- Department of Nuclear Medicine, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai, China
Epidermal growth factor receptor 2 (ERBB2) is commonly over-expressed in advanced or metastatic tissues of papillary thyroid cancer (PTC) with poor prognosis, while it remains unknown whether ERBB2 plays a role in the progression of PTC. Thus, we analyzed the data derived from online repositories, including TCGA, KEGG, GO, GeneMANIA, and STRING, to explore the relationship between ERBB2 expression and prognosis, tumor phenotypes of interest, and immune infiltrates in PTC. Compared to normal thyroid tissue, ERBB2 was up-regulated in PTC samples (p < 0.001); In comparison with the group with low expression of ERBB2, the group with high expression of ERBB2 had poorer progression-free interval in stage III/IV patients (p = 0.008) and patients aged >45 years (p = 0.019). The up-regulated ERBB2 was associated with iodine metabolism dysfunction, proliferation, metastasis, angiogenesis, and drug resistance. The expression of ERBB2 negatively correlated with enrichment scores of B cells (r = −0.176, p < 0.001), CD8+ T cells (r = −0.160, p < 0.001), cytotoxic cells (r = −0.219, p < 0.001), NK CD56dim cells (r = −0.218, p < 0.001), plasmacytoid dendritic cells (r = −0.267, p < 0.001), T cells (r = −0.164, p < 0.001), T follicular helper cells (r = −0.111, p = 0.012), gamma delta T cells (r = −0.105, p = 0.017), and regulatory T cells (r = −0.125, p = 0.005). In conclusion, ERBB2 may serve as a prognostic biomarker and an immunotherapeutic target in PTC, deserving further exploration.
Introduction
Thyroid cancer is one of the most common endocrine neoplasms, and the incidence rate is overgrowing, with 53,815 new cases expected in the US (Siegel et al., 2021) and 224,023 in China (Xia et al., 2022) in 2022. Papillary thyroid cancer (PTC) is the most widely recognized thyroid tumor, accounting for around 84% of all thyroid cancer patients (Fagin and Wells, 2016). Alarmingly, the incidence of invasive and metastatic PTC has risen 1.5–5-fold over the past 30 years (Lim et al., 2017). Within 10 years after initial surgery and radioiodine therapy, approximately 1/10 to 1/5 of PTCs developed local recurrence and distant metastasis; Once distant metastases of PTC occurred, the 5-year survival rate of patients would decrease from 98% to 63–75% (Cancer Facts 2022). The high rates of recurrence, metastasis, and radioiodine refractoriness have become the key bottleneck, stymying the cure of PTC (Jin et al., 2018).
Epidermal growth factor receptor 2 (ERBB2), a well-known oncogene of multiple cancers, such as breast cancer and ovarian cancer (Baxevanis et al., 2004), was found to be firmly connected with cancer occurrence, proliferation, metastasis, drug resistance, immune escape and poor prognosis (Dawood et al., 2009; Bates et al., 2018). Owe to the research on ERBB2, multiple diagnostic and therapeutic methods were established for tumor management (Mitri et al., 2012). In thyroid cancer, however, the deep-going research on ERBB2 is limited. Although ERBB2 is over-expressed in progressive and metastatic PTC cases (Kremser et al., 2003), and high expression of ERBB2 was found to be responsible for resistance to mitogen-activated extracellular signal-regulated kinase inhibitors (MEKi) (Montero-Conde et al., 2013), the ERBB2 upregulation-related unfavorable phenotypes, such as tumor occurrence, proliferation, metastasis, and poor progression-free interval (PFI), have not been comprehensively explored in PTC, and the relations between ERBB2 expression and the immune microenvironment in PTC remains unknown.
Given that the application of ERBB2-related management approaches, such as imaging probes and immunotherapeutic drugs, is limited by lacking knowledge of the association between ERBB2 expression and oncological features of PTC; we, therefore, conducted the bioinformatic study with integrated data from online repositories, including TCGA, KEGG, GO, GeneMANIA, and STRING to explore the potency and translational value of ERBB2 as a prognostic biomarker and immunotherapeutic target for PTC management.
Materials and methods
Data collection
The mRNA data and clinical characteristics of PTC patients were downloaded from the TCGA database (https://www.cancer.gov/tcga/), an open data portal that compiled clinical information and RNA-Seq data of 33 cancers. The thyroid cancer (THCA) project of the TCGA database includes 568 samples (510 PTC and 58 paracancerous normal samples). Clinical data collection for the THCA project was mainly completed in 2014; thus, PTC staging was adopted in AJCC 7th edition (Agrawal et al., 2014). The mRNA sequencing data of all samples were converted to the format of transcripts per million (TPM) for subsequent analysis.
OncoPrint analysis online
OncoPrint is a method for visualizing samples by integrating gene expression heat map and gene variant distribution map. The gene expression and gene mutation data in OncoPrint analysis were derived from the TCGA-THCA dataset and analyzed online (https://www.cbioportal.org). The OncoPrint analysis was utilized to show the association between ERBB2 expression and BRAF and RAS mutant PTC sample distribution.
Progression-free interval analysis
The Kaplan-Meier Progression-free interval (PFI) curve compared prognosis differences between patients with high and low expression of ERBB2. Since the death reports of PTC in the TCGA database were few (n = 16), the clinical prognosis data included in this study were PFI. Due to the limited number of patients with distant metastases in the database (n = 9), we could not analyze PFI in the subgroup of patients with distant metastases.
Co-expression heat map
Co-expression heat map assesses the correlation between the expression of ERBB2 and gene sets, including the iodine metabolism-related gene set of TSHR, SLC5A8, SLC26A4, and TPO (Portulano et al., 2013), the tumor angiogenesis gene set of VEGFA, FLT1, KDR, FLT4, PECAM1, VWF, TIE1, TEK, ANGPT1, ANGPT2, CDH5, and CLDN5 (Smith et al., 2010; Haibe et al., 2020), Lymph node metastasis gene set of EVA1A, TIMP1, SERPINA1, FAM20A, FN1, TNC, and MXRA8 (Wu et al., 2021), distant metastatic gene sets of MMP2, PTTG1, VEGFC, CXCR4, and FGF2, tumor cell proliferation set of MKI67, PCNA, and MCM2) (Liang et al., 2011), and MEKi resistance marker gene set of SPRY2, SPRY4, ETV4, ETV5, DUSP4, DUSP6, CCND1, EPHA2, and EPHA4 (Mazzoni et al., 2019; Degirmenci et al., 2021).
Analysis of immune infiltrates
The enrichment scores of 24 immune cells were based on the reported literature (Bindea et al., 2013). The correlation analysis between ERBB2 expression and enrichment scores of immune infiltration was tested with a single sample gene set enrichment analysis (ssGSEA) with R packages of clusterProfiler (version 3.8.0).
Analysis of ERBB2-related differentially expressed genes (DEGs)
To further understand the role of ERBB2 in the progression of PTC, we screened the DEGs between samples with high and low expression of ERBB2. We applied Log2(fold change) > 1.5 and adjusted p < 0.001 to select DEGs. The analysis is performed using the R package of DESeq2 (version 3.8). All the DEGs were graphed in a volcano plot.
Enrichment analysis of KEGG and GO terms
GO analysis is a widely applied bioinformatics tool for determining ERBB2-related biological processes, cellular components, and molecular functions. KEGG analysis is used to analyze ERBB2- related signal path changes. We applied GO and KEGG to analyze the biological function of ERBB2 in PTC. To understand how ERBB2 is involved in tumorigenesis, we used KEGG and GO online tools to analyze the signaling pathways and gene functions in which ERBB2-related DEGs participated. Gene set enrichment analysis (GSEA) with R package clusterProfiler (version 3.8.0) evaluated the ERBB2- related DEGs contributing to annotated gene functions, cell phenotypes, signal pathways, and diseases.
GeneMANIA and string analysis online
The network of gene-gene relations was constructed with the online tool GeneMANIA (https://genemania.org/). The website extensively integrated data on gene-gene interactions, gene co-expression networks, and gene function enrichment. To understand the ERBB2-involved mechanisms, the STRING tool (version 11.5, https://string-db.org/) was utilized to analyze the protein-protein interaction (PPI) network of corresponding proteins of ERBB2-related DEGs. The PPI network was displayed with Cytoscape software (version 3.9.1).
Statistical analysis
All statistical analyses and graphs were performed using R software (version 3.6.3). The ERBB2 expression in unpaired and paired samples was analyzed with Wilcoxon rank-sum test and Wilcoxon signed-rank test, respectively. Kruskal-Wallis test was tested to assess the relationship between clinical/cytogenetic features and ERBB2 expression. The differences in enrichment scores of immune infiltrate were analyzed using the Wilcoxon rank sum tests. The Spearman statistical method was adopted for the correlation analysis between ERBB2 expression and the gene expression of interest or enrichment scores of 24 immune cells. Hazard ratio, log-rank tests, and the Kaplan-Meier curve were applied to assess the role of ERBB2 as a prognostic biomarker. P < 0.05 was considered statistically significant.
Results
Aberrantly upregulated ERBB2 expression in PTC samples
By analyzing the RNA-Seq data downloaded from TCGA data, we found that the ERBB2 expression in PTC samples was generally higher than that in normal thyroid tissue (unpaired test: p < 0.001, Figure 1A; paired test: p < 0.001, Supplementary Figure S1). OncopPrint plot displayed that PTC tissues with high expression of ERBB2 were mainly distributed in BRAF mutation samples rather than RAS mutation samples (Figure 1B). Subgroup analysis showed that the ERBB2 expression in BRAF mutant PTC, classic PTC, and PTC with bilateral foci were significantly higher than those in corresponding subgroups of RAS mutant PTC (p < 0.001), follicular variant PTC (FVPTC) (p < 0.001), and PTC with unilateral foci (p = 0.02), respectively (Figure 2). To determine the ERBB2-related clinical value and the underlying mechanism, PTC samples with high and low expression of ERBB2 were divided based on the median TPM value. The characteristics of patients with the high and low expression of ERBB2 were summarized in Supplementary Table S1.
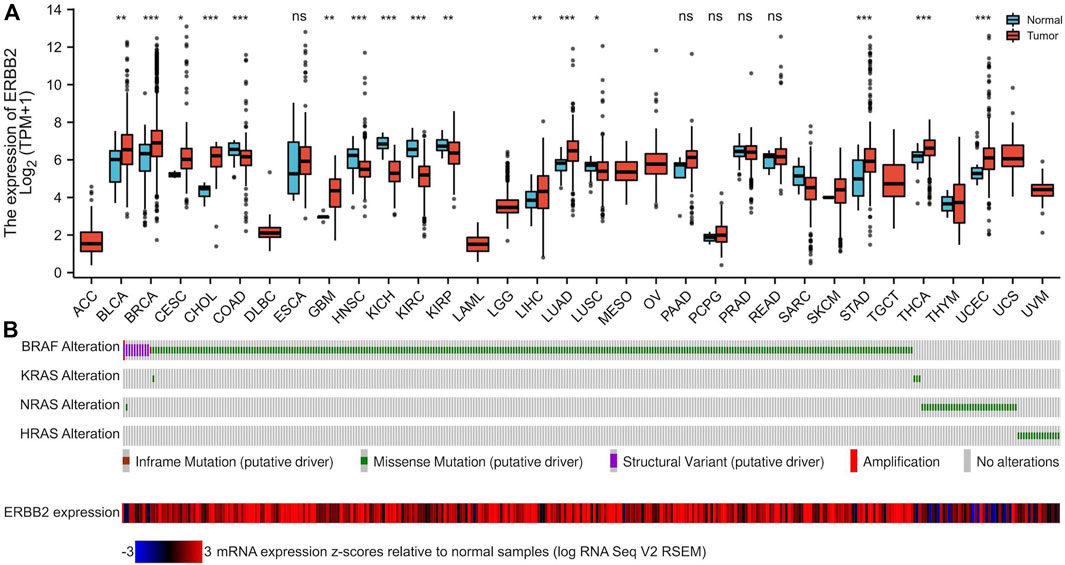
FIGURE 1. The mRNA expression of ERBB2 in pan-cancers and tumor-adjacent normal samples. (A) Compared with paired normal tissues, mRNA expression of ERBB2 was upregulated in 16 cancer datasets: BLCA (p = 0.007), BRCA (p < 0.001), CESC (p = 0.034), CHOL (p < 0.001), GBM (p = 0.002), LIHC (p = 0.008), LUAD (p < 0.001), STAD (p < 0.001), and THCA (p < 0.001) and UCEC (p < 0.001). (B) The OncoPrint plot displays an overview of gene alterations of the BRAF, KRAS, NRAS, and HRAS, and the expression of ERBB2 in papillary thyroid cancer samples from TCGA. *p < 0.05, **p < 0.01, ***p < 0.001.
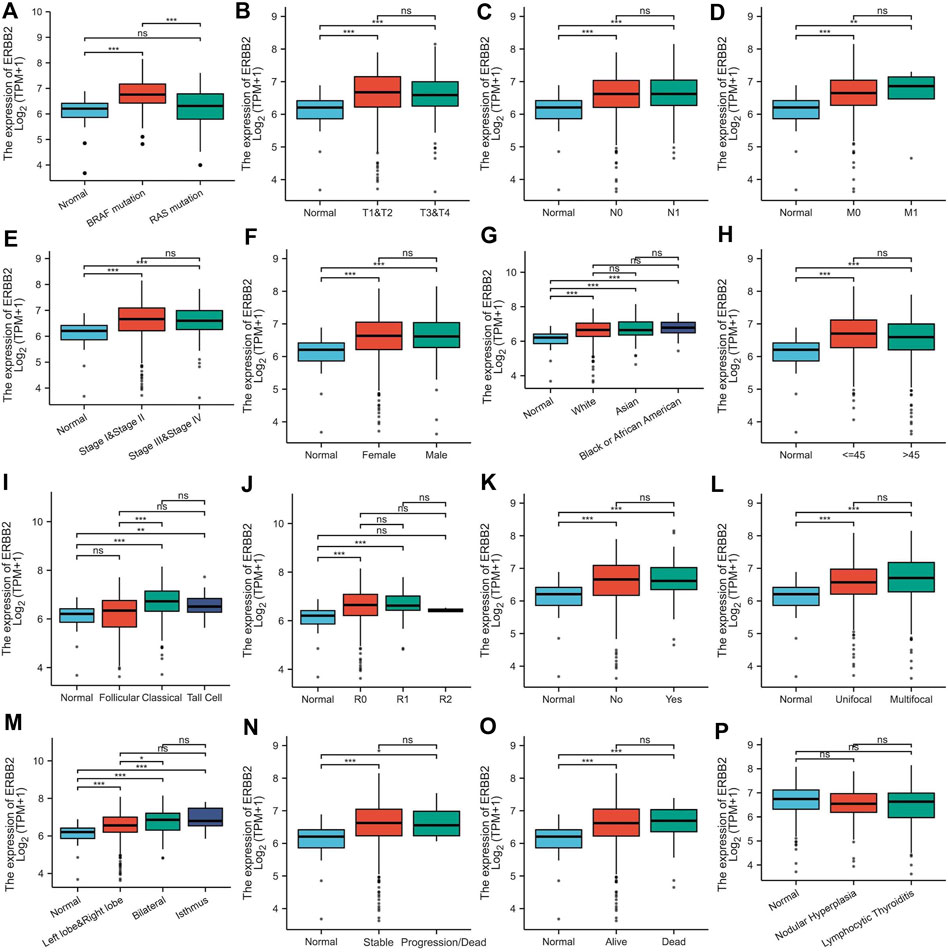
FIGURE 2. ERBB2 expression in papillary thyroid cancer classified by characteristics: (A) mutation, (B) T stage, (C) N stage, (D) M stage, (E) pathological stage, (F) gender, (G) race, (H) age, (I) histological type, (J) residual tumor, (K) extrathyroidal extension, (L) primary tumor focus type, (M) tumor location, (N) overall survival event, (O) progression-free interval event, and (P) thyroid gland disorder history. *p < 0.05, **p < 0.01, ***p < 0.001.
Elevated ERBB2 expression was associated with the poor prognosis
The relations between ERBB2 expression and PFI were analyzed using the Kaplan-Meier curve. As is shown in Figure 3, the group with high expression of ERBB2 had a significantly poorer prognosis than that with low expression of ERBB2 in stage III/IV patients (HR: 2.7, CI: 1.29–5.66, p = 0.008) and patients aged >45 years (HR: 2.22, CI: 1.09–4.54, p = 0.019) (Figure 3); the detailed subgroup analyses of PFI Kaplan-Meier curves were plotted in Supplementary Figures S2–4.
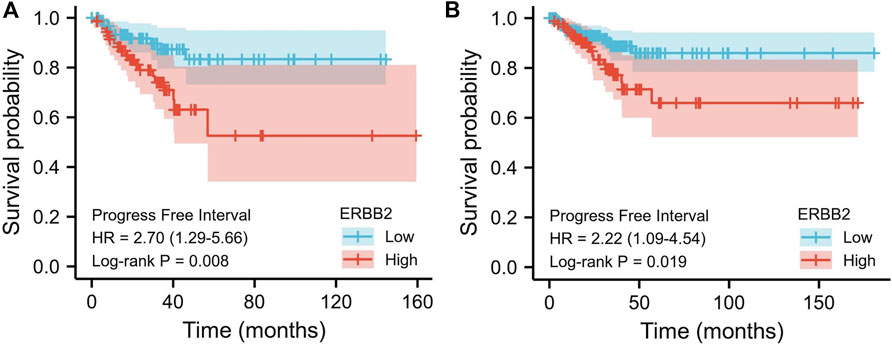
FIGURE 3. Kaplan-Meier curves of progression-free interval comparing the high and low expression of ERBB2 in thyroid cancer patients. (A) In stage III/IV patients, the progression-free interval of the high ERBB2 expression group was poorer than that of the low expression group (p = 0.008). (B) In patients aged >45 years, the progression-free survival of the high ERBB2 expression group was poorer than that of the low expression group (p = 0.019).
Highly expressed ERBB2 was correlated to unfavorable tumor phenotypes
The association between ERBB2 expression and clinical phenotypes was performed with correlation analysis. The co-expression heat map showed that the expression of ERBB2 positively correlated with TSHR but negatively correlated with SLC5A8, SLC26A4, and TPO (Figure 4A); In addition, the expression of ERBB2 positively correlated with gene sets of tumor angiogenesis (VEGFA, FLT1, KDR, FLT4, PECAM1, VWF, TIE1, TEK, ANGPT1, ANGPT2, CDH5, and CLDN5) (Figure 4B), lymph node metastasis (Gene sets: EVA1A, TIMP1, SERPINA1, FAM20A, FN1, TNC, and MXRA8) (Figure 4C), distant metastases (Gene sets: MMP2, VEGFC, CXCR4, and FGF2) (Figure 4D), tumor cell proliferation (Gene sets: MKI67, PCNA, and MCM2) (Figure 4E), and MEKi resistance (Gene sets: SPRY2, SPRY4, ETV4, ETV5, DUSP4, DUSP6, CCND1, EPHA2, and EPHA4) (Figure 4F).
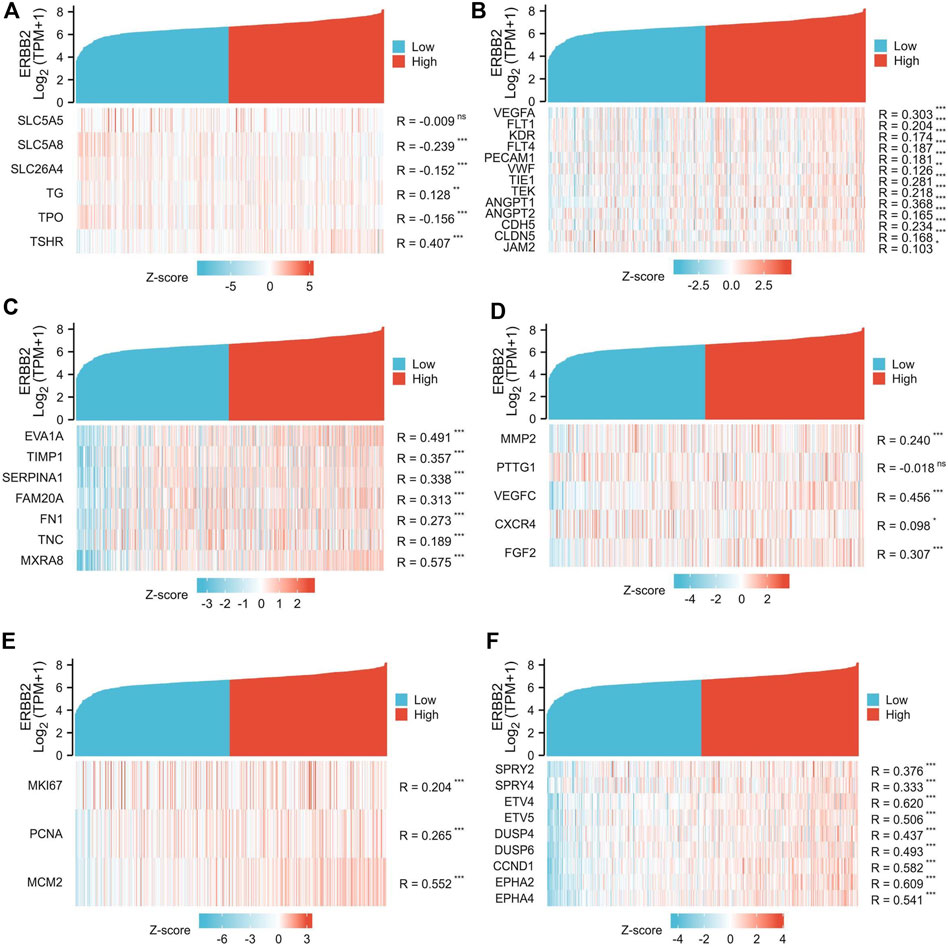
FIGURE 4. Co-expression analysis of ERBB2 and genes related to unfavorable thyroid cancer phenotypes. (A) the expression of ERBB2 is negatively correlated with iodine metabolism genes of SLC5A8, SLC26A4, and TPO. (B) the expression of ERBB2 was positively correlated with the tumor angiogenesis gene set. (C) the expression of ERBB2 is positively correlated with the gene set of lymph node metastasis. (D) the expression of ERBB2 was positively correlated with the distant metastasis gene set. (E) the expression of ERBB2 was positively correlated with the gene set of cell proliferation. (F) the expression of ERBB2 was positively correlated with the MEK inhibitor resistance gene set. *p < 0.05, **p < 0.01, ***p < 0.001.
Up-regulated ERBB2 was associated with suppressed tumor-infiltration of immune cells
The tumor-infiltrating immune cells were quantified by ssGSEA. Compared to the samples with low expression of ERBB2, the enrichment scores of T cells, CD8+ T cells, cytotoxic cells, NK CD56dim cells, plasmacytoid dendritic (pDC) cells, gamma delta T cells (Tgd), and regulatory T cells (TReg) were lower in samples with high expression of ERBB2 (Figure 5A). The correlation analysis showed that ERBB2 expression was negatively correlated with the enrichment scores of B cells, CD8+ T cells, cytotoxic cells, NK CD56dim cells, pDC, T cells, follicular helper T cells (TFH), Tgd cells, and TReg cells (Figure 5B, Supplementary Table S2).
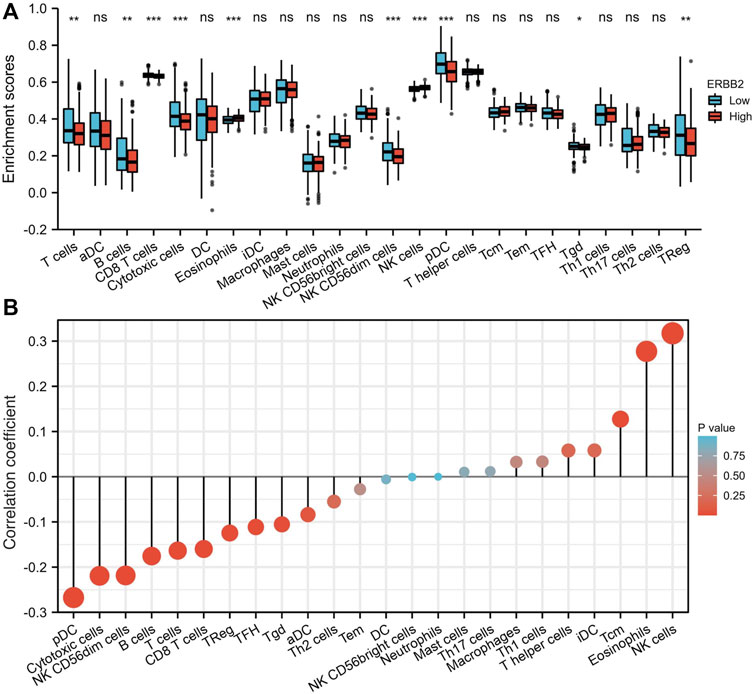
FIGURE 5. Immune cell enrichment scores in thyroid cancer tissues with different ERBB2 expression. (A) The enrichment scores of T cells, B cells, CD8+ T cells, cytotoxic cells, NK CD56dim cells, pDCs, Tgd, and TReg in the ERBB2 high expression group were lower than those in the ERBB2 low expression group. (B) Correlation analysis between the expression of ERBB2 and the enrichment scores of immune infiltrating cells in the tumor. ERBB2 expression negatively correlated with the enrichment scores of CD8+ T cell, cytotoxic cell, NK CD56dim cell, pDC, T cell, TFH, Tgd, and TReg. DC, dendritic cell; aDC, activated dendritic cell; pDC, plasmacytoid dendritic cell; iDC, interdigitating dendritic cell; Tcm, central memory T cell; Tem, effector memory T cell; TFH, follicular helper T cell; Tgd, T gamma delta, γδ; Th1, T helper type 1; Th17, T helper type 17; Th2, T helper type 2; Treg, regulatory T cells; NK, natural killer. *p < 0.05, **p < 0.01, ***p < 0.001.
Identification of DEGs between samples with high and low expression of ERBB2
A total of 146 DEGs between samples with high and low expression of ERBB2 were yielded (Supplementary Table S3). The five most up-regulated genes and the five most downregulated genes were shown in the volcano plot (Figure 6). Spearman correlation analysis found that the expression of ERBB2 negatively correlated with KLK15, KLK1, ARSF, and FGF21; and positively correlated with TAGLN3, GLRA1, RSPO1, SPAG11B, and SPAG11A (Supplementary Figure S5).
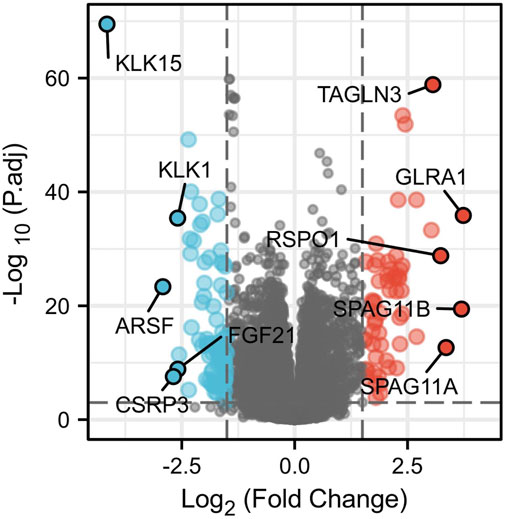
FIGURE 6. Volcano plot of differentially expressed genes (DEGs) between high and low ERBB2 expression groups. The significantly up-regulated genes are red dots; the significantly down-regulated genes are blue dots. The dots marked with gene names are the most up-regulated five DEGs and the most down-regulated five DEGs.
ERBB2-related DEGs participated in the primary immunodeficiency signaling pathway
Annotations from KEGG and GO were enriched with 146 ERBB2-related DEGs, showing that the ERBB2-related DEGs were involved in the KEGG term of primary immunodeficiency and GO term of the humoral immune response, regulation of execution phase of apoptosis, and negative regulation of execution phase of apoptosis (Figure 7A). Thereinto, the majority of ERBB2-related DEGs (n = 23) were enriched into the primary immunodeficiency signaling pathway, attracting more attention. GSEA analysis adds more insight into the 23 ERBB2-related DEGs and primary immunodeficiency signaling pathway, showing that ERBB2-related DEGs associated with primary immunodeficiency were mainly down-regulated ERBB2-related DEGs, such as CD79A, CD19, CD3D, CD3E, CD8A, and CD4 (Figure 7B).
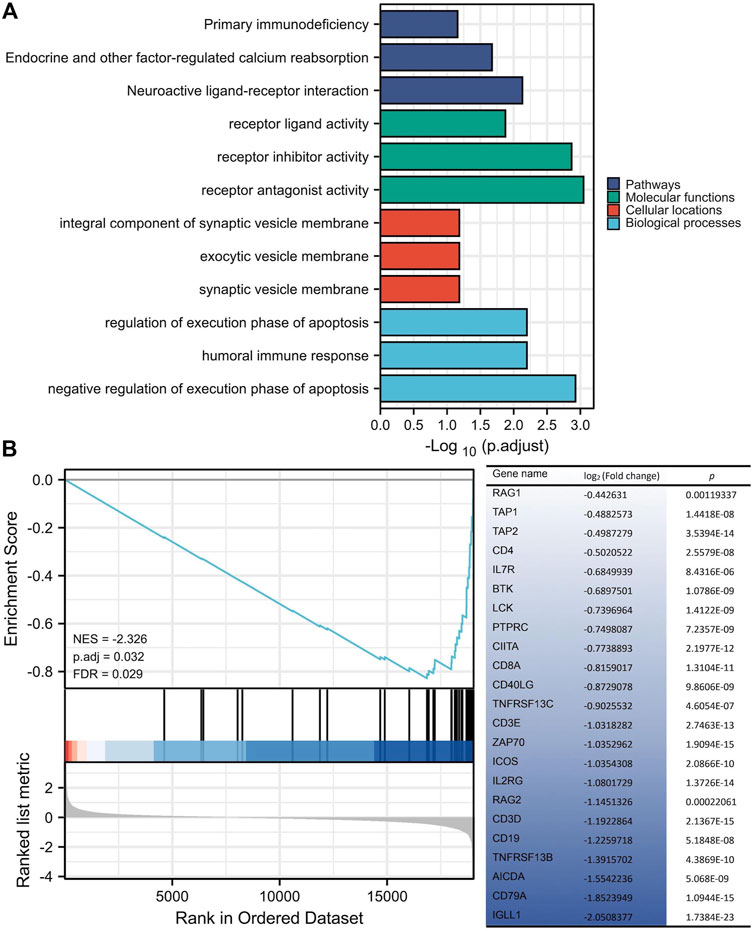
FIGURE 7. KEGG and GO enrichment analysis of differentially expressed genes between high and low expression of ERBB2 groups. (A) 146 ERBB2-related differentially expressed genes were enriched in KEGG signaling pathways and GO annotations, such as the primary immunodeficiency. (B) GSEA analysis of 19433 protein-coding genes and 146 ERBB2-related differentially expressed genes showed that the genes involved in the primary immunodeficiency pathway were mainly 23 down-regulated genes in the ERBB2 high expression group; Gene expression was sorted by log2 (fold change).
ERBB2-related DEGs constructed the core PPI networks
Network analysis showed that the 23 ERBB2-related DEGs had gene interaction and co-expression relationships with each other; the functional enrichment found that the 23 ERBB2-related DEGs involved in the function of a variety of immune cells, including the functions of B cells, monocytes, and T cells (Supplementary Figure S6). The PPI network of corresponding proteins of the ERBB2-related DEGs was screened with the STRING tool with a confidence threshold of 0.4. In total, 84 nodes and 127 edges were connected, and the networks with nodes ≥3 were displayed with Cytoscape (Supplementary Figure S7A). The core network was analyzed with the Cytoscape-MCODE function, showing the networks with an MCODE score of 4.815 (Supplementary Figure S7B) and 4.612 (Supplementary Figure S7C); the proteins of NPY and ESR1 were located in the center of the PPI network (Supplementary Figure S7).
Discussion
Despite ERBB2 being a well-known oncogene and regarded as a crucial diagnostic/prognostic biomarker and therapeutic target in multiple tumors, such as breast cancer (Wynn and Tang, 2022), the exact function of ERBB2 in tumors remains largely unknown. Only a few data are available in PTC, showing an 18%–100% rate of positive expression of ERBB2 protein via immunohistochemistry in 43–45 cases (Kremser et al., 2003; Ruggeri et al., 2016). Thus, expression profiles of the ERBB2 gene in the PTC need to be comprehensively investigated in the context of a large-scale study. In the present study, we took advantage of a large-scale analysis of gene expression profiles in 510 PTC patients in the TCGA database. The bioinformatic analysis adds the knowledge of detailed relations between ERBB2 expression and the clinical characteristics, prognosis, and immunoenvironment of PTC. Moreover, it is a novel finding that the expression levels of ERBB2 were significantly correlated to the markers of the immune cells in the PTC tissues, suggesting that ERB2 may play a crucial role in regulating the tumor-immunoenvironment. The present study uncovered the biological and clinical roles of ERBB2 in PTC, which might help imply novel ERBB2-based management strategies to improve the prognosis of PTC patients.
ERBB2 is up-regulated in multiple tumors located in breast, bladder, pancreas, ovary, and esophagus, especially in tumors with poor prognostic characteristics; in breast cancer, high expression of ERBB2 has become one of the hallmarks of poor prognosis (Dawood et al., 2009). Similarly, our study showed that the expression of ERBB2 in PTC was aberrantly up-regulated. A step further, we found that ERBB2 expression in patients with bilateral PTCs was higher than that in those with unilateral foci, which may be related to the fact that bilateral PTC is more aggressive and metastatic than the PTC with unilateral foci (Qu et al., 2016). Meanwhile, we found that the expression of ERBB2 in classic PTC was higher than that in FVPTC, which may be related to the poorer prognosis of PTC than that of FVPTC (Liu et al., 2018). The prognosis analysis in our study uncovered that the high expression of ERBB2 is a risk factor for poor prognosis in PTC.
The highly expressed ERBB2 was deemed to participate in cancer progression, promoting tumor proliferation, invasion, and metastasis (Mitri et al., 2012). PTC patients with high expression of ERBB2 were prone to suffer from distant metastasis (Kremser et al., 2003). In our study, the expression of ERBB2 was found to be positively correlated with VEGF-associated genes of VEGFA, FLT, KDR, and FLT4 (Shibuya, 2011), vascular endothelial cell markers of PECAM1 and VWF (Bauer et al., 2015), and vascular support-related genes of TEK, ANGPT1, ANGPT2, CDH5, CLDN5, and JAM2 (Oshi et al., 2021). Furthermore, the expression of ERBB2 was positively correlated with lymph node metastatic signature genes such as EVA1A, TIMP1, SERPINA1, FAM20A, FN1, TNC, and MXRA8 (Wu et al., 2021), and distant metastatic signature genes of MMP2, PTTG1, VEGFC, CXCR4, and FGF2 (Liang et al., 2011). Besides, ERBB2 expression was also positively correlated with proliferation-related genes, such as MKI67, PCNA, and MCM2 (Juríková et al., 2016). Collectively, highly expressed ERBB2 is involved in progressive oncological behaviors of PTC, which might be responsible for the poor prognosis.
Interestingly, high expression of ERBB2 was found to be associated with dysfunction of iodine metabolism of PTC in our study. The expression of ERBB2 was negatively correlated with SLC5A8, SCL26A4, and TPO, which relate to iodine absorption, transport, and organization. The downregulated SLC5A8 was associated with impaired resorption of organic iodine (Porra et al., 2005), making it challenging to transport iodine across the basal membrane to cell cytoplasm in thyroid follicular cells (Anekpuritanang et al., 2021). Meanwhile, TPO is the crucial protein to organize iodine with the participation of H2O2, and TPO deficiency would prevent iodine from taking part in the organization process (Kessler et al., 2008). Although the expression of ERBB2 was positively correlated with that of TSHR, the role of TSHR in radioiodine refractoriness remains debated because the highly expressed TSHR is not only associated with high radioiodine uptake (Hou et al., 2010) but also causes fast thyroid tumor growth (Lu et al., 2010). Therefore, the high expression of ERBB2 is likely to associate with the poor efficacy of radioactive iodine to some extent, and this mechanism needs to be further studied.
In recent years, targeted drugs, including commonly applied receptor tyrosine kinase inhibitors (RTKi) and novel MEKi, have become valuable antitumor drugs for advanced or progressive thyroid cancer (Jin et al., 2018). Notwithstanding the broad application of RTKi, the occurrence of RTKi-related severe adverse events is frequent, and drug resistance eventually develops in almost all PTC patients (Cheng et al., 2020). Compared to RTKi, the MEKi has the additional capability of inducing tumor differentiation, activating immune recognition, and possesses the characteristic of a lower incidence of severe adverse events (Neuzillet et al., 2014), attracting more attentions and expectations in the field of anti-PTC. Still, MEKi resistance is the typical scenario, often calling for a combination of MEKi and other therapeutics; nevertheless, the improvement of efficacy by the combination is generally limited (Shah et al., 2017), requiring a crucial key to solving the most pressing challenge. MEKi resistance has been proven to be closely related to the high expression of ERBB2 (Sa et al., 2022). For instance, high ERBB2 expression is one characteristic phenotype of thyroid cancer resistance to MEKi selumetinib (Montero-Conde et al., 2013). Our study further explored the correlation between the expression of ERBB2 and MEKi resistance genes of SPRY2, SPRY4, ETV4, ETV5, DUSP4, DUSP6, CCND1, EPHA2, and EPHA4 (Wagle et al., 2018). In BRAF or RAS mutant tumor cells, DUSP4, DUSP6, SPRY2, and SPRY4 tend to be highly expressed, allowing tumors to evade regular MAPK signaling pathway feedback (Pratilas et al., 2009). Other genes, such as ETV4, ETV5, CCND1, EPHA2, and EPHA4, also play a role in activating the MAPK pathway, promoting tumor resistance to MEKi (Chesnokov et al., 2022). Together with the previous study (Montero-Conde et al., 2013), our study confirmed the relationship between highly expressed ERBB2 and MEKi resistance in PTC.
Over the past decade, immune-related diagnostic techniques have successfully become new tools in tumor management. For instance, Michael D Farwell et al. conducted a phase I trial of PET/CT scan with 89Zr-labeled IAB22M2C minibody targeting CD8+ cells, which visualized the biodistribution of tumor-infiltrating CD8+ T cells and predicted early response to immunotherapy (Farwell et al., 2021). In the present research, ERBB2 expression was inversely correlated with the enrichment scores of CD8+ T cells. Therefore, radioactive or non-radioactive probes, such as 68Ga-labeled anti-ERBB2 Nanobody (Keyaerts et al., 2016) or polyethylene glycol-conjugated anti-ERBB2 peptides targeting ERBB2 protein (Guan et al., 2018) might be of value for predicting the response to immunotherapy in PTCs. In addition, ERBB2-targeted imaging would overcome the false positive uptake of 89Zr-labeled IAB22M2C in CD8+ normal tissues, such as the bone marrow and lymphnodes.
The finding, ERBB2 overexpression related to suppressed T cell infiltration in tumors, is not unique to PTC. Liu et al. (2021) also found that ERBB2 expression was negatively correlated with the infiltration of B cells and CD8+ T cells in cutaneous melanoma, similar to our findings. In breast cancer, ERBB2 is related to low expression of MHC class I surface antigen, causing impaired recognition between tumor cells and CD8+ T cells (Herrmann et al., 2004; Seliger and Kiessling, 2013). Besides, the PD-1/PD-L1 antibodies have been used in clinical trials to treat advanced, progressive, or metastatic thyroid cancer. However, the results were unsatisfactory; most patients still have an unfavorable response to PD-1 antibody therapy (Mehnert et al., 2019), related to the limited T-cell infiltration in PTC (Bastman et al., 2016). DEG analysis showed that in PTC samples with high expression of ERBB2, KLK15 and KLK1 were downregulated. The down-regulated KLKs were previously reported to be responsible for impaired immune microenvironment reprogramming during an antitumor immune response (Srinivasan et al., 2022). In our research, KEGG and GO annotation analysis found that the DEGs were mainly associated with the primary immunodeficiency pathway, which is closely related to tumor occurrence and the failure of antitumor immunotherapy (Tangye et al., 2020). Therefore, increasing tumor-infiltrating immune cells by applying bispecific antibodies or adding cytokines, e.g., PD-L1×CD3 bispecific antibody (Yang et al., 2021) or interleukin-17 (Nagaoka et al., 2020), in ERBB2 highly expressed tumors may be beneficial to improving the response to immunotherapy in PTC patients.
In addition, we found that NPY and ESR1 were located at the center of the ERBB2-related PPI network. NPY is a pleiotropic gene initially thought to be an endogenous anxiolytic peptide whose expression can be regulated by stress; however, NPY has been found to promote the growth and migration of breast cancer cells in recent years (Lin et al., 2021). ESR1 encodes an estrogen receptor, plays a crucial role in the occurrence and progression of breast and endometrial cancer, and is the main reason for resistance to estrogen suppression therapy (Piscuoglio et al., 2018). These results suggest that ERBB2 may be involved in the occurrence and progression of PTC together with NPY and ESR1; the mechanism needs further investigation.
Despite the fact that our data analysis enhanced the understanding of the roles of ERBB2 associated with PTC progression, there remain some limitations. First, the role of ERBB2 in follicular thyroid cancer, poorly differentiated thyroid cancer, and anaplastic thyroid cancer could not be investigated because those pathological types or endpoint information were lacking in online databases. Second, despite large sample studies in public databases prone to possess the superiority of strong evidence, public databases remain to lack some clinical information on PTC tissue around the prognostic tumor status of radioiodine uptake, angiogenesis, proliferation, drug resistance, and serum thyroglobulin levels; Therefore, we cannot perform a clinically based analysis comparing those prognostic factors above, and directly investigating their relations with ERBB2 expression. Thus, a more comprehensive prospective study is of value in the future to detail the role of ERBB2 in thyroid cancer. Lastly, this research mainly relied on mRNA data from the TCGA database; therefore, it is necessary to further experimentally investigate the direct evidence of ERBB2-related mechanisms in PTC tumorigenesis and progression in vitro or in vivo.
Because of the complex interaction between ERBB2 and mutant genes, we didn't separate poor prognosis contributed by ERBB2 overexpression and gene mutations. To step further, we have planned a more profound analysis in the subsequent study to separate their contributions to poor prognosis. As is known, BRAF mutation is a driver gene for tumorigenesis, which causes ERBB2 overexpression and poor prognosis of PTC (Kebebew et al., 2007; Caria et al., 2016). It remains unknown whether ERBB2 overexpression independently contributes to the poor prognosis of PTC, which need further validation.
The present work, the first to document, provides a comprehensive study of the relationship between the ERBB2 expression and clinical characteristics of a large-scale PTC cohort. We found that ERBB2 was highly expressed in PTC, and elevated ERBB2 expression was associated with iodine metabolism dysfunction, tumor proliferation, metastasis, angiogenesis, MEKi resistance, and poor prognosis. Meanwhile, ERBB2 expression was inversely correlated with the infiltration of immune cells in PTC tissues. It seems that ERBB2 might be a prognostic biomarker and an immunotherapeutic target in PTC, warranting further clinical validation. The study might also be a new start to expect future investigations on the latent mechanisms that bridge ERBB2 expression, clinical characteristics, and immunosuppressive environment in PTC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization, YJ, XQ, and ZH; methodology, YJ; software, YJ; validation, XQ, and ZH; formal analysis, YJ; investigation, YJ and ZH; resources, LC; data curation, ZH, and RS; writing—original draft preparation, YJ; writing—review and editing, XQ, ZH, JW, and RS; visualization, YJ; supervision, LC; project administration, LC; funding acquisition, LC. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the National Natural Science Foundation of China, grant numbers 81671711 and 82171981.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.966365/full#supplementary-material
References
Agrawal, N., Akbani, R., Aksoy, B. A., Ally, A., Arachchi, H., Asa, S. L., et al. (2014). Integrated genomic characterization of papillary thyroid carcinoma. Cell. 159, 676–690. doi:10.1016/j.cell.2014.09.050
Anekpuritanang, T., Uataya, M., Claimon, A., Laokulrath, N., Pongsapich, W., and Pithuksurachai, P. (2021). The association between radioiodine refractory in papillary thyroid carcinoma, sodium/iodide symporter expression, and BRAFV600E mutation. Onco. Targets. Ther. 14, 3959–3969. doi:10.2147/ott.s308910
Bastman, J. J., Serracino, H. S., Zhu, Y., Koenig, M. R., Mateescu, V., Sams, S. B., et al. (2016). Tumor-infiltrating T cells and the PD-1 checkpoint pathway in advanced differentiated and anaplastic thyroid cancer. J. Clin. Endocrinol. Metab. 101, 2863–2873. doi:10.1210/jc.2015-4227
Bates, J. P., Derakhshandeh, R., Jones, L., and Webb, T. J. (2018). Mechanisms of immune evasion in breast cancer. Bmc Cancer 18, 556. doi:10.1186/s12885-018-4441-3
Bauer, A. T., Suckau, J., Frank, K., Desch, A., Goertz, L., Wagner, A. H., et al. (2015). von Willebrand factor fibers promote cancer-associated platelet aggregation in malignant melanoma of mice and humans. Blood 125, 3153–3163. doi:10.1182/blood-2014-08-595686
Baxevanis, C. N., Sotiropoulou, P. A., Sotiriadou, N. N., and Papamichail, M. (2004). Immunobiology of HER-2/neu oncoprotein and its potential application in cancer immunotherapy. Cancer Immunol. Immunother. 53, 166–175. doi:10.1007/s00262-003-0475-7
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795. doi:10.1016/j.immuni.2013.10.003
Cancer Facts (2022). Cancer-facts-statistics. Available at: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html (Accessed May 3, 2022).
Caria, P., Cantara, S., Frau, D. V., Pacini, F., Vanni, R., and Dettori, T. (2016). Genetic heterogeneity of HER2 amplification and telomere shortening in papillary thyroid carcinoma. Int. J. Mol. Sci. 17, 1759. doi:10.3390/ijms17101759
Cheng, L., Fu, H., Jin, Y., Sa, R., and Chen, L. (2020). Clinicopathological features predict outcomes in patients with radioiodine-refractory differentiated thyroid cancer treated with sorafenib: A real-world study. Oncologist 25, e668–e678. doi:10.1634/theoncologist.2019-0633
Chesnokov, M. S., Yadav, A., and Chefetz, I. (2022). Optimized transcriptional signature for evaluation of MEK/ERK pathway baseline activity and long-term modulations in ovarian cancer. Biorxiv. doi:10.1101/2022.03.21.485160
Dawood, S., Broglio, K., Buzdar, A. U., Hortobagyi, G. N., and Giordano, S. H. (2009). Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: An institutional-based review. J. Clin. Oncol. 28, 92–98. doi:10.1200/jco.2008.19.9844
Degirmenci, U., Yap, J., Sim, Y. R. M., Qin, S., and Hu, J. (2021). Drug resistance in targeted cancer therapies with RAF inhibitors. Cancer Drug resist. 4, 665–683. doi:10.20517/cdr.2021.36
Fagin, J. A., and Wells, S. A. (2016). Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 375, 1054–1067. doi:10.1056/nejmra1501993
Farwell, M. D., Gamache, R. F., Babazada, H., Hellmann, M. D., Harding, J. J., Korn, R., et al. (2021). CD8-Targeted PET imaging of tumor-infiltrating T cells in patients with cancer: A phase I first-in-humans study of 89Zr-Df-IAB22M2C, a radiolabeled anti-CD8 minibody. J. Nucl. Med. 63, 720–726. doi:10.2967/jnumed.121.262485
Guan, S. S., Wu, C. T., Chiu, C. Y., Luo, T. Y., Wu, J. Y., Liao, T. Z., et al. (2018). Polyethylene glycol-conjugated HER2-targeted peptides as a nuclear imaging probe for HER2-overexpressed gastric cancer detection in vivo. J. Transl. Med. 16, 168. doi:10.1186/s12967-018-1550-3
Haibe, Y., Kreidieh, M., Hajj, H. E., Khalifeh, I., Mukherji, D., Temraz, S., et al. (2020). Resistance mechanisms to anti-angiogenic therapies in cancer. Front. Oncol. 10, 221. doi:10.3389/fonc.2020.00221
Herrmann, F., Lehr, H.-A., Drexler, I., Sutter, G., Hengstler, J., Wollscheid, U., et al. (2004). HER-2/neu-Mediated regulation of components of the MHC class I antigen-processing pathway. Cancer Res. 64, 215–220. doi:10.1158/0008-5472.can-2522-2
Hou, P., Bojdani, E., and Xing, M. (2010). Induction of thyroid gene expression and radioiodine uptake in thyroid cancer cells by targeting major signaling pathways. J. Clin. Endocrinol. Metab. 95, 820–828. doi:10.1210/jc.2009-1888
Jin, Y., Nostrand, D. V., Cheng, L., Liu, M., and Chen, L. (2018). Radioiodine refractory differentiated thyroid cancer. Crit. Rev. Oncol. Hematol. 125, 111–120. doi:10.1016/j.critrevonc.2018.03.012
Juríková, M., Danihel, Ľ., Polák, Š., and Varga, I. (2016). Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 118, 544–552. doi:10.1016/j.acthis.2016.05.002
Kebebew, E., Weng, J., Bauer, J., Ranvier, G., Clark, O. H., Duh, Q.-Y., et al. (2007). The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann. Surg. 125, 466–470. doi:10.1097/sla.0b013e318148563d
Kessler, J., Obinger, C., and Eales, G. (2008). Factors influencing the study of peroxidase-generated iodine species and implications for thyroglobulin synthesis. Thyroid 18, 769–774. doi:10.1089/thy.2007.0310
Keyaerts, M., Xavier, C., Heemskerk, J., Devoogdt, N., Everaert, H., Ackaert, C., et al. (2016). Phase I study of 68Ga-HER2-Nanobody for PET/CT assessment of HER2 expression in breast carcinoma. J. Nucl. Med. 57, 27–33. doi:10.2967/jnumed.115.162024
Kremser, R., Obrist, P., Spizzo, G., Erler, H., Kendler, D., Kemmler, G., et al. (2003). Her2/neu overexpression in differentiated thyroid carcinomas predicts metastatic disease. Virchows Arch. 442, 322–328. doi:10.1007/s00428-003-0769-3
Liang, H., Zhong, Y., Luo, Z., Huang, Y., Lin, H., Zhan, S., et al. (2011). Diagnostic value of 16 cellular tumor markers for metastatic thyroid cancer: An immunohistochemical study. Anticancer Res. 31, 3433–3440.
Lim, H., Devesa, S. S., Sosa, J. A., Check, D., and Kitahara, C. M. (2017). Trends in thyroid cancer incidence and mortality in the United States, 1974-2013. Jama 317, 1338–1348. doi:10.1001/jama.2017.2719
Lin, S., Li, Y., Sun, X., Chen, Q., Huang, S., Lin, S., et al. (2021). Update on the role of neuropeptide Y and other related factors in breast cancer and osteoporosis. Front. Endocrinol. 12, 705499. doi:10.3389/fendo.2021.705499
Liu, S., Geng, R., Lin, E., Zhao, P., and Chen, Y. (2021). ERBB1/2/3 expression, prognosis, and immune infiltration in cutaneous melanoma. Front. Genet. 12, 602160. doi:10.3389/fgene.2021.602160
Liu, Z., Zeng, W., Huang, L., Wang, Z., Wang, M., Zhou, L., et al. (2018). Prognosis of FTC compared to PTC and FVPTC: Findings based on SEER database using propensity score matching analysis. Am. J. Cancer Res. 8, 1440–1448.
Lu, C., Zhao, L., Ying, H., Willingham, M. C., and Cheng, S.-Y. (2010). Growth activation alone is not sufficient to cause metastatic thyroid cancer in a mouse model of follicular thyroid carcinoma. Endocrinology 151, 1929–1939. doi:10.1210/en.2009-1017
Mazzoni, M., Mauro, G., Erreni, M., Romeo, P., Minna, E., Vizioli, M. G., et al. (2019). Senescent thyrocytes and thyroid tumor cells induce M2-like macrophage polarization of human monocytes via a PGE2-dependent mechanism. J. Exp. Clin. Cancer Res. 38, 208. doi:10.1186/s13046-019-1198-8
Mehnert, J. M., Varga, A., Brose, M. S., Aggarwal, R. R., Lin, C.-C., Prawira, A., et al. (2019). Safety and antitumor activity of the anti–PD-1 antibody pembrolizumab in patients with advanced, PD-L1–positive papillary or follicular thyroid cancer. Bmc Cancer 19, 196. doi:10.1186/s12885-019-5380-3
Mitri, Z., Constantine, T., and O’Regan, R. (2012). The HER2 receptor in breast cancer: Pathophysiology, clinical use, and new advances in therapy. Chemother. Res. Pract. 2012, 743193. doi:10.1155/2012/743193
Montero-Conde, C., Ruiz-Llorente, S., Dominguez, J. M., Knauf, J. A., Viale, A., Sherman, E. J., et al. (2013). Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 3, 520–533. doi:10.1158/2159-8290.cd-12-0531
Nagaoka, K., Shirai, M., Taniguchi, K., Hosoi, A., Sun, C., Kobayashi, Y., et al. (2020). Deep immunophenotyping at the single-cell level identifies a combination of anti-IL-17 and checkpoint blockade as an effective treatment in a preclinical model of data-guided personalized immunotherapy. J. Immunother. Cancer 8, e001358. doi:10.1136/jitc-2020-001358
Neuzillet, C., Tijeras-Raballand, A., Mestier, L., de Cros, J., Faivre, S., and Raymond, E. (2014). MEK in cancer and cancer therapy. Pharmacol. Ther. 141, 160–171. doi:10.1016/j.pharmthera.2013.10.001
Oshi, M., Satyananda, V., Angarita, F. A., Kim, T. H., Tokumaru, Y., Yan, L., et al. (2021). Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients. Am. J. Cancer Res. 11, 1659–1671.
Piscuoglio, S., Ng, C. K. Y., Weigelt, B., Chandarlapaty, S., and Reis-Filho, J. S. (2018). ESR1 and endocrine therapy resistance: More than just mutations. Ann. Oncol. 29, 787–789. doi:10.1093/annonc/mdy081
Porra, V., Ferraro-Peyret, C., Durand, C., Selmi-Ruby, S., Giroud, H., Berger-Dutrieux, N., et al. (2005). Silencing of the tumor suppressor gene SLC5A8 is associated with BRAF mutations in classical papillary thyroid carcinomas. J. Clin. Endocrinol. Metab. 90, 3028–3035. doi:10.1210/jc.2004-1394
Portulano, C., Paroder-Belenitsky, M., and Carrasco, N. (2013). The Na+/I− symporter (NIS): Mechanism and medical impact. Endocr. Rev. 35, 106–149. doi:10.1210/er.2012-1036
Pratilas, C. A., Taylor, B. S., Ye, Q., Viale, A., Sander, C., Solit, D. B., et al. (2009). V600EBRAF is associated with disabled feedback inhibition of RAF–MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. U. S. A. 106, 4519–4524. doi:10.1073/pnas.0900780106
Qu, N., Zhang, L., Wu, W. L., Ji, Q. H., Lu, Z. W., Zhu, Y. X., et al. (2016). Bilaterality weighs more than unilateral multifocality in predicting prognosis in papillary thyroid cancer. Tumour Biol. 37, 8783–8789. doi:10.1007/s13277-015-4533-5
Ruggeri, R. M., Campennì, A., Giuffrè, G., Giovanella, L., Siracusa, M., Simone, A., et al. (2016). HER2 analysis in sporadic thyroid cancer of follicular cell origin. Int. J. Mol. Sci. 17, 2040. doi:10.3390/ijms17122040
Sa, R., Liang, R., Qiu, X., He, Z., Liu, Z., and Chen, L. (2022). IGF2BP2-dependent activation of ERBB2 signaling contributes to acquired resistance to tyrosine kinase inhibitor in differentiation therapy of radioiodine-refractory papillary thyroid cancer. Cancer Lett. 527, 10–23. doi:10.1016/j.canlet.2021.12.005
Seliger, B., and Kiessling, R. (2013). The two sides of HER2/neu: Immune escape versus surveillance. Trends Mol. Med. 19, 677–684. doi:10.1016/j.molmed.2013.08.003
Shah, M. H., Wei, L., Wirth, L. J., Daniels, G. A., Souza, J. A. D., Timmers, C. D., et al. (2017). Results of randomized phase II trial of dabrafenib versus dabrafenib plus trametinib in BRAF-mutated papillary thyroid carcinoma. J. Clin. Oncol. 35, 6022. doi:10.1200/jco.2017.35.15_suppl.6022
Shibuya, M. (2011). Vascular endothelial growth factor (vegf) and its receptor (vegfr) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes. Cancer 2, 1097–1105. doi:10.1177/1947601911423031
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. Ca. Cancer J. Clin. 71, 7–33. doi:10.3322/caac.21654
Smith, N. R., Baker, D., James, N. H., Ratcliffe, K., Jenkins, M., Ashton, S. E., et al. (2010). Vascular endothelial growth factor receptors VEGFR-2 and VEGFR-3 are localized primarily to the vasculature in human primary solid cancers. Clin. Cancer Res. 16, 3548–3561. doi:10.1158/1078-0432.ccr-09-2797
Srinivasan, S., Kryza, T., Batra, J., and Clements, J. (2022). Remodelling of the tumour microenvironment by the kallikrein-related peptidases. Nat. Rev. Cancer 22, 223–238. doi:10.1038/s41568-021-00436-z
Tangye, S. G., Al-Herz, W., Bousfiha, A., Chatila, T., Cunningham-Rundles, C., Etzioni, A., et al. (2020). Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 40, 24–64. doi:10.1007/s10875-019-00737-x
Wagle, M.-C., Kirouac, D., Klijn, C., Liu, B., Mahajan, S., Junttila, M., et al. (2018). A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis. Oncol. 2, 7. doi:10.1038/s41698-018-0051-4
Wu, L., Zhou, Y., Guan, Y., Xiao, R., Cai, J., Chen, W., et al. (2021). Seven genes associated with lymphatic metastasis in thyroid cancer that is linked to tumor immune cell infiltration. Front. Oncol. 11, 756246. doi:10.3389/fonc.2021.756246
Wynn, C. S., and Tang, S.-C. (2022). Anti-HER2 therapy in metastatic breast cancer: Many choices and future directions. Cancer Metastasis Rev. 41, 193–209. doi:10.1007/s10555-022-10021-x
Xia, C., Dong, X., Li, H., Cao, M., Sun, D., He, S., et al. (2022). Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 135, 584–590. doi:10.1097/cm9.0000000000002108
Keywords: ERBB2, prognosis, biomarker, immune microenviroment, papillary thyroid cancer
Citation: Jin Y, Qiu X, He Z, Wang J, Sa R and Chen L (2022) ERBB2 as a prognostic biomarker correlates with immune infiltrates in papillary thyroid cancer. Front. Genet. 13:966365. doi: 10.3389/fgene.2022.966365
Received: 10 June 2022; Accepted: 28 October 2022;
Published: 09 November 2022.
Edited by:
Amitabha Chaudhuri, SciGenom Labs, IndiaReviewed by:
Laura Sterian Ward, State University of Campinas, BrazilWei Zhou, Cancer Hospital, Chongqing University, China
Copyright © 2022 Jin, Qiu, He, Wang, Sa and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Libo Chen, lbchen@sjtu.edu.cn
†These authors have contributed equally to this work
 Yuchen Jin
Yuchen Jin