerase chain reaction (PCR) amplification was carried out in a 20 μL reaction following Zhang et al. (2021). The PCR products were examined using electrophoresis on a 1.0% TAE agarose gel and sequenced by Sangon Biotech (Shanghai, China).
Annotation, gene order, and genomic structure analyses
The overlapping fragments were assembled into a linear mitochondrial DNA sequence using SeqMan (DNASTAR), and the assembled sequences were manually checked (Jin and Sun, 2018). The complete mitochondrial genome sequences of Xenocyprinae species included all the Xenocyprinae fish that have been reported so far, representing four genera (species and accession numbers are listed in Table 1), except for Xenocyprioides. The complete mitochondrial genome sequences were downloaded from the GenBank with their features and structures. All the sequences were annotated using the MITOS web server (Bernt et al., 2013) and determined in MitoFish (http://mitofish.aori.utokyo.ac.jp/) to compare the genomic structure and gene order (Sato et al., 2018). Transferred RNA genes and their secondary structures were predicted using the tRNAscan SE web server (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Chan, 2016). The composition of amino acids, nucleotide bases, and relative synonymous codon usage (RSCU) was calculated using MEGA X software (Kumar et al., 2018). Variable sites among the sequences were also detected using MEGA software. Nucleotide composition skew was calculated using the following formulas: AT-skew = (A—T)/(A + T) and GC-skew = (G—C)/(G + C) (Perna and Kocher 1995).
Phylogenetic analysis
Phylogenetic analyses were reconstructed using maximum likelihood (ML) and Bayesian inference (BI) methods, based on concatenated nucleotide sequences of 13 PCGs from 12 individuals belonging to the subfamily Xenocyprinae, with C. carpio, H. molitrix, and A. nobilis as outgroups. The best-fit partition model of nucleotide evolution of PCGs was identified using PartitionFinder v2 (Lanfear et al., 2012) in the PhyloSuite platform (Zhang et al., 2020), and it was GTR + I + G according to the Akaike information criterion (Bozdogan 1987). ML and BI analyses were performed using MrBayes v3.2.7 and RaxML v8.2.12 programs following their manuals, respectively (Ronquist et al., 2012; Stamatakis 2014). Bootstrap ML analysis was implemented under the GTRGAMMAI model, and 1000 replications were used to evaluate the bootstrap support values and search for the best ML tree. BI analysis was run as two simultaneous Markov chain Monte Carlo (MCMC) chains for 10 million generations, with sampling of every 1000 generations, using a burn-in rate of 25%. Phylogenetic trees were visualized through the online tool Interactive Tree Of Life v5 (iTOL, https://itol.embl.de/) (Letunic and Bork 2021).
Divergence time estimation
Divergence time among the subfamilies was estimated using the amino acid sequences of 13 PCGs with a relaxed clock log-normal model in BEAST v.1.10 (Drummond and Rambaut, 2007). A model of the Yule process was selected for the tree prior. To estimate the divergence time calibration, two calibration points were used as priors for the divergence time of the corresponding splits. The most recent common ancestor of Xenocyprinae is dated to 15 million years ago (Mya), as described by Chang et al. (1996) and Chen et al. (2005). Also, the 18.80 Mya point calibration was set as the divergence time of Hypophthalmichthyinae from cyprinids (Wang et al., 2012). The final Markov chain was run twice for 100 million generations, with sampling of every 10,000 generations. The first 10% of generations were discarded as burn-in, according to the convergence of chains checked using Tracer v. 1.7, and the ESS values of all parameters were above 200. The maximum clade credibility tree was generated using TreeAnnotator v. 2.4.1 (part of the Beast package) and visualized in the iTOL web tool.
Results and Discussion
Mitogenome structure and organization
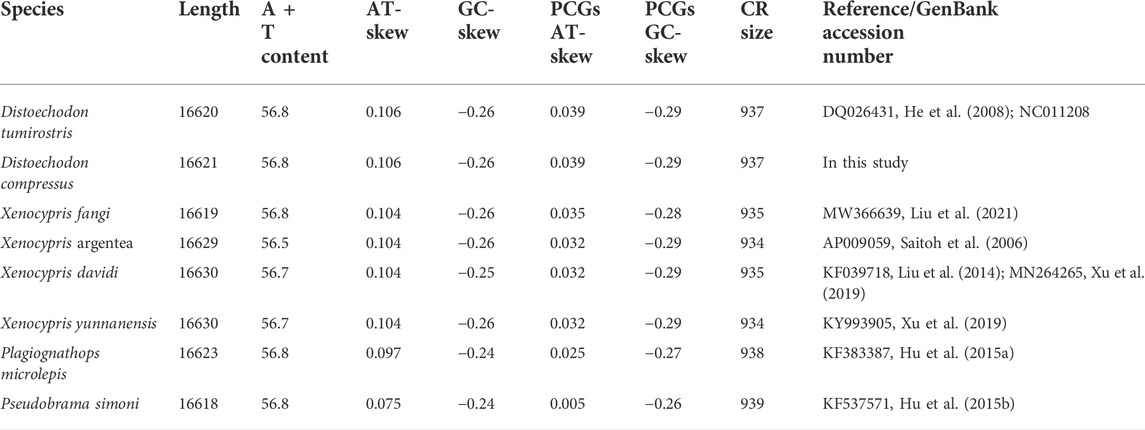
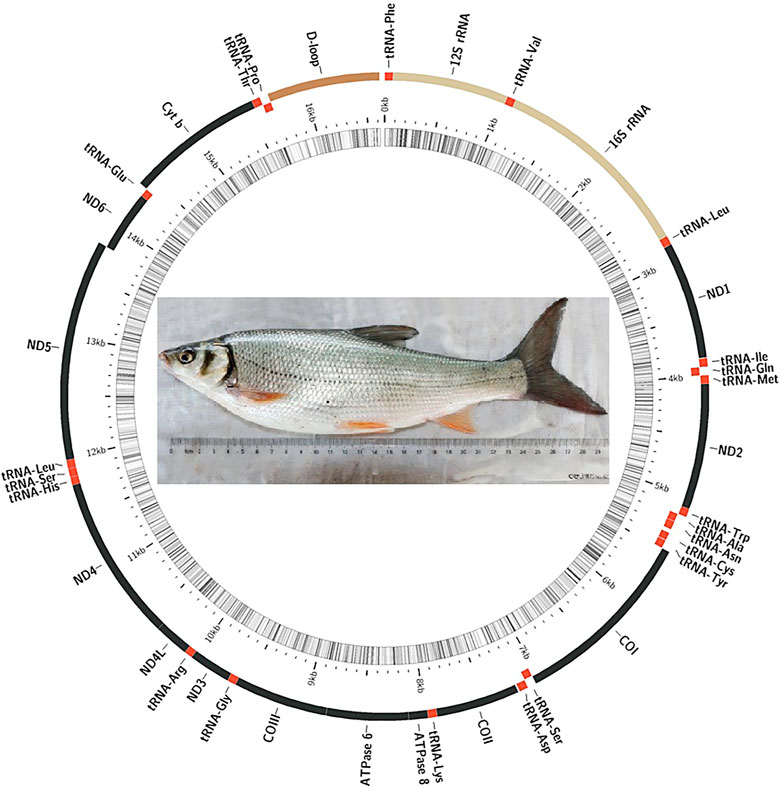
In the present study, the complete mitogenome of D. compressus was successfully sequenced and annotated. It was deposited in the GenBank under accession number OM994436. All the reported mitogenomes of Xenocyprinae were compared to analyze the similarities and differences in structure and organization between the sequences. The mitogenomes of nine xenocyprinid fish, ranging from 16623 bp to 16630 bp length, have 2511 variable sites, where 698 variable sites are singletons among them. The gene content and order were as expected for a typical vertebrate mitogenome which comprises 13 PCGs, 22 tRNA genes, 2 rRNA genes, and 1 non-coding control region. More than half of the genes were encoded on the heavy (H-) strand: 9 PCGs and 14 tRNAs (Figure 1). The bias toward high AT content (55.8%–58.8%) is consistent with general findings in teleost mitogenomes (Sharma et al., 2020; Sun et al., 2021; Yu et al., 2021). The average nucleotide composition of all sequences is A (31.2%), T (25.5%), G (16.2%), and C (27.1%). For the complete mitogenomes, AT-skew was all positive, ranging from 0.075 (P. simoni) to 0.106 (D. compressus and D. tumirostris), and GC-skew was slightly negative (−0.24 ∼ −0.26) (Table 1).
Protein-coding genes
Similar to other vertebrates, ATG is the regular initiation codon in almost all PCGs, except for Cox1, which uses GTG. However, many PCGs were not terminated with TAG or TAA but possessed an incomplete stop codon (six or seven genes), which was more than most of the mitogenomes of teleosts (Sun et al., 2021; Yu et al., 2021; Zhang et al., 2021). This can be corrected by posttranscriptional polyadenylation (Jondeung et al., 2007; Shi et al., 2018). Ranging from 11422 to 11429 bp, there was a slightly positive AT-skew (0.005–0.039) and negative GC-skew (−0.26 ∼ −0.29) (Table 1). This is consistent with the bias of using A rather than T in the PCGs of most teleosts, suggesting that there was no unique selective pressure or processes of the xenocyprinid species detected in several fish (Yu et al., 2021; Zhang et al., 2021). Amino acid codon usage of 11 mitogenomes was assessed by relative synonymous codon usage (RSCU) values. Leucine 2 (Leu2), threonine (Thr), and proline (Pro) were the most frequently translated amino acids, whereas glutamine (Glu) and aspartate (Asp) were the least used amino acids. CUA (Leu), AUU (Ile), and AAU (Asn) were codons commonly used in Xenocyprinae mitogenomes. The over-usage of A and T at the third position indicates the possibility of genome bias, optimal selection of tRNA, or DNA repair efficiency, referring to other teleosts (Fischer et al., 2013; Zhang et al., 2021).
Non-coding genes
The sizes of the 22 tRNA genes ranged from 68 bp to 76 bp, which comprised 9.4% of the complete mitogenomes. Most secondary structures of tRNAs were recovered as ordinal clover-leaf structures (21 of the 22 tRNAs), which included the amino acid-accepting stem (AAS), dihydrouridine stem and loop (DSL), anticodon stem and loop (ASL), thymidine stem and loop (TSL), and variable stem and loop (VSL). However, tRNASer (AGN) lacks the DHU arm, which is commonly observed in many metazoan mitogenomes (Frazen-Abel and Hagerman, 2008; Watanabe et al., 2014). The two rRNA genes were separated by tRNAVal, with lengths of 961–964 bp and 1688–1693 bp. Similarly, a strong AT-bias was also detected in rRNAs, with a strongly positive AT-skew (0.253–0.269) and a slightly negative GC-skew (−0.06 ∼ −0.08).
The length of the CR ranged from 930 to 938 bp, with a significantly higher A + T content (65.5%) and relatively low AT-skew (0.0046). The control region contains three domains: a termination-associated sequence (TAS), central conserved domain (CD), and conserved sequence block (CSB). The identified conserved sequences also exist in other Cypriniformes (Liu, 2002b; Xiao and Zhang, 2000; Shi et al., 2018). Miya and Nishida (2015) proposed that CR plays a crucial role in the initiation and regulation of transcription and replication. The TAS may terminate the synthesis of the heavy strand (Fischer et al., 2013), and the central conserved domains (CSB-F, CSB-E, and CSB-D) are thought to be associated with the positioning of RNA polymerase for both priming replication and transcription (Castresana, 2000). All the conserved blocks may maintain the function of the control region. We found that CSB2 was the most conserved. However, some domains (such as CSB-E) experienced rapid evolution 5 to 10 times that of the complete mtDNA. In this study, the variation in CR may be an important marker for distinguishing different species of Xenocyprinae.
Genetic distance and variation in mitochondrial sequences
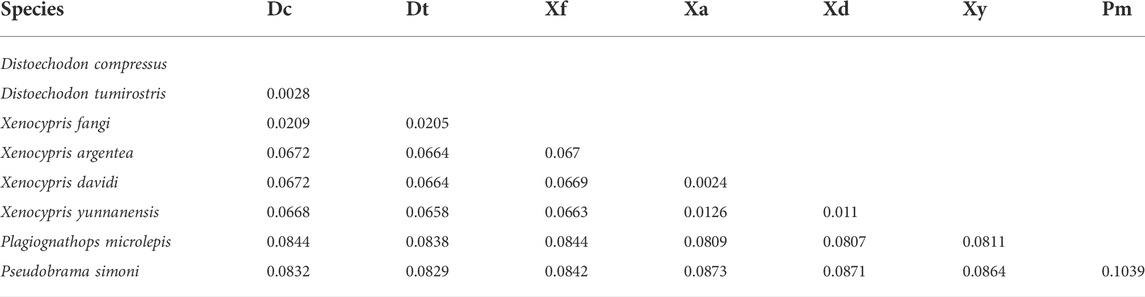
The pairwise genetic distances ranged from 0.002 (X. davidi and X. argentea) to 0.104 (P. microlepis and P. simoni) with an average of 0.064 ± 0.029 (Table 2). In the present study, the variation sites between the two Distoechodon species were only 0.28% of the complete mitogenomes, far less than other intra-species studies. However, morphological differences between these two Distoechodon species have been widely reported (Cao and Meng, 1992; Zhao et al., 2009; Lin, 2020; Chen et al., 2021). The amount of nucleotide variation in the mitogenomes that can be used to distinguish species or genera levels is still not clarified. The main differences in nucleotide composition included two parts. First, the length of each part showed three differences in 16s rRNA, ND2, and the gaps after ND2. The length of the 16 S rRNA ranged from 1688 bp to 1693 bp. ND2, X. davidi, X. argentea, and X. yunnanensis had a complete stop codon (TAA), whereas the remaining sequences had an incomplete stop codon (T++). Six nucleotides were identified between ND2 and tRNATrp in X. davidi, X. argentea, and X. yunnanensis more than others. Hence, these two differences can be used as loci to distinguish Xenocypris from other genera. In addition, we examined the number and percentage of variation in all PCGs. ND2 and ND4 were the most conserved PCGs, with variation sites accounting for 5%. Alternatively, the variation sites of ND1, ATP6, ND3, ND5, and ND6 exceeded 20% among the nine xenocyprinid fish. CR also showed a high variation within less than 1 kb sequence, but most of the variation sites were concentrated in the CD and CSB domains.
Phylogenetic relationship and updated classification of the subfamily Xenocyprinae
Phylogenetic trees were constructed using the BI and ML methods, based on concatenated nucleotide sequences of 13 PCGs. The phylogenetic trees exhibited a congruent topology according to both methods. As shown in Figure 2, the subfamily Xenocyprinae was monophyletic within Cyprinidae corroborating the result of previous studies on the phylogeny of Cyprinidae (Saitoh et al., 2006; Wang et al., 2007; He et al., 2008; Yang et al., 2015; Tao et al., 2019; Cheng et al., 2022). Within the subfamily Xenocyprinae, there were three clades representing different genera and their phylogenetic relationships. Plagiognathops was located at the base of the phylogenetic tree with a pairwise genetic distance of more than 0.08 between all the other xenocyprinid fish, which are completely different from those of previous studies based on partial mitochondrial genes (He et al., 2004; Xiao et al., 2012; Cheng et al., 2022). For the remaining two clades, it mainly consisted of genera Xenocypris and (Distoechodon + Pseudobrama). X. fangi is closely related to Distoechodon with a genetic distance of less than 0.02. We suspect that sampling or sequencing in Liu et al. (2021) contributed to this unusual result. All previous phylogenetic analyses using partial mtDNA genes revealed that X. fangi clustered to the Xenocypris clade (He et al., 2004; Xiao et al., 2012; Zhang et al., 2022). Therefore, more sequences of X. fangi are needed to understand their accurate phylogenetic position.
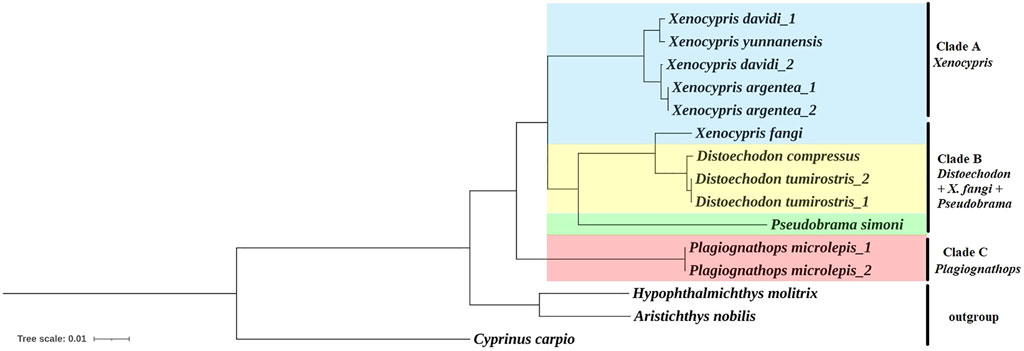
FIGURE 2. Phylogenetic relationship within the subfamily Xenocyprinae using BI and ML methods based on the 13 PCGs. Note: All the support values for the ML/Bayesian method are 1.00 or 100 so that it is not labeled at the nodes in the figure.
The current results clarified some controversies in the systematics of the subfamily Xenocyprinae. All of the aforementioned results support the validity of genus Plagiognathops. The key characteristics of Plagiognathops are lateral line scales of less than 50 and gill rakers on the first gill arch of more than 100 (Zhao et al., 2009). Second, strong evidence supports the validity of D. compressus. Previous studies based on partial mitochondrial markers could not accurately distinguish D. compressus and D. tumirostris (Cheng et al., 2022; Zhang et al., 2022). In the present study, using complete mitogenomes, the two species could be separated from each other by phylogenetic analysis. Although the genetic distance between the two species is very low, we also found the main variation between two Distoechodon species in Cox1 and ND5 genes, so that species classification can be identified by specific loci.
Divergence time estimation
The two clusters (Xenocypris and (Distoechodon + Pseudobrama)) diverged 10.0 Mya (95% CI: 7.4–12.6 Mya) (Figure 3). The divergence time in this study was earlier than that of previous studies on cyprinid fish, which were based on nuclear recombination activating gene 2 and the mitochondrial 16 S rRNA and cytochrome b genes (6.7 Mya, Wang et al., 2012) but later than that reported for East Asian opsariichthyin-xenocyprinin-cultrin fish (14.4 Mya) by Cheng et al. (2022). Plagiognathops was reconfirmed as a valid genus and began to diverge from Xenocypris and Distoechodon approximately 12.1 Mya. The two Distoechodon species (D. compressus and D. tumirostris) separated from each other 0.6 Mya and were still at an early stage of differentiation. Xenoprinins, as an important part of East Asian freshwater fish, were recently shown to be related to monsoon-driven climatic conditions and formation of river and river–lake environments in East Asia (Cheng et al., 2022). With a close relationship to the Tibetan Plateau uplift from 25 to 20 Mya (Xiao et al., 2012), the Asian monsoon climate was rebuilt in the Early Neogene (23 Mya) (Guo et al., 2002; Favre et al., 2015). The East Asian monsoon was further strengthened in the late Early Miocene, from the Middle Miocene (approximately 15–13 Mya) to the Late Miocene (approximately 10–7 Mya), and the Middle Pliocene (approximately 3.5 Mya) (Clift et al., 2008). In this study, the major radiation of Xenocyprinae species possibly occurred in the Middle to Late Miocene and Late Pliocene, which also confirmed that Xenocyprinae fish were the vital case of Asian monsoon climate resulting in speciation and diversification.

FIGURE 3. Divergence time estimation of Xenocyprinae utilized two fossil clocks in BEAST based on the best-scoring maximum-likelihood tree. Note: Node bars show 95% CIs of the divergence time estimate.
Data availability statement
The mitochondrial genomes for this study can be found in the GenBank with accession number OM994436.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Minjiang University.
Author contributions
ZZ and JC designed the research. ZZ performed the experiments and analyzed the data. BL, JL, and XZ collected the samples. ZZ and JC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Natural Science Foundation of Fujian Province of China (2022J011136), Youth Foundation of East China Sea Bureau of Ministry of Natural Resources (202112), and Fujian Middle-Aged and Young Teacher Education Research Project (JAT200445).
Conflict of interest
Author JL is the boss of Liancheng Jiming Fish Farming Company, who provided samples for this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.966633/full#supplementary-material
References
Aljanabi, S. M., and Martinez, I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25 (22), 4692–4693. doi:10.1093/nar/25.22.4692
Avise, J. C. (2000). Phylogeography: The history and formation of species. Cambridg: Harvard University Press.
Bănărescu, P. (1970). Remarks on the genus Xenocypris (Pisces, Cyprinidae) with description of a new subspecies. Rev. Roum. Biol. Zool. 15, 395–402.
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69 (2), 313–319. doi:10.1016/j.ympev.2012.08.023
Bleidorn, C., Podsiadlowski, L., Zhong, M., Eeckhaut, I., Hartmann, S., Halanych, K. M., et al. (2009). On the phylogenetic position of myzostomida: Can 77 genes get it wrong? BMC Evol. Biol. 9 (1), 150. doi:10.1186/1471-2148-9-150
Bozdogan, H. (1987). Model selection and akaike's information criterion (AIC): The general theory and its analytical extensions. Psychometrika 52, 345–370. doi:10.1007/BF02294361
Cao, L., and Meng, Q. (1992). Studies on isozymes, skeleton characters of the Xenocyprinae fishes of China and discussion on systematic evolution. Acta Zootax Sin. 17 (3), 366–376. (In Chinese with English Abstract).
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 (4), 540–552. doi:10.1093/oxfordjournals.molbev.a026334
Chaban, Y., Boekema, E. J., and Dudkina, N. V. (2014). Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim. Biophys. Acta 1837 (4), 418–426. doi:10.1016/j.bbabio.2013.10.004
Chang, M., Chen, Y., and Tong, H. (1996). A new Miocene xenocyprinine (Cyprinidae) from heilongjiang Province northeast China and succession of late cenozoic fish faunas of East Asia. Vertebr. Palasiat. 34 (3), 165–183.
Chen, G., Fang, F., and Chang, M. M. (2005). A new cyprinid closely related to cultrins+ xenocyprinins from the Mid-Tertiary of South China. J. Vertebrate Paleontology 25 (3), 492–501. doi:10.1671/0272-4634(2005)025
Chen, X. L., Yue, P. Q., and Lin, E. D. (1984). Major groups within the family Cyprinidae and their phylogenetic relationships. Acta Zootaxonomica Sin. 9 (4), 424–438.
Chen, Y., Lin, B., Wang, W., Tan, K., Zhang, X., Song, X., et al. (2021). Morphological and biochemical genetic characteristics of Distoechodon compressus. J. Anhui Agric. Sci. 49 (2), 77–80. (In Chinese with English Abstract). doi:10.3969/j.issn.0517-6611.2021.02.023
Chen, Y. Y. (1982). Description of a new genus and species of Cyprinid fish. Acta Zootax Sin. 7, 425–427.
Cheng, P., Yu, D., Tang, Q., Yang, J., Chen, Y., and Liu, H. (2022). Macro-evolutionary patterns of East Asian opsariichthyin-xenocyprinin-cultrin fishes related to the formation of river and river-lake environments under monsoon climate. Water Biol. Secur. 1, 100036. doi:10.1016/j.watbs.2022.100036
Chu, Y. T. (1935). Comparative studies on the scales and on the pharyngeal and their teeth in Chinese cyprinids, with particular reference to taxonomy and evolution. Biol. Bull. St. John's Univ. Shanghai 2, 1–225.
Clift, P. D., Hodges, K. V., Heslop, D., Hannigan, R., Van Long, H., and Calves, G. (2008). Correlation of Himalayan exhumation rates and Asian monsoon intensity. Nat. Geosci. 1 (12), 875–880. doi:10.1038/ngeo351
Drummond, A. J., and Rambaut, A. (2007). Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7 (1), 214–218. doi:10.1186/1471-2148-7-214
Ellegren, H. (2014). Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 29 (1), 51–63. doi:10.1016/j.tree.2013.09.008
Fang, F., Noren, M., Liao, T. Y., Källersjö, M., and Kullander, S. O. (2009). Molecular phylogenetic interrelationships of the south Asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae). Zool. Scr. 38 (3), 237–256. doi:10.1111/j.1463-6409.2008.00373.x
Favre, A., Päckert, M., Pauls, S. U., Jähnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 90 (1), 236–253. doi:10.1111/brv.12107
Fischer, C., Koblmüller, S., Gülly, C., Schlötterer, C., Sturmbauer, C., and Thallinger, G. G. (2013). Complete mitochondrial DNA sequences of the threadfin cichlid (Petrochromis trewavasae) and the blunthead cichlid (Tropheus moorii) and patterns of mitochondrial genome evolution in cichlid fishes. PLoS One 8, e67048. doi:10.1371/journal.pone.0067048
Frazer-Abel, A. A., and Hagerman, P. J. (2008). Core flexibility of a truncated metazoan mitochondrial tRNA. Nucleic Acids Res. 36 (17), 5472–5481. doi:10.1093/nar/gkn529
Günther, A. C. L. G. (1868). Catalogue of the Fishes in the British museum. England: Wheldon & Wesley.
Guo, Z. T., Ruddiman, W. F., Hao, Q. Z., Wu, H. B., Qiao, Y. S., Zhu, R. X., et al. (2002). Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416 (6877), 159–163. doi:10.1038/416159a
He, M. J. (1989). “Xenocyprininae,” in Fishes of zhujiang river. Editor C. Y. Zheng (Beijing: Science), 112–115.
He, S., Gu, X., Mayden, R. L., Chen, W. J., Conway, K. W., and Chen, Y. (2008). Phylogenetic position of the enigmatic genus psilorhynchus (ostariophysi: Cypriniformes): Evidence from the mitochondrial genome. Mol. Phylogenet. Evol. 1 (47), 419–425. doi:10.1016/j.ympev.2007.10.012
He, S. P., Liu, H. Z., Chen, Y. Y., Masayuki, K., Tsuneo, N., and Zhong, Y. (2004). Molecular phylogenetic relationships of Eastern Asian Cyprinidae (Pisces: Cypriniformes) inferred from cytochrome b sequences. Sci. China. C Life Sci. 47 (2), 130–138. doi:10.1360/03yc0034
Howes, G. J. (1981). Anatomy and phylogeny of the Chinese major carps ctenopharygodon steind., 1866 and Hypophthalmichthys bleeker, 1860. Bull. Br. Mus. Nat. Hist. (Zool.) 41 (1), 1–52.
Hu, Y. X., Song, Y., Zhou, Q., Xue, Y., Xu, H., Luo, X., et al. (2015b). Complete mitochondrial genome of the pseudobream, Pseudobrama simoni (teleostei: Cypriniformes: Cyprinidae). Mitochondrial DNA 26 (5), 694–695. doi:10.3109/19401736.2013.843078
Hu, Y. X., Zhou, Q., Song, Y., Chen, D. Q., and Li, Y. (2015a). Complete mitochondrial genome of the smallscale yellowfin, Plagiognathops microlepis (Teleostei: Cypriniformes: Cyprinidae). Mitochondrial DNA 26 (3), 463–464. doi:10.3109/19401736.2013.830298
Jin, J. Q., and Sun, Y. B. (2018). AutoSeqMan: Batch assembly of contigs for sanger sequences. Zool. Res. 39 (2), 123–126. doi:10.24272/j.issn.2095-8137.2018.027
Jondeung, A., Sangthong, P., and Zardoya, R. (2007). The complete mitochondrial DNA sequence of the Mekong giant catfish (Pangasianodon gigas), and the phylogenetic relationships among Siluriformes. Gene 387, 49–57. doi:10.1016/j.gene.2006.08.001
Kobayashi, G., Itoh, H., Fukuda, H., and Kojima, S. (2021). The complete mitochondrial genome of the sand bubbler crab Scopimera globosa and its phylogenetic position. Genomics 113 (1), 831–839. doi:10.1016/j.ygeno.2020.10.014
Koepfli, K. P., Paten, B., and O'Brien, S. J. (2015). Genome 10K community of scientists, & O’brien, S. JThe genome 10K Project: A way forward. Annu. Rev. Anim. Biosci. 3 (1), 57–111. doi:10.1146/annurev-animal-090414-014900
Kong, L., Li, Y., Kocot, K. M., Yang, Y., Qi, L., Li, Q., et al. (2020). Mitogenomics reveals phylogenetic relationships of Arcoida (Mollusca, Bivalvia) and multiple independent expansions and contractions in mitochondrial genome size. Mol. Phylogenet. Evol. 150, 106857. doi:10.1016/j.ympev.2020.106857
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Lanfear, R., Calcott, B., Ho, S. Y., and Guindon, S. (2012). Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29 (6), 1695–1701. doi:10.1093/molbev/mss020
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi:10.1093/nar/gkab301
Lin, B. (2020). Analysis of main morphological characteristic of Distoechodon compressus in Liancheng. J. Fish. Res. 42 (6), 629–634. doi:10.14012/j.cnki.fjsc.202006011
Lin, B., Li, J., Zhang, X., and Zhang, Z. (2021). Genetic diversity of aquacultural populations of Distoechodon compressus in Liancheng. China Fish. 11, 91–93.
Liu, H. Z. (2002a). Mitochondrial DNA sequence variations and systematics of the genus Distoechodon (Teleostei: Cyprinidae). J. Appl. Ichthyol. 18, 181–184. doi:10.1046/j.1439-0426.2002.00325.x
Liu, H. Z. (2002b). The structure and evolution of mitochondrial DNA control region of fish: A case study to bitterlings. Prog. Nat. Sci. 12 (3), 266–271. (In Chinese with English Abstract). doi:10.3321/j.issn:1002-008X.2002.03.008
Liu, H. Z. (1998). “Xenocyprinae,” in Fauna Sinica, OsteichthyesCypriniformes II. Editor Y. Y. Chen (Beijing: Science), 208–223.
Liu, N., Gu, H., Cheng, X., Wang, Y., and Wang, Z. (2021). The complete mitochondrial genome sequence of Xenocypris fangi. Mitochondrial DNA. B Resour. 6 (3), 1200–1201. doi:10.1080/23802359.2021.1903361
Liu, Y. (2014). The complete mitochondrial genome sequence of Xenocypris davidi (Bleeker). Mitochondrial DNA 25 (5), 374–376. doi:10.3109/19401736.2013.809429
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 (W1), W54–W57. doi:10.1093/nar/gkw413
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14 (1), 91–104. doi:10.1093/oxfordjournals.molbev.a025706
Miya, M., and Nishida, M. (2015). The mitogenomic contributions to molecular phylogenetics and evolution of fishes: A 15-year retrospect. Ichthyol. Res. 62, 29–71. doi:10.1007/s10228-014-0440-9
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41 (3), 353–358. doi:10.1007/BF00186547
Ronquist, F., Teslenko, M., Mark, P. V. D., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi:10.1093/sysbio/sys029
Saitoh, K., Sado, T., Mayden, R. L., Hanzawa, N., Nakamura, K., Nishida, M., et al. (2006). Mitogenomic evolution and interrelationships of the Cypriniformes (actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J. Mol. Evol. 63 (6), 826–841. doi:10.1007/s00239-005-0293-y
Satoh, T. P., Miya, M., Mabuchi, K., and Nishida, M. (2016). Structure and variation of the mitochondrial genome of fishes. BMC genomics 17 (1), 719–720. doi:10.1186/s12864-016-3054-y
Shan, X. H. (1998). Phylogeny and dispersal-vicariance explanation of the extant fishes of Xenocyprinae (Teleostei: Cyprinidae). Acta Zootax Sin. 23, 56–66.
Sharma, A., Siva, C., Ali, S., Sahoo, P. K., Nath, R., Laskar, M. A., et al. (2020). The complete mitochondrial genome of the medicinal fish, Cyprinion semiplotum: Insight into its structural features and phylogenetic implications. Int. J. Biol. Macromol. 164, 939–948. doi:10.1016/j.ijbiomac.2020.07.142
Shi, L. X., Zhang, C., Wang, Y. P., Tang, Q. Y., Danley, P. D., and Liu, H. Z. (2018). Evolutionary relationships of two balitorids (Cypriniformes, Balitoridae) revealed by comparative mitogenomics. Zool. Scr. 47 (3), 300–310. doi:10.1111/zsc.12282
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. doi:10.1093/bioinformatics/btu033
Sun, C. H., Liu, H. Y., Xu, N., Zhang, X. L., Zhang, Q., and Han, B. P. (2021). Mitochondrial genome structures and phylogenetic analyses of two tropical characidae fishes. Front. Genet. 12, 627402. doi:10.3389/fgene.2021.627402
Tang, K. L., Agnew, M. K., Hirt, M. V., Lumbantobing, D. N., Raley, M. E., Sado, T., et al. (2013). Limits and phylogenetic relationships of East asian fishes in the subfamily oxygastrinae (teleostei: Cypriniformes: Cyprinidae). Zootaxa 3681 (2), 101–135. doi:10.11646/zootaxa.3681.2.1
Tao, W., Yang, L., Mayden, R. L., and He, S. (2019). Phylogenetic relationships of Cypriniformes and plasticity of pharyngeal teeth in the adaptive radiation of cyprinids. Sci. China. Life Sci. 62 (4), 553–565. doi:10.1007/s11427-019-9480-3
Wang, X., Gan, X., Li, J., Mayden, R. L., and He, S. (2012). Cyprinid phylogeny based on bayesian and maximum likelihood analyses of partitioned data: Implications for Cyprinidae systematics. Sci. China. Life Sci. 55 (9), 761–773. doi:10.1007/s11427-012-4366-z
Wang, X., Li, J., and He, S. (2007). Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 42 (1), 157–170. doi:10.1016/j.ympev.2006.06.014
Wang, Y., Yang, Y., Liu, H., Kong, L., Yu, H., Liu, S., et al. (2021). Phylogeny of Veneridae (Bivalvia) based on mitochondrial genomes. Zool. Scr. 50 (1), 58–70. doi:10.1111/zsc.12454
Watanabe, Y. L., Suematsu, T., and Ohtsuki, T. (2014). Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 5, 109. doi:10.3389/fgene.2014.00109
Xiao, G., Guo, Z., Dupont-Nivet, G., Lu, H., Wu, N., Ge, J., et al. (2012). Evidence for northeastern Tibetan Plateau uplift between 25 and 20 ma in the sedimentary archive of the xining basin, northwestern China. Earth Planet. Sci. Lett. 317, 185–195. doi:10.1016/j.epsl.2011.11.008
Xu, H., Zhu, Y., Zheng, D., and Yang, S. (2019). Molecular identification and phylogenetic analysis of mitogenome of the Xenocypris davidi from Cao’e River. Mitochondrial DNA. B Resour. 4 (2), 3998–3999. doi:10.1080/23802359.2019.1688099
Xue, S., Lin, J., Li, H., Ma, K., and Han, Y. (2019). The complete mitochondrial genome of Xenocypris yunnanensis (Nichols, 1925), threatened fishes of the world. Mitochondrial DNA Part B 4 (1), 428–429. doi:10.1080/23802359.2018.1547138
Yang, G. R. (1964). “Xenocyprininae,” in The Cyprinid fishes of China. Editor X. W. Wu (Beijing: Science), 128–132.
Yang, L., Sado, T., Hirt, M. V., Pasco-Viel, E., Arunachalam, M., Li, J., et al. (2015). Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (teleostei: Cypriniformes). Mol. Phylogenet. Evol. 85, 97–116. doi:10.1016/j.ympev.2015.01.014
Yu, P., Zhou, L., Yang, W. T., Miao, L. J., Li, Z., Zhang, X. J., et al. (2021). Comparative mitogenome analyses uncover mitogenome features and phylogenetic implications of the subfamily Cobitinae. BMC genomics 22 (1), 50–19. doi:10.1186/s12864-020-07360-w
Yu, P., Zhou, L., Yang, X. L., Li, L. L., Jin, H., Li, Z., et al. (2022). Comparative mitogenomic analyses unveil conserved and variable mitogenomic features and phylogeny of Chedrinae fish. Zool. Res. 43 (1), 30–32. doi:10.24272/j.issn.2095-8137.2021.243
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20 (1), 348–355. doi:10.1111/1755-0998.13096
Zhang, Y., Zhou, C., Gao, Y., Wang, L., Yang, F., Liu, H., et al. (2015). Purification effect on eutrophic water body with combination of Xenocyprinae and Hyriopsis cumingii under different hydrodynamic conditions. Chin. J. Environ. Eng. 9 (3), 1109–1116. (In Chinese with English Abstract). doi:10.12030/j.cjee.20150320
Zhang, Z., Li, J., Zhang, X., Lin, B., and Chen, J. (2022). Genetic diversity of Distoechodon compressus populations and discussion of phylogenetic relationships within subfamily Xenocyprinae. Biot. Resour. 44 (5). In press, (In Chinese with English Abstract).
Zhang, Z., Li, S., Zhang, J., Song, W., Yang, J., and Mu, J. (2021). The complete mitochondrial genome of an endangered minnow Aphyocypris lini (Cypriniformes: Xenocyprididae): Genome characterization and phylogenetic consideration. Biologia 76 (11), 3311–3321. doi:10.1007/s11756-021-00811-z
Keywords: Xenocyprinae, mitogenome, phylogeny, divergence time, taxonomy
Citation: Zhang Z, Li J, Zhang X, Lin B and Chen J (2022) Comparative mitogenomes provide new insights into phylogeny and taxonomy of the subfamily Xenocyprinae (Cypriniformes: Cyprinidae). Front. Genet. 13:966633. doi: 10.3389/fgene.2022.966633
Received: 11 June 2022; Accepted: 10 October 2022;
Published: 25 October 2022.
Edited by:
Claudio Oliveira, São Paulo State University, BrazilReviewed by:
Larry Croft, Deakin University, AustraliaImran Parvez, Hajee Mohammad Danesh Science & Technology University, Bangladesh
Copyright © 2022 Zhang, Li, Zhang, Lin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Zhang, emhpemhhbmdAbWp1LmVkdS5jbg==; Jianming Chen, Y2hlbmptQG1qdS5lZHUuY24=
 Zhi Zhang
Zhi Zhang Jiming Li2,3
Jiming Li2,3 Jianming Chen
Jianming Chen