- 1Department of Nephrology, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 2Laboratory of Basic Medical Sciences, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 3Department of General Practice, Qilu Hospital of Shandong University, Jinan, Shandong, China
Diabetic nephropathy (DN), a frequent microvascular complication of diabetes, has been recognized as a primary cause of chronic kidney disease (CKD) and end-stage renal disease (ESRD). Previous studies found that autophagy of renal tubular epithelial cells plays an important role in DN pathogenesis. Our research aimed to investigate the differentially expressed autophagy-related genes (DEARGs) between DN and healthy renal tubule samples and identify a novel autophagy-related biomarker associated with tubulointerstitial injury in DN. In this study, gene expression profiles of renal tubules from 10 DN patients and 24 healthy controls in the GSE30122 dataset were analyzed, and 43 DEARGs were identified by bioinformatics analysis. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and correlation analysis were performed on DEARGs, and the hub gene prolyl 4-hydroxylase subunit beta (P4HB) was screened by protein–protein interaction and verified by utilizing other datasets and stimulating HK-2 cells under high glucose concentration. We found that the expression of P4HB in renal tubules was correlated with renal function. In summary, our research provided novel insights for comprehension of DN molecular mechanisms and identified P4HB as a novel autophagy-related biomarker of DN.
Introduction
Diabetic nephropathy (DN), a common microvascular complication of diabetes, has been recognized as a primary cause of chronic kidney disease (CKD) and end-stage renal disease (ESRD) in many developed and developing countries (Bikbov et al., 2020; Johansen et al., 2021). According to recent reports, DN has accounted for 20 %–40 % of patients requiring kidney replacement therapy worldwide, contributing to more than 950,000 deaths worldwide each year (Tang and Yiu, 2020). DN has become a serious global healthcare problem.
Although glomerular injury is a major pathological manifestation of DN, there is growing evidence suggesting that tubulointerstitial pathological changes, such as tubular atrophy, interstitial fibrosis, autophagy, and apoptosis of tubular epithelial cells, play a crucial role in DN development (Brezniceanu et al., 2010; C. Yang et al., 2021a; Liu et al., 2022). Apart from all other reasons, non-enzymatic glycation is considered one of the major reasons behind diabetes, including DN. It was found that DNA-AGEs and auto-antibodies against glycated DNA are related to diabetic microvascular complications, such as DN (Ahmad et al., 2014). At the same time, it was also reported that some commonly used drugs like ezetimibe and rosuvastatin can show strong antidiabetic and renal protective effects by targeting AGE/RAGE-associated signaling (Nabi et al., 2021a; Nabi et al., 2021b). In addition, some common components in plants such as ellagic acid and lycopene play a protective role in diabetes and DN by reducing the formation of AGEs (Tabrez et al., 2015; Ahmad et al., 2022).
Autophagy is a cellular process in which damaged organelles, protein aggregates, and other macromolecules are degraded in the cytoplasm (Galluzzi et al., 2017). Autophagy dysfunction is associated with pathogenesis of various diseases (Dikic and Elazar, 2018; C. Zhang et al., 2020). Several pathways affect the biological function of DN by influencing autophagy. Li et al. (2021) reported that activation of the epidermal growth factor receptor (EGFR) signaling pathway can exacerbate kidney damage by inhibiting autophagy. However, autophagy-related genes (ARGs) in DN are still largely unknown and require further exploration. Exploration and illumination of differentially expressed autophagy-related genes (DEARGs) in DN will provide us with novel biomarkers for treatment of DN. Bioinformatics is a method for efficiently and accurately processing large quantities of data, providing valuable information to patients. Nevertheless, studies on the investigation of the expression of diabetic renal tubular interstitial genes and autophagy through bioinformatics are still lacking.
Woroniecka et al. completed the DN-related dataset GSE30122 for analysis of gene expression differences between DN patients and healthy controls (Woroniecka et al., 2011). In this study, we re-analyzed their dataset from other perspectives to explore DEARGs in DN vs. normal human renal tubular interstitial genes. Subsequently, we performed Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and correlation analysis on DEARGs. Protein–protein interaction (PPI) showed that prolyl 4-hydroxylase subunit beta (P4HB) was identified as an autophagy-related hub gene for DN. Finally, we further validated the upregulated expression of P4HB in DN tubulointerstitium by exploiting another database (GSE104954) (Grayson et al., 2018) and the European Renal cDNA Bank (ERCB) cohort, as well as establishing an in vitro model, and the expression of P4HB in renal tubules was correlated with renal function. The experiments’ schematic workflow is displayed in Figure 1. Our study suggested that P4HB is a potential key biomarker in the pathogenesis of renal tubular injury in DN.
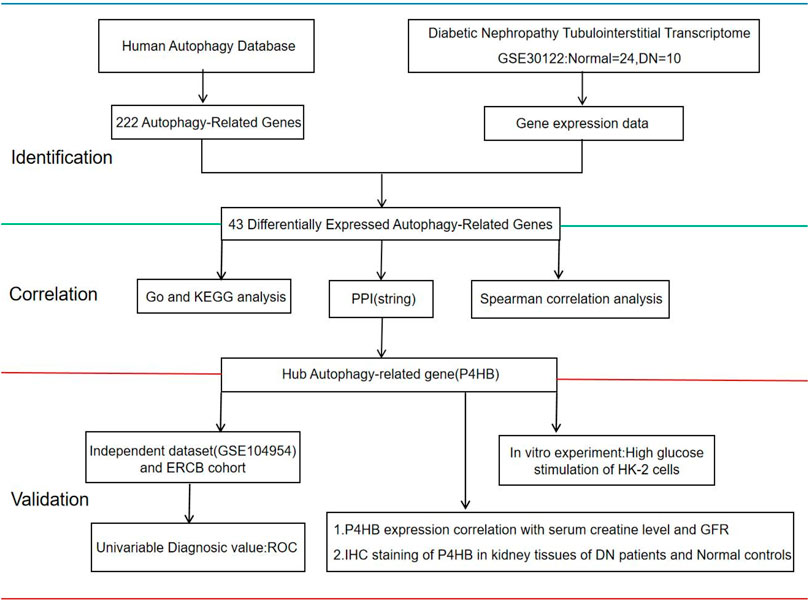
FIGURE 1. Workflow of this study. The gene expression of human renal tubulointerstitial data was extracted from the GEO database (GSE30122). GEO, Gene Expression Omnibus; Normal, healthy controls; DN, patients with diabetic nephropathy; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein Interaction; ERCB: European Renal cDNA Bank; ROC, receiver operating curve; GFR, glomerular filtration rate; IHC, immunohistochemistry.
Materials and methods
Data download and preprocessing
The Human Autophagy Database (HADb; http://www.autophagy.lu/index.html) was used to obtain genes involved in autophagy. Genomic and transcriptomic datasets of DN and healthy renal tubule samples were obtained using Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Data from GSE30122 (GPL571 platform, Affymetrix Human Genome U133A 2.0 Array) included the data of 10 patients with DN (GSM757014–GSM757023) and 24 control human kidney tubules (GSM757024–GSM757035, GSM758498–GSM758509). The basic characteristics of 10 DN patients are summarized in Supplementary Table S1, and the histological evaluation of the 10 DN patients showed that renal tubular atrophy, interstitial fibrosis, vascular sclerosis, and mesangial matrix dilatation increased significantly (Woroniecka et al., 2011). Data from GSE104954 (GPL22, 945 platform, Affymetrix Human Genome U133 Plus 2.0 Array, and GPL 24,120 platform, Affymetrix Human Genome U133A Array) were used for validation. The “sva” (Parker et al., 2014) and “limma” (Ritchie et al., 2015) packages in R software were used to normalize raw data in batches.
Differentially expressed autophagy-related gene analysis
The reproducibility of the GSE30122 data was examined using the principal component analysis (PCA) method. The “limma” package was utilized to investigate the differential expression of genes related to autophagy. Genes were identified as differentially expressed genes based on an adjusted p-value of <0.05 and an absolute fold-change value of >1.5. Heatmaps were created using the “pheatmap” package in R, and the volcano plots and box plots were performed using the Sangerbox tools, a free online platform for data analysis (http://vip.sangerbox.com/home.html).
Gene functions and correlation analysis
Enrichment analyses were conducted for DEARGs. GO and KEGG pathway enrichment analyses were performed using DAVID version 6.8 (https://david.ncifcrf.gov/conversion.jsp), a commonly used tool for detailed analysis and classification of genes and protein functions in bioinformatics research. The GO analysis included cellular composition (CC), biological process (BP), and molecular function (MF). Enrichment results and correlation analysis were drawn by https://www.bioinformatics.com.cn, a free online platform for data analysis and visualization.
Protein–protein interaction network construction
The PPI network was constructed on the basis of the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online analysis (https://string-db.org/). PPI network visualization and analyses were performed with Cytoscape (Version 3.9.1). Thirty hub genes related to autophagy were identified by the Density of Maximum Neighborhood Component (DMNC), and P4HB was screened as a hub gene. An online platform (https://www.bioinformatics.com.cn) was also used to show the receiver operating curve (ROC) to evaluate the ability of P4HB to discriminate between DN patients from healthy controls.
P4HB validation and correlation analysis with renal function
The differential gene expression of P4HB between DN and healthy renal tubule samples was verified in the GSE104954 dataset and ERCB cohort (31 healthy controls and 17 DN patients) (Ju et al., 2015). Then, the association analysis of P4HB expression and clinical characteristics was validated in ERCB using Pearson’s correlation analysis by using the Nephroseq v5 online database (http://v5.nephroseq.org).
Cell culture and treatments
The human proximal tubular cell line HK-2 was purchased from the National Collection of Authenticated Cell Cultures and cultured in Dulbecco’s modified Eagle’s medium (c11885500BT, Gibco) by adding 10 % fetal bovine serum (10270-106, Gibco) and 1 % penicillin–streptomycin (p1400, Solarbio) at 37°C and 5 % CO2 in a humidified environment. High glucose concentration (30 mM, G8644, Sigma) for 24 h (Zhan et al., 2015) was used to cause HK-2 cell damage, and the addition of 5.5 mmol/L glucose served as the control.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from cultured HK-2 cells using the RNAfast200 Kit (Fastagen), and cDNA was synthesized using the SureScriptTM First-Stand cDNA Synthesis Kit (QP056T, GeneCopoeia). Real-time quantitative polymerase chain reaction (RT-qPCR) was conducted using SYBR Green reagent (1725201, Bio-Rad) on a Bio-Rad CFX PCR System (Bio-Rad). The procedure was repeated three times for each sample. The primers are given in Table 1. Gene expression analysis was performed using the 2−ΔΔCt method, and expression levels were normalized to those of GAPDH.
Protein extraction and Western blotting analysis
Total proteins were extracted by the incubation of cultured HK-2 cells with radioimmunoprecipitation assay (RIPA) buffer (P0013D, Beyotime) and adding 1 % phenylmethylsulfonyl fluoride (PMSF) (329-98-6, Solarbio) and quantified by Bio-RAD assays. The same amount of protein was separated by 10 % sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were then blocked with 5 % skim milk and subsequently incubated with primary antibodies against P4HB (1:1,000 dilution, Cat.137,110, Abcam) and GAPDH (1:6,000 dilution, Cat. 60004-1-Ig, ProteinTech) overnight at 4°C. After washing, the proteins were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:6,000 dilution, Cat. SA00001-2, ProteinTech) for 1 h at room temperature. The bands were visualized using an enhanced chemiluminescence (ECL) system, and the signal intensity was quantitatively processed by ImageJ software.
Immunohistochemistry and immunofluorescent staining
The paraffin-embedded kidney sections of the kidney tissues of three DN patients diagnosed by renal biopsy from the pathology department of Qilu Hospital affiliated with Shandong University and the paraffin-embedded kidney sections of healthy adjacent kidney tissues of three individuals who underwent tumor nephrectomy (no diabetes or other kidney diseases) were immunohistochemically stained by P4HB, as approved by the Research Ethics Committee Qilu Hospital of Shandong University (NO. KYLL-2020(KS)-030). These sections were incubated with an anti-P4HB antibody (1:1,000 dilution, Cat.137110, Abcam) at 4°C overnight. The general two-step method was used for detection and incubation, and the 3, 3′-diaminobenzidine (DAB) chromogenic kit was used for immunohistochemical staining. Images were collected and analyzed using the NIS Element software and Nikon microscope imaging system, and ImageJ software was used for quantitative analysis.
For immunofluorescent staining of cells, the cells were fixed with 4 % paraformaldehyde, incubated with TritonX-100 (1 %) for 20 min, and then incubated with the P4HB primary antibody (1:200 dilution, Cat.137110, Abcam) overnight at 4°C, Then, the cells were incubated with goat anti-rabbit immunoglobulin (Ig)G DyLight 594 (1:500, Cat. A23420, Abbkine Scientific Company) coupled with a fluorescent probe in the dark at room temperature for 1 h, and the nuclei were observed by DAPI (AR1176, Boster Bio) staining. Images were captured by a fluorescence microscope (Olympus).
Statistical analysis
GraphPad Prism 7 and R software version 4.1.3 were used for statistical analysis. A two-tailed unpaired t test was used for comparisons between the two groups. Differences were considered statistically significant at p <0.05. (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Results
Identification of differentially expressed autophagy-related genes between the DN and healthy tubule samples
The renal tubular transcriptome data GSE10322 were used for further analysis to investigate the role of ARGs in DN pathogenesis. The distribution of DN and normal samples was shown by the PCA results (Figure 2A). The expression of 222 ARGs in 10 DN patients and 24 normal samples was subsequently analyzed, and 43 ARGs showed differential expression in DN with an adjusted p value <0.05 and an absolute fold-change value of >1.5, including 38 upregulated and 5 downregulated genes (Table 2; Figures 2B–D).
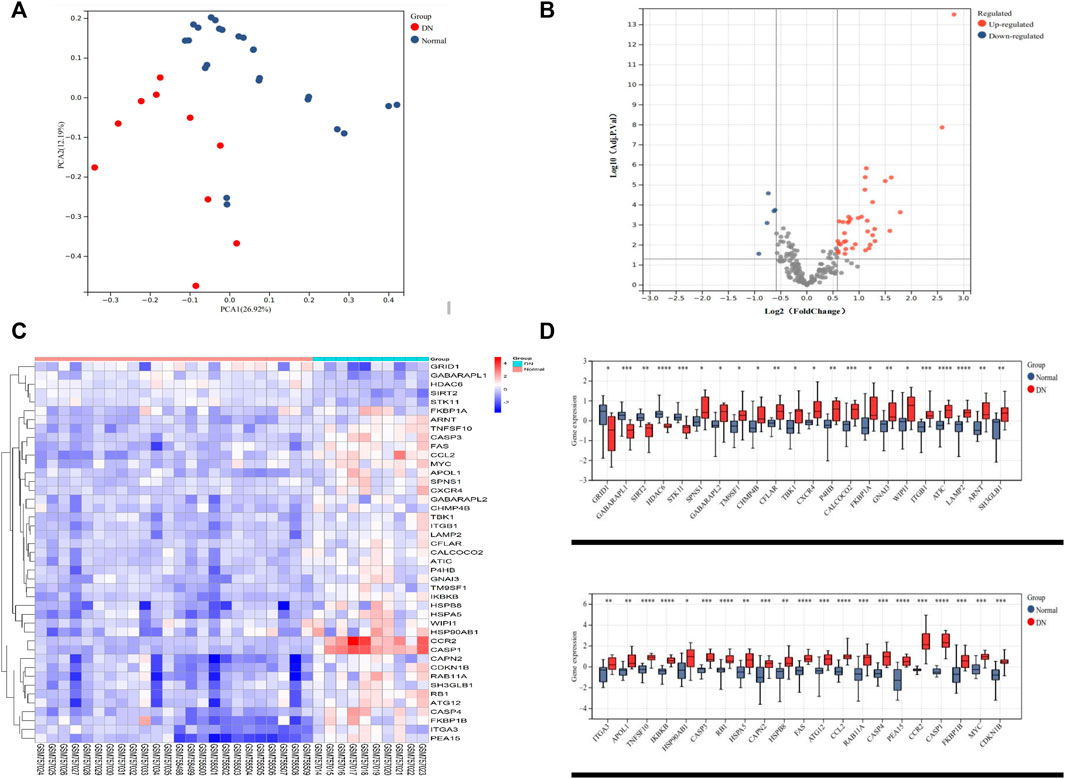
FIGURE 2. Differentially expressed autophagy-related genes in the tubulointerstitium between DN and normal renal tissue. (A) PCA for GSE30122. (B) Volcano plot of 222 DEARGs in DN and normal samples. (C) Heatmap of the 43 DEARGs in DN and normal samples. (D) Boxplot of the 43 DEARGs in DN and normal samples. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. PCA, principal component analysis; DEARG, differentially expressed autophagy-related gene; DN, diabetic nephropathy.
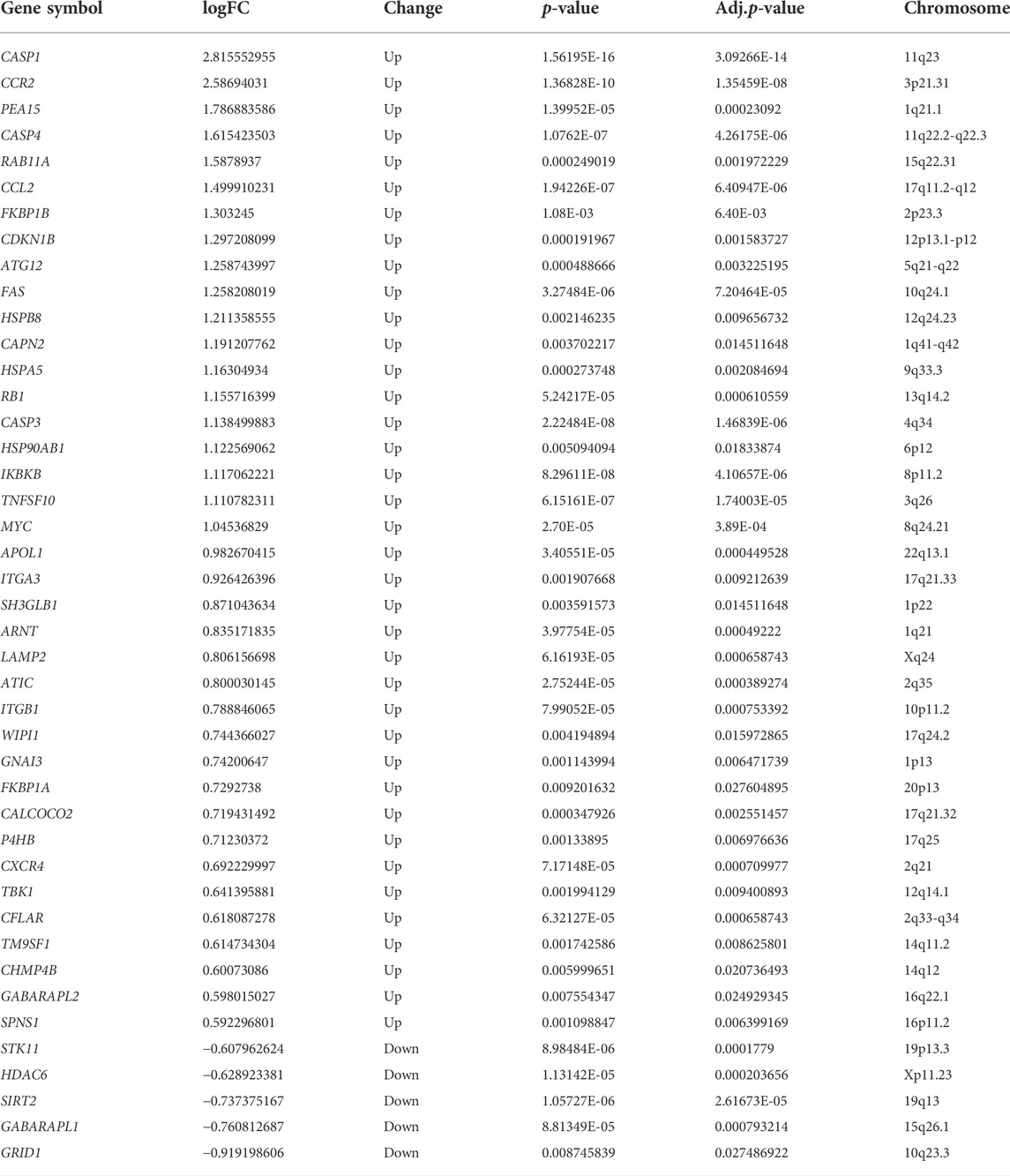
TABLE 2. 43 differentially expressed autophagy-related genes in the tubulointerstitium between DN and normal renal tissue.
Functional enrichment analysis of the differentially expressed autophagy-related genes
GO and KEGG pathway enrichment analyses were performed to further explore the biological functions of these DEARGs. The most involved processes or components in the GO included autophagy and apoptotic process (biological process), autophagosome membrane and membrane raft (cell component), and cysteine-type endopeptidase activity involved in the apoptotic signaling pathway and ubiquitin protein ligase binding (molecular function) (Figures 3A, B; Supplementary Table S2). The DEARGs mainly involved in the autophagy-associated process were shown by the KEGG pathway enrichment analysis (Figures 3C, D; Supplementary Table S3).
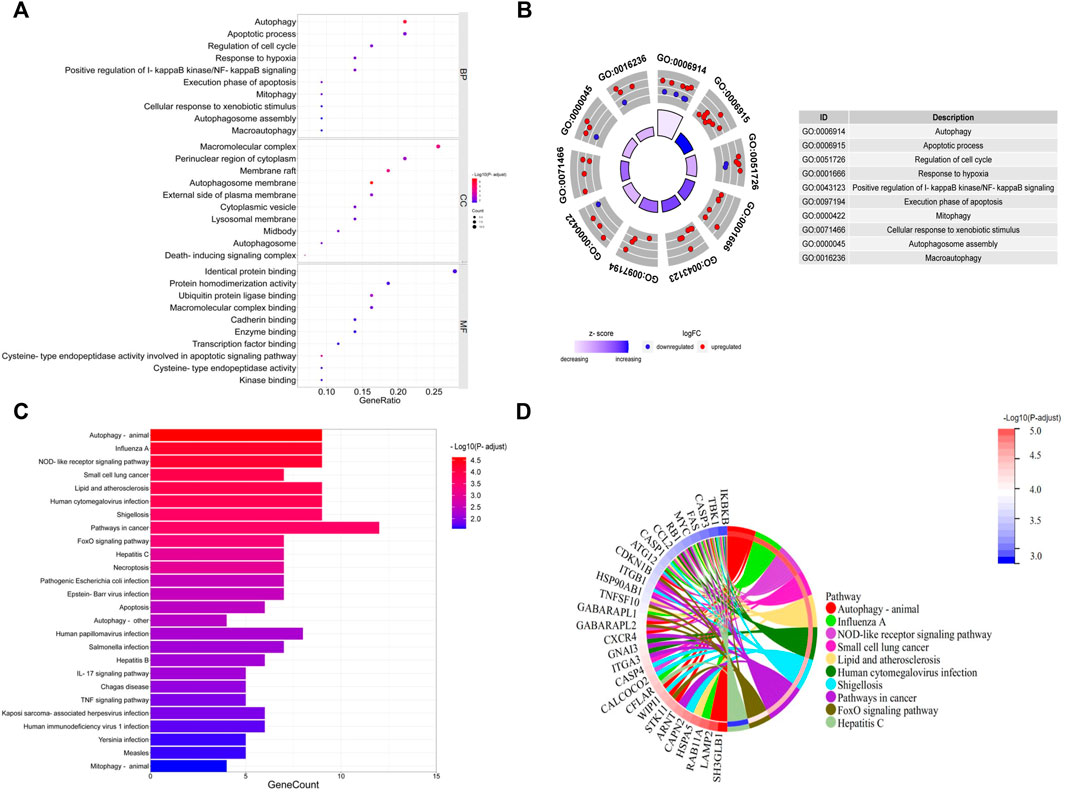
FIGURE 3. GO and KEGG enrichment analyses of 43 differentially expressed autophagy-related genes. (A, B) Bubble plot and circle plot of GO enrichment terms. BP, biological process; CC, cellular component; MF, molecular function. (C, D) Bar plot and Chord plot of KEGG enrichment pathways. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Hub gene identification and validation
A correlation analysis was conducted to investigate the expression relevance of these DEARGs. The relationship of 43 DEARGs in the GSE30122 dataset is shown in Figure 4A. The human protein interaction database (String) was chosen to identify the interactions among DEARGs, and 30 hub genes were calculated by the DMNC algorithm. P4HB was screened as a hub DEARG in DN (Figure 4B). We utilized another independent dataset GSE104954 and an ERCB cohort to further verify the change in P4HB expression, showing that P4HB expression was significantly upregulated in DN tubule samples (Figures 4C–F). Additionally, the ROC analysis demonstrated that the expression of P4HB showed an excellent diagnostic value for DN patients and healthy controls (Figures 4C–F).
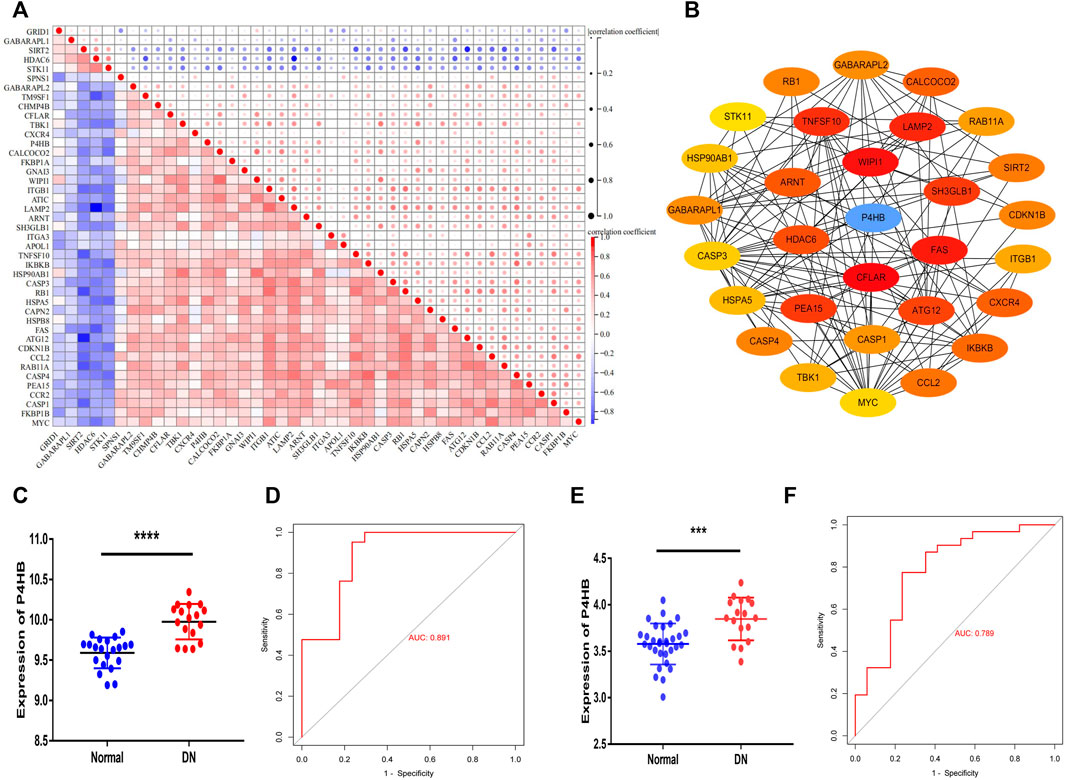
FIGURE 4. Identifying P4HB as a hub autophagy-related gene in DN and validating in the GSE104954 database and ERCB cohort. (A) Spearman’s correlation analysis of the 43 DEARGs. (B) Top 30 hub DEARGs identified via the PPI network. (C, D) Validation of P4HB in GSE104954. (C) Significantly upregulated expression of P4HB in DN patients (n = 17) compared to healthy samples (n = 21) (p < 0.0001) (D) ROC curve of P4HB expression in DN (AUC = 89.1%). (E, F) Validation of P4HB in the ERCB cohort. (E) Significantly upregulated expression of P4HB in DN patients (n = 17) compared to healthy samples (n = 31) (p < 0.0001). (F) ROC curve of P4HB expression in DN (AUC = 78.9%). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. DEARG, differentially expressed autophagy-related gene; DN, diabetic nephropathy; PPI, protein–protein Interaction; ROC, receiver operating curve; AUC, area under the curve; ERCB: European Renal cDNA Bank.
Validation of P4HB expression under high glucose stimulation
After determining from other datasets that the expression of P4HB was significantly elevated in DN patients, the transcriptional and protein levels of P4HB were explored after high glucose stimulation and normal glucose treatment of HK-2 cells. RT-qPCR (Figure 5A) and Western blot analysis (Figures 5B, C) further validated that P4HB expression was elevated in HK-2 cells under high glucose (30 mM) stimulation, accompanied by increased expression of NGAL, a marker for renal tubular injury, indicating that there was significant damage in HK-2 cells. Simultaneously, cell immunofluorescence showed that the expression of P4HB in HK-2 cells increased significantly after high glucose stimulation (Figure 5D).
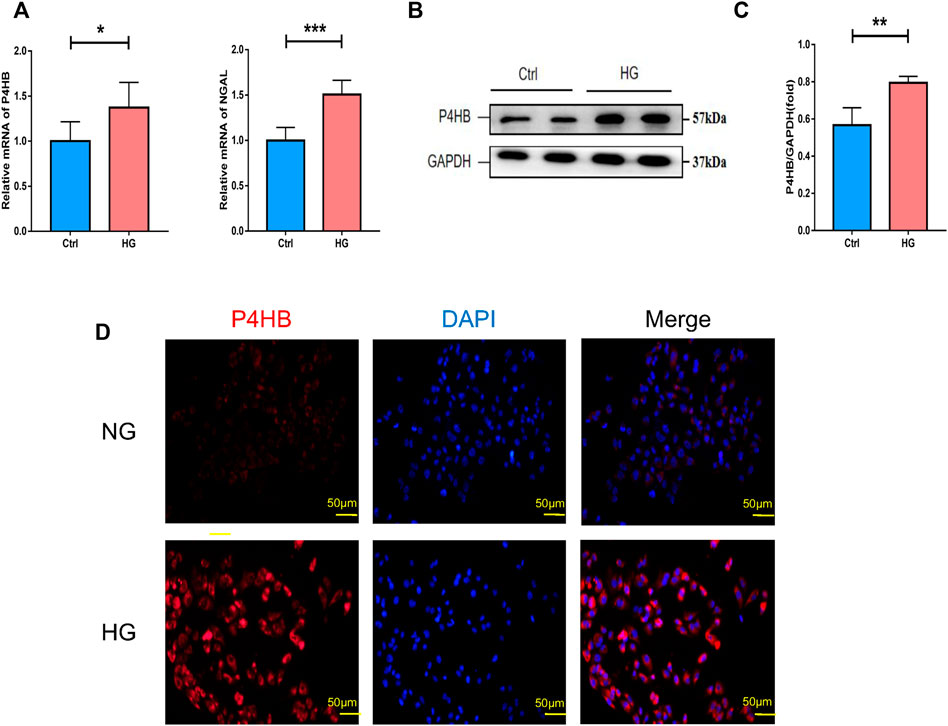
FIGURE 5. Validation of P4HB expression in vitro. (A) RT-qPCR analysis of P4HB and NGAL in HK-2 cells after HG stimulation and NG treatment. (B) Western blot analysis of P4HB in HK-2 cells after HG stimulation and NG treatment. (C) Densitometric quantification of P4HB in HK-2 cells after HG stimulation and NG treatment. Results are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. (D) Immunofluorescence showing the expression of P4HB in HK-2 after HG stimulation and NG treatment. Scale bars, 50 μm. RT-qPCR, real-time quantitative polymerase chain reaction; SEM, standard error of the mean; HG, high glucose; NG, normal glucose.
Clinical relevance of P4HB expression and immunohistochemical validation of P4HB expression
The correlation analysis of the ERCB cohort was performed to validate the relationship between the expression of P4HB in renal tubular and renal function. P4HB expression was positively correlated with serum creatinine levels (r = 0.351, p = 0.028) and negatively correlated with glomerular filtration rate (GFR) (r = −0.472, p = 0.002) (Figures 6A,B). Additionally, we performed immunohistochemical staining of P4HB in kidney tissues of healthy controls and DN patients, and found that the expression of P4HB in the renal tubules of DN patients was significantly higher than that in normal kidneys (Figures 6C,D).
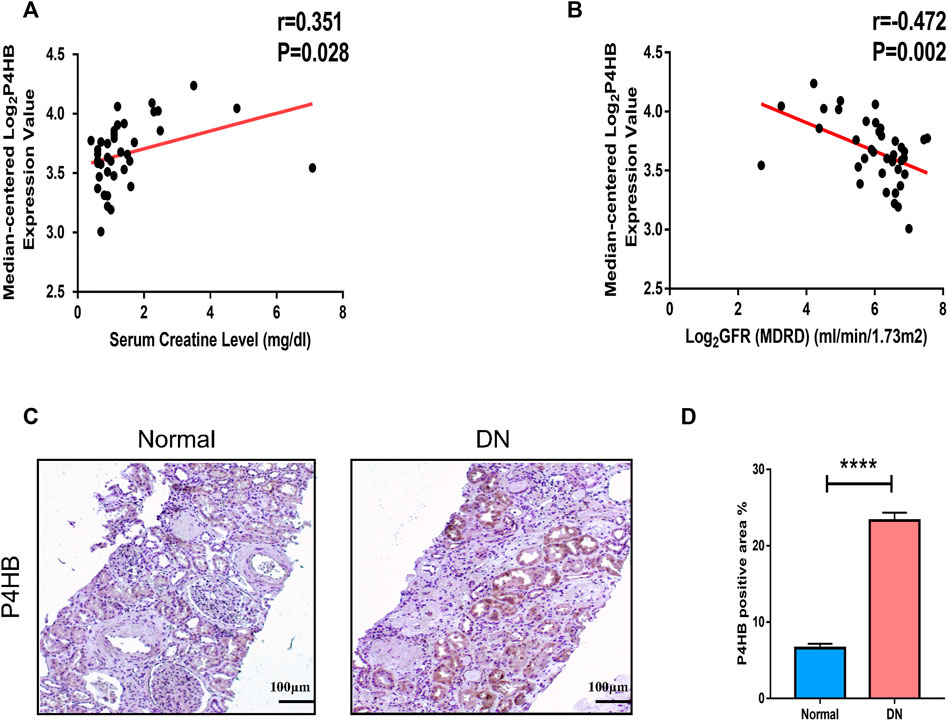
FIGURE 6. Clinical relevance of P4HB and immunohistochemical validation of P4HB expression. (A) Relevance of the expression of P4HB and serum creatinine level. (B) Relevance of the expression of P4HB and GFR. (C,D) Typical images and statistical charts of immunohistochemical staining of P4HB in normal renal tissues and DN renal tissues. Scale bars, 100 μm. Results are expressed as the mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001, and ****p < 0.0001. GFR, glomerular filtration rate; DN, diabetic nephropathy; SEM, standard error of the mean.
Discussion
DN is one of the most serious microvascular complications of diabetes, accounting for approximately 30 %–40 % of end-stage kidney disease patients worldwide (Maezawa, Takemoto and Yokote., 2015; L. Zhang et al., 2016). Poor prognosis and low quality of life are distinctive characteristics of patients with DN (Slieker et al., 2021). Therefore, novel and satisfactory strategies are urgently needed to treat DN. There is growing evidence that multiple biological functions are engaged in the pathogenesis of DN, such as immunity, inflammation, and autophagy (Yang et al., 2019; Yang M. et al., 2021; Yu et al., 2021; Li et al., 2022). In our research, starting from autophagy, for the first time, we identified DEARGs in DN vs. normal human renal tubular interstitial cells by bioinformatics analysis, as well as determined a new autophagy-related biomarker for DN, thus providing novel insights into the tubulointerstitial pathogenesis of DN and contributing to the identification of novel therapeutic targets.
P4HB, a member of the protein disulfide isomerase (PDI) family, is a multifunctional protein capable of catalyzing the generation and reorganization of disulfide bonds (Noiva, 1999). P4HB, as an autophagy-related gene, can be detected in various diseases that involve inflammation and apoptosis, including cancer, endocrine diseases, and skin diseases. Elevated expression of P4HB has been reported in several solid tumors, such as ovarian cancer (Bonome et al., 2008), bladder cancer (Lyu et al., 2020; Wang et al., 2020), and prostate cancer (Welsh et al., 2001; Singh et al., 2002). Additionally, Ding et al. (2020) reported that targeting P4HB can reduce inflammation and melanogenesis of the skin. Furthermore, previous studies found that PDIA1 contributes to oxidative maturation of proinsulin in the endoplasmic reticulum to support insulin production and ß-cell health in diet-induced obesity (Jang et al., 2019), indicating that P4HB could indirectly influence insulin production and ß-cell health.
One study has determined that P4HB was substantially elevated as an ARG in kidney renal clear cell carcinoma (KIRC) and showed high diagnostic and prognostic ability (Xie et al., 2020). There are also reports demonstrating that P4HB overexpression was associated with poor prognosis in human KIRC (Zhu et al., 2019; Wu et al., 2021), indicating that overexpression of P4HB is an adverse prognostic factor. Additionally, Fu et al. (2021) reported that the self-antigen P4HB located on the cell membrane of kidney cells could be cross-recognized by anti-HU1 (a conserved peptide derived from DNABII proteins) and induce lupus nephritis (LN). Hence, evidence suggests that P4HB might be a key biomarker and therapeutic target for human kidney diseases.
In this study, transcriptomic variations of ARGs in the gene expression profiles of 10 DN and 24 healthy renal tubule samples were analyzed, and 43 DEARGs were identified in DN samples compared with healthy samples. The GO and KEGG enrichment analyses of DEARGs were subsequently performed. These genes were mostly enriched in autophagy-related biological processes, such as autophagy, mitophagy, autophagosome assembly, and macroautophagy. These processes may be associated with various infections and diseases that were inferred from the enrichment analysis of pathways, including influenza A, lipid dysfunction and atherosclerosis, and cancer. Previous studies have also shown that DN was associated with various autophagy-related biological functions (Yang M. et al., 2021; Liu et al., 2022). Then, correlation analysis and the PPI network were constructed to further explore the correlation between the DEARG expression. According to the PPI network, DMNC scores from cytoHubba confirmed that P4HB was a key hub gene in DN tubular injury.
The elevation of renal tubular P4HB expression in DN was verified in other datasets and in vitro experiments. We first verified the elevated gene expression by other databases, and the result was consistent with that of our previous study. The ROC analysis indicated that P4HB expression showed an excellent diagnostic value for DN patients and healthy controls. Through in vitro experiments, we found that the expression of P4HB was significantly elevated after high glucose stimulation of HK-2, accompanied by increased expression of NGAL, a marker for kidney tubular injury, indicating that there was significant damage in HK-2 cells (Satirapoj, 2018). In the meantime, we found that P4HB expression was positively correlated with serum creatinine levels and negatively correlated with GFR, and immunohistochemistry staining showed that the expression of P4HB in the renal tubules of DN patients was significantly higher than that of normal kidneys. Therefore, P4HB might serve as an autophagy-related biomarker for DN.
This study can provide novel insights and potential targets for further studies on the connection between DN and autophagy. However, our analysis was limited by the number of samples included, as transcriptomic data on DN tubuleinterstitium are restricted and most data were tested with different platforms. More combined samples and clinical information are required to clarify the potential mechanisms of P4HB in DN.
Conclusion
We identified DEARGs in DN vs. normal human renal tubular interstitial cells by bioinformatics analysis for the first time, and P4HB was found and confirmed as an autophagy-related biomarker for DN, thereby providing new insights and potential targets for further studies on the correlation between DN and autophagy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below:, GSE30122; further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
FB carried out the assays, made relevant analysis, and wrote the manuscript. FB, KY, YY, YZ, LD, and XA conducted this research. FF, NS, and JF provided materials and skills and assisted in building cell models. LL, HY, and XY designed the experiment and reviewed and approved the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 82070746) and the National Natural Science Fund for Outstanding Youth Fund (Grant No. 82000692).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.965816/full#supplementary-material
References
Ahamd, S., Alouffi, S., Khan, S., Khan, M., Akasha, R., Ashraf, J. M., et al. (2022). Physicochemical characterization of in vitro LDL glycation and its inhibition by ellagic acid (EA): An in vivo approach to inhibit diabetes in experimental animals. Biomed. Res. Int. 2022, 5583298. doi:10.1155/2022/5583298
Ahamd, S., MmoinuddinShahab, U., Habisb, S., Khan, M. S., Alam, K., and Ali, A. (2014). Glycoxidative damage to human DNA: Neo-antigenic epitopes on DNA molecule could be a possible reason for autoimmune response in type 1 diabetes. Glycobiology 24 (3), 281–291. doi:10.1093/glycob/cwt109
Bikbov, B., Purcell, C., Levey, A. S., Smith, M., Abdoli, A., Abebe, M., et al. (2020). Global, regional, and national burden of chronic kidney disease, 1990-2017: A systematic analysis for the global burden of disease study 2017. Lancet 395 (10225), 709–733. doi:10.1016/s0140-6736(20)30045-3
Bonome, T., Levine, D. A., Shih, J., Randonovich, M., Pise-Masison, C. A., Bogomolniy, F., et al. (2008). A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 68 (13), 5478–5486. doi:10.1158/0008-5472.Can-07-6595
Brezniceanu, M.-L., Lau, C. J., Godin, N., Chenier, I., Duclos, A., Ethier, J., et al. (2010). Reactive oxygen species promote caspase-12 expression and tubular apoptosis in diabetic nephropathy. J. Am. Soc. Nephrol. 21 (6), 943–954. doi:10.1681/asn.2009030242
Dikic, I., and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19 (6), 349–364. doi:10.1038/s41580-018-0003-4
Ding, X.-J., Zhang, Z.-Y., Jin, J., Han, J.-X., Wang, Y., Yang, K., et al. (2020). Salidroside can target both P4HB-mediated inflammation and melanogenesis of the skin. Theranostics 10 (24), 11110–11126. doi:10.7150/thno.47413
Fu, W., Liu, Y., Liu, F., Liu, C., Li, J., Niu, J., et al. (2021). A novel autoantibody induced by bacterial biofilm conserved components aggravates Lupus nephritis. Front. Immunol. 12, 656090. doi:10.3389/fimmu.2021.656090
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Pedro, J. M. B.-S., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. Embo J. 36 (13), 1811–1836. doi:10.15252/embj.201796697
Grayson, P. C., Eddy, S., Taroni, J. N., Lightfoot, Y. L., Mariani, L., Parikh, H., et al. (2018). Metabolic pathways and immunometabolism in rare kidney diseases. Ann. Rheum. Dis. 77 (8), 1226–1233. doi:10.1136/annrheumdis-2017-212935
Jang, I., Pottekat, A., Poothong, J., Yong, J., Lagunas-Acosta, J., Charbono, A., et al. (2019). PDIA1/P4HB is required for efficient proinsulin maturation and ß cell health in response to diet induced obesity. Elife 8, e44528. doi:10.7554/eLife.44528
Johansen, K. L., Chertow, G. M., Foley, R. N., Gilbertson, D. T., Herzog, C. A., Ishani, A., et al. (2021). US renal data system 2020 annual data report: Epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 77, A7–A8. doi:10.1053/j.ajkd.2021.01.002
Ju, W., Nair, V., Smith, S., Zhu, L., Shedden, K., Song, P. X. K., et al. (2015). Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci. Transl. Med. 7 (316), 316ra193. doi:10.1126/scitranslmed.aac7071
Li, A., Yi, B., Han, H., Yang, S., Hu, Z., Zheng, L., et al. (2022). Vitamin D-VDR (vitamin D receptor) regulates defective autophagy in renal tubular epithelial cell in streptozotocin-induced diabetic mice via the AMPK pathway. Autophagy 18 (4), 877–890. doi:10.1080/15548627.2021.1962681
Li, Y., Pan, Y., Caouasasaki, S. K., Wang, Y., Niu, A., Fan, X, et al. (2021). Podocyte EGFR inhibits autophagy through upregulation of rubicon in type 2 diabetic nephropathy. Diabetes 70 (2), 562–576. doi:10.2337/db20-0660
Liu, L., Bai, F., Song, H., Xiao, R., Wang, Y. Z., Yang, H. M., et al. (2022). Upregulation of TIPE1 in tubular epithelial cell aggravates diabetic nephropathy by disrupting PHB2 mediated mitophagy. Redox Biol. 50, 102260. doi:10.1016/j.redox.2022.102260
Lyu, L., Xiang, W., Zheng, F., Huang, T., Feng, Y., Yuan, J., et al. (2020). Significant prognostic value of the autophagy-related gene P4HB in bladder urothelial carcinoma. Front. Oncol. 10, 1613. doi:10.3389/fonc.2020.01613
Maezawa, Y., Takemoto, M., and Yokote, K. (2015). Cell biology of diabetic nephropathy: Roles of endothelial cells, tubulointerstitial cells and podocytes. J. Diabetes Investig. 6 (1), 3–15. doi:10.1111/jdi.12255
Nabi, R., Alvi, S. S., Alouffi, S., Khan, S., Ahmad, A., Khan, M., et al. (2021a). Amelioration of neuropilin-1 and RAGE/matrix metalloproteinase-2 pathway-induced renal injury in diabetic rats by rosuvastatin. Arch. Biol. Sci. 73 (2), 265–278. doi:10.2298/abs210316021n
Nabi, R., Alvi, S. S., Shah, A., Chaturvedi, C. P., Faisal, M., Alat, A. A., et al. (2021b). Ezetimibe attenuates experimental diabetes and renal pathologies via targeting the advanced glycation, oxidative stress and AGE-RAGE signalling in rats. Arch. Physiol. Biochem. 2021, 1–16. doi:10.1080/13813455.2021.1874996
Noiva, R. (1999). Protein disulfide isomerase: The multifunctional redox chaperone of the endoplasmic reticulum. Semin. Cell Dev. Biol. 10 (5), 481–493. doi:10.1006/scdb.1999.0319
Parker, H. S., Leek, J. T., Favorov, A. V., Considine, M., Xia, X., Chavan, S., et al. (2014). Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction. Bioinformatics 30 (19), 2757–2763. doi:10.1093/bioinformatics/btu375
Ritcheie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. doi:10.1093/nar/gkv007
Satirapoj, B. (2018). Tubulointerstitial biomarkers for diabetic nephropathy. J. Diabetes Res. 2018, 2852398. doi:10.1155/2018/2852398
Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola, J., Ladd, C., et al. (2002). Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1 (2), 203–209. doi:10.1016/s1535-6108(02)00030-2
Slieker, R. C., Van Der Heijden, A. A. W. A., Siddiqui, M. K., Langendoen-Gort, M., Nijpels, G., Herings, R., et al. (2021). Performance of prediction models for nephropathy in people with type 2 diabetes: Systematic review and external validation study. Bmj-British Med. J. 374, n2134. doi:10.1136/bmj.n2134
Tabrez, S., Al-Shali, K. Z., and Ahmad, S. (2015). Lycopene powers the inhibition of glycation-induced diabetic nephropathy: A novel approach to halt the AGE-RAGE axis menace. Biofactors 41 (5), 372–381. doi:10.1002/biof.1238
Tang, S. C. W., and Yiu, W. H. (2020). Innate immunity in diabetic kidney disease. Nat. Rev. Nephrol. 16 (4), 206–222. doi:10.1038/s41581-019-0234-4
Wang, X., Bai, Y., Zhang, F., Yang, Y., Feng, D., Li, A., et al. (2020). Targeted inhibition of P4HB promotes cell sensitivity to gemcitabine in urothelial carcinoma of the bladder. Onco. Targets. Ther. 13, 9543–9558. doi:10.2147/ott.S267734
Welsh, J. B., Sapinoso, L. M., Su, A. I., Kern, S. G., Wang-rodriguez, J., Moskaluk, C. A., et al. (2001). Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 61 (16), 5974–5978.
Woroniecka, K. I., Park, A. S. D., Mohtat, D., Thomas, D. B., Pullman, J. M., and Susztak, K. (2011). Transcriptome analysis of human diabetic kidney disease. Diabetes 60 (9), 2354–2369. doi:10.2337/db10-1181
Wu, P., Xiang, T., Wang, J., Lv, R., Ma, S., Yuan, L., et al. (2021). Identification of immunization-related new prognostic biomarkers for papillary renal cell carcinoma by integrated bioinformatics analysis. BMC Med. Genomics 14 (1), 241. doi:10.1186/s12920-021-01092-w
Xie, L., Li, H., Zhang, L., Ma, X., Dang, Y., Guo, J., et al. (2020). Autophagy-related gene P4HB: A novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging 12 (2), 1828–1842. doi:10.18632/aging.102715
Yang, C., Chen, X.-C., Li, Z.-H., Wu, H.-L., Jing, K.-P., Huang, X.-R., et al. (2021a). SMAD3 promotes autophagy dysregulation by triggering lysosome depletion in tubular epithelial cells in diabetic nephropathy. Autophagy 17 (9), 2325–2344. doi:10.1080/15548627.2020.1824694
Yang, H., Xie, T., Li, D., Du, X., Wang, T., Li, C., et al. (2019). Tim-3 aggravates podocyte injury in diabetic nephropathy by promoting macrophage activation via the NF-kappa B/TNF-alpha pathway. Mol. Metab. 23, 24–36. doi:10.1016/j.molmet.2019.02.007
Yang, M., Luo, S., Jiang, N., Wang, X., Han, Y., Zhao, H., et al. (2021b). DsbA-L ameliorates renal injury through the AMPK/NLRP3 inflammasome signaling pathway in diabetic nephropathy. Front. Physiol. 12, 659751. doi:10.3389/fphys.2021.659751
Yu, K., Li, D., Xu, F., Guo, H., Feng, F., Ding, Y., et al. (2021). Ido1 as a new immune biomarker for diabetic nephropathy and its correlation with immune cell infiltration. Int. Immunopharmacol. 94, 107446. doi:10.1016/j.intimp.2021.107446
Zhan, M., Usman, I. M., Sun, L., and Kanwar, Y. S. (2015). Disruption of renal tubular mitochondrial quality control by myo-inositol oxygenase in diabetic kidney disease. J. Am. Soc. Nephrol. 26 (6), 1304–1321. doi:10.1681/asn.2014050457
Zhang, C., Jiang, J., Wang, L., Zheng, L., Xu, J., Qi, X., et al. (2020). Identification of autophagy-associated biomarkers and corresponding regulatory factors in the progression of colorectal cancer. Front. Genet. 11, 245. doi:10.3389/fgene.2020.00245
Zhang, L., Long, J., Jiang, W., Shi, Y., He, X., Zhou, Z., et al. (2016). Trends in chronic kidney disease in China. N. Engl. J. Med. 375 (9), 905–906. doi:10.1056/NEJMc1602469
Keywords: autophagy, P4HB, bioinformatics analysis, diabetic nephropathy, biomarker
Citation: Bai F, Yu K, Yang Y, Zhang Y, Ding L, An X, Feng F, Sun N, Fan J, Liu L, Yang H and Yang X (2022) Identification and validation of P4HB as a novel autophagy-related biomarker in diabetic nephropathy. Front. Genet. 13:965816. doi: 10.3389/fgene.2022.965816
Received: 10 June 2022; Accepted: 25 August 2022;
Published: 26 September 2022.
Edited by:
Saheem Ahmad, University of Hail, Saudi ArabiaReviewed by:
Xiaoyan Bai, Guangdong Provincial People’s Hospital, ChinaHao Zhang, Central South University, China
Copyright © 2022 Bai, Yu, Yang, Zhang, Ding, An, Feng, Sun, Fan, Liu, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangdong Yang, eXhkQGVtYWlsLnNkdS5lZHUuY24=
 Fang Bai
Fang Bai Kuipeng Yu
Kuipeng Yu Yanjiang Yang1
Yanjiang Yang1 Feng Feng
Feng Feng Huimin Yang
Huimin Yang